Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
14-Residue peptaibol velutibol A from Trichoderma velutinum: its structural and cytotoxic evaluation†
Varun Pratap Singhab,
Anup Singh Pathaniac,
Manoj Kushwahad,
Samsher Singhe,
Vandana Sharmadg,
Fayaz A. Malikc,
Inshad A. Khanef,
Anil Kumarb,
Deepika Singh *ad and
Ram A. Vishwakarma*a
*ad and
Ram A. Vishwakarma*a
aMedicinal Chemistry Division, CSIR-Indian Institute of Integrative Medicine, Canal Road, Jammu 180 001, India. E-mail: dsingh@iiim.res.in; deeps.csir@gmail.com; ram@iiim.res.in
bDepartment of Biotechnology, Faculty of Sciences, Shri Mata Vaishno Devi University, Katra, Jammu and Kashmir 182320, India
cPharmacology Division, CSIR-Indian Institute of Integrative Medicine, Canal Road, Jammu 180 001, India
dQuality Control & Quality Assurance Division, CSIR-Indian Institute of Integrative Medicine, Canal Road, Jammu 180 001, India
eClinical Microbiology Division, CSIR-Indian Institute of Integrative Medicine, Canal Road, Jammu 180 001, India
fDepartment of Microbiology, Central University, Rajasthan-305 817, India
gAcademy of Scientific and Innovative Research, Jammu 180001, India
First published on 24th August 2020
Abstract
Velutibol A (1), a new 14-residue peptaibol was isolated from the Himalayan cold habitat fungus Trichoderma velutinum. The structural characterization was carried out by 1D and 2D NMR studies, and tandem mass studies, and Marfey's method aided in determining the stereochemistry of the amino acids. The CD analysis revealed folding of the peptide in a 310-helical conformation. The intramolecular H-bonding was determined by an NMR-VT experiment. Cytotoxic evaluation was carried out against a panel of cancer cell lines. The cell cycle assay was carried out on human myeloid leukaemia (HL-60) cells and revealed the formation of apoptotic bodies and DNA damage in a dose-dependent manner. Three other peptaibols namely velutibol B (2), velutibol C (3), and velutibol D (4) were also isolated in trace amounts from the psychotropic fungus and characterized through tandem mass spectroscopy and Marfey's analysis.
Introduction
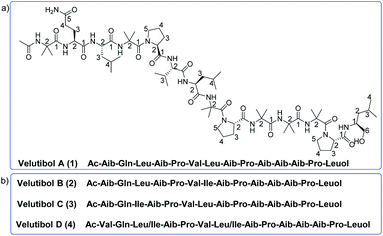
Peptaibols belong to the class of non-ribosomal peptides comprising non-proteinogenic amino acid residues including α-aminoisobutyric acid (Aib), ethyl norvaline, isovaline, hydroxylproline etc. The N-terminus in these peptides is acetylated and the C-terminus residue is an amino alcohol.1–7 They commonly exhibit various biological activities owing to their ion-channel formation,8–10 and membranolytic11 properties in modifying biological membranes. This includes activities like cytotoxic,2,12 neuroleptic,13–15 anthelmintic,16 hemolytic,17 anti-tubercular,18 antifungal,19–21,56 and antibacterial20–22 activity and has also shown significant effects against various phytopathogens.23,24 Their amino acid length varies from 5–20 amino acids which are classified into 9 subfamilies (SF1–SF9) depending upon their sequence, substitution pattern, and structure.8 Among these subfamilies, the SF1 (18–20 residues), SF4 (11 or 14 residues), SF5 (lipopeptaibols) and SF9 (7 residues) group of peptaibols are produced mainly from fungal genus Trichoderma.25 With more than 250 species of fungal genus Trichoderma,26,27 the majority of them are peptaibol producers,28 which include strains obtained from soil,4,29 and endophytic,30 and marine sources.31,32 Recently, we have reported lipopeptaibols from fungus T. velutinum from the soils of Himalayan cold habitat.33 There are almost 1000 peptaibols reported to date.28,34,35 They display microheterogeneity in character i.e. containing closely related peptides with limited sequence variation in the natural mixture.36,37 Herein, we found few 14-residue peptaibols of SF4 group with very little sequence variation. To the best of our knowledge, there are more than 70 sequences of peptaibols with 14 residues are reported belonging mainly to SF4 group except bergofungin D8,38 a peptaibol of SF2 group. All the 14 residue peptaibols of SF4 group contains three Aib-LPro dipeptide motifs at 4th, 8th and 12th positions with no aromatic amino acid in the sequence39–44 whereas the SF2 group does not have the Aib-LPro dipeptide motifs but contains phenylalaninol at C-terminus.8 These peptaibols with rich Aib motifs have been reported to fold into α-helix,45 310-helix46,47 and β-bend ribbon spiral conformation.48 These 14-residue peptaibols are reported to have antibacterial,38,39 antifungal,39 and amyloid β-peptide formation inhibitor41 activities.In our previous report, four lipopeptaibols namely lipovelutibols A–D from fungus T. velutinum were isolated from soils of Himalayan cold habitat.33 In continuing the exploration of peptaibiotics from T. velutinum, we herein, report isolation, structure elucidation, conformational analysis along with cytotoxic and anti-tubercular evaluation of peptaibol velutibol A (1) (Fig. 1a). As per our knowledge, this study represents the first report of 14-residue peptaibol having cytotoxic and weak anti-tubercular activity. In addition to 1, we herein, also provide probable structures of three other 14-residue peptaibols namely velutibol B–D (2–4) as shown in Fig. 1b, isolated in a trace amount and thus characterized through HRESIMS/MS studies and Marfey's analysis using LCMS.
Results and discussion
The chromatographic purification of the ethyl acetate extract was carried out through RP-HPLC, which gave 15 fractions (Fig. S1, ESI†). The re-purification of fraction-6, 7, 8 and 10 gave four lipopeptaibols as described previously.33 The Fr-6 and Fr-7 also contained mixtures of peptides with higher m/z values, which were further purified through RP-HPLC to get the peptaibols (1–4) as discussed in the Experimental section (Fig. S2 and S3, ESI†).The High Resolution Mass Spectroscopy (HRMS) of velutibol A (1) gave the molecular formula as C69H119N15O16 showing m/z 1414.9031 [M + H]+ (calcd 1414.9032) and 1436.8857 [M + Na]+ (calcd 1436.8851) (Fig. S4, ESI†) indicating eighteen double bond equivalents. The 1D proton NMR spectrum showed 13 NH protons in the region ranging at δH 6.5–9.5 ppm, 8 α-H ranging at δH 3.7–4.5 ppm and presence of acetyl protons at δH 1.9 ppm. It also showed the presence of 36 protons signals for 12 methyls in region δH 1.2–1.5 ppm and 24 protons signals corresponding to 8 methyls at δH 0.7–1.0 ppm. The close observation of 1D carbon NMR experiments including 13C and DEPT spectra revealed 69 carbon signals, which were assigned for 15 carbonyls, 6 carbons (α,α-disubstituted carbon), 12 methines (distinguished in DEPT-90), 15 methylene (including 3 methylene merged in DMSO signal as observed in DEPT-135) and 21 methyl carbons (including signal at δC 23.66 ppm for 2 carbons and one methyl carbon at δC 25.61 ppm as observed in DEPT-135) (Table 1) (Fig. S5–S8, ESI†).
| Residue | Type | 13C (δ) | 1H (δ), (mult. J in Hz) | Residue | Type | 13C (δ) | 1H (δ), (mult. J in Hz) | ||
|---|---|---|---|---|---|---|---|---|---|
| a From DEPT-135.b Signal overlap. | |||||||||
| Ac | 1 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
171.2 | — | 5 | CH3 | 21.5 | 0.79 (m)b | |
| 2 | CH3 | 22.9 | 1.90, (s) | 6 | CH3 | 22.8 | 0.85 (d, 6.8) | ||
| Aib-1 | 1 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
176.3 | — | NH | 7.21 (d, 8.4) | |||
| 2 | C | 55.9 | — | Aib-8 | 1 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
172.1 | ||
| 3 | CH3 | 23.6 | 1.34 (s) | 2 | C | 55.6 | |||
| 4 | CH3 | 26.5 | 1.36 (s)b | 3 | CH3 | 23.7b | 1.40 (s) | ||
| NH | 8.73 (s) | 4 | CH3 | 25.6 | 1.29 (s) | ||||
| Gln-2 | 1 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
172.6 | NH | 7.63 (s) | ||||
| 2 | CH | 55.0 | 3.96 (m) | Pro-9 | 1 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
172.9 | ||
| 3 | CH2 | 25.0 | 1.97 (m) | 2 | CH | 63.2 | 4.13 (t, 8.4) | ||
| 4 | CH2 | 31.1 | 2.29 (m), 2.18 (m) | 3 | CH2 | 28.5 | 2.20(m), 1.61 (m) | ||
| 5 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
174.5 | 4 | CH2 | 25.1 | 1.86 (m), 1.74(m) | |||
| NH2 | 7.48 (br s), 6.95 (br s) | 5 | CH2 | 48.1 | 3.68(m), 3.58 (m) | ||||
| NH | 8.93 (d, 5.2) | Aib-10 | 1 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
173.6 | ||||
| Leu-3 | 1 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
171.7 | 2 | C | 56.1 | |||
| 2 | CH | 51.2 | 4.18 (m) | 3 | CH3 | 25.6a | 1.36 (s)b | ||
| 3 | CH2 | 39.1a | 1.78 (m), 1.47 (m) | 4 | CH3 | 23.6 | 1.36 (s)b | ||
| 4 | CH | 24.2 | 1.57 (m) | NH | 7.84 (s) | ||||
| 5 | CH3 | 22.8 | 0.89 (d, 6.8) | Aib-11 | 1 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
175.5 | ||
| 6 | CH3 | 20.7 | 0.79 (m)b | 2 | C | 56.0 | |||
| NH | 7.88 (d, 8.4) | 3 | CH3 | 26.5 | 1.36 (s)b | ||||
| Aib-4 | 1 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
173.6 | 4 | CH3 | 23.6 | 1.44 (s) | ||
| 2 | C | 55.8 | NH | 7.34 (s) | |||||
| 3 | CH3 | 23.4 | 1.49 (s) | Aib-12 | 1 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
171.6 | ||
| 4 | CH3 | 25.4 | 1.36 (s)b | 2 | C | 55.7 | |||
| NH | 7.93 (s) | 3 | CH3 | 23.7b | 1.39 (s) | ||||
| Pro-5 | 1 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
172.7 | 4 | CH3 | 25.0 | 1.36 (s)b | ||
| 2 | CH | 63.1 | 4.23 (m) | NH | 7.58 (s) | ||||
| 3 | CH2 | 28.7 | 2.18 (m), 2.08 (m) | Pro-13 | 1 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
170.9 | ||
| 4 | CH2 | 25.4a | 1.86 (m), 1.74 (m) | 2 | CH | 61.9 | 4.23 (m) | ||
| 5 | CH2 | 48.4 | 3.68 (m), 3.58 (m) | 3 | CH2 | 28.1 | 2.18(m), 2.08 (m) | ||
| Val-6 | 1 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
171.4 | 4 | CH2 | 25.6a | 1.86(m), 1.74 (m) | ||
| 2 | CH | 60.1 | 3.85 (t, 8.0) | 5 | CH2 | 48.4 | 3.68(m), 3.58 (m) | ||
| 3 | CH | 28.8 | 2.18 (m) | Leuol | 1 | CH | 48.4 | 3.77 (m) | |
| 4 | CH3 | 19.0 | 0.97 (d, 6.4) | 2 | CH2 | 39.3a | 1.37 (m) | ||
| 5 | CH3 | 18.9 | 0.91 (d, 6.8) | 3 | CH | 23.8 | 1.67 (m) | ||
| NH | 7.28 (d, 8.4) | 4 | CH3 | 22.8 | 0.82 (m)b | ||||
| Leu-7 | 1 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
172.2 | 5 | CH3 | 20.2 | 0.79 (m)b | ||
| 2 | CH | 51.2 | 4.29 (m) | 6 | CH2OH | 63.9 | 3.27(m), 3.18 (m) | ||
| 3 | CH2 | 39.6a | 1.54 (m) | NH | 7.15 (d, 9.6) | ||||
| 4 | CH | 24.4 | 1.67 (m) | OH | 4.23 (m) | ||||
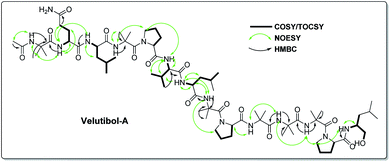
The detailed examination of 1D along with 2D experiments such as COSY, TOCSY, NOESY and HMBC data elucidated the structure as 1, which comprises 13 amino acids viz. six α-aminoisobutyric acids (Aib), two leucines (Leu), one valine (Val), three prolines (Pro) and one glutamine (Gln) and an amino alcohol leucinol (Leuol) at the C-terminus. The 14-residue sequence was found to have acetylation at the N-terminus (Fig. 2). For instance, the 2D experiments such as COSY and TOCSY showed three Pro, two Leu, one Leuol, and one Val moieties in the molecule. The amino acid Gln was recognized by the presence of correlation between two NHs having broad singlets at δH 6.95 and 7.48 in COSY and TOCSY experiments along with the correlations obtained from α-H, β-H2, and γ-H2. The presence of six Aib was revealed by HMBC correlations as shown in Fig. 2 (Fig. S9–S13, ESI†).
The peptide primary sequence was determined using NOESY correlations. The NOE experiment revealed acetyl moiety at N-terminus of Aib-1 i.e. α-H3 (δH 1.90 acetyl)/NH-1 (δH 8.73, Aib-1). Thus, the sequential assignment was observed through β-H3 (δH 1.36, Aib-1)/NH-2 (δH 8.93, Gln-2), NH-2/NH-3 (δH 7.88, Leu-3), NH-4 (δH 7.93, Aib-4)/δ-H2-5 (δH 3.68, 3.58, Pro-5), δ-H2-5/NH-5 (δH 7.28, Val-6), β-H-6 (δH 2.18, Val-6)/NH-6 (δH 7.21, Leu-7), NH-6/NH-7 (δH 7.63, Aib-8), NH-8/δ-H2-9 (δH 3.68, 3.58, Pro-9), α-H-9 (δH 4.13, Pro-9)/NH-9 (δH 7.84, Aib-10), NH-9/NH-10 (δH 7.34, Aib-11), NH-10/NH-11 (δH 7.58, Aib-12), NH-11/δ-H2-13 (δH 3.68, 3.58, Pro-13), δ-H2-13/NH-12 (δH 7.15, Leuol). These observations along with HMBC correlations as shown in Fig. 2 (Tables S1 and S2, ESI†) gave the primary sequence of 1 as Ac-Aib-Gln-Leu-Aib-Pro-Val-Leu-Aib-Pro-Aib-Aib-Aib-Pro-Leuol. The MS/MS studies for [M + H]+at m/z 1414.90 showed prominent b-ions at m/z 1200.74 (b12), 848.52 (b8) and 454.26 (b4), an indication of easily labile Aib–Pro bond in ESI fragmentation, along with other b-ions at 1115.68 (b11), 1030.62 (b10), 763.47 (b7) and 369.22 (b3). Further, MS/MS of 1200.74 (b12) gave daughter ions at 650.39 (b6), 551.31 (b5) and 256.13 (b2). The daughter ion 128.07 (b1) was observed via MS/MS of 848.53 (b8) (Fig. 3). The MS/MS data in combination with the NOESY and HMBC correlations established the sequence of compound 1 as Ac-Aib-Gln-Leu-Aib-Pro-Val-Leu-Aib-Pro-Aib-Aib-Aib-Pro-Leucinol.
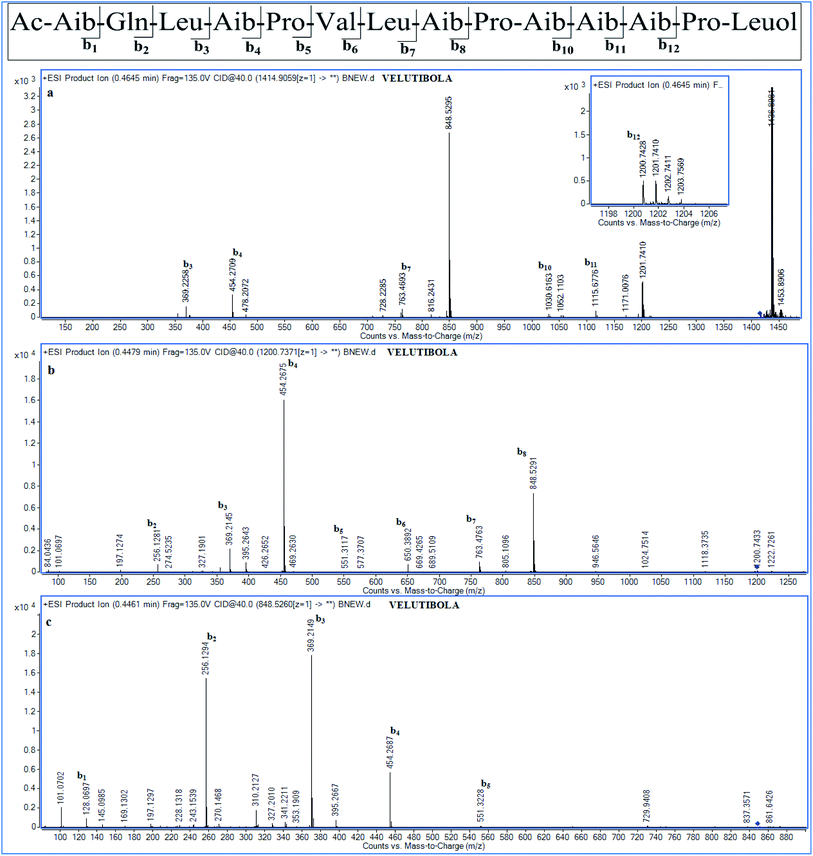 | ||
| Fig. 3 MSn studies of compound 1 (a) MS2 at m/z 1414.9059 [M + H]+; (b) MS3 for daughter ion b12 at m/z 1200.7371 (c) MS3 for daughter ion b8 at m/z 848.526. | ||
The advanced Marfey's method49 was used for the determination of stereochemistry. Acid hydrolysis of the compound 1 was done and derivatized with 1-fluoro-2,4-dinitrophenyl-5-L-alaninamide (LFDAA). Likewise, both the D- and L-amino acid standards were derivatized with LFDAA. The amino acid glutamine gets converted into glutamic acid on acid hydrolysis,35 thus LGlu and DGlu was derivatized for stereochemical determination of Gln. The amino acid standards thus obtained were compared with L-FDAA derivatives of acid hydrolyzed compounds using LCMS instrument. The retention times for standard amino acid-LFDAA derivatives were found to be LGlu/DGlu (21.0/21.8), LPro/DPro (23.6/24.4), LVal/DVal (26.9/29.5), LLeu/DLeu (29.8/32.3) and LLeuol/DLeuol (29.4/32.6) and for the acid hydrolyzed-LFDAA derivatives were 21.0, 23.5, 27.0, 29.4 and 29.8 for LGlu/LGln, LPro, LVal, LLeuol, and LLeu respectively. Thus, the advanced Marfey's analysis carried out for 1 indicated the L-form of Glu/Gln, Leu, Val, Pro and Leuol (Fig. S14, ESI†).
As the compounds velutibol B–D (2–4) were isolated in very low amounts (0.3–0.1 mg) the structural characterization could not be established through NMR. The characterization of velutibol B–D (2–4) was established through tandem mass spectrometry utilizing HRMS. The HRMS of 2 and 3 gave the molecular formula as C69H119N15O16 showing m/z 1414.9060 [M + H]+ and 1414.9022 [M + H]+ respectively (calcd 1414.9032). The probable primary sequence of these peptides has been given in Table 2. The compound 2 and 3 have similar m/z values as that of 1. The sequence of 2 was elucidated by tandem mass spectroscopy. The MS/MS scan of 1414.903 [M + H]+ which gave b-ions at m/z 256.13 (b2), 369.21 (b3), 454.27 (b4), 551.31 (b5), 650.38 (b6), 763.47 (b7), 848.52 (b8), 945.57 (b9), 1030.63 (b10), 1115.67 (b11), and 1200.73 (b12). The b1 ion (m/z 128.07) was found from the MS/MS scan for the peak of m/z 454.27 (Fig. S20a and b, ESI†). This gave the sequence as Ac-Aib-Gln-Lxx-Aib-Pro-Vxx-Lxx-Aib-Pro-Aib-Aib-Aib-Pro-Lxxol. The similar sequence was observed for compound 3. The daughter ions obtained from tandem mass spectroscopy of [M + H]+ ion of 3 are given in Table 2 (Fig. S23a–c, ESI†). The molecular formula for compound 4 was found to be C70H121N15O16 showing m/z 1428.9188 [M + H (calcd 1428.9188). The tandem mass studies for compound 4 revealed a difference of 14 Da for b1 ion as m/z of 142.09 corresponding for Ac-Vxx moiety. The daughter ions obtained from the fragmentation of compound 4 having m/z 1428.9191 [M + H]+ were 468.28 (b4), 862.54 (b8), 1044.65 (b10), 1129.71 (b11), and 1214.75 (b12). Further, MS/MS for m/z 1214.7532 (b12) gave daughter ions as 142.09 (b1), 270.14 (b2), 383.23 (b3), 565.34 (b5), 644.40 (b6), 777.49 (b7) and 969.60 (b9) along with other b ions (Fig. S26a and b, ESI†). This gave the sequence as Ac-Vxx-Gln-Lxx-Aib-Pro-Vxx-Lxx-Aib-Pro-Aib-Aib-Aib-Pro-Lxxol. These compounds were eluted at different retention times during purification and subsequently showed different retention times on further HPLC analysis (Fig. S27, ESI†), which reflected that these compounds were different with similar m/z values.
| Compounds | Sequence | m/za | tR (min) | Respective b-ions |
|---|---|---|---|---|
| a HRMS observed values for [M + H]+ and [M + Na]+, tR = retention time. | ||||
| Velutibol B (2) | Ac-Aib-Gln-Lxx-Aib-Pro-Vxx-Lxx-Aib-Pro-Aib-Aib-Aib-Pro-Lxxol | 1414.9060, 1436.8882 | 43.5 | 1200.73, 1115.67, 1030.63, 945.57, 848.52, 763.47, 650.38, 551.31, 454.27, 369.21, 256.13 and 128.07 |
| Velutibol C (3) | Ac-Aib-Gln-Lxx-Aib-Pro-Vxx-Lxx-Aib-Pro-Aib-Aib-Aib-Pro-Lxxol | 1414.9022, 1436.8835 | 44.3 | 1200.74, 1115.68, 1030.62, 848.52, 763.47, 650.38, 551.31, 454.27, 369.21, 256.13 and 128.07 |
| Velutibol D (4) | Ac-Vxx-Gln-Lxx-Aib-Pro-Vxx-Lxx-Aib-Pro-Aib-Aib-Aib-Pro-Lxxol | 1428.9188, 1450.9013 | 45.0 | 1214.75, 1129.71, 1044.65, 959.60, 862.54, 777.49, 644.40, 565.34, 468.28, 383.23, 270.14 and 142.09 |
The Marfey's analysis of the compounds 2–4 were also carried at a scale of 20 μg owing to their low quantities. It revealed presence of LGln/LGlu, LPro, LVal, LIle, LLeu and LLeuol. The extracted ion chromatogram (EIC) for m/z 368.0, 370.0, 384.0 and 400.0 gave one, two, two and one peaks respectively for compound 2–4. The respective amino acid derivatives were found to be in-line with the EIC obtained for compounds 2–4 (Fig. S28–S33, ESI†). The amino acid LIle was distinguished from L-allo-isoleucine (Lallo-Ile) via using chiral LCMS. The three peptaibols studied here were found to have LIle in their respective sequence (Fig. S34, ESI†). Moreover, the compound 2 and 3 having similar primary sequence with same amino acids asserted that the amino acid Ile and Leu were probably exchanged in respective position. The close observation of MS/MS for 2 (Fig. S20b, ESI†) gave a m/z 327.20 for fragmentation of –CH(CH3)2 corresponding to Leu-3 similarly a m/z 341.22 for fragment –CH2CH3 corresponding to Ile-3 for 3 (Fig. S23b, ESI†). Thus, combining Marfey's analysis with tandem mass spectral analysis gave the sequence of 2, 3 and 4 as Ac-Aib-Gln-Leu-Aib-Pro-Val-Ile-Aib-Pro-Aib-Aib-Aib-Pro-Leuol, Ac-Aib-Gln-Ile-Aib-Pro-Val-Leu-Aib-Pro-Aib-Aib-Aib-Pro-Leuol and Ac-Val-Gln-Leu/Ile-Aib-Pro-Val-Leu/Ile-Aib-Pro-Aib-Aib-Aib-Pro-Leuol respectively.
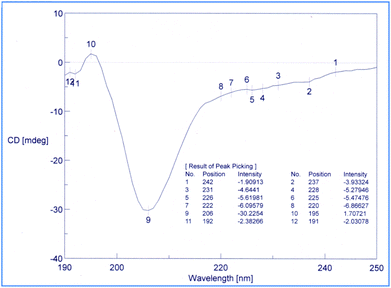
Conformational studies were performed through CD technique50,51 and NMR-VT (variable temperature).52,53 The CD spectra of the 1 in methanol were analyzed as shown in Fig. 4. The shapes of the CD pattern showed a −ive maxima at 206 nm and a −ive shoulder at 220 nm (in the vicinity of 222 nm). The ellipticity ratio R [θT220]/[θT206] was found to be 0.23. This clearly indicated that the compound 1 folds in a 310-helix in methanol as the value for R was less than 0.50, which is known for the high propensity of 310-helix.
The NMR-VT examined the manner in which the NH groups in the peptide behaved as a function of temperature.52,53 1H NMR of peptaibol 1 was recorded at four different temperatures i.e. 298 K, 308 K, 318 K, 328 K in DMSO-d6. Out of the thirteen NH protons in the putative peptide, the chemical shifts of NH-1, NH-2, NH-5 along with amide NH2 of Gln (NH-8 and NH-13) were sensitive to heating while the other NH groups had shown very little change in chemical shift values as shown in Fig. 5. On the basis of these 1H NMR-VT experiment results (Fig. S35, ESI†), it was evident that NH-3 (Leu-3), NH-4 (Aib-4), NH-6 (Leu-7), NH-7 (Aib-8), NH-9 (Aib-10), NH-10 (Aib-11), NH-11 (Aib-12), and NH-12 (Leuol) were involved in intramolecular hydrogen bonding in peptaibol 1.
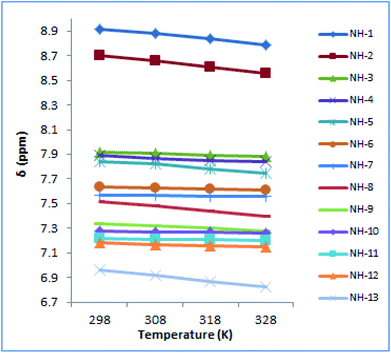 | ||
| Fig. 5 NMR-VT experiment performed for compound 1 at four different temperatures i.e. 298 K, 308 K, 318 K and 328 K. | ||
The cytotoxicity testing of compound 1 was performed against various cell lines, which includes human lung cancer (A549), human colon cancer (LS-180), human myeloid leukemia (HL-60) and human breast cancer (MDA-MB-231) using MTT (3-(4,5-dimethylthiazole-2-yl)-2,5-diphenyltetrazolium bromide). Herein, paclitaxel was used as positive control for cytotoxic assay. The MTT assay showed inhibitory activity of compound 1 against MDA-MD-231 and HL-60 cancer cell lines having IC50 values of 7 μM and 4 μM respectively (Table 3). The IC50 for positive control was found to be in range of 2.0 to 6.8 nM against four cancer cell lines tested. Further, the cell-cycle assay was performed on HL-60 cell lines. The compound 1 was treated against HL-60 cells at concentrations 5, 10, 20 and 30 μM and incubated for 24 h. The compound 1 showed DNA damage in a dose-dependent manner. The HL-60 cells, with no treatment, showed non-significant 2% apoptotic DNA whereas the cells treated with 1 showed 70%, 85% and 99% apoptotic DNA population at 10, 20 and 30 μM concentrations respectively (Fig. 6a). Apoptotic bodies are small DNA fragments wrapped around the single membrane vesicles mainly formed when cells undergo apoptosis. These apoptotic bodies can be visualized under a fluorescent microscope by using DNA staining dyes such as 4′,6-diamidino-2-phenylindole (DAPI). The HL-60 cells were incubated with the compound 1 at different concentrations (5, 10, 20 and 30 μM) for 24 h. The apoptotic bodies (indicated by white arrows in Fig. 6b) were seen in cells treated with tested compound whereas healthy and round nuclei with no DNA fragmentation was observed with the untreated cells. Formation of apoptotic bodies increased in a dose-dependent manner. Similarly, the phase contrast microscopy of cells treated with 1 showed bled and distorted cells while the control cells were observed as healthy (Fig. 6c). The % apoptosis is also represented graphically in Fig. 7.
| IC50a ± standard deviation (μM) | ||||
|---|---|---|---|---|
| A549 | LS-180 | HL-60 | MDA-MB-231 | |
| a IC50 = half maximal inhibitory concentration. The experiments were conducted three times and data represented as mean ± standard deviation, with p value less than 0.05 calculated using Student t-test. | ||||
| 1 | 23 ± 0.8 | 26 ± 1.1 | 4 ± 0.1 | 7 ± 0.2 |
| Paclitaxel (nM) | 6.2 ± 0.1 | 2.2 ± 0.1 | 1.8 ± 0.1 | 3.3 ± 0.1 |
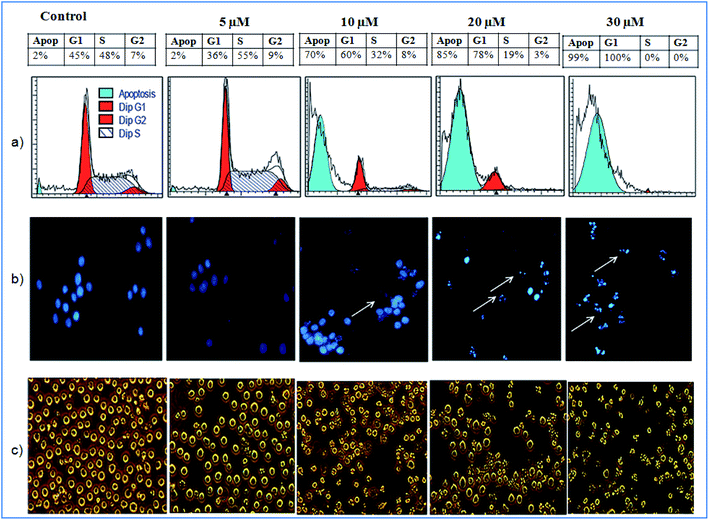 | ||
| Fig. 6 Effect of 1 (a) on cell cycle phase distribution; (b) on the morphology and (c) on the nuclear morphology of HL-60 cells. | ||
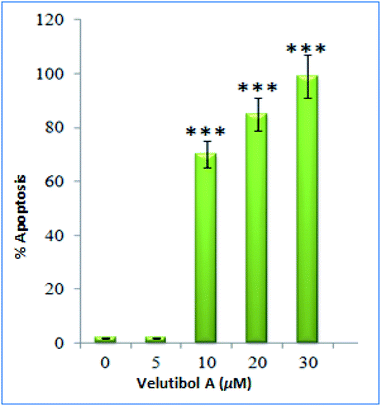 | ||
| Fig. 7 Graphical representation of cells undergoing apoptosis after treatment with compound 1. Columns, mean; bars, SD with ***p < 0.001 versus control. | ||
The compound 1 was also tested for anti-tubercular potential. The Minimum Inhibitory Concentration (MIC) of compound 1 was found to be 32 μg mL−1 against Mycobacterium tuberculosis. Rifampicin was used as a positive control with MIC 0.06 μg mL−1 (Fig. S36, ESI†).
Experimental
Materials
The chemicals used in the study such as Marfey's reagent (LFDAA), all the standard L and D-amino acids (except Lallo-Ile), propidium iodide (PI), RPMI-1640, 4′,6-diamidino-2-phenylindole (DAPI), and 3-(4,5-dimethylthiazole-2-yl)-2,5 diphenyltetrazolium bromide (MTT) were purchased from Sigma-Aldrich, USA. The amino acid Lallo-Ile was purchased from TCI Chemicals, Japan. The Middlebrook 7H9 broth was purchased from Difco Laboratories, Detroit, MI, USA, Tween 80 from Himedia, Mumbai, India and albumin dextrose catalase (ADC) from Becton Dickinson, Sparks, MD, USA. The HPLC grade solvents such as water, methanol and acetonitrile were purchased from Merck-India. The HL-60 cancer cell lines were purchased from ECACC, England. The cells were grown at 37 °C with 95% humidity in a CO2 incubator (Thermocon Electron Corporation, Houston, TX) in RPMI-1640 growth medium supplemented with 10% FCS and 1× antibiotic/antimycotic solution obtained from Gibco, USA. The M. tuberculosis H37Rv (ATCC 27294) was obtained from American Type Culture Collection, Manassas, VA, USA. The FT-IR spectroscopy was carried out in chloroform on PerkinElmer instrument. The optical rotation of compound 1 was recorded on a Perkin-Elmer 241 polarimeter.Fungal cultivation
The isolation, microbial identification, characterization and cultivation for Trichoderma velutinum has been described in previous reports.33,54Isolation of compounds (1–4)
The semi-preparative HPLC purification was performed using reverse-phase (RP) column (Reprosil Gold C18, 10 × 250 mm, 5 μm, Dr Maisch GmbH) on ThermoFinnigan HPLC system. The purification details were same as provided in the previous report.33 The Fr-6 on further isolation gives three fractions (Fig. S2, ESI†), one was lipovelutibol A, the other two being velutibol A (1) and velutibol B (2) in 3.5 mg and 0.3 mg respectively. Similarly, Fr-7 gave two fractions, the first being lipovelutibol B, and the second fraction (0.4 mg) was re-purified through-HPLC using RP column (LiChrospher RP18e, 125 × 4 mm, 5 μm, Merck) on Shimadzu HPLC system through a gradient program comprising 0.1% TFA in water as mobile phase A and acetonitrile as mobile phase B. The method used was initiated with 10% B for 5 min then gradually increased to 60% B in next 30 min, kept at 60% B for 10 min, then gradually decreased to 10% B in 5 min and maintained at 10% B for next 10 min, with an overall run time of 60 min at 1 mL min−1 flow rate of and observed at 214 nm wavelength. This gave two fractions as velutibol C (3) (0.2 mg approx.) and velutibol D (4) (0.1 mg approx.) (Fig. S3, ESI†).HPLC purity analysis
The HPLC analysis was carried out on Shimadzu HPLC system coupled with a PDA detector, thermostat and quaternary pump using a RP column (LiChrospher RP18e, 5 μm, 250 × 4 mm, Merck). The mobile phase A comprises of 0.1% formic acid in water and mobile phase B as acetonitrile. The gradient program was started initially by keeping 10% B for 5 min, gradually increased from 10–60% B in next 30 min, remain at 60% B for next 10 min, followed by gradual decrease from 60–10% B in 5 min and finally remained at 10 for next 13 min with an overall run time of 63 min. It was detected at 214 nm with 1 mL min−1 flow rate.LCHRMS analysis
The HRMS and tandem mass spectroscopy was carried out on Agilent HRMS 6450 4HD Q-TOF Mass Spectrometer coupled with LC system. The instrument was maintained at capillary voltage 3500 V, source temperature 350 °C, 7.0 L min−1 gas flow and spray voltage of 4.5 KV with a resolution of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 were used for HRMS analysis. The tandem mass spectroscopy (MSn) was performed for the range of 100–2000 amu at fragmentor voltage at 135.0 V. 20 ions per cycle was selected for precursor ion with the minimum intensity of 5000 and collision induced dissociation (CID) at 30 and 40 V.
000 were used for HRMS analysis. The tandem mass spectroscopy (MSn) was performed for the range of 100–2000 amu at fragmentor voltage at 135.0 V. 20 ions per cycle was selected for precursor ion with the minimum intensity of 5000 and collision induced dissociation (CID) at 30 and 40 V.
NMR analysis
The 1D and 2D NMR experiments were recorded on a Bruker 400 MHz NMR instrument. The data was recorded in ppm scale using tetramethylsilane or residual NMR solvent resonance as a reference. All the 1D and 2D NMR spectra were processed via MestReNova. The NMR-VT experiment was carried out at Bruker 500 MHz spectrometer coupled with a thermostat. The data was acquired at four different temperatures i.e. at 298 K, 308 K, 318 K and 328 K.CD analysis
The circular dichroism (CD) experiment was performed on a Jasco J-1500 CD spectrophotometer using 2 mm cuvette. The solution of compound 1 in methanol was prepared at 0.1 mg mL−1 concentration. The spectrum was recorded from 260 to 180 nm with 50 nm min−1 scanning speed at a response time of 1 s and 1 nm bandwidth. The data obtained was accumulation of two scans.Cytotoxic assay (MTT-assay)
The cytotoxic effect of compound 1 on cancer cells was assayed by using MTT dye. The HL-60 cell lines were seeded for 4 h in round bottom 96 well plates. It was then treated with different concentrations of compound 1 dissolved in DMSO and incubated for 48 h. With 3 h earlier to experiment termination, the MTT dye (20 μL) from the stock solution of 2.5 mg mL−1 was added to each well. The MTT-formazan crystals thus formed were dissolved in DMSO (160 μL) and observed at 570 nm. The untreated control culture was treated with DMSO (<0.2%).Cell cycle analysis
The cell cycle phase distribution analysis in HL-60 cells was carried out by propidium iodide (PI) staining. The HL-60 cells were treated with different concentration (5, 10, 20 and 30 μM) of 1. After 24 h, the cells were centrifuged and collected at 400 × g and washed two times with PBS followed by fixing with ice-cold ethanol (70%). It was incubated overnight at 4 °C. Next day, cells were centrifuged at 4000 rpm, collected and washed with PBS followed by incubation with RNase (0.2 mg mL−1) for 90 min at 37 °C. It was then stained with PI (10 μg mL−1) and kept for 30 min in dark. The analysis of cells were carried out through flow-cytometer (FACS Calibur, Becton Dickinson) using list mode on 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events for FL2-A vs. FL2-W. The ModFit software was used for the calculation of fluorescence intensity of the sub-G1 cell fraction representing the apoptotic cell population.
000 events for FL2-A vs. FL2-W. The ModFit software was used for the calculation of fluorescence intensity of the sub-G1 cell fraction representing the apoptotic cell population.
Hoechst staining
HL-60 cells were incubated with compound 1 in a 12 well plate. After 24 h, cells were centrifuged at 400 × g, collected and washed two times with PBS. The cells were fixed using 1 mL solution of cold acetic acid in methanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) and incubated overnight at 4 °C. Further, the cells were centrifuged at 400 × g, collected and re-suspended in fixing solution (100 μL). Further, the cells were spread on a slide and dried for 4–6 h at room temperature. Slides were then stained with DAPI in PBS containing 0.05% Tween 20 (5 μg mL−1) and incubated for 30 min followed by washing with water and PBS. The slide covered by a coverslip was mounted with fluid containing 50 μL glycerol and PBS (1
3) and incubated overnight at 4 °C. Further, the cells were centrifuged at 400 × g, collected and re-suspended in fixing solution (100 μL). Further, the cells were spread on a slide and dried for 4–6 h at room temperature. Slides were then stained with DAPI in PBS containing 0.05% Tween 20 (5 μg mL−1) and incubated for 30 min followed by washing with water and PBS. The slide covered by a coverslip was mounted with fluid containing 50 μL glycerol and PBS (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and sealed with white nail polish. Nuclei were observed using microscope (20×) and photographed by Olympus IX70 microscope. The treated and untreated control cells were simply photographed using a microscope.
1) and sealed with white nail polish. Nuclei were observed using microscope (20×) and photographed by Olympus IX70 microscope. The treated and untreated control cells were simply photographed using a microscope.
Anti-tubercular assay
The MIC of compound 1 was determined by the micro-broth dilution method against M. tuberculosis H37Rv.55 The bacterial cells was grown in Middlebrook 7H9 broth containing glycerol (0.5%), Tween 80 (0.05%), and ADC (10%) under constant shaking for 10 to 15 days at 37 °C in 5% CO2. A suspension of M. tuberculosis cells was prepared in normal saline solution containing Tween 80 (0.5%). The turbidity of the resulting suspension was attuned by 1 McFarland (McF) standard (≈1.0 × 107 CFU mL−1). The bacterial suspension was further diluted 10 folds with the medium. The compound 1 was serially diluted in Middlebrook 7H9 to 2-folds in 100 μL per well in 96-well U bottom microtitre plates. A 100 μL of the diluted suspension was added to each well resulting in a final inoculum of 1.0 × 106 CFU mL−1 in the well, and the compound 1 was finally in range from 0.5 μg mL−1 to 128 μg mL−1. The plates were incubated at 37 °C for seven days in 5% CO2. The Resazurin Microtiter Assay (REMA) method was used for evaluation of the results. After incubation, resazurin (15 μL, 0.04%) and Tween 80 (12.5 μL, 20%) was added in each well of the plate. The plates were incubated for 48 h and were read visually to ascertain the MIC from the minimum concentration of the compound showing no change in color.Conclusions
Herein, we report isolation of four 14-residue peptaibols from a psychrotrophic fungus T. velutinum of the Himalayan cold habitat. The complete structural characterization of 1 was established by intensive 1D, 2D NMR, and tandem mass spectroscopic studies. Whereas, the probable primary sequence for compound 2–4 was established using MS/MS studies. The stereochemical determination by Marfey's analysis revealed L-form of the amino acids in these putative peptides. The conformational studies carried out using CD spectroscopy revealed that compound 1 folds in a 310-helix in methanol. The variable temperature experiment performed through NMR showed that NH-3 (Leu-3), NH-4 (Aib-4), NH-6 (Leu-7), NH-7 (Aib-8), NH-9 (Aib-10), NH-10 (Aib-11), NH-11 (Aib-12) and NH-12 (Leuol) were involved in intramolecular hydrogen bonding in peptaibol 1. These experiments also gave an insight into its secondary structure and may correlate to the biological activities asserted by 1. The cytotoxic assay showed compound 1 having IC50 values of 7 μM and 4 μM against MDA-MD-231 and HL-60 cancer cells respectively. Further, the cell cycle assay revealed DNA damage and formation of apoptotic bodies in a dose-dependent manner against HL-60 cancer cells. The compound 1 also showed weak MIC of 32 μg mL−1 against M. tuberculosis.Conflicts of interest
There are no conflicts to declare.Acknowledgements
VPS, ASP, MK and SS are grateful to CSIR for their Senior Research Fellowship (SRF). The support of Er. Rajneesh Anand and Mr Vikram Bhardwaj of Instrumentation Division is greatly acknowledged. Authors are indebted to Dr Asha Chaubey and Dr Richa Sharma from Fermentation division for providing the culture. The manuscript bears Institutional IPR reference number CSIR-IIIM/IPR/00116.Notes and references
- R. D. Sussmuth and A. Mainz, Angew. Chem., Int. Ed., 2017, 56, 3770–3821 CrossRef PubMed.
- D. Liu, H. Lin, P. Proksch, X. Tang, Z. Shao and W. Lin, Org. Lett., 2015, 17, 1220–1223 CrossRef CAS PubMed.
- W. H. Jiao, Z. Khalil, P. Dewapriya, A. A. Salim, H. W. Lin and R. J. Capon, J. Nat. Prod., 2018, 81, 976–984 CrossRef CAS PubMed.
- J. F. Daniel and E. R. Filho, Nat. Prod. Rep., 2007, 24, 1128–1141 RSC.
- R. Mikkola, M. A. Andersson, L. Kredics, P. A. Grigoriev, N. Sundell and M. S. Salkinoja-Salonen, FEBS J., 2012, 279, 4172–4190 CrossRef CAS PubMed.
- M. Y. Summers, F. Kong, X. Feng, M. M. Siegel, J. E. Janso, E. I. Graziani and G. T. Carter, J. Nat. Prod., 2007, 70, 391–396 CrossRef CAS PubMed.
- T. Degenkolb and H. Bruckner, Chem. Biodivers., 2008, 5, 1817–1843 CrossRef CAS PubMed.
- J. K. Chugh and B. A. Wallace, Biochem. Soc. Trans., 2001, 29, 565–570 CrossRef CAS PubMed.
- T. Salditt, C. Li and A. Spaar, Biochim. Biophys. Acta, 2006, 1758, 1483–1498 CrossRef CAS PubMed.
- M. S. P. Q. Sansom, Annu. Rev. Biophys., 1993, 26, 365–421 CAS.
- G. Gabernet, A. T. Müller, J. A. Hiss and G. Schneider, RSC Med. Chem, 2016, 7, 2232–2245 RSC.
- M. Shi, H. N. Wang, S. T. Xie, Y. Luo, C. Y. Sun, X. L. Chen and Y. Z. Zhang, Mol. Canc., 2010, 9, 26 CrossRef PubMed.
- M. Ritzau, S. Heinze, K. Dornberger, A. Berg, W. Fleck, B. Schlegel, A. Hartl and U. Grafe, J. Antibiot., 1997, 50, 722–728 CrossRef CAS PubMed.
- M. Kronen, P. Kleinwachter, B. Schlegel, A. Hartl and U. Grafe, J. Antibiot., 2001, 54, 175–178 CrossRef CAS PubMed.
- T. V. Ovchinnikova, N. G. Levitskaya, O. G. Voskresenskaya, Z. A. Yakimenko, A. A. Tagaev, A. Y. Ovchinnikova, A. N. Murashev and A. A. Kamenskii, Chem. Biodivers., 2007, 4, 1374–1387 CrossRef CAS PubMed.
- S. Ayers, B. M. Ehrmann, A. F. Adcock, D. J. Kroll, E. J. Carcache de Blanco, Q. Shen, S. M. Swanson, J. O. Falkinham III, M. C. Wani, S. M. Mitchell, C. J. Pearce and N. H. Oberlies, J. Pept. Sci., 2012, 18, 500–510 CrossRef CAS PubMed.
- M. Dathe, C. Kaduk, E. Tachikawa, M. F. Melzig, H. Wenschuh and M. Bienert, Biochim. Biophys. Acta, 1998, 1370, 175–183 CrossRef CAS.
- I. Kavianinia, L. Kunalingam, P. W. R. Harris, G. M. Cook and M. A. Brimble, Org. Lett., 2016, 18, 3878–3881 CrossRef CAS PubMed.
- I. Touati, N. Ruiz, O. Thomas, I. S. Druzhinina, L. Atanasova, O. Tabbene, S. Elkahoui, R. Benzekri, L. Bouslama, Y. F. Pouchus and F. Limam, World J. Microbiol. Biotechnol., 2018, 34, 98 CrossRef PubMed.
- D. Ishiyama, T. Satou, H. Senda, T. Fujimaki, R. Honda and S. Kanazawa, J. Antibiot., 2000, 53, 728–732 CrossRef CAS PubMed.
- M. Y. Summers, F. Kong, X. Feng, M. M. Siegel, J. E. Janso, E. I. Graziani and G. T. Carter, J. Nat. Prod., 2007, 70, 391–396 CrossRef CAS PubMed.
- C. Wilhelm, H. Anke, Y. Flores and O. Sterner, J. Nat. Prod., 2004, 67, 466–468 CrossRef CAS PubMed.
- A. Otto, A. Laub, L. Wendt, A. Porzel, J. Schmidt, G. Palfner, J. Becerra, D. Kruger, M. Stadler, L. Wessjohann, B. Westermann and N. Arnold, J. Nat. Prod., 2016, 79, 929–938 CrossRef CAS PubMed.
- M. Shi, L. Chen, X. W. Wang, T. Zhang, P. B. Zhao, X. Y. Song, C. Y. Sun, X. L. Chen, B. C. Zhou and Y. Z. Zhang, Microbiology, 2012, 158, 166–175 CrossRef CAS PubMed.
- T. Neuhof, R. Dieckmann, I. S. Druzhinina, C. P. Kubicek and H. von Dohren, Microbiology, 2007, 153, 3417–3437 CrossRef CAS PubMed.
- J. Bissett, W. Gams, W. Jaklitsch and G. J. Samuels, IMA Fungus, 2015, 6, 263–295 CrossRef PubMed.
- T. Marik, C. Tyagi, G. Racic, D. Rakk, A. Szekeres, C. Vagvolgyi and L. Kredics, Microorganisms, 2018, 6, 85 CrossRef CAS PubMed.
- N. K. Neumann, N. Stoppacher, S. Zeilinger, T. Degenkolb, H. Bruckner and R. Schuhmacher, Chem. Biodivers., 2015, 12, 743–751 CrossRef CAS PubMed.
- J. P. Brito, M. H. Ramada, M. T. de Magalhães, L. P. Silva and C. J. Ulhoa, SpringerPlus, 2014, 3, 600 CrossRef PubMed.
- L. Maddau, A. Cabras, A. Franceschini, B. T. Linaldeddu, S. Crobu, T. Roggio and D. Pagnozzi, Microbiology, 2009, 155, 3371–3381 CrossRef CAS PubMed.
- M. Mohamed-Benkada, M. Montagu, J.-F. Biard, F. Mondeguer, P. Verite, M. Dalgalarrondo, J. Bissett and Y. F. Pouchus, Rapid Commun. Mass Spectrom., 2006, 20, 1176–1180 CrossRef CAS PubMed.
- J. Ren, C. Xue, L. Tian, M. Xu, J. Chen, Z. Deng, P. Proksch and W. Lin, J. Nat. Prod., 2009, 72, 1036–1044 CrossRef CAS PubMed.
- V. P. Singh, N. Yedukondalu, V. Sharma, M. Kushwaha, R. Sharma, A. Chaubey, A. Kumar, D. Singh and R. A. Vishwakarma, J. Nat. Prod., 2018, 81, 219–226 CrossRef CAS PubMed.
- N. Stoppacher, N. K. Neumann, L. Burgstaller, S. Zeilinger, T. Degenkolb, H. Bruckner and R. Schuhmacher, Chem. Biodivers., 2013, 10, 734–743 CrossRef CAS PubMed.
- J. Rivera-Chávez, H. A. Raja, T. N. Graf, J. M. Gallagher, P. Metri, D. Xue, C. J. Pearce and N. H. Oberlies, RSC Adv., 2017, 7, 45733–45741 RSC.
- G. Pocsfalvi, A. Ritieni, P. Ferranti, G. Randazzo, K. Vekey and A. Malorni, Rapid Commun. Mass Spectrom., 1997, 11, 922–930 CrossRef CAS PubMed.
- G. Pócsfalvi, F. Scala, M. Lorito, A. Ritieni, G. Randazzo, P. Ferranti, K. Vékey and A. Malorni, J. Mass Spectrom., 1998, 33, 154–163 CrossRef.
- A. Berg, B. Schlegel, W. Ihn, U. Demuth and U. Grafe, J. Antibiot., 1999, 52, 666–669 CrossRef CAS PubMed.
- S. Rebuffat, C. Goulard and B. Bodo, J. Chem. Soc., Perkin Trans. 1, 1995, 1849–1855 RSC.
- G. Leclerc, S. Rebuffat and B. Bodo, J. Antibiot., 1998, 51, 178–183 CrossRef CAS PubMed.
- N. Hosotani, K. Kumagai, S. Honda, A. Ito, T. Shimatani and I. Saji, J. Antibiot., 2007, 60, 184–190 CrossRef CAS PubMed.
- T. Degenkolb, R. Dieckmann, K. F. Nielsen, T. Gräfenhan, C. Theis, D. Zafari, P. Chaverri, A. Ismaiel, H. Brückner, H. von Döhren, U. Thrane, O. Petrini and G. J. Samuels, Mycol. Prog., 2008, 7, 177–219 CrossRef.
- P. K. Mukherjee, A. Wiest, N. Ruiz, A. Keightley, M. E. Moran-Diez, K. McCluskey, Y. F. Pouchus and C. M. Kenerley, J. Biol. Chem., 2011, 286, 4544–4554 CrossRef CAS PubMed.
- T. Degenkolb, R. Karimi Aghcheh, R. Dieckmann, T. Neuhof, S. E. Baker, I. S. Druzhinina, C. P. Kubicek, H. Bruckner and H. von Dohren, Chem. Biodivers., 2012, 9, 499–535 CrossRef CAS PubMed.
- G. R. Marshall, E. E. Hodgkin, D. A. Langs, G. D. Smith, J. Zabrocki and M. T. Leplawy, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 487–491 CrossRef CAS PubMed.
- M. Lucaciu, S. Rebuffat, C. Goulard, H. Duclohier, G. Molle and B. Bodo, Biochim. Biophys. Acta, 1997, 1323, 85–96 CrossRef CAS.
- R. Gessmann, D. Axford, R. L. Owen, H. Brückner and K. Petratos, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2012, 68, 109–116 CrossRef CAS PubMed.
- H. F. Christoffersen, S. K. Hansen, B. S. Vad, E. H. Nielsen, J. T. Nielsen, T. Vosegaard, T. Skrydstrup and D. E. Otzen, Biochim. Biophys. Acta, 2015, 1854, 882–889 CrossRef CAS PubMed.
- R. Bhushan and H. Bruckner, Amino Acids, 2004, 27, 231–247 CrossRef CAS PubMed.
- F. Formaggio, M. Crisma, P. Rossi, P. Scrimin, B. Kaptein, Q. B. Broxterman, J. Kamphuis and C. Toniolo, Chem. - Eur. J., 2000, 6, 4498–4504 CrossRef CAS.
- C. Toniolo, A. Polese, F. Formaggio, M. Crisma and J. Kamphuis, J. Am. Chem. Soc., 1996, 118, 2744–2745 CrossRef CAS.
- S. C. Yasui, T. A. Keiderling, F. Formaggio, G. M. Bonora and C. Toniolo, J. Am. Chem. Soc., 1986, 108, 4988–4993 CrossRef CAS.
- C. Toniolo, G. M. Bonora, A. Bavoso, E. Benedetti, B. Di Blasio, V. Pavone and C. Pedone, Macromolecules, 1986, 19, 472–479 CrossRef CAS.
- R. Sharma, V. P. Singh, D. Singh, F. Yusuf, A. Kumar, R. A. Vishwakarma and A. Chaubey, Appl. Microbiol. Biotechnol., 2016, 100, 9091–9102 CrossRef CAS PubMed.
- G. Munagala, K. R. Yempalla, S. Singh, S. Sharma, N. P. Kalia, V. S. Rajput, S. Kumar, S. D. Sawant, I. A. Khan, R. A. Vishwakarma and P. P Singh, Org. Biomol. Chem., 2015, 13, 3610–3624 RSC.
- M. Katoch, D. Singh, K. K. Kapoor, R. A. Vishwakarma, BMC Microbiol., 2019, 19(98), 1–10 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra05780k |
| This journal is © The Royal Society of Chemistry 2020 |