Open Access Article
Open Access ArticleParallel G-quadruplex-mediated protein dimerization and activation†
Tuom Tinh Thi Truonga,
Cuong Caob and
Dung Thanh Dang *ac
*ac
aUniversity of Science, Vietnam National University, Ho Chi Minh City, Vietnam. E-mail: dung.dthanh@ou.edu.vn
bSchool of Biological Sciences, Queen's University Belfast, UK
cHo Chi Minh City Open University, Ho Chi Minh City, Vietnam
First published on 13th August 2020
Abstract
We studied parallel G4-mediated protein dimerization and activation by incorporating a RHAU peptide with a fluorescent protein FRET pair CFP/YFP and an apoptotic casp9. Occurrence of energy tranfer (from donor CFP to acceptor YFP) and enhancement of 60-fold cleavage efficiency of casp9 were observed in the presence of parallel G4, which indicated that parallel G4 can induce dimerization and activation of proteins. This novel approach holds a great promise for studying G4-targeting functional dimeric proteins in celllular biology.
G-quadruplex (G4) is a four-stranded structure formed by G-rich sequences stacking multiple G-tetrads.1–3 The G4 structure is highly polymorphic, and depends on orientation of the strand loops that can adopt parallel and nonparallel structure topologies.4,5 Computational analysis of the human genome identified more than 700
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 sequences that have potential to form G4 structure.6 G4 highly localizes at telomeres and promoter regions of genes.7,8 They have also been found in 5′-UTR of the encoded RNA9 and the long G-rich RNA transcripts of telomeric DNA.10 G4 location is not randomly distributed, correlating with functional genomic regions. The formation of G4 structure correlates with many cellular processes such as replication, transcription, translation and telomere maintenance that regulate cell proliferation.11,12 Therefore, G4 has recently emerged as a potential target for anti-cancer drug design.13–15
000 sequences that have potential to form G4 structure.6 G4 highly localizes at telomeres and promoter regions of genes.7,8 They have also been found in 5′-UTR of the encoded RNA9 and the long G-rich RNA transcripts of telomeric DNA.10 G4 location is not randomly distributed, correlating with functional genomic regions. The formation of G4 structure correlates with many cellular processes such as replication, transcription, translation and telomere maintenance that regulate cell proliferation.11,12 Therefore, G4 has recently emerged as a potential target for anti-cancer drug design.13–15
Specific recognition and stabilization of G4 by peptide or protein is a promising approach for regulation of various biological processes. In cells, several helicases such as BLM, FANCJ, PIF1 and RHAU were showed to selectively bind and resolve G4 structure.16–19 A G4-specific binding motif was also identified in N-terminus of RHAU (RNA helicase associated with AU rich element) protein. RHAU peptide selectively binds and stabilizes only parallel G4 structure including DNA and RNA G4s.20,21 NMR structure solution of a complex between an 18-residue peptide fragment (RHAU18) consisting a G4-specific binding motif and a parallel DNA G4 has showed that the RHAU18 peptide forms an α-helix that specifically recognizes the G-tetrad platform of G4. Interestingly, the parallel DNA G4 can selectively bind two RHAU peptides at the 3′ and 5′ end G-tetrads.20 The peptide covers the G-tetrad and clamps the G4 with three-anchor-point electrostatic interactions between negatively charged phosphate groups of the G4 and three positively charged amino acids of the peptide.20 Recently, specific recognition of G4 by RHAU peptide has been applied for chemical biology applications, i.e. incorporating the RHAU peptide with a fluorescent protein provided a useful protein probe for distinguishing different G4 topologies.21 Generation of new ribonuclease by incorporating the RHAU peptide with the catalytic domain of RNase HI that can target G4 and efficiently cleave the single-stranded RNA in a site-specific manner.22 However, G4 has not been applied as a target molecule for dimerization and activation of protein. Herein, we studied on characterization of G4-mediated dimerization by incorporating a RHAU peptide with a fluorescent protein pair: cyan fluorescent protein/yellow fluorescent protein (CFP/YFP) that was physically detected by the fluorescence resonance energy transfer (FRET) technique (Fig. 1). In addition, we also introduced a RHAU peptide in an apoptotic casp9 to study G4-enhanced enzymatic activity. Such approach holds a great promise for inactive monomeric proteins to specifically target G4 and play a function at dimeric form in cellular biology.
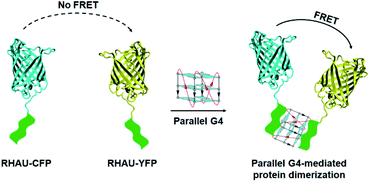 | ||
| Fig. 1 Schematic representation of parallel G4-mediated protein dimerization that can be physically detected by hetero-FRET with excitation at 400 nm and emission at 527 nm. | ||
Protein dimerization is a ubiquitous mechanism to regulate activity of protein in a broad range of cellular processes including receptor clustering, signal transduction, and apoptosis.23 Therefore, control over protein dimerization is highly desirable. Casp9 is an apoptotic cystein protease which is primarily present in its inactive monomeric form under normal physiological conditions.24,25 It becomes active upon induced dimerization by auxiliary factors and plays a key role in the apoptosis pathway, cleaving proteins at specific aspartate residues. Engineering of the casp9 dimerization interface by using specific mutations could enhance enzymatic activity in cell.26 Principle limitations of this engineered approach are the unknown effects of the point mutations on the conformation of the active site and lack of controllable reversibility.27 In addition, introduction of an N-terminal phenylalanine–glycine–glycine (FGG) motif in casp9 allowed cucurbit[8]uril to induce dimerization of casp9, resulting in an enhancement of cleavage enzyme activity.27 However, poor water solubility and permeability of cucurbit[8]uril limited applications of molecule-induced dimerization and activation of casp9 in cell. Therefore, G4 with high solubility and permeability would be a potential target molecule for inducing dimerization and activation of protein in cellular processes.
It has been evident that a short 16-aa RHAU peptide (aa 53–68) is sufficient for specific recognition of a parallel G4, the length of the RHAU peptides significantly influences binding affinity.21 Herein, we designed a fluorescent protein FRET pair RHAU–CFP/RHAU–YFP for characterization of G4-mediated protein dimerization by incorporating an engineered RHAU peptide 30-aa (consisting of RHAU specific binding motif) with the CFP and YFP, respectively. In addition, G4-mediated activation of protein was also proofed by incorporating this RHAU peptide with inactive monomeric casp9. DNA sequences coding RHAU–CFP, RHAU–YFP and RHAU–casp9 were confirmed by DNA sequencing. All proteins were expressed in E.coli BL21 (DE3) under regulation of IPTG. Proteins consisting His-tag at N-terminus were purified by the His-tag chromatography column. Subsequently, the purified proteins were evaluated by SDS-PAGE (Fig. S2, ESI†).
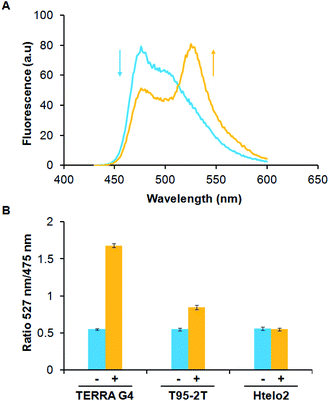
FRET technology has emerged as a powerful tool for determining protein–protein interaction and molecules-induced protein dimerization.28 Physically, FRET involves the excitation of an acceptor molecule by the emission of a donor molecule within a distance range of 1–10 nm. Herein, we used a hetero-FRET system (energy transfer from donor CFP to acceptor YFP) to elucidate G4-mediated dimerization of RHAU–CFP and RHAU–YFP. Addition of parallel G4s (1 μM) to an equimolar mixture of RHAU–CFP and RHAU–YFP (both at 1 μM) resulted in a strong hetero-FRET signal (Fig. 2A and S3, ESI†). The strong FRET is more notable, considering that G4-induced protein heterodimerization in this case is probably accompanied by 50% homodimerization of protein, which does not contribute to the hetero-FRET. The addition of TERRA (parallel RNA G4) and T95-2T (parallel DNA G4) (Table 1) to a solution of RHAU–CFP/RHAU–YFP mixture resulted in an increase of the peak ratio 527 nm/475 nm from 0.55 to 1.68 and 0.55 to 0.8, respectively (Fig. 2B). The different increase of the peak ratio showed energy transfer of CFP/YFP in the system of TERRA-induced protein dimerization was greater than that of T95-2T-induced protein dimerization. That may explain that RHAU binds RNA G4 with a somewhat greater affinity than DNA G4 (ref. 29) or TERRA-induced protein dimerization may be more optimal for position and orientation of protein fluorescence which may impact the FRET signal. In contrast, addition of Htelo2 (nonparallel DNA G4, Table 1) (1 μM) to a mixture of RHAU–CFP and RHAU–YFP (both at 1 μM) did not result in an increase of the peak ratio 527 nm/475 nm (from 0.55 to 0.54) (Fig. 2B and S3, ESI†). These results showed parallel G4s (TERRA and T95-2T) are capable of selectively binding and inducing protein dimerization.
| Name | Sequences (5′–3′) | Structure21 |
|---|---|---|
| TERRA | r(UAGGGUUAGGGUUAGGGUUAGGGUU) | Parallel G4 |
| T95-2T | d(TTGGGTGGGTGGGTGGGT) | Parallel G4 |
| Htelo2 | d(TAGGGTTAGGGTTAGGGTTAGGGTT) | Non-parallel G4 |
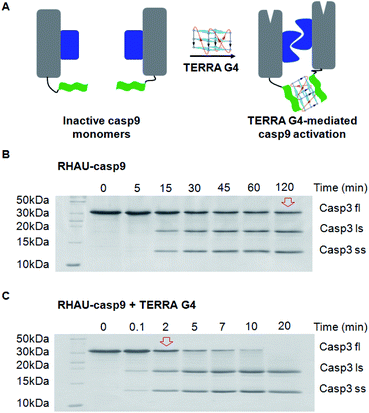
Parallel G4-mediated dimerization of protein approach was applied for dimer-driven activation of casp9 (Fig. 3A). Casp9 exits primarily inactive monomeric form and becomes active dimeric form under biophysical condition, resulting in apoptosis. In the pathway of apoptosis, casp9 catalyzes, amongst others, the activation of caspase3 (casp3) through cleavage into a small and a large subunit. The catalytic efficiency of the TERRA G4-induced casp9 dimerization and activation was therefore determined using casp3 as a natural substrate.27 In the absence of TERRA the RHAU–casp9 monomer requires approximately 120 min to cleave half of the casp3 substrate (Fig. 3B). Simple addition of TERRA results in a strong >60-fold enhancement of catalytic efficiency towards the natural substrate, resulting in a half-time of cleavage of approximately only 2 min (Fig. 3C). The catalytic efficiency of the reference construct casp9 is similar to that of the RHAU–casp9 without RNA G4, but in contrast does not increase upon addition of the TERRA (Fig. S4, ESI†), thus again showing the high specificity of the parallel G4 and RHAU interaction.
DNA and RNA G4 play key role in regulation of cellular processes such as replication, transcription, translation and telomere maintenance. Therefore, specific recognition of G4 by peptides and proteins provides a promising approach for regulation of many biological processes. Parallel G4s capable of selectively binding two RHAU peptides have been characterized by hetero-FRET. A set of fluorescent protein FRET pair CFP/YFP was used owing to their simple expression and stability. Hetero-FRET only occurs in the mixture of RHAU–CFP/RHAU–YFP in the presence of parallel G4 that indicates parallel G4-mediated protein dimerization via a specific interaction between RHAU and parallel G4 following the binding mode of 1G4 to 2RHAUs. Parallel G4-mediated protein dimerization system is applied for activation of inactive monomeric casp-9, an apoptotic enzyme. Results show that enzymatic activity of casp-9 significantly increase in the presence of TERRA (parallel RNA G4). It is notable that high activity of casp-9 depends on (i) dimerization of protein and (ii) rearrangement of active site.27 Therefore, TERRA can play as a target molecule for inducing both dimerization and rearrangement of the active site of RHAU–casp9. Such approach would be used to promote apoptosis of cancer cells by G4-mediated dimerization and activation of casp9. Induction of protein dimerization by G4 is crucial for studies in function of protein and interplay between protein oligomerization state and activation, not only casp-9, but also many other protein homodimerization events.
In conclusion, the results show that parallel G4 can act as a target-inducer of protein dimerization and activation, thereby leading to energy transfer from donor RHAU–CFP to acceptor RHAU–YFP and optimal protein reorganization for enzymatic activity of RHAU–casp9. Specific recognition of parallel G4 by two RHAU peptides allows inactive monomeric proteins fusing with RHAU peptide to specifically target parallel G4 and play a dimer-driven activation of proteins. We believe that parallel G4-mediated protein dimerization and activation hold great promises for studying not only caspases, but also many other protein homodimerization events such as dimerizing enzymes and membrane receptor proteins in cellular processes.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research is funded by Vietnam National Foundation for Science and Technology Development (NAFOSTED) under grant number 108.02-2017.305. We would like to thank Prof. Anh Tuân Phan (Nanyang Technological University, Singapore) for scientific discussion and kindly giving plasmids.Notes and references
- M. Gellert, M. N. Lipsett and D. R. Davies, Proc. Natl. Acad. Sci. U.S.A., 1962, 48, 2013–2018 CrossRef CAS PubMed.
- D. Sen and W. Gilbert, Nature, 1988, 334, 364–366 CrossRef CAS PubMed.
- C. Weldon, I. Behm-Ansmant, L. H. Hurley, G. A. Burley, C. Branlant, I. C. Eperon and C. Dominguez, Nat. Chem. Biol., 2017, 13, 18–20 CrossRef CAS PubMed.
- S. Burge, G. N. Parkinson, P. Hazel, A. K. Todd and S. Neidle, Nucleic Acids Res., 2006, 34, 5402–5415 CrossRef CAS PubMed.
- D. J. Patel, A. T. Phan and V. Kuryavyi, Nucleic Acids Res., 2007, 35, 7429–7455 CrossRef CAS PubMed.
- V. S. Chambers, G. Marsico, J. M. Boutell, M. Di Antonio, G. P. Smith and S. Balasubramanian, Nat. Biotechnol., 2015, 33, 877–881 CrossRef PubMed.
- S. Balasubramanian, L. H. Hurley and S. Neidle, Nat. Rev. Drug Discov., 2011, 10, 261–275 CrossRef CAS PubMed.
- G. Biffi, D. Tannahill, J. McCafferty and S. Balasubramanian, Nat. Chem., 2013, 5, 182–186 CrossRef CAS PubMed.
- J. D. Beaudoin and J. P. Perreault, Nucleic Acids Res., 2010, 38, 7022–7036 CrossRef CAS PubMed.
- A. T. Phan, FEBS J., 2010, 277, 1107–1117 CrossRef CAS PubMed.
- N. Maizels and L. T. Gray, PLoS Genet., 2013, 9(4), e1003468 CrossRef CAS PubMed.
- D. Rhodes and H. J. Lipps, Nucleic Acids Res., 2015, 43, 8627–8637 CrossRef CAS PubMed.
- S. M. Kerwin, B. Mamiya, C. Brian, T. Fletcher, J. T. Kern and P. W. Thomas, Abstracts of Papers of the American Chemical Society, 2000, vol. 219, p. U6 Search PubMed.
- H. Y. Han and L. H. Hurley, Trends Pharmacol. Sci., 2000, 21, 136–142 CrossRef CAS PubMed.
- J. L. Mergny and C. Helene, Nature Medicine, 1998, 4, 1366–1367 CrossRef CAS PubMed.
- P. Mohaghegh, J. K. Karow, R. M. Brosh Jr, V. A. Bohr and I. D. Hickson, Nucleic Acids Res., 2001, 29, 2843–2849 CrossRef CAS PubMed.
- J. P. Vaughn, S. D. Creacy, E. D. Routh, C. Joyner-Butt, G. S. Jenkins, S. Pauli, Y. Nagamine and S. A. Akman, J. Biol. Chem., 2005, 280, 38117–38120 CrossRef CAS PubMed.
- C. Ribeyre, J. Lopes, J. B. Boule, A. Piazza, A. Guedin, V. A. Zakian, J. L. Mergny and A. Nicolas, PLoS Genet., 2009, 5, e1000475 CrossRef PubMed.
- O. Mendoza, A. Bourdoncle, J. B. Boule, R. M. Brosh Jr and J. L. Mergny, Nucleic Acids Res., 2016, 44, 1989–2006 CrossRef CAS PubMed.
- B. Heddi, V. V. Cheong, H. Martadinata and A. T. Phan, Proc. Natl. Acad. Sci. U.S.A., 2015, 112, 9608–9613 CrossRef CAS PubMed.
- D. T. Dang and A. T. Phan, Chembiochem, 2016, 17, 42–45 CrossRef CAS PubMed.
- D. T. Dang and A. T. Phan, Sci. Rep., 2019, 9, 7432 CrossRef PubMed.
- N. J. Marianayagam, M. Sunde and J. M. Matthews, Trends Biochem. Sci., 2004, 29, 618–625 CrossRef CAS PubMed.
- C. Pop, J. Timmer, S. Sperandio and G. S. Salvesen, Mol. Cell, 2006, 22, 269–275 CrossRef CAS PubMed.
- S. B. Bratton and G. S. Salvesen, J. Cell Sci., 2010, 123, 3209–3214 CrossRef CAS PubMed.
- Y. Chao, E. N. Shiozaki, S. M. Srinivasula, D. J. Rigotti, R. Fairman and Y. Shi, PLoS Biol., 2005, 3, e183 CrossRef PubMed.
- D. T. Dang, H. D. Nguyen, M. Merkx and L. Brunsveld, Angew. Chem. Int. Ed., 2013, 52, 2915–2919 CrossRef CAS PubMed.
- H. D. Nguyen, D. T. Dang, J. L. van Dongen and L. Brunsveld, Angew. Chem. Int. Ed., 2010, 49, 895–898 CrossRef CAS PubMed.
- S. D. Creacy, E. D. Routh, F. Iwamoto, Y. Nagamine, S. A. Akman and J. P. Vaughn, J. Biol. Chem., 2008, 283, 34626–34634 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra06173e |
| This journal is © The Royal Society of Chemistry 2020 |