Open Access Article
Open Access ArticleEfficient polymerase chain reaction assisted by metal–organic frameworks†
Chunli
Sun
 a,
Yong
Cheng
a,
Yong
Cheng
 *b,
Yong
Pan
a,
Juliang
Yang
c,
Xudong
Wang
a and
Fan
Xia
*b,
Yong
Pan
a,
Juliang
Yang
c,
Xudong
Wang
a and
Fan
Xia
 *ac
*ac
aHubei Key Laboratory of Bioinorganic Chemistry & Materia Medica, School of Chemistry and Chemical Engineering, Huazhong University of Science and Technology, Wuhan 430074, China. E-mail: xiafan@hust.edu.cn
bSchool of Materials Science and Engineering, Huazhong University of Science and Technology, Wuhan 430074, China. E-mail: chengy@hust.edu.cn
cEngineering Research Center of Nano-Geomaterials of Ministry of Education, Faculty of Materials Science and Chemistry, China University of Geosciences, Wuhan 430074, China
First published on 6th December 2019
Abstract
As a powerful tool for obtaining sufficient DNA from rare DNA resources, polymerase chain reaction (PCR) has been widely used in various fields, and the optimization of PCR is still in progress due to the dissatisfactory specificity, sensitivity and efficiency. Although many nanomaterials have been proven to be capable of optimizing PCR, their underlying mechanisms are still unclear. So far, the scientifically compelling and functionally evolving metal–organic framework (MOF) materials with high specific surface area, tunable pore sizes, alterable surface charges and favourable thermal conductivity have not been used for PCR optimization. In this study, UiO-66 and ZIF-8 were used to optimize error-prone two round PCR. The results demonstrated that UiO-66 and ZIF-8 not only enhanced the sensitivity and efficiency of the first round PCR, but also increased the specificity and efficiency of the second round PCR. Moreover, they could widen the annealing temperature range of the second round PCR. The interaction of DNA and Taq polymerase with MOFs may be the main reason. This work provided a candidate enhancer for PCR, deepened our understanding on the enhancement mechanisms of nano-PCR, and explored a new application field for MOFs.
Introduction
As the gold standard of in vitro nucleic acid amplification, polymerase chain reaction (PCR) has been extensively used in many fields related to medical diagnostics and molecular biology.1 However, due to the lack of correction mechanisms similar to in vivo amplification, nonspecific amplification and primer mismatch might occur occasionally in the PCR, resulting in unsatisfactory specificity, sensitivity and efficiency. Until now, many sophisticated methods have been developed to make up for these drawbacks, such as the development of PCR machines with rapid heating–cooling responses,2 adopting unique PCR strategies,3 optimization of reaction conditions and adding additives to the PCR mixture.4 In the last two decades, nanomaterials as effective enhancers of PCR have attracted more attention (termed nano-PCR). So far, the reported nanomaterials contain gold nanoparticles, graphene oxide, quantum dots, ZnO, TiO2 and nanocomposites.5 However, most of them could only optimize one or two characteristics of PCR including specificity, sensitivity and efficiency, and few materials could enhance these three characteristics together. Besides, the limited optimization effect, complex preparation methods and high price would limit their widespread application. Therefore, looking for new materials with comprehensive optimization effects and simple preparation methods with low prices will be highly desirable.Metal–organic frameworks (MOFs) are special organic–inorganic hybrid porous solids with extraordinarily high surface areas, tunable pore sizes, adjustable internal surface properties and extraordinary degree of variable structures. These features endow them with potential applications in gas or liquid adsorption/storage, drug delivery, polymerization, catalysis and biosensors.6 As far as we know, the combination of MOFs and DNA has been frequently studied.7 However, MOFs have not yet been used in the improvement of PCR.
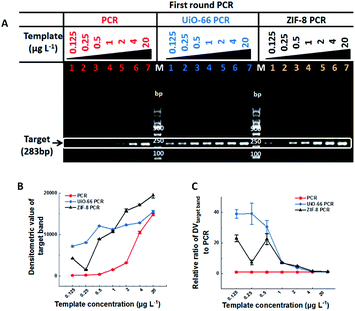
In this study, two kinds of MOFs with exceptional chemical robustness and thermal stability, ZIF-8 and UiO-66, were used to optimize PCR. They were synthesized according to the relative references,8,9 and their characterization results are shown in Fig. S1, S2 and Table S1† respectively. The stock solutions (10 g L−1) were stable aqueous solutions formed by sonication and autoclaving. The two-round error-prone PCR system with λ-DNA (48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 502 bp) as template was employed as the model, and a single 283-bp band will be amplified after 35 cycles of the first round PCR, while many nonspecific bands will be accumulated after the second round 35-cycle PCR (Fig. S3†). The effect of MOFs on PCR is directly proportional to the densitometric value (DV) of PCR products on agarose gel. Specifically, the sensitivity of PCR could be reflected by the minimum template concentration that produces visual target bands, and the efficiency and specificity of PCR can be reflected by the DV of target bands (DVtarget band) and the proportion of target bands in the total bands (DVtarget band/DVtotal band), respectively. The outline of this study is illustrated in Table 1.
502 bp) as template was employed as the model, and a single 283-bp band will be amplified after 35 cycles of the first round PCR, while many nonspecific bands will be accumulated after the second round 35-cycle PCR (Fig. S3†). The effect of MOFs on PCR is directly proportional to the densitometric value (DV) of PCR products on agarose gel. Specifically, the sensitivity of PCR could be reflected by the minimum template concentration that produces visual target bands, and the efficiency and specificity of PCR can be reflected by the DV of target bands (DVtarget band) and the proportion of target bands in the total bands (DVtarget band/DVtotal band), respectively. The outline of this study is illustrated in Table 1.
| First round PCR (283bp) | Second round PCR (283bp) | |||
|---|---|---|---|---|
| PCR | MOF–PCR (UiO-66 or ZIF-8) | PCR | MOF–PCR (UiO-66 or ZIF-8) | |
| Sensitivity | 0.5 μg L−1 | 0.125 μg L−1 | Inapplicable | Inapplicable |
| Specificity | Inapplicable | Inapplicable | 1 | 29/8 |
| Efficiency | 1 | 39/23 | 1 | 24/2 |
| Annealing temperature | Unverified | Unverified | Narrow | Wide |
Results and discussion
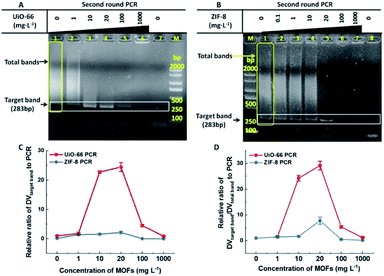
In most cases, one round conventional PCR with a suitable copy template was sufficient to obtain the desired amount of DNA, while fewer copy templates often result in random amplification, false negative or failure to obtain enough products. Therefore, improving the sensitivity of the first round PCR is a critical issue. Our study found that UiO-66 and ZIF-8 could improve the sensitivity and efficiency of the first round PCR in the absence of MOFs with good stability (Fig. 1A and S4†). And no band could be detected when the concentration of template was less than 0.5 μg L−1 (Fig. 1A, left lane 1–3). By contrast, in the presence of UiO-66 (Fig. 1A, middle lane 1–7) or ZIF-8 (Fig. 1A, right lane 1–7), obvious target bands can still be detected even at a template concentration as low as 0.125 μg L−1 (Fig. 1A). Importantly, UiO-66 showed much better PCR efficiency than ZIF-8 at special and lower template concentrations from 0.125 to 0.5 μg L−1 (Fig. 1B). Moreover, the normalized analysis of DVtarget band showed that UiO-66 and ZIF-8 could increase the PCR efficiency about 39- and 23-fold compared to conventional PCR, respectively (Fig. 1C). In all, this experiment suggested that UiO-66 and ZIF-8 could improve the sensitivity and efficiency of the first round PCR.The effect of UiO-66 and ZIF-8 on the second round PCR is shown in Fig. 2, in which the 100-fold diluted product of the first round PCR was used as template. In the absence of UiO-66 or ZIF-8, nonspecific bands accumulated as manifested by smear bands (Fig. 2A lane 1, Fig. 2B lane 1 and Fig. S2† lane 1), while gradually diminishing smear bands were observed with the increase of UiO-66 or ZIF-8 concentration from 1 to 20 mg L−1 (Fig. 2A lane 2–4 and Fig. 2B lane 2–5). And a single predominant target band was observed at a concentration of 20 mg L−1 (Fig. 2A lane 4 and Fig. 2B lane 5). Specifically, the metal ions, specific ligand and excess addition of UiO-66 or ZIF-8 can significantly inhibit PCR as manifested by the attenuation or disappearance of target bands (Fig. S5,†Fig. 2A lane 5–6 and Fig. 2B lane 6–7). Furthermore, the analysis of DV showed that the specificity and efficiency of PCR increased first and then decreased with the increase of concentrations of UiO-66 or ZIF-8 (Fig. 2C and D). This phenomenon happened in the first round PCR (Fig. S6†). When the optimal concentration of 20 mg L−1 MOFs was used, UiO-66 and ZIF-8 could increase the specificity of PCR by 29 times and 8 times and increase the efficiency of PCR by 24 times and 2 times, respectively (Fig. 2C and D). It is worth noting that the performance of UiO-66 is much better than that of ZIF-8. In summary, these experiments confirmed that both UiO-66 and ZIF-8 can improve the specificity and efficiency of the second round PCR at appropriate concentrations.
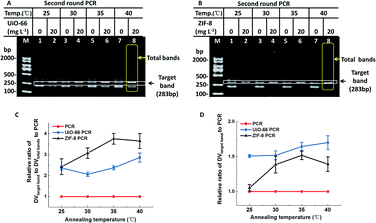
As we know, the annealing temperature is important for efficiency and specificity of PCR. Generally speaking, a low annealing temperature facilitates the efficiency of PCR and a high annealing temperature reduces the specificity of PCR.10 Therefore, much more sophisticated optimization of annealing temperature was further investigated. Like several other nanomaterials,11 UiO-66 and ZIF-8 exhibited reliable PCR results at low annealing temperatures (Fig. 3). When non-optimal annealing temperatures (25–40 °C) were employed in PCR, conventional PCR produced a predominant band corresponding to 200-bp with numerous non-specific bands and few target bands. However, in the presence of UiO-66/ZIF-8 (20 mg L−1), most non-specific bands disappeared, and target bands were amplified as the main band with an obviously higher intensity than the conventional PCR. More strikingly, the predominant band corresponding to 200-bp produced in conventional PCR totally disappeared in the presence of ZIF-8 (Fig. 3B). The relative DV analysis showed that both UiO-66 and ZIF-8 could improve the specificity of PCR at all tested non-optimal annealing temperatures, and this effect became more obvious with the increase of the annealing temperatures (Fig. 3C). Furthermore, ZIF-8 had a better effect than UiO-66 (Fig. 3C). In addition, at almost all the test temperatures, UiO-66 and ZIF-8 showed more improved PCR efficiency than conventional PCR. With the increase of annealing temperatures, the improvement extent of the PCR efficiency also slightly increased, which was especially evident in ZIF-8 (Fig. 3D). Moreover, the four different kinds of PCR experiments showed similar results (Fig. S7†). In short, UiO-66 and ZIF-8, especially ZIF-8, could widen the ideal range of annealing temperature from 40–65 °C to below 40 °C. It not only could obviate the time-consuming screening process, but also could improve the efficiency at much lower annealing temperatures.
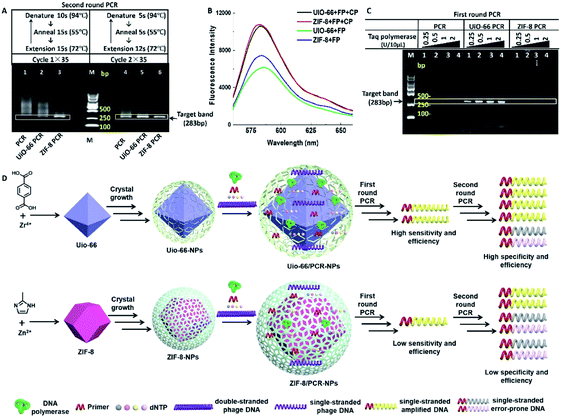
Although some studies related to nano-PCR attributed the widened annealing temperature to the superior thermal conductivity of nanomaterials,12 both UiO-66 and ZIF-8 are thermally inert MOFs, and their thermal conductivity is about 0.11 and 0.165 W m−1 K−1 respectively,13 which are far lower than the thermal conductivity of reported nano-PCR materials.14 Nevertheless, PCR was performed with two different cycles, in which denaturation, annealing and extension times were reduced. It showed that smear bands amplified in the second round PCR (Fig. 4A, lane 1 and lane 4) were only slightly reduced in the presence of UiO-66 (Fig. 4A, lane 2 and lane 5), but they were obviously reduced in the presence of ZIF-8 (Fig. 4A, lane 3 and 6). Besides, the intensity of target bands obviously increased in the presence of MOFs. These results suggested that the reduction of reaction time weakened rather than blocking the effect of UiO-66 and ZIF-8 on the specificity and efficiency of PCR. It means that the thermal conductivity of MOFs may contribute to the improvement of PCR. The better effect of ZIF-8 on PCR than UiO-66 might be explained by the higher thermal conductivity of ZIF-8.
The most well-known application of MOFs is their adsorption/storage ability of ions, nucleic acids, proteins and other substances.15 And the conjugated π-electron system of MOFs allows their binding of ssDNA through π–π stacking.16 Using this property, UiO-66 and ZIF-8 have been used in biosensors to test some special single stranded DNA (ssDNA).8,17 Therefore, it is reasonable to speculate that UiO-66 and ZIF-8 selectively attract or adsorb ssDNA rather than double stranded DNA (dsDNA) to improve the efficiency of PCR. To confirm this speculation, we compared the relative fluorescence intensity of different solutions containing UiO-66 (or ZIF-8) and a fluorescence labelled nucleic acid probe (FP) in the absence or presence of a complementary probe (CP). For the sequences of the FP and CP, refer to Table S2.† As shown in Fig. 4B, the solutions containing the FP and CP have obviously higher fluorescence intensity than the FP alone, which means that the attraction or adsorption to ssDNA contribute to the optimization effect of UiO-66 and ZIF-8.
Another advantage of MOFs is their potential as an enzyme immobilization platform.18 It could be inferred that DNA polymerase is immobilized by UiO-66 or ZIF-8 materials, and the efficiency of DNA polymerase is improved greatly, leading to the improved PCR effect. To test this possibility, we performed the first round PCR with a lower template concentration (0.2 μg L−1) that could cause amplification failure in conventional PCR. It could not be reversed by increasing the concentration of Taq polymerase (lane 2–4) (Fig. 4C lane 1). However, the addition of UiO-66 (20 mg L−1) reversed the amplification failure, and with the increase of the Taq polymerase concentration, the intensity of target bands and some non-specific bands increased (Fig. 4C lane 5–8), while ZIF-8 (20 mg L−1) did not show the same effect as UiO-66, and only some non-specific bands were observed with the highest concentration of Taq polymerase (Fig. 4C lane 9–12). This difference may be caused by their different pore sizes and total pore volumes, just as shown in the inset of Fig. S1, S2 and Table S1.† This suggests that ZIF-8 contains microporous particles, while UiO-66 contains tridimensional mesoporous particles. Besides, UiO-66 has a larger total pore volume than ZIF-8. Maybe these characters make UiO-66 much more suitable for enzyme immobilization than ZIF-8.
Another possible explanation may lie in their different zeta potentials; ZIF-8 (about 15 mV) is more positively charged than UiO-66 (about −5 mV) (Fig. S8†), which means that they prefer to bind with negatively charged nucleic acids. The SEM and TEM morphology and elemental analysis of MOFs before and after the addition of a high concentration of Taq polymerase or template also supports this explanation (Fig. S9–S16†). As revealed in the SEM results, UiO-66 showed significant morphological change after adding a high concentration of the Taq enzyme, while ZIF-8 had indistinct morphological changes after adding a high concentration of λ-DNA (Fig. S9†). Moreover, the TEM results indicated that the adsorption capacity of UiO-66 was stronger than that of ZIF-8 (Fig. S10–S15†). The results showed that both surface charges and pore sizes have certain roles in the MOFs' enhancement effect on PCR, and pores may play a more important role for UiO-66 rather than ZIF-8. And the relationship among templates, Taq polymerase and UiO-66 (or ZIF-8) during the extension process of PCR is illustrated in Fig. 4D.
Conclusions
In summary, our work preliminarily demonstrated that for the error-prone two round PCR system, UiO-66 and ZIF-8 could remarkably improve the sensitivity and efficiency of the first round PCR and they also could remarkably enhance the specificity and efficiency of the second round PCR, even at a very low annealing temperature. The possible mechanism might be attributable to their selective adsorption to ssDNA and the ability of immobilizing Taq polymerase. This work provides a new insight into the understanding of the nano-PCR effects and their enhancement mechanism for PCR. It also opened up a new field for the application of MOFs. MOFs could provide a variety of materials with remarkable geometrical shapes and chemical properties of the internal surface of nanoporous materials. Further investigation into the effect of MOFs on PCR should be focused on the influence of shape, particle size, hydrophilicity, colloidal stability, surface charge, pore sizes and other factors on PCR.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This research is supported by the National Natural Science Foundation of China (21525523, 21574048, 21375042, 21405054), the National Basic Research Program of China (the 973 program, 2015CB932600, 2013CB933000), the Special Fund for Strategic New Industry Development of Shenzhen, China (grant no. JCYJ20150616144425376) and 1000 Young Talent (to Fan Xia).Notes and references
- (a) Y. L. Jung, C. Jung, H. Parab, D. Y. Cho and H. G. Park, ChemBioChem, 2011, 12, 1387–1390 CrossRef CAS PubMed; (b) R. K. Saiki, D. H. Gelfand, S. Stoffel, S. J. Scharf, R. Higuchi, G. T. Horn, K. B. Mullis and H. A. Erlich, Science, 1988, 239, 487–491 CrossRef CAS PubMed; (c) R. K. Saiki, S. Scharf, F. Faloona, K. B. Mullis, G. T. Horn, H. A. Erlich and N. Arnheim, Science, 1985, 230, 1350–1354 CrossRef CAS PubMed; (d) Y. Wang, C. Liu, X. Zhang, W. Yang, F. Wu, G. Zou, X. Wen and X. Zhou, Chem. Sci., 2018, 9, 3723–3728 RSC.
- (a) J. Berg, V. Nagl, G. Mühlbauer and H. Stekel, J. Clin. Virol., 2001, 20, 71–75 CrossRef CAS PubMed; (b) M. S. Ibrahim, R. S. Lofts, P. B. Jahrling, E. A. Henchal, V. W. Weedn, M. A. Northrup and P. Belgrader, Anal. Chem., 1998, 70, 2013–2017 CrossRef CAS PubMed.
- (a) J. H. Van, M. M. Ehlers, W. Z. Van and W. O. Grabow, Water Res., 2003, 37, 3704–3708 CrossRef; (b) N. Paul, J. Shum and T. Le, Hot start PCR, Humana Press, Totowa, NJ, 2010, vol. 630, pp. 301–318 Search PubMed; (c) R. H. Don, P. T. Cox, B. J. Wainwright, K. Baker and J. S. Mattick, Nucleic Acids Res., 1991, 19, 4008 CrossRef CAS; (d) A. Roloff and O. Seitz, Chem. Sci., 2013, 4, 432–436 RSC.
- (a) W. Henke, K. Herdel, K. Jung, D. Schnorr and S. A. Loening, Nucleic Acids Res., 1997, 25, 3957–3958 CrossRef CAS; (b) M. Musso, R. Bocciardi, S. Parodi, R. Ravazzolo and I. Ceccherini, J. Mol. Diagn., 2006, 8, 544–550 CrossRef CAS PubMed; (c) H. A. Erlich, D. Gelfand and J. J. Sninsky, Science, 1991, 252, 1643–1651 CrossRef CAS PubMed; (d) R. T. D'Aquila, L. J. Bechtel, J. J. Videler, J. J. Eron, P. Gorczyca and L. C. Kaplan, Nucleic Acids Res., 1991, 19, 3749 CrossRef.
- (a) H. Li, J. Huang, J. Lv, H. An, X. Zhang, Z. Zhang, C. Fan and J. Hu, Angew. Chem., Int. Ed., 2005, 44, 5100–5103 CrossRef CAS; (b) M. Li, Y. Lin, C. Wu and H. Liu, Nucleic Acids Res., 2005, 33, e184 CrossRef PubMed; (c) J. Jia, L. Sun, N. Hu, G. Huang and J. Weng, Small, 2012, 8, 2011–2015 CrossRef CAS; (d) Y. Liang, F. Luo, Y. Lin, Q. Zhou and G. Jiang, Carbon, 2009, 47, 1457–1465 CrossRef CAS; (e) X. Cao, J. Chen, S. Wen, C. Peng, M. Shen and X. Shi, Nanoscale, 2011, 3, 1741–1747 RSC; (f) F. Sang, Z. Zhang, L. Yuan and D. Liu, Analyst, 2018, 143, 1259–1267 RSC; (g) A. Khaliq, P. J. Sonawane, B. K. Sasi, B. S. Sahu, T. Pradeep, S. K. Das and N. R. Mahapatra, Nanotechnology, 2010, 21, 255704 CrossRef PubMed; (h) X. Cao, X. Shi, W. Yang, X. Zhang, C. Fan and J. Hu, Analyst, 2009, 134, 87–92 RSC.
- (a) G. Maurin, C. Serre, A. Cooper and G. Ferey, Chem. Soc. Rev., 2017, 46, 3104–3107 RSC; (b) J. Navarro-Sánchez, N. Almora-Barrios, B. Lerma-Berlanga, J. J. Ruiz-Pernía, V. A. Lorenz-Fonfria, I. Tuñón and C. Martí-Gastaldo, Chem. Sci., 2019, 10, 4082–4088 Search PubMed; (c) W. Wang, S. Wu, J. Wang, Z. Li, H. Cui, S. Lin, J. Zhu and Q. Chen, Chem. Sci., 2019, 10, 4476–4485 RSC.
- (a) S. Wang, C. M. McGuirk, M. B. Ross, S. Wang, P. Chen, H. Xing, Y. Liu and C. A. Mirkin, J. Am. Chem. Soc., 2017, 139, 9827–9830 CrossRef CAS; (b) W. Morris, W. E. Briley, E. Auyeung, M. D. Cabezas and C. A. Mirkin, J. Am. Chem. Soc., 2014, 136, 7261–7264 CrossRef CAS; (c) J. S. Kahn, L. Freage, N. Enkin, M. A. A. Garcia and I. Willner, Adv. Mater., 2017, 29, 1602782 CrossRef; (d) B. V. Schmidt and . Macromol, Rapid Commun. Mass Spectrom., 2019, 1900333 CrossRef PubMed; (e) Y. Liu, W. Hou, L. Xia, C. Cui, S. Wan, Y. Jiang, Y. Yang, Q. Wu, L. Qiu and W. Tan, Chem. Sci., 2018, 9, 7505–7509 RSC.
- Y. Pan, S. Zhan and F. Xia, Anal. Biochem., 2018, 546, 5–9 CrossRef CAS PubMed.
- J. H. Cavka, S. Jakobsen, U. Olsbye, N. Guillou, C. Lamberti, S. Bordiga and K. P. Lillerud, J. Am. Chem. Soc., 2008, 130, 13850–13851 CrossRef PubMed.
- W. Rychlik, W. J. Spencer and R. E. Rhoads, Nucleic Acids Res., 1990, 18, 6409–6412 CrossRef CAS PubMed.
- (a) H. Li, J. Huang, J. Lv, H. An, X. Zhang, Z. Zhang, C. Fan and J. Hu, Angew. Chem., 2005, 44, 5100–5103 CrossRef CAS PubMed; (b) H. R. Kim, A. Baek, I. J. Lee and D. E. Kim, ACS Appl. Mater. Interfaces, 2016, 8, 33521–33528 CrossRef CAS PubMed; (c) L. Wang, Y. Zhu, Y. Jiang, R. Qiao, S. Zhu, W. Chen and C. Xu, J. Phys. Chem. B, 2009, 113, 7637–7641 CrossRef CAS PubMed.
- (a) M. Li, Y. C. Lin, C. C. Wu and H. S. Liu, Nucleic Acids Res., 2005, 33, e184 CrossRef PubMed; (b) Z. Xun, X. Zhao and Y. Guan, Nanotechnology, 2013, 24, 355504 CrossRef PubMed.
- (a) H. Babaei, A. J. McGaughey and C. E. Wilmer, Chem. Sci., 2017, 8, 583–589 RSC; (b) B. Cui, C. O. Audu, Y. Liao, S. T. Nguyen, O. K. Farha, J. T. Hupp and M. Grayson, ACS Appl. Mater. Interfaces, 2017, 9, 28139–28143 CrossRef CAS PubMed; (c) J. Huang, X. Xia, X. Hu, S. Li and K. Liu, Int. J. Heat Mass Transfer, 2019, 138, 11–16 CrossRef CAS.
- (a) A. A. Balandin, S. Ghosh, W. Bao, I. Calizo, D. Teweldebrhan, F. Miao and C. N. Lau, Nano Lett., 2008, 8, 902–907 CrossRef CAS PubMed; (b) A. Li, B. Zhou, C. S. Alves, B. Xu, R. Guo, X. Shi and X. Cao, ACS Appl. Mater. Interfaces, 2016, 8, 25808–25817 CrossRef CASM. Fujii, X. Zhang, H. Xie, H. Ago, K. Takahashi, T. Ikuta and T. Shimizu, Phys. Rev. Lett., 2005, 95, 065502 CrossRef PubMed.
- (a) M. Eddaoudi, J. Kim, N. Rosi, D. Vodak, J. Wachter, M. O'Keeffe and O. M. Yaghi, Science, 2002, 295, 469–472 Search PubMed; (b) O. M. Yaghi, M. O'Keeffe, N. W. Ockwig, H. K. Chae, M. Eddaoudi and J. Kim, Nature, 2003, 423, 705–714 Search PubMed; (c) G. Ferey, Chem. Soc. Rev., 2008, 37, 191–214 RSC.
- (a) Y. J. Cui, Y. F. Yue, G. D. Qian and B. L. Chen, Chem. Rev., 2012, 112, 1126 CrossRef CAS PubMed; (b) X. Zhu, H. Zheng, X. Wei, Z. Lin, L. Guo, B. Qiu and G. Chen, Chem. Commun., 2013, 49, 1276–1278 RSC.
- (a) Y. Cui, Y. Yue, G. Qian and B. Chen, Chem. Rev., 2012, 112, 1126–1162 CrossRef CAS PubMed; (b) H. T. Zhang, J. W. Zhang, G. Huang, Z. Y. Du and H. L. Jiang, Chem. Commun., 2014, 50, 12069–12072 RSC.
- (a) X. Lian, Y. Fang, E. Joseph, Q. Wang, J. Li, S. Banerjee, C. Lollar, X. Wang and H. C. Zhou, Chem. Soc. Rev., 2017, 46, 3386–3401 RSC; (b) M. Ding, X. Cai and H. L. Jiang, Chem. Sci., 2019, 10, 10209–10230 RSC; (c) P. Li, J. A. Modica, A. J. Howarth, E. Vargas, P. Z. Moghadam, R. Q. Snurr, M. Mrksich, J. T. Hupp and O. K. Farha, Chem, 2016, 1, 154–169 CrossRef CAS; (d) J. Liang, F. Mazur, C. Tang, X. Ning, R. Chandrawati and K. Liang, Chem. Sci., 2019, 10, 7852–7858 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9sc03202a |
| This journal is © The Royal Society of Chemistry 2020 |