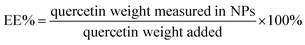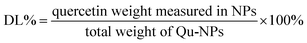Open Access Article
Open Access ArticleImproved therapeutic efficacy of quercetin-loaded polymeric nanoparticles on triple-negative breast cancer by inhibiting uPA†
Yang Zhou‡
a,
Dan Chen‡a,
Guangpu Xuea,
Shujuan Yua,
Cai Yuanb,
Mingdong Huang *a and
Longguang Jiang
*a and
Longguang Jiang *ac
*ac
aCollege of Chemistry, National & Local Joint Biomedical Engineering Research Center on Photodynamic Technologies, Fuzhou University, Fuzhou, Fujian 350116, China. E-mail: jianglg@fzu.edu.cn
bCollege of Biological Science and Engineering, Fuzhou University, Fuzhou, Fujian 350116, China
cFujian Key Laboratory of Marine Enzyme Engineering, Fuzhou University, Fuzhou, Fujian 350116, P.R. China
First published on 17th September 2020
Abstract
Triple negative breast cancer (TNBC) is one kind of breast cancer that demonstrates highly aggressive tumor biology. The high heterogeneity of TNBC makes its individual clinical treatment extremely blind and limited, which also introduces more challenges into the diagnosis and treatment of diseases. Urokinase-type plasminogen activator (uPA) is a high level marker for breast cancer, which mediates tumor growth and metastasis. Quercetin is a plant-derived flavonoid in many plants, which inhibits uPA and has low bioavailability and mediocre pharmaceutical efficacy. Thus, we herein developed polymeric nanoparticulate systems from PLGA-TPGS (Qu-NPs) for quercetin oral delivery and evaluated the anticancer effect of this formulation on TNBC in vitro and in vivo. Qu-NPs have a uniform spherical morphology with a mean diameter of 198.4 ± 7.8 nm and good drug loading capacity (8.1 ± 0.4%). Moreover, Qu-NPs exhibited significantly improved inhibition on the growth and metastasis in TNBC cells. Following oral gavage, a remarkable antitumor effect of Qu-NPs on 4T1-bearing mice was observed with a tumor inhibition ratio of 67.88% and fewer lung metastatic colonies. Furthermore, the inhibitory effect of quercetin on the migration of uPA knockdown MDA-MB231 cells was greatly attenuated. Together, Qu-NPs improved the significant antitumor and antimetastatic effects by inhibiting uPA, which provides a new strategy for the treatment of TNBC.
1. Introduction
A steep increase in the incidence of breast cancer has been noted globally in recent years. Breast cancer, the most common malignancy in women, is a heterogeneous group of tumors with diverse histology, behavior and response to therapies, and remains a major public health problem.1 Among breast cancer subtypes, triple negative breast cancer (TNBC) accounts for approximately 15–20%. TNBC is defined by the lack of estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (HER 2).2–4 Therefore, TNBC lacks specific molecular targets and is extremely aggressive, making its treatment a daunting task and leading to higher mortality than the other two subtypes.2 Conventional treatments, including chemotherapy, radiation, and surgical resection, are often limited by pain and resistance. Some new targets have been reported, such as epidermal growth factor receptor (EGFR) and androgen receptor (AR), but they have not made encouraging progress in clinical trials.5 Therefore, TNBC still requires an effective targeted therapy and the treatment for this disease should continue to improve.Urokinase-type plasminogen activator (uPA), a biomarker in breast cancer,6 is an extracellular serine proteolytic enzyme, which is involved in many physiological and pathological processes such as wound healing, fibrin degradation, migration of inflammatory cells, dissipation of extravascular hematoma, and invasion and metastasis of tumor cells.7–9 According to the exciting results, some tumor cells have high uPA expression to activate plasminogen around the cells to degrade the extracellular matrix and basement membrane of tumor cells, and promotes the invasion and metastasis of tumor cells. Therefore, the study of uPA inhibitors is of great significance for the clinical prognosis of cancer and anti-cancer metastasis.
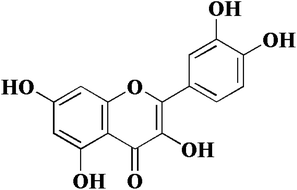
In view of the important role of uPA in cancer, we have been focusing on the research of uPA and a series of serine protease inhibitors for a long time including flavonoids.10–12 Flavonoids have a wide range of biological activities and have been used in many diseases and health care since ancient times as a component of Chinese herbal medicine.13–16 Quercetin (Fig. 1), a plant-derived flavonoid from many plants, binds to uPA with a general inhibition (IC50 = 7 μM).17 In our previous work, we determined the crystal structure of uPA in a complex with quercetin, and the inhibition mechanism of quercetin was preliminarily understood. We found that quercetin interacts with uPA in a different way from traditional uPA inhibitors (e.g. phenylguanidine). The main pharmacophore of quercetin is the catechol group (Fig. 1), which interacts with the Asp189 in the uPA substrate recognition pocket.10 However, there are still some issues of quercetin that need to be addressed, such as its poor water solubility and susceptibility to oxidation leading to the loss of efficacy.18,19 Hence, we not only focused on inhibiting potential targets, but also enhancing the anticancer potential of existing drugs by designing new drug delivery systems. For most drugs, oral administration is the most common form, which has the advantages of economy, safety, easy dose control, and convenience. At the same time, it can effectively improve patient compliance. However, quercetin has poor bioavailability, which is a major obstacle to oral administration due to poor water solubility and easy oxidation. Therefore, we sought to develop a quercetin-loaded nanoparticle oral delivery system.
PLGA shows advantageous performance compared with most other types of delivery systems due to its biodegradability, sustained-release efficacy and long-term stability of entrapped bioactive molecules.20 TPGS is another well-known biocompatible and amphiphilic polymer extensively used in nanoparticle surface coatings to improve encapsulation efficiency, bioavailability and circulation time in vivo.21 Hence, in this study, we encapsulated quercetin in PLGA/TPGS nanoparticles (Qu-NPs) to obtain uniform and stable nanoparticles with high drug loading. On this basis, we established an orthotopic model of 4T1 breast cancer with oral administration of Qu-NPs. The results showed that Qu-NPs had improved quercetin's inhibition on the proliferation, migration and invasion of TNBC cells much better both in vitro and in vivo compared with quercetin alone. Mouse oral gavage showed a remarkable antitumor effect of Qu-NPs with a good tumor inhibition ratio (67.88%) and few lung metastatic colonies. Furthermore, the inhibitory effect of quercetin on the migration of uPA knockdown MDA-MB231 cells was greatly attenuated. This study indicates uPA may serve as an effective target of TNBC and provides a new strategy for drug development towards the treatment for breast cancer.
2. Materials and methods
2.1 Materials and cell lines
Poly(lactic-co-glycolic acid) (PLGA) with a 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 monomer ratio and viscosity of 0.7 dL g−1 was purchased from Jinan Dai Gang Biological Technology Co. (Jinan, China). Quercetin (CAS number: 117-39-5), D-α-tocopherol polyethylene glycol 1000 succinate (TPGS, CAS number: 9002-96-4) and all reagents were purchased either from Sigma-Aldrich (St Louis, MO, USA) or from Sinopharm Chemical Reagent Co. Ltd. (Shanghai, China) unless otherwise stated. Human breast cancer cells (MDA-MB-231), human embryo lung fibroblasts (HELF), and mouse breast cancer cells (4T1) were purchased from American Type Culture Collection (ATCC, Rockville, MD, USA). Cells were cultured in Dulbecco's Modified Eagle's Medium-high glucose (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotics (penicillin–streptomycin solution). All cells were kept at 37 °C in a humidified incubator with 5% CO2 atmosphere and maintained in logarithmic phase with viability >95%.22
25 monomer ratio and viscosity of 0.7 dL g−1 was purchased from Jinan Dai Gang Biological Technology Co. (Jinan, China). Quercetin (CAS number: 117-39-5), D-α-tocopherol polyethylene glycol 1000 succinate (TPGS, CAS number: 9002-96-4) and all reagents were purchased either from Sigma-Aldrich (St Louis, MO, USA) or from Sinopharm Chemical Reagent Co. Ltd. (Shanghai, China) unless otherwise stated. Human breast cancer cells (MDA-MB-231), human embryo lung fibroblasts (HELF), and mouse breast cancer cells (4T1) were purchased from American Type Culture Collection (ATCC, Rockville, MD, USA). Cells were cultured in Dulbecco's Modified Eagle's Medium-high glucose (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% antibiotics (penicillin–streptomycin solution). All cells were kept at 37 °C in a humidified incubator with 5% CO2 atmosphere and maintained in logarithmic phase with viability >95%.22
2.2 Preparation and characterization of Qu-NPs
Qu-loaded PLGA/TPGS nanoparticles were prepared by a single-step nanoprecipitation approach.23 Briefly, PLGA was first dissolved in acetone with a concentration of 9.6 mg mL−1, and then quercetin was dissolved in the above PLGA solution with a concentration of 0.8 mg mL−1. Additionally, TPGS was dissolved and stirred in a 4% ethanol aqueous solution with a concentration of 5.76 mg mL−1 in a 37 °C water bath. Qu-NPs were prepared by adding 10 mL of the preheated aqueous solution dropwise into 2 mL of PLGA acetone solution under gentle stirring. After stirring for 30 minutes, 8 mL of ultrapure water was added drop-wise into the system. Qu-NPs were allowed to self-assemble for another 30 minutes, and the remaining organic solvent and non-encapsulated free drugs were removed by washing the Qu-NPs using Amicon Ultra-4 centrifugal filters (Millipore, MA) with a MWCO of 10 kDa. Qu-NPs were used immediately or stored at −4 °C.24,25 The size distribution and diameter of quercetin nanoparticles and PLGA/TPGS nanoparticles were measured by a dynamic light scattering (DLS) instrument (Nano ZS ZEN 3600, Malvern Instruments, Malvern, UK) at room temperature. Meanwhile, the zeta potential of two nanoparticles was also detected by DLS. Next, the sample was placed on a 300-mesh copper grid coated with carbon after concentration exchange, dried at 65 °C for 4 h, the morphology of Qu-NPs was observed by a transmission electron microscope (TEM, Hitachi H-7650) operated at an acceleration voltage of 80 kV and an ultra-high resolution field emission scanning electron microscope (SEM, Verios G4 UC) operated at an acceleration voltage of 3 kV.22,26 The content of quercetin in Qu-NPs was determined by UV spectroscopy. The absorbance of quercetin was measured at the determined maximum absorption wavelength (370 nm). Encapsulation efficiency (EE%) and drug-loading efficiency (DL%) were calculated as following:272.3 In vitro drug release
The in vitro drug release study of Qu-NPs formulations was studied by the dialysis bag diffusion method. Drug-loaded nanoparticles (2 mL) were dispersed into a dialysis bag and the dialysis bag was then kept in a beaker containing 20 mL of pH 7.4 phosphate buffer, 0.1% SDS (v/v) and 2.5% tween 80 (v/v). The beaker was placed over a magnetic stirrer and the temperature of the assembly was maintained at 37 ± 1 °C throughout the experiment. During the experiment rpm was maintained at 100 rpm. Samples (100 μL) were withdrawn at a definite time intervals and replaced with equal amounts of fresh dialysate buffer. Afterwards, the samples were analyzed using a UV-visible spectrophotometer at 370 nm.2.4 Cytotoxicity of quercetin and Qu-NPs determined by MTT assay
The cells were seeded into 96-well plates (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well), and 24 h later, breast cancer cells were treated with a series of concentrations of free quercetin and Qu-NPs, respectively. After 48 hours, the medium in the 96-well plate was removed and washed with PBS. Then 100 μL of MTT was added to each well. After 4 hours, the MTT solution in the 96-well plate was aspirated, and 100 μL of DMSO solution was added to each well. After incubating for 30 min on a shaker, the absorbance was measured at 490 nm with a microplate reader.28
000 cells per well), and 24 h later, breast cancer cells were treated with a series of concentrations of free quercetin and Qu-NPs, respectively. After 48 hours, the medium in the 96-well plate was removed and washed with PBS. Then 100 μL of MTT was added to each well. After 4 hours, the MTT solution in the 96-well plate was aspirated, and 100 μL of DMSO solution was added to each well. After incubating for 30 min on a shaker, the absorbance was measured at 490 nm with a microplate reader.28
2.5 Anti-migration effect of quercetin and Qu-NPs determined by wound healing assay
The cells were seeded at a density of 5 × 105 cells per well in a 12-well plate. A confluent monolayer of cells was scraped with a white pipette tip after approximately 24 hours and washed with PBS to remove floating cells. The cells were then incubated with different concentrations (0, 5, 10, and 50 μM) of quercetin and Qu-NPs, respectively, and images of the migration of cells into the wound were captured at 0, 12 and 24 hours.28 For each cell type, the wound area was measured using Image J software at the beginning of the experiment (0 h) and after 24 h. The data were expressed as the change in the wound area over time and calculated as a percentage of the original wound area. Experiments were performed in duplicates and repeated twice.2.6 Anti-invasion effect of quercetin and Qu-NPs determined by transwell chamber invasion assay
For migration experiments, cells in 5 × 104 serum-free medium were placed in the upper chamber of a Transwell permeable support with an 8 μm-well cell culture insert. For the invasion assay, the upper chamber of the insert was coated with 100 μL of 50-fold diluted BD matrigel and allowed to cure in the incubator. Next, basement membrane hydration was performed on Matrigel, and 5 × 104 cells in serum-free DMEM medium were added to the upper chamber and treated with different concentrations of quercetin and Qu-NPs. DMEM medium which contained 10% FBS was added to the lower chamber of a 24-well plate. The cells were incubated for 24 h in a carbon dioxide cell incubator (37 °C, 5% CO2). Then, the cells remaining on the upper side of the membrane were removed with a cotton swab, and the cells that migrated or invaded were fixed in methanol for 20 min, stained with crystal violet for 20 min, photographed under a microscope, and finally decolorized with 33% acetic acid. The OD value of the decolorizing solution was measured by using a microplate reader at 570 nm.282.7 Antitumor and antimetastatic efficacy in orthotopic 4T1 breast tumor model
We established an orthotopic 4T1 breast tumor model to assess the anti-proliferative and anti-metastatic efficacy of nanomedicine in vivo. 4T1 cancer cells (4 × 106) were inoculated into a fat pad under the fourth mammary gland of female Balb/c mice (weighing approximately 20 g). Mice were randomly divided into 2 groups (n = 5) when the tumor volume reached 50–100 mm3. Mice in the control group received saline (10 μL g−1 by gavage). Mice in the Qu-NPs group were treated with Qu-NPs orally at a Qu dose of 30 mg kg−1 once a day for consecutive 10 days. The tumor size and body weight were recorded daily. The mice were sacrificed the day after the final administration, and the tumor and lung were harvested immediately and weighed. Nodules of the metastatic tumor on the surface of the lungs were counted. After that, the tumor and lung were fixed in formalin, embedded in paraffin, sectioned into 5 μm sections and stained with H&E for further histologic analysis.29 The protocols and handling of the animals were conducted in accordance with the Guidelines for Care and Use of Laboratory Animals of Fuzhou University as approved by the Animal Ethics Committee in Fuzhou University.2.8 Construction of stable uPA knockdown cell lines
Small interfering oligonucleotides specific for uPA (CCGGGGGCGAACGACAATAGCTTTACTCGAGTAAAGCTATTGTCGTTCGCCCTTTTTG) were synthesized and annealed. The designed sequences of the primes are shown in ESI Table S2.† A uPA RNAi plasmid vector that expresses shRNA for uPA under the control of a human U6 promoter was constructed by inserting pairs of the annealed DNA oligonucleotides specific for uPA at the Age I site and EcoR I site sequentially into the pLKO.1 puro vector (sh-uPA). Then we transfected the shRNA construct. Before transfection, the cells were grown and subcultured to a six-well plate at a density of 2.5 × 105 cells per well until they reached a confluency of 60–80% (24 h). The experiment was divided into a blank group, control group and sh-uPA experimental group. The blank group was transfected with the negative control shRNA expression vector (1 μg), and the experimental group was transfected with the sh-uPA expression vector (1 μg). Cells were transfected with LIPOFECTAMINE™ 2000 reagent at 37 °C for 24 h. With increased concentrations of puromycin, we obtained a stable uPA knockdown cell line.30–322.9 The quantitative real-time polymerase chain reaction (qPCR) analysis
Total RNA was first extracted from the control and transfected cells using TRIzol reagent (Invitrogen). The isolated RNA was reverse transcribed using the Verso cDNA synthesis kit (Thermo Scientific) according to the manufacturer's protocol. PCR amplification was then performed using the SYBR green method with the help of CFX96™ Optics Module machine (BIO-RAD). GAPDH was used as a reference gene for normalization.33 The PCR conditions consisted of 1 cycle of initial activation at 95 °C for 10 minutes, 40 cycles of denaturation at 95 °C for 10 seconds, annealing at 50 °C for 20 seconds and then extension at 72 °C for 15 seconds, followed by a melting reaction step of 65 °C and 95 °C for 1 cycle. Relative changes in the gene expression were analyzed using the 2−ΔΔCT method.34–36 The sequences of the primers used are presented in ESI Table S2.†2.10 Statistics
All data were presented as mean ± standard deviation (SD). All experiments were performed at least three times unless otherwise indicated. The experimental data in vitro were analyzed using the one-way ANOVA. Differences at the 95% confidence level were considered to be statistically significant (P < 0.05).3. Results
3.1 Characterization of quercetin-loaded nanoparticles (Qu-NPs)
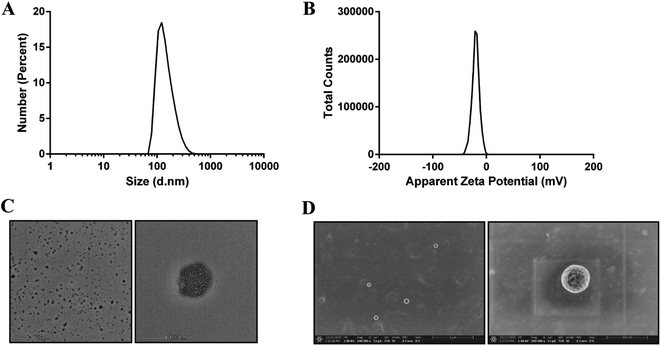
The Qu-NPs were prepared as described previously,23 and were observed to be opalescent and yellow with a particle size of 198.4 ± 7.8 nm with a polydispersity index (PDI) of 0.095, demonstrating a good homogeneity. The zeta potential of Qu-NPs was −22.5 ± 2.5 mV (Table 1, Fig. 2A and B). After quercetin entrapment, nanoparticles showed a slight size increase compared to drug-free nanoparticles (approximately 173 nm) (Fig. S1A and B†). The particle size and zeta potential of the Qu-NPs did not change significantly during storage at 4 °C for one week, showing good storage stability (Table S1†). Qu-NPs were uniform in size with a spherical shape and dense structure as revealed by TEM and SEM (Fig. 2C and D). Furthermore, we developed a simple method to determine the concentration of quercetin by UV spectroscopy. The free quercetin solution had a maximum absorption peak at 370 nm, and the maximum absorption peak of the Qu-NPs solution (Qu-NPs) also appeared at 370 nm; however, the PLGA-TPGS NPs had no obvious absorption peak at 370 nm. These results demonstrated that quercetin had already been packed into the PLGA-TPGS NPs (Fig. S2A†). Furthermore, we built the standard curve determined by UV spectroscopy at 370 nm (Fig. S2B†). The drug loading of this nano-formulation was calculated to be 8.1 ± 0.4%, and the encapsulation efficiency was 82.3 ± 5.7% (Table 1). In addition, in vitro release experiments showed that the release rate of Qu-NPs is much slower than that of free quercetin, and achieves a sustained release effect and improves the medicinal value of quercetin (Fig. S3†). Based on the above-mentioned in vitro characteristic results, we concluded that this nano-formulation was a stable nanoparticulate system with high quercetin loading.| Formulation | Size (nm) | Zeta potential | DL (%) | EE (%) |
|---|---|---|---|---|
| Qu-NPs | 198.4 ± 7.8 | −22.5 ± 2.5 | 8.1 ± 0.4 | 82.3 ± 5.7 |
3.2 In vitro cytotoxic effects of Qu-NPs on TNBC cell lines
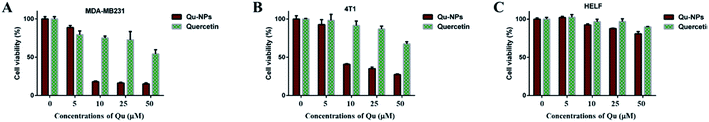
Cytotoxic effects of Qu-NPs in vitro were evaluated on two TNBC cell lines (mouse 4T1 cells and human MDA-MB231 cells), which have high-level uPA expression.37 Human embryonic lung fibroblast (HELF) cells were also used as a control. Cells were treated with different concentrations of quercetin, and MTT assays were performed at the indicated time points. The results showed that free quercetin at high concentrations (>100 μM) has weak cytotoxic effects against both of the TNBC cell lines, which is consistent with the result reported before38 and limits the practical applications of quercetin (Fig. S4†). Therefore, we investigated the cytotoxic effect of Qu-NPs on the above mentioned cells. Studies have shown that PLGA-TPGS formulations have no toxic effect on breast cancer and exhibit relatively slow drug release.39–42 Compared with quercetin alone, it can be easily observed that the inhibitory effects of Qu-NPs on MDA-MB231 and 4T1 are greatly improved at lower concentrations (10 μM) (Fig. 3A and B). Moreover, the cytotoxic effect of Qu-NPs to HELF cells is not significantly increased (Fig. 3C). These results not only fully demonstrated that nano-formulations have an enhanced impact on quercetin to increase the inhibitory effects on TNBC cells proliferation, but also showed that Qu-NPs had a better specific inhibitory effect for TNBC cells.3.3 Inhibitory effects of Qu-NPs on the migration of TNBC cells
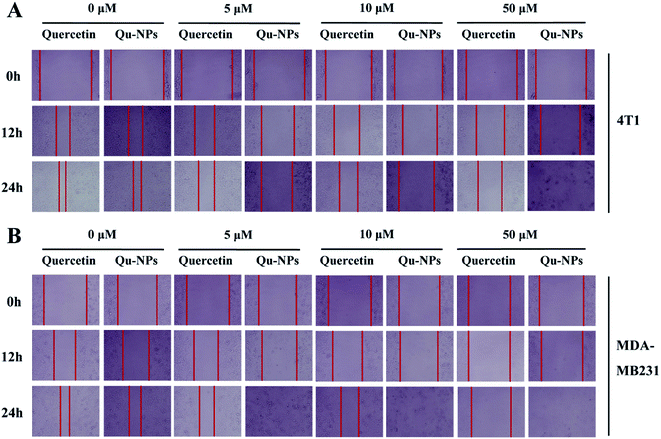
In addition to uncontrolled growth, malignant tumors are capable of invading surrounding normal tissues and then spreading throughout the body via lymphatic or circulatory systems. To investigate the effects of quercetin and Qu-NPs on cell motility, the confluent cell layers were scraped with a pipette tip and treated with different concentrations of quercetin and Qu-NPs, respectively. In order to avoid the strong cytotoxic effects of Qu-NPs in 48 h, we decided to record the images of the cell migration at different time points in 24 h (0 h, 12 h, and 24 h). As shown in Fig. 4, compared with the untreated groups, both quercetin and Qu-NPs significantly inhibited the migration of 4T1 and MDA-MB231 cells along with the increased concentration of quercetin. The comparison of the effects of quercetin and Qu-NPs at the same concentration and time point revealed that the inhibition effect of Qu-NPs on the migration of 4T1 and MDA-MB231 cells was much stronger than that of quercetin alone. Furthermore, quercetin and Qu-NPs inhibited the migration of MDA-MB231 cells more strongly than that of 4T1 cells, which is consistent with the results of the inhibition on breast cancer cell proliferation. These results demonstrated that nano-encapsulation greatly enhanced the inhibition efficacy of quercetin on the migration of TNBC cells.3.4 Inhibitory effects of Qu-NPs on the invasion of TNBC cells
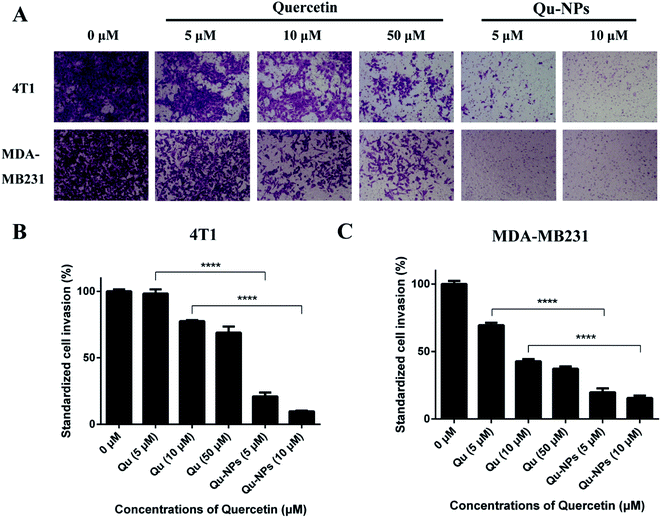
We further measured the invasion of TNBC cells using a transwell invasion assay in 4T1 and MDA-MB231 cells. For the same reason in the assay of inhibitory effects of Qu-NPs on cell migration, we treated the cells with different concentrations of Qu and Qu-NPs for 24 hours, respectively. As shown in Fig. 5A, the amount of 4T1 and MDA-MB231 cells penetrating the matrigel and entering the lower chamber gradually decreased with increasing concentrations of free quercetin. In addition, it is noted that only a small number of both 4T1 and MDA-MB231 cells were invaded when treated with 5 μM of Qu-NPs, and the quantity of invaded cells was further reduced when treated with 10 μM of Qu-NPs. The results indicated a concentration-dependent inhibitory effect of free quercetin and Qu-NPs on tumor cell invasion. Moreover, the invasion rate of cells treated with Qu-NPs was significantly reduced compared with that treated with the same concentration of free quercetin (Fig. 5B and C). For 4T1 cells, compared with the control group (0 μM quercetin), 10 μM of free quercetin reduced the cell invasion rate to 77.5%, while treatment with 10 μM of Qu-NPs decreased the cell invasion rate to 9.8%, indicating that Qu-NPs have a better capability to prevent 4T1 cell invasion. For MDA-MB231 cells, incubation with 10 μM of free quercetin reduced the cell invasion rate to 42.9% compared with the control group, while incubation with 5 μM of Qu-NPs decreased the cell invasion rate to 18.1%. Therefore, these results showed that the inhibitory effect of Qu-NPs on 4T1 and MDA-MB231 cell invasion is much stronger than that of free quercetin.3.5 Qu-NPs exerted antitumor and antimetastatic effect in an orthotopic 4T1 breast tumor model
Furthermore, we investigated the inhibitory effect of Qu-NPs on the proliferation and metastasis of triple-negative 4T1 mammary carcinoma in vivo. After oral gavage for 10 days, Qu-NPs significantly inhibited tumor growth with much lower tumor weight and smaller tumor volume at the end of a once-daily dosing compared with saline treatment (Fig. 6A–C). The H&E staining of the excised tumor and lung sections showed that the tumors in the control group treated with saline grew well, while the tumors in the Qu-NPs treatment group showed partial necrosis and apoptosis. Metastatic foci were seen in all of the lungs of mice in the control group, while no metastatic foci were observed in the mice treated by Qu-NPs (Fig. 6D). This indicated that Qu-NPs greatly inhibited the proliferation of orthotopic 4T1 mammary carcinoma and hindered their metastasis in vivo, which is consistent with the in vitro results. Moreover, we also found that there was no significant decrease in body weight in the experimental animals, indicating that Qu-NPs did not have severe toxicity to mice (Fig. 6E).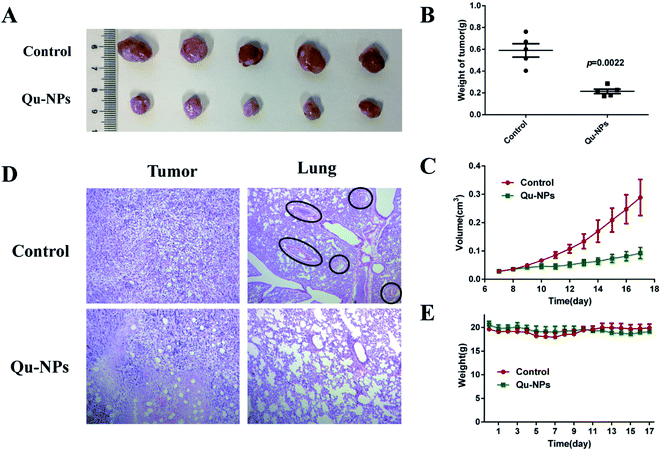 | ||
| Fig. 6 The anti-tumor and anti-metastatic efficacies of Qu-NPs in vivo. The suppression of the tumor growth in Balb/c mice orthotopically implanted with 4T1 cells at the daily dosage of 30 mg kg−1 Qu-NPs for 10 days. Representative graphics (A) and tumor weights (B) of resected tumors with and without treatments. (C) The tumor volume of Balb/c mice was recorded from the seventh day of tumor seeding, and the data were processed with 0.5 × length × width.2 The values were represented as mean ± SEM. (D) The graphics of the H&E-stained histological sections of the tumor and lungs with and without the treatments. (E) The changes of the body weights of mice orthotopically implanted with 4T1 cells with the treatment of Qu-NPs. PBS buffer as a control, the values were represented as mean ± SEM. | ||
3.6 Quercetin inhibits migration of triple-negative breast cancer cells by inhibiting uPA
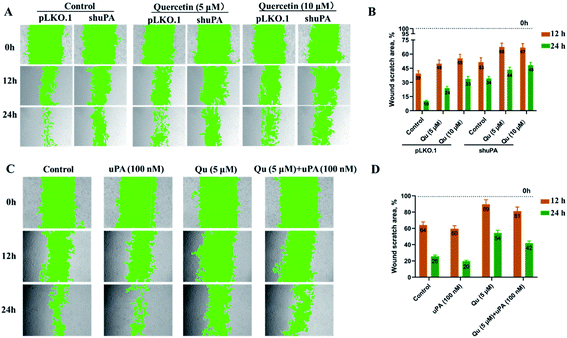
To study if the inhibition of quercetin is mediated by uPA on triple-negative breast tumors, we used small hairpin RNAs to knock down the expression of endogenous uPA genes in the human breast cancer cell line MDA-MB231 (Table S2†). The real-time quantitative PCR showed that we successfully constructed a stable uPA knockdown cell line (MDA-MB231 shuPA) with 70% knockdown efficiency (Fig. S5†). One of the key functions of uPA is the promotion of tumor metastasis. Some studies have shown that knocking down uPA inhibited the invasion and metastasis of cancer cells.43,44 To evaluate the impact of uPA knockdown on the migration of TNBC cells, we performed wound healing assays using MDA-MB231 pLKO.1 cells and MDA-MB231 shuPA cells. As shown in Fig. 7, without quercetin treatments, the migration rate of shuPA cells was significantly slower compared with pLKO.1 cells. However, under the condition of quercetin treatment, the migration rate of MDA-MB231 pLKO.1 cells was greatly reduced, and the inhibitory effect of quercetin on the migration of MDA-MB231 shuPA cells was also greatly attenuated. Moreover, in order to verify the relationship between quercetin and uPA, we also performed wound healing experiments in MDA-MB231 shuPA cells by adding additional uPA. Compared with the control, the migration ability of MDA-MB231 shuPA cells co-cultured with uPA was significantly improved. After incubation with 5 μM of quercetin, the migration ability of MDA-MB231 shuPA cells, which still have 30% uPA expression, was significantly reduced compared with the control. However, after co-cultivation with quercetin and uPA, its migration ability was slightly restored. The above experimental results show that knockdown of uPA expression by RNAi can inhibit the migration of MDA-MB231 cells to a certain degree. Supplemental uPA can restore the migration of uPA knockdown cells (MDA-MB231 shuPA), and then treating with quercetin again inhibited the migration of uPA knockdown cells. Combined with our previous findings on the crystal structure of quercetin and uPA complex, we conclude that quercetin inhibits the migration of TNBC cells mediated by uPA.4. Discussion
Quercetin is different from the traditional uPA inhibitors, which usually contain a guanidine or amidine as the P1 group. Although these basic groups can help uPA inhibitors target the S1 pocket of the uPA active site,10,45,46 they are prone to protonation at physiological pH, hindering uPA inhibitors from penetrating the cell membrane into the interior of the cell, thus resulting in poor oral availability.18 Quercetin is one of the most important flavonoids in the human diet with a wide range of biological activities. Previous studies have shown that quercetin can bind to uPA with moderate inhibition at the micromolar level;14 however, there is no basic group in the structure of quercetin that might be a replacement to overcome the poor oral availability of traditional uPA inhibitors. In addition, the toxic effects showed that the inhibition of quercetin alone on a series of breast cancer cells is not obvious, and the specificity is not good enough due to being easily oxidized (Fig. S4†). In order to solve the above-mentioned disadvantages as well as improve the inhibition efficacy and specificity of quercetin on triple-negative breast cancer, we packaged quercetin with various materials, but only PLGA/TPGS nanoparticles exhibited good encapsulation. Qu-NPs prepared in this work have good uniformity and stability (Fig. 2). The cytotoxicity test showed that Qu-NPs had a much stronger inhibition effect on TNBC cells than quercetin alone (Fig. 3). A large number of studies and our study have shown that PLGA-TPGS formulations not only have no toxicity or side effects on many cancers but also have relatively slow drug release, which may prevent the Qu from being oxidized quickly.39–42Tumor metastasis is the main reason for the low survival rate of patients with malignant tumors, and it is the bottleneck of clinical treatment of malignant tumors. In this study, wound healing and matrigel invasion experiments were performed to verify the effect of Qu-NPs on the metastasis of TNBC cells. The results demonstrated that Qu-NPs have a much stronger inhibitory effect on TNBC metastasis than quercetin alone (Fig. 4 and 5). Du et al. reported that quercetin only slowed tumor growth but failed to reduce the initial tumor size and cure tumor-bearing mice.47 We further tested the effect of Qu-NPs by mouse orthotopic tumor models. The results showed that Qu-NPs significantly inhibit the growth of TNBC tumors in vivo. The results of cell experiments were further confirmed from lung sections. As a result, the transfer of TNBC cells to the lungs of mice treated with Qu-NPs were greatly reduced after treating with Qu-NPs, demonstrating that Qu-NPs effectively inhibit the growth and metastasis of TNBC tumors in vivo (Fig. 6).
While we did not measure the biodistribution of free quercetin and Qu-NPs in vivo, the biodistribution of free quercetin and its nanoparticle formulations have been reported in related literature, which revealed that quercetin can be significantly accumulated in the lung, kidney and liver after oral administration.48–51 However, it is still unclear whether quercetin exerts its anti-TNBC effect by interacting with uPA. To illuminate this issue, we established a stable TNBC cell line that knocked down uPA (MDA-MB231 shuPA). Wound healing experiments were performed on this cell line to reveal the role of uPA in cell migration. As results have shown, the migration capabilities were significantly reduced after MDA-MB231 knocked down the expression of uPA. However, the migration capabilities were rescued after supplementation with uPA and co-culturing with it, indicating that uPA has a strong relationship with the migration and invasion of TNBC cells. Moreover, the migration decreased slightly when adding quercetin by inhibiting uPA (Fig. 7).
5. Conclusion
In this study, quercetin-loaded PLGA/TPGS nanoparticles were prepared with high uniformity and stability, and was characterized by high drug loading capacity. After nano-encapsulation, compared with quercetin alone, Qu-NPs had much stronger inhibition not only on the proliferation but also on the migration and invasion of breast cancer cells. In the mechanism of action, we demonstrated that quercetin inhibited the migration and invasion of triple-negative breast cancer cells by inhibiting the activity of uPA. In general, our results demonstrated that quercetin-loaded PLGA/TPGS nanoparticles are more effective in treating breast cancer than free quercetin, indicating that uPA may serve as an effective target of TNBC and provide a new strategy for drug development towards treatment for breast cancer.Author contributions
Y. Z. and D. C. contributed equally to this work. Y. Z. and D. C. performed most of the experiments and prepared the draft, L. J. and C. Y. participated in the experimental guidance; S. Y. and G. X. assisted in preparing and operating experiments; M. H. and L. J. conceived and led the project and finalized the manuscript.Abbreviations
| EGFR | Epidermal growth factor receptor |
| AR | Androgen receptor |
| PLGA | Poly(lactic-co-glycolic acid) |
| TPGS | D-α-Tocopherol polyethylene glycol 1000 succinate |
| uPA | uPA-type plasminogen activator |
| TNBC | Triple negative breast cancer |
| Qu | Quercetin |
| Qu-NPs | Quercetin-loaded nanoparticles |
Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
Our research work is financially supported by grants from Natural Science Foundation of Fujian Province (2018J01897 and 2018J01729), National Key R&D Program of China (2017YFE0103200), and Natural Science Foundation of China (31370737, 31400637, 31570745, 31670739, U1405229).References
- X. Dai, T. Li, Z. Bai, Y. Yang, X. Liu, J. Zhan and B. Shi, Am. J. Cancer Res., 2015, 5(10), 2929–2943 CAS.
- A. M. Brewster, M. Chavez-MacGregor and P. Brown, Lancet Oncol., 2014, 15(13), e625–e634 CrossRef.
- S. Sharma, M. Barry, D. J. Gallagher, M. Kell and V. Sacchini, Surg. Oncol., 2015, 24(3), 276–283 CrossRef.
- M. Lippman, G. Bolan and K. Huff, Cancer Res., 1976, 36(12), 4595–4601 CAS.
- C. I. Herold and C. K. Anders, Oncology, 2013, 27(9), 846–854 Search PubMed.
- M. J. Duffy, P. M. McGowan, N. Harbeck, C. Thomssen and M. Schmitt, Breast Cancer Res., 2014, 16(4), 428 CrossRef.
- F. Blasi, J. D. Vassalli and K. Dano, J. Cell Biol., 1987, 104(4), 801–804 CrossRef CAS.
- D. M. Evans, K. Sloan-Stakleff, M. Arvan and D. P. Guyton, Clin. Exp. Metastasis, 1998, 16(4), 353–357 CrossRef CAS.
- P. Mikus, T. Urano, P. Liljestrom and T. Ny, Eur. J. Biochem., 1993, 218(3), 1071–1082 CrossRef CAS.
- G. Xue, L. Gong, C. Yuan, M. Xu, X. Wang, L. Jiang and M. Huang, Food Funct., 2017, 8(7), 2437–2443 RSC.
- L. Jiang, E. Oldenburg, T. Kromann-Hansen, P. Xu, J. K. Jensen, P. A. Andreasen and M. Huang, Biochim. Biophys. Acta, Gen. Subj., 2018, 1862(9), 2017–2023 CrossRef CAS.
- L. Jiang, X. Zhang, Y. Zhou, Y. Chen, Z. Luo, J. Li, C. Yuan and M. Huang, RSC Adv., 2018, 8(49), 28189–28197 RSC.
- T. Maliar, A. Jedinak, J. Kadrabova and E. Sturdik, Eur. J. Med. Chem., 2004, 39(3), 241–248 CrossRef CAS.
- M. Cuccioloni, M. Mozzicafreddo, L. Bonfili, V. Cecarini, A. M. Eleuteri and M. Angeletti, Chem. Biol. Drug Des., 2009, 74(1), 1–15 CrossRef CAS.
- L. Liu, H. Y. Ma, N. Y. Yang, Y. P. Tang, J. M. Guo, W. W. Tao and J. A. Duan, Thromb. Res., 2010, 126(5), E365–E378 CrossRef CAS.
- B. H. Havsteen, Pharmacol Ther., 2002, 96(2–3), 67–202 CrossRef CAS.
- N. V. Gorlatova, H. Elokdah, K. Fan, D. L. Crandall and D. A. Lawrence, J. Biol. Chem., 2003, 278(18), 16329–16335 CrossRef CAS.
- J. C. Ngo, L. Jiang, Z. Lin, C. Yuan, Z. Chen, X. Zhang, H. Yu, J. Wang, L. Lin and M. Huang, Curr. Drug Targets, 2011, 12(12), 1729–1743 CrossRef CAS.
- J. Barrenetxe, P. Aranguren, A. Grijalba, J. M. Martinez-Penuela, F. Marzo and E. Urdaneta, Br. J. Nutr., 2006, 95(3), 455–461 CrossRef CAS.
- E. Perez-Herrero and A. Fernandez-Medarde, Eur. J. Pharm. Biopharm., 2015, 93, 52–79 CrossRef CAS.
- Z. Zhang, S. Huey Lee and S. S. Feng, Biomaterials, 2007, 28(10), 1889–1899 CrossRef CAS.
- S. Li, C. Yuan, J. Chen, D. Chen, Z. Chen, W. Chen, S. Yan, P. Hu, J. Xue, R. Li, K. Zheng and M. Huang, Theranostics, 2019, 9(3), 884–899 CrossRef CAS.
- W. Tao, X. Zeng, T. Liu, Z. Wang, Q. Xiong, C. Ouyang, L. Huang and L. Mei, Acta Biomater., 2013, 9(11), 8910–8920 CrossRef CAS.
- P. V. Farago, R. P. Raffin, A. R. Pohlmann, S. S. Guterres and S. F. Zawadzki, J. Braz. Chem. Soc., 2008, 19(7), 1298–1305 CrossRef CAS.
- S. Desgouilles, C. Vauthier, D. Bazile, J. Vacus, J. L. Grossiord, M. Veillard and P. Couvreur, Langmuir, 2003, 19(22), 9504–9510 CrossRef CAS.
- A. Pawar and P. Prabhu, Biomed. Pharmacother., 2019, 110, 319–341 CrossRef CAS.
- J. Liu, Z. Qiu, S. Wang, L. Zhou and S. Zhang, Biomed. Mater., 2010, 5(6), 065002 CrossRef.
- S. Mander, D. J. You, S. Park, D. H. Kim, H. J. Yong, D. S. Kim, C. Ahn, Y. H. Kim, J. Y. Seong and J. I. Hwang, Arch. Pharmacal Res., 2018, 41(2), 229–242 CrossRef CAS.
- D. Wang, Y. S. Yang, L. G. Jiang, Y. Wang, J. Y. Li, P. A. Andreasen, Z. Chen, M. D. Huang and P. Xu, J. Med. Chem., 2019, 62(4), 2172–2183 CrossRef CAS.
- S. M. Pulukuri, C. S. Gondi, S. S. Lakka, A. Jutla, N. Estes, M. Gujrati and J. S. Rao, J. Biol. Chem., 2005, 280(43), 36529–36540 CrossRef CAS.
- H. Raghu, C. S. Gondi, D. H. Dinh, M. Gujrati and J. S. Rao, Mol. Cancer, 2011, 10, 130 CrossRef CAS.
- R. Subramanian, C. S. Gondi, S. S. Lakka, A. Jutla and J. S. Rao, Int. J. Oncol., 2006, 28(4), 831–839 CAS.
- D. Alfano, G. Votta, A. Schulze, J. Downward, M. Caputi, M. P. Stoppelli and I. Iaccarino, Mol. Cell. Biol., 2010, 30(7), 1838–1851 CrossRef CAS.
- A. Moirangthem, B. Bondhopadhyay, M. Mukherjee, A. Bandyopadhyay, N. Mukherjee, K. Konar, S. Bhattacharya and A. Basu, Sci. Rep., 2016, 6, 21903 CrossRef CAS.
- L. X. Yan, Q. N. Wu, Y. Zhang, Y. Y. Li, D. Z. Liao, J. H. Hou, J. Fu, M. S. Zeng, J. P. Yun, Q. L. Wu, Y. X. Zeng and J. Y. Shao, Breast Cancer Res., 2011, 13(1), R2 CrossRef CAS.
- H. Y. Huang, Z. F. Jiang, Q. X. Li, J. Y. Liu, T. Wang, R. Zhang, J. Zhao, Y. M. Xu, W. Bao, Y. Zhang, L. T. Jia and A. G. Yang, Cancer Invest., 2010, 28(7), 689–697 CrossRef CAS.
- V. Pavet, Y. Shlyakhtina, T. He, D. G. Ceschin, P. Kohonen, M. Perala, O. Kallioniemi and H. Gronemeyer, Cell Death Discovery, 2014, 5, e1043 CrossRef CAS.
- G. Du, H. Lin, M. Wang, S. Zhang, X. Wu, L. Lu, L. Ji and L. Yu, Cancer Chemother. Pharmacol., 2010, 65(2), 277–287 CrossRef CAS.
- X. Tang, Y. Liang, X. Feng, R. Zhang, X. Jin and L. Sun, Mater. Sci. Eng., C, 2015, 49, 348–355 CrossRef CAS.
- W. Tao, X. Zeng, T. Liu, Z. Wang, Q. Xiong, C. Ouyang, L. Huang and L. Mei, Acta Biomater., 2013, 9(11), 8910–8920 CrossRef CAS.
- L. Mu and S. S. Feng, J. Controlled Release, 2003, 86(1), 33–48 CrossRef CAS.
- R. H. Gaonkar, S. Ganguly, S. Dewanjee, S. Sinha, A. Gupta, S. Ganguly, D. Chattopadhyay and M. Chatterjee Debnath, Sci. Rep., 2017, 7(1), 530 CrossRef.
- D. Meng, M. Lei, Y. Han, D. Zhao, X. Zhang, Y. Yang and R. Liu, OncoTargets Ther., 2018, 11, 7733–7743 CrossRef CAS.
- M. C. Huber, R. Mall, H. Braselmann, A. Feuchtinger, S. Molatore, K. Lindner, A. Walch, E. Gross, M. Schmitt, N. Falkenberg and M. Aubele, BMC Cancer, 2016, 16, 615 CrossRef.
- S. Sperl, U. Jacob, N. A. de Prada, J. Sturzebecher, O. G. Wilhelm, W. Bode, V. Magdolen, R. Huber and L. Moroder, Proc. Natl. Acad. Sci. U. S. A., 2000, 97(10), 5113–5118 CrossRef CAS.
- E. Krissinel and K. Henrick, J. Mol. Biol., 2007, 372(3), 774–797 CrossRef CAS.
- G. Du, H. Lin, Y. Yang, S. Zhang, X. Wu, M. Wang, L. Ji, L. Lu, L. Yu and G. Han, Int. Immunopharmacol., 2010, 10(7), 819–826 CrossRef CAS.
- J. Bieger, R. Cermak, R. Blank, V. C. de Boer, P. C. Hollman, J. Kamphues and S. Wolffram, J. Nutr., 2008, 138(8), 1417–1420 CrossRef CAS.
- W. Gang, W. J. Jie, Z. L. Ping, S. Ming du, L. J. Ying, W. Lei and Y. Fang, Expert Opin. Drug Delivery, 2012, 9(6), 599–613 CrossRef.
- L. Liu, Y. Tang, C. Gao, Y. Li, S. Chen, T. Xiong, J. Li, M. Du, Z. Gong, H. Chen, L. Liu and P. Yao, Colloids Surf., B, 2014, 115, 125–131 CrossRef CAS.
- K. A. Khaled, Y. M. El-Sayed and B. M. Al-Hadiya, Drug Dev. Ind. Pharm., 2003, 29(4), 397–403 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra04231e |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2020 |