Open Access Article
Open Access ArticleNovel four-disulfide insulin analog with high aggregation stability and potency†
Xiaochun
Xiong‡
 a,
Alan
Blakely‡
a,
Alan
Blakely‡
 a,
Prasoona
Karra
b,
Michael A.
VandenBerg
c,
Gabrielle
Ghabash
a,
Frank
Whitby
a,
Yi Wolf
Zhang
a,
Matthew J.
Webber
a,
Prasoona
Karra
b,
Michael A.
VandenBerg
c,
Gabrielle
Ghabash
a,
Frank
Whitby
a,
Yi Wolf
Zhang
a,
Matthew J.
Webber
 c,
William L.
Holland
b,
Christopher P.
Hill
c,
William L.
Holland
b,
Christopher P.
Hill
 *a and
Danny Hung-Chieh
Chou
*a and
Danny Hung-Chieh
Chou
 *a
*a
aDepartment of Biochemistry, University of Utah, Salt Lake City UT 84112, USA. E-mail: chris@biochem.utah.edu; dchou@biochem.utah.edu
bDepartment of Nutrition and Integrative Physiology, University of Utah, Salt Lake City UT 84112, USA
cDepartment of Chemical & Biomolecular Engineering, University of Notre Dame, IN 46556, USA
First published on 5th November 2019
Abstract
Although insulin was first purified and used therapeutically almost a century ago, there is still a need to improve therapeutic efficacy and patient convenience. A key challenge is the requirement for refrigeration to avoid inactivation of insulin by aggregation/fibrillation. Here, in an effort to mitigate this problem, we introduced a 4th disulfide bond between a C-terminal extended insulin A chain and residues near the C-terminus of the B chain. Insulin activity was retained by an analog with an additional disulfide bond between residues A22 and B22, while other linkages tested resulted in much reduced potency. Furthermore, the A22-B22 analog maintains the native insulin tertiary structure as demonstrated by X-ray crystal structure determination. We further demonstrate that this four-disulfide analog has similar in vivo potency in mice compared to native insulin and demonstrates higher aggregation stability. In conclusion, we have discovered a novel four-disulfide insulin analog with high aggregation stability and potency.
Introduction
Insulin is a peptide hormone that regulates blood glucose metabolism and is an indispensable therapeutic for treating diabetes for millions of people worldwide.1 Human native insulin comprises two chains: A chain with 21 amino acid residues (A1-21) and B chain with 30 amino acid residues (B1-30).2 The two chains are connected by two inter-chain disulfide bonds (A7-B7 and A20-B19) and one intra-A-chain disulfide (A6-A11). Human insulin forms hexamers in the presence of zinc ions through coordination of B10 His. In all commercially available insulin formulations, the hexamer, which is relatively stable, is the dominant form. However, even with the presence of three disulfides and the hexameric form, human insulin is still prone to formation of aggregated fibrils, which makes continuous refrigeration necessary to ensure quality.3 Thus, there is a need to further increase insulin stability to provide an analog that does not require continuous refrigeration.All known vertebrate insulins have a conserved three-disulfide skeleton.4 There are some examples of insulin-like peptides from invertebrates with four disulfides, although their aggregation stability was not well characterized.5,6 Introduction of additional disulfides into a protein may lead to higher stability,7 and an insulin analog with an additional disulfide between A10 and B4 has been described.8 This A10-B4 linkage was chosen because the two Cα were in close (5.4 Å) proximity, and crystal structure determination of the A10-B4-4SS-Ins hexamer revealed that the extra disulfide has only minor impacts on tertiary structure. Importantly, A10-B4-4SS-Ins has a similar binding affinity as native insulin but has a higher aggregation and thermal stability, which is a promising feature for insulin therapeutics.8,9 In an analogous study, the DiMarchi group recently synthesized another four-disulfide insulin-like peptide, Con-Ins G2, with the extra disulfide between the human A4 position and an N-terminal extended B chain (human B′′-6′′ position) and showed that Con-Ins G2 is inactive.6 They further performed disulfide scanning to identify a number of other additional disulfide bonds that result in reduced bioactivity.10 These observations indicate that an additional disulfide may lead to higher insulin stability but that the extra disulfide may compromise bioactivity.
Guided by our studies of novel venom insulin molecules from fish-hunting cone snails,11 we hypothesized that an additional disulfide may be inserted into native insulin between an extended A-chain and B-chain residues B21-B23. Here, we report the chemical synthesis and characterization of 4 insulin analogs with additional disulfide bonds in this region. We further evaluated the properties of A22-B22-4SS-Ins, and identified it as a novel four-disulfide insulin analog with high stability and activity.
Results and discussion
Design of 4SS-Ins molecules
Efforts to incorporate an extra disulfide between C-terminally extended A chain residues and B21-B23 have not been reported. Indeed, the extended A-chain residues of the insulin relatives IGF-1 and IGF-2 point away from the B chain.12,13 However, our studies of cone snail venom insulin molecules suggested that this might be a viable approach. Specifically, venom insulins Con-Ins-K1 and K2 have a C-terminally extended A chain (24 residues) and truncated B chain (22 residues), and modeling studies suggested that the extended A chain may point toward the C-terminus of the B chain.11 We therefore designed insulin analogs that include a fourth disulfide bond in this region.Using two independent insulin crystal structures (PDB 1EVR and 1ZNJ),14 we found that the distance of the two Cα for both A21-B22 and A21-B23 is ∼6 Å, which is borderline for the introduction of a disulfide bond (Fig. 1). Our designs therefore included analogs with an extra C-terminal residue, A22, to provide more flexibility for the additional disulfide. Based on the modeling, we decided to investigate the structure–activity relationship of four analogs that displayed the additional disulfides: A22-B21, A22-B22, A22-B23 and A21-B22.
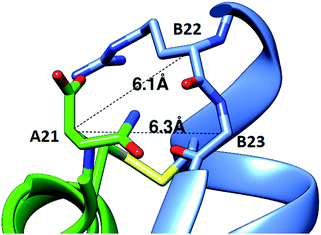 | ||
| Fig. 1 Modeling of a fourth disulfide bond in insulin. The crystal structure of R6 insulin (PDB: 1EVR) is shown with Cα distances between A21 and nearby residues on the B-chain indicated. | ||
Synthesis of 4SS-Ins molecules
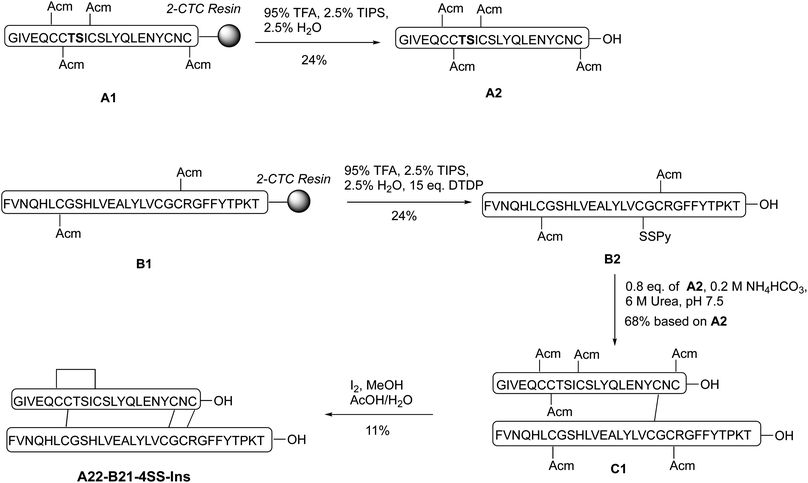
Our synthetic route toward four-disulfide insulin molecules is described in Scheme 1. First, the protected A chain [A6, 7, 11, 22-Acm] A1 was synthesized on 2-chlorotrityl chloride (2-CTC) resin through solid phase peptide synthesis (SPPS). Note that the A8-A9 dipeptide was synthesized as an isoacylpeptide to improve A chain solubility (Bold residues in Scheme 1).15A1 was then treated with a cocktail of TFA/TIPS/H2O to yield the corresponding four Acm-protected product A2 in 24% yield. A chain [A6, 7, 11, 21-Acm] A3 was obtained in 26% yield using the same route (Scheme 2). Next, protected B chain [B7, 21-Acm] B1 was generated through SPPS and activated with DTDP during acidic cleavage to give the [B7, 21-Acm, B19-SPy] chain B2 in 24% yield. B chains [B7, 22-Acm, B19-Spy] B3 and [B7, 23-Acm, B19-Spy] B4 were synthesized in 25% and 21% yield respectively (Scheme 2).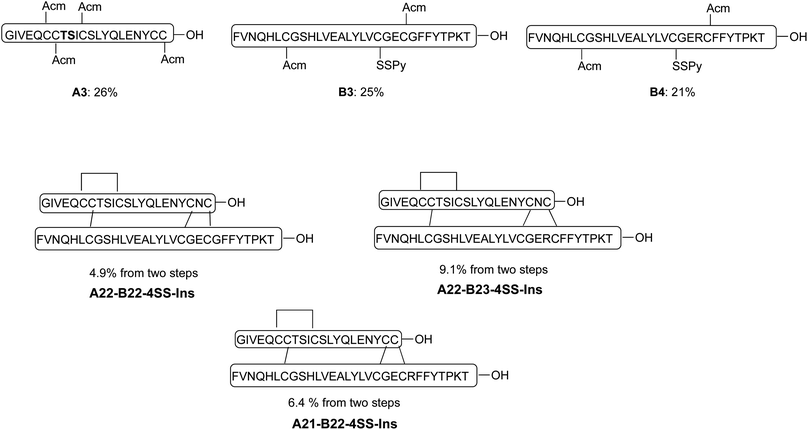 | ||
| Scheme 2 Synthesis of three additional analogs using the same synthetic route described in scheme 1. | ||
With both A and B chains in hand, we attempted to form the four disulfide bonds through a two-step process. First, A2 and B2 were dissolved and stirred in 6 M urea containing 0.2 M NH4HCO3 (pH 7.5) buffer solution. The reaction reached complete conversion of A2 in 10 min and yielded a heterodimer C1 in 68% yield based on the amount of A2 after HPLC purification. Conversion of C1 to A22-B21-4SS-Ins was smoothly achieved by treatment with excess amount of iodine in methanol, and the A6-A11, A7-B7 and A22-B21 disulfide bonds were formed through in situ Acm deprotection/disulfide formation. The product A22-B21-4SS-Ins was obtained in 11% yield after purification (Scheme 1). Three additional analogs A22-B22-4SS-Ins, A22-B23-4SS-Ins and A21-B22-4SS-Ins were obtained in 4.9%, 9.1% and 6.4% yield, respectively, over two steps (Scheme 2).
A22-B22-4SS-Ins displays native activity and resistance to aggregation
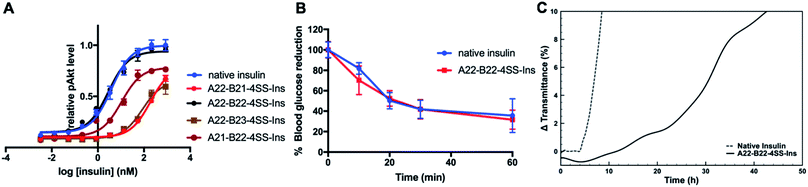
A phospho-AKT (Ser 473) insulin signaling stimulation assay was used to evaluate insulin bioactivity of the four analogs (Fig. 2A). Native insulin and A22-B22-4SS-Ins displayed similar potency (EC50 3.86 nM vs. 2.87 nM, respectively). In contrast, both A22-B21-4SS-Ins and A22-B23-4SS-Ins have much reduced potency (50- and 20-fold reduction respectively), and A21-B22-4SS-Ins is 4-fold less potent than native insulin. These results indicate that the relatively close proximity between A21 and B22 can lead to partially preserved bioactivity, and the extra flexibility provided by the A22 residue results in close to native potency for the A22-B22-4SS-Ins analog. Consistent with the cell-based assay, an insulin tolerance test in mice showed that A22-B22-4SS-Ins and native insulin display similar in vivo activity following subcutaneous injection (Fig. 2B).We further evaluated propensity for aggregation using a stressed aging assay of continuous agitation in pH 7.4 buffer at 37 °C. As shown in Fig. 2C, native insulin behaves similarly to previous reports as it aggregated in less than 10 hours of shaking,16 while A22-B22-4SS-Ins did not reach an equivalent level of aggregation until at least 42 hours of shaking. This result demonstrates that A22-B22-4SS-Ins has a superior effect over native insulin in preventing aggregation, a key factor for insulin formulation.
A22-B22-4SS-Ins crystal structure and comparison to native insulin
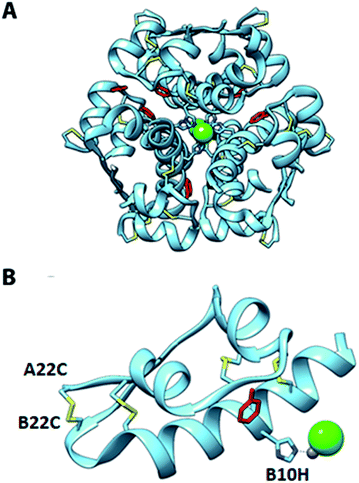
To investigate the structural basis of the high stability and potency, crystals of A22-B22-4SS-Ins were grown by sitting-drop vapor diffusion. The crystals contained one insulin hexamer in the asymmetric unit. The hexamer is bound by seven phenol molecules occupying the six classical phenolic binding sites, and one additional site. Also present are two Zn2+ ions that, together with the phenol, stabilize the hexameric R6 conformation of the insulin (Fig. 3). Phenolic compounds have been well documented for their use in stabilizing insulin hexamers,17,18 and under our screen conditions the addition of phenol was found to be essential for the formation of crystals. Electron density for the fourth disulfide bond connecting the C-terminus of the A chain to the type-I β-turn on the B-chain is well defined (Fig. 4), and the overall structure reveals that the insulin backbone is able to accommodate the new disulfide linkage without appreciable conformational changes. The average root mean square deviation (RMSD) between equivalent Cα atoms of our structure and two wild-type human insulin R6 hexamers, PDB entries 1ZNJ and 1EVR,14 is only 0.40 Å and 0.51 Å across 21 residues of the A chain, and 0.39 Å and 0.36 Å across 29 residues of the B chain, respectively.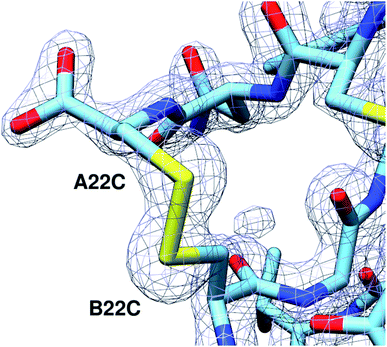 | ||
| Fig. 4 The A22-B22 disulfide bond is clearly defined. 2Fo-Fc electron density map of the A22-B22 disulfide bond at a contour level of 1.8 Å RMSD. | ||
An important structural element of insulin is the type-I β-turn formed by residues B20-B22. Biochemical and structural experiments have shown that this β-turn is critical for the stability and activity of insulin.19 For instance, a modification that ablates this turn through the incorporation of (α,β)-dehydrophenylalanine results in an aggregation-prone analog, while the converse modification of rigidifying the turn by substituting the B20 and B22 glycines with D-alanine to reinforce the positive φ-angles at those positions diminishes activity. These results are consistent with structures of insulin in complex with its receptor, which reveal an induced-fit binding mechanism in which the C-terminal β-strand of the insulin B-chain hinges away from the hydrophobic core of the hormone.19–21 These observations underscore the refractory nature of insulin to mutations in this region of the B-chain. In agreement with the native-like activity of A22-B22-4SS-Ins, alignment of our structure with wild-type insulin shows that residues B20-B23 closely match the corresponding backbone conformation of wildtype insulin despite the incorporation of a new disulfide connecting the C-terminus of the A-chain to the loop of the β-turn (Fig. 5 and S1†). Further, and consistent with wildtype insulin structures, residues comprising the β-turn have locally elevated thermal B-factors, indicating that the flexibility of the β-turn is not hindered by the A22-B22 disulfide bond.
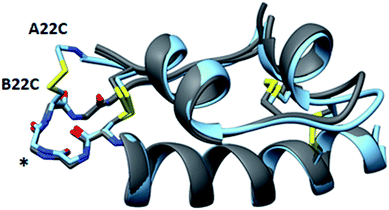 | ||
| Fig. 5 Structural similarity of A22-B22-4SS-Ins and native insulins. Overlay of A22-B22-4SS-Ins (cyan) with wild-type structures (PDB 1EVR and 1ZNJ; both gray) with the backbone type-1 beta-turn depicted on the left side of the structure (asterisk). | ||
Because residues near the C-terminus of the B chain are important for insulin dimerization and for binding to the insulin receptors, we characterized each of these properties for A22-B22-4SS-Ins. We used isothermal calorimetry to measure the dimer forming tendency, and found that A22-B22-4SS-Ins has a similar dimer formation tendency as native insulin (Fig. S2†). As noted above, A22-B22-4SS-Ins and native insulin give equivalent phosphorylation of Atk (Fig. 2A). We also determined signaling through each of the IR-A and IR-B insulin receptor isoforms by quantification of receptor autophosphoryation. As for native insulin,22 signaling was essentially identical for the two isoforms (Fig. S3†). In summary, we have demonstrated that the C-terminal of B chain in A22-B22-4SS-Ins is structurally and functionally similar to that of native insulin.
Comparison of A22-B22-4SS-Ins with A10-B4-4SS-Ins
In addition to the A22-B22-4SS-Ins characterized by this work, one other fully-active, four disulfide human insulin has been reported in the literature, with cysteine mutations connecting A10 and B4.8 This analog (A10-B4-4SS-Ins) has improved stability relative to native insulin but displayed altered structural characteristics. The crystal structure of A10-B4-4SS-Ins displays a register shift that results in the formation of a new β-sheet between B2-B4 and A10-A12 in one monomer in the insulin dimer. Additionally, CD spectroscopy indicated resistance of A10-B4-4SS-Ins to adoption of the phenol-induced R-state conformation. In contrast, the structure of A22-B22-4SS-Ins shows fewer structural alterations resulting from the new disulfide bond, a property that could be favorable for the development of novel, ultra-stable therapeutic insulins.Conclusion
Our results demonstrate that tethering between the C-terminal A chain and the B21-B23 region can lead to bioactivity variation ranging from retained potency to 50-fold reduction. Further, we report A22-B22-4SS-Ins as a novel insulin analog with retained potency, increased aggregation stability, and a crystal structure that is closely similar to native insulin. Our findings thereby complement recent efforts in generating four-disulfide insulin9,10 and single-chain insulin,23,24 and advance efforts to develop a much more stable therapeutic insulin. Further investigations regarding insulin signaling pathway activation in vitro and in vivo are underway.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank NIDDK (R01DK120430) and JDRF (5-CDA-2018-572 A-N) for funding support. X. X. is a JDRF postdoctoral fellow. Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences. UCSF Chimera, which was used for molecular representations and analyses, was developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311.References
- A. N. Zaykov, J. P. Mayer and R. D. DiMarchi, Pursuit of a perfect insulin, Nat. Rev. Drug Discovery, 2016, 15(6), 425–439 CrossRef CAS.
- M. A. Weiss, The structure and function of insulin: decoding the TR transition, Vitam. Horm., 2009, 80, 33–49 CAS.
- Q. X. Hua, S. H. Nakagawa, W. Jia, K. Huang, N. B. Phillips, S. Q. Hu and M. A. Weiss, Design of an active ultrastable single-chain insulin analog: synthesis, structure, and therapeutic implications, J. Biol. Chem., 2008, 283(21), 14703–14716 CrossRef CAS.
- J. M. Conlon, Evolution of the insulin molecule: insights into structure–activity and phylogenetic relationships, Peptides, 2001, 22(7), 1183–1193 CrossRef CAS.
- H. Safavi-Hemami, J. Gajewiak, S. Karanth, S. D. Robinson, B. Ueberheide, A. D. Douglass, A. Schlegel, J. S. Imperial, M. Watkins, P. K. Bandyopadhyay, M. Yandell, Q. Li, A. W. Purcell, R. S. Norton, L. Ellgaard and B. M. Olivera, Specialized insulin is used for chemical warfare by fish-hunting cone snails, Proc. Natl. Acad. Sci. U. S. A., 2015, 112(6), 1743–1748 CrossRef CAS PubMed.
- F. Wu, J. P. Mayer, V. M. Gelfanov, F. Liu and R. D. DiMarchi, Synthesis of Four-Disulfide Insulin Analogs via Sequential Disulfide Bond Formation, J. Org. Chem., 2017, 82(7), 3506–3512 CrossRef CAS.
- J. Mansfeld, G. Vriend, B. W. Dijkstra, O. R. Veltman, B. Van den Burg, G. Venema, R. Ulbrich-Hofmann and V. G. Eijsink, Extreme stabilization of a thermolysin-like protease by an engineered disulfide bond, J. Biol. Chem., 1997, 272(17), 11152–11156 CrossRef CAS.
- T. N. Vinther, M. Norrman, U. Ribel, K. Huus, M. Schlein, D. B. Steensgaard, T. A. Pedersen, I. Pettersson, S. Ludvigsen, T. Kjeldsen, K. J. Jensen and F. Hubalek, Insulin analog with additional disulfide bond has increased stability and preserved activity, Protein Sci., 2013, 22(3), 296–305 CrossRef CAS.
- T. N. Vinther, I. Pettersson, K. Huus, M. Schlein, D. B. Steensgaard, A. Sorensen, K. J. Jensen, T. Kjeldsen and F. Hubalek, Additional disulfide bonds in insulin: Prediction, recombinant expression, receptor binding affinity, and stability, Protein Sci., 2015, 24(5), 779–788 CrossRef CAS.
- F. M. Brunel, J. P. Mayer, V. M. Gelfanov, A. N. Zaykov, B. Finan, D. Perez-Tilve and R. D. DiMarchi, A disulfide-scan of insulin by [3 + 1] methodology exhibits site-specific influence on bioactivity, ACS Chem. Biol., 2019, 14(8), 1829–1835 CrossRef CAS.
- P. Ahorukomeye, M. M. Disotuar, J. Gajewiak, S. Karanth, M. Watkins, S. D. Robinson, P. Florez Salcedo, N. A. Smith, B. J. Smith, A. Schlegel, B. E. Forbes, B. Olivera, D. Hung-Chieh Chou and H. Safavi-Hemami, Fish-hunting cone snail venoms are a rich source of minimized ligands of the vertebrate insulin receptor, Elife, 2019, 8, e41574 CrossRef.
- A. M. Brzozowski, E. J. Dodson, G. G. Dodson, G. N. Murshudov, C. Verma, J. P. Turkenburg, F. M. de Bree and Z. Dauter, Structural origins of the functional divergence of human insulin-like growth factor-I and insulin, Biochemistry, 2002, 41(30), 9389–9397 CrossRef CAS.
- D. T. Dransfield, E. H. Cohen, Q. Chang, L. G. Sparrow, J. D. Bentley, O. Dolezal, X. Xiao, T. S. Peat, J. Newman, P. A. Pilling, T. Phan, I. Priebe, G. V. Brierley, N. Kastrapeli, K. Kopacz, D. Martik, D. Wassaf, D. Rank, G. Conley, Y. Huang, T. E. Adams and L. Cosgrove, A human monoclonal antibody against insulin-like growth factor-II blocks the growth of human hepatocellular carcinoma cell lines in vitro and in vivo, Mol. Cancer Ther., 2010, 9(6), 1809–1819 CrossRef CAS PubMed.
- G. D. Smith, E. Ciszak, L. A. Magrum, W. A. Pangborn and R. H. Blessing, R6 hexameric insulin complexed with m-cresol or resorcinol, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2000, 56(12), 1541–1548 CrossRef CAS.
- F. Liu, E. Y. Luo, D. B. Flora and A. R. Mezo, A synthetic route to human insulin using isoacyl peptides, Angew. Chem., Int. Ed. Engl., 2014, 53(15), 3983–3987 CrossRef CAS.
- M. J. Webber, E. A. Appel, B. Vinciguerra, A. B. Cortinas, L. S. Thapa, S. Jhunjhunwala, L. Isaacs, R. Langer and D. G. Anderson, Supramolecular PEGylation of biopharmaceuticals, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(50), 14189–14194 CrossRef CAS.
- S. Rahuel-Clermont, C. A. French, N. C. Kaarsholm, M. F. Dunn and C. I. Chou, Mechanisms of stabilization of the insulin hexamer through allosteric ligand interactions, Biochemistry, 1997, 36(19), 5837–5845 CrossRef CAS.
- H. Berchtold and R. Hilgenfeld, Binding of phenol to R6 insulin hexamers, Biopolymers, 1999, 51(2), 165–172 CrossRef CAS.
- J. G. Menting, Y. Yang, S. J. Chan, N. B. Phillips, B. J. Smith, J. Whittaker, N. P. Wickramasinghe, L. J. Whittaker, V. Pandyarajan, Z. L. Wan, S. P. Yadav, J. M. Carroll, N. Strokes, C. T. Roberts Jr, F. Ismail-Beigi, W. Milewski, D. F. Steiner, V. S. Chauhan, C. W. Ward, M. A. Weiss and M. C. Lawrence, Protective hinge in insulin opens to enable its receptor engagement, Proc. Natl. Acad. Sci. U. S. A., 2014, 111(33), E3395–E3404 CrossRef CAS.
- G. Scapin, V. P. Dandey, Z. Zhang, W. Prosise, A. Hruza, T. Kelly, T. Mayhood, C. Strickland, C. S. Potter and B. Carragher, Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis, Nature, 2018, 556(7699), 122–125 CrossRef CAS.
- F. Weis, J. G. Menting, M. B. Margetts, S. J. Chan, Y. Xu, N. Tennagels, P. Wohlfart, T. Langer, C. W. Muller, M. K. Dreyer and M. C. Lawrence, The signalling conformation of the insulin receptor ectodomain, Nat. Commun., 2018, 9(1), 4420 CrossRef.
- R. Slaaby, Specific insulin/IGF1 hybrid receptor activation assay reveals IGF1 as a more potent ligand than insulin, Sci. Rep., 2015, 5, 7911 CrossRef CAS.
- M. D. Glidden, K. Aldabbagh, N. B. Phillips, K. Carr, Y. S. Chen, J. Whittaker, M. Phillips, N. P. Wickramasinghe, N. Rege, M. Swain, Y. Peng, Y. Yang, M. C. Lawrence, V. C. Yee, F. Ismail-Beigi and M. A. Weiss, An ultra-stable single-chain insulin analog resists thermal inactivation and exhibits biological signaling duration equivalent to the native protein, J. Biol. Chem., 2018, 293(1), 47–68 CrossRef CAS.
- M. Y. Jeong, J. Rutter and D. H. Chou, Display of Single-Chain Insulin-like Peptides on a Yeast Surface, Biochemistry, 2019, 58(3), 182–188 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Detailed synthesis procedure and mass spectrometry data along with biological experiment protocol. See DOI: 10.1039/c9sc04555d |
| ‡ These authors contribute equally. |
| This journal is © The Royal Society of Chemistry 2020 |