Open Access Article
Open Access ArticleSuper-capacity information-carrying systems encoded with spontaneous Raman scattering†
Yuchen
Tang
*ab,
Caili
He
ab,
Xingxing
Zheng
ab,
Xuqi
Chen
ab and
Tingjuan
Gao
 *ab
*ab
aCollege of Chemistry, Central China Normal University, Wuhan 430079, China. E-mail: ytang@mail.ccnu.edu.cn; tingao@mail.ccnu.edu.cn
bChina Key Laboratory of Pesticide and Chemical Biology of Ministry of Education, Wuhan 430079, China
First published on 2nd March 2020
Abstract
Optical multiplex barcode systems have been significantly boosting the throughput of scientific discovery. A high volume of barcodes can be made from combinations of distinct spectral bands and intensity levels. However, the practical capacity often reaches a ceiling due to the overlaps of signal frequencies or intensities when massive information is written on individual carriers. In this paper, we built super-capacity information-carrying systems by tuning vibrational signals into octal numeral intensities in multiple bands of Raman-silent regions. This novel approach experimentally yielded the largest capacity of distinct optical barcodes to date. The experiments of encoding ASCII and Unicode systems to write and read languages indicate that the Raman coding method provides a new strategy for super-capacity data storage. In addition, multiplex screening of a cell-binding ligand was implemented to demonstrate the feasibility of this technology for fast and in situ high-throughput bio-discovery. These information-carrying systems may open new scenarios for the development of high-throughput screening, diagnostics and data storage.
Introduction
Recent advances in the advent of lasers, luminescent materials, and high-resolution microscopy offer opportunities to discover optical systems carrying multiplex information that boost the throughput of scientific discovery.1–5 For such optical barcode systems, in order to enhance the information capacity, new distinct optical dimensions of a medium material are being discovered for encoding. Among the many selections of optical properties, the combination of wavelength and intensity is the most widely explored. Theoretically, 106 codes can be made from a combination of 10 distinguishable intensities at each wavelength of 6 distinct peaks. However, practically a large coding capacity is never achieved due to the problems of spectral overlaps, difficulties in precisely controlling the fabrication process of information carriers, signal instability, etc.Specifically, organic fluorescent dyes inevitably encounter problems of photobleaching and cross talking caused by broad emission spectra. Inorganic nano-materials, e.g. semiconductor quantum dots and upconversion nanoparticles, are subtly dependent on the processes of fabrication and/or energy transfer which affect the stability of the information carriers and the subsequent coding signals (Fig. 1).6–9 In contrast, Raman scattering as vibrational signatures of molecules, provides much narrower and more stable spectra, thus allowing a large number of different peaks to be placed inside the spectral range. However, Raman scattering has very low efficiency and is difficult to be detected. With the strategy of Surface Enhanced Raman Spectroscopy (SERS), the current achievable coding capacities increase but are still restricted, due to the fact that the signaling peaks of the commonly used SERS reporters are located in the fingerprint region (500–2000 cm−1) where multiple Raman peaks crowd and interfere.10–12 Recently, methods were invented using the intense spontaneous Raman scattering of alkyne molecules in the Raman silent region (1800–2600 cm−1). Higher coding capacities can be achieved using this strategy to generate clean combinatorial spectra.13–15 These current methods of employing SERS and alkyne reporters focus on discovering more distinct spectral wavelengths to increase the coding capacities, but they encounter a bottleneck of exploring more diversities within the dimension of intensity. The key issues are the difficulties in precisely controlling the fabrication process, when the goal for large capacity requires encoding an increasing number of involved agents into the carriers with distinguishable ratios.15–17 Therefore, the total number of codes that can be experimentally made is highly restricted.
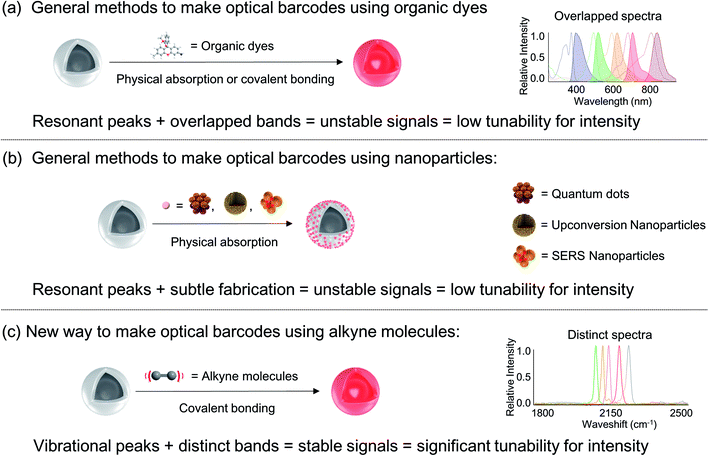 | ||
| Fig. 1 Concept of optical barcode fabrication including the typical methods using fluorescent dyes (a) and nanoparticles (b), as well as the new method using alkyne molecules (c). | ||
Besides aiming for simple ways to edit and write code units to carriers and achieve large coding capacities, effective information-carrying systems also require maintaining the stability of carriers after writing as well as retrieving the information fast and accurately. Specifically, when the systems are used for high-throughput bio-discovery, specific binding sites have to be generated at the interfaces via chemical reactions. The encoded information has to survive these reactions and the subsequent screening conditions. In addition, it is also important for the encoded information to be insusceptible to the ambient environment, e.g. light and temperature, thus allowing the system to act as the same platform for variable uses in scientific and technological discovery.
In this study, we proposed and achieved super-capacity information-carrying systems, by tuning the intensities of spontaneous Raman scattering of the alkyne reporters in 4 distinct spectral bands. The stock solutions of individual reporters were made as normalized code units, and then edited to obtain octal code units by combinatorial mixing. These octal code units were simply written on 2-D surfaces through spotting, or on microbeads through chemical bonding, creating information-carrying systems with super coding capacities. The coding capacities on the carriers of 2-D surfaces and 3-D beads were experimentally found to reach ∼500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 200
000 and 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, respectively. They can increase to a higher level when more Raman reporters are discovered to maintain relevant optical properties. Decoding the systems does not require multiple excitation sources, and is performed using conventional Raman microscopes that are easily accessible. The decoding results were obtained in short turn-around times. We demonstrated a quartz surface as the information carrier for encoding ASCII and Unicode systems, through which we were able to write/read the authors' affiliation “Central China Normal University” in both English and Chinese. We also applied the encoded resin beads for a multiplex screening of a specific target binding to cancer cells. The encoded beads were stable during the chemical reactions and biological interactions, and the encoded information was kept unchanged in the ambient environment for more than 5 months.
000, respectively. They can increase to a higher level when more Raman reporters are discovered to maintain relevant optical properties. Decoding the systems does not require multiple excitation sources, and is performed using conventional Raman microscopes that are easily accessible. The decoding results were obtained in short turn-around times. We demonstrated a quartz surface as the information carrier for encoding ASCII and Unicode systems, through which we were able to write/read the authors' affiliation “Central China Normal University” in both English and Chinese. We also applied the encoded resin beads for a multiplex screening of a specific target binding to cancer cells. The encoded beads were stable during the chemical reactions and biological interactions, and the encoded information was kept unchanged in the ambient environment for more than 5 months.
Results and discussion
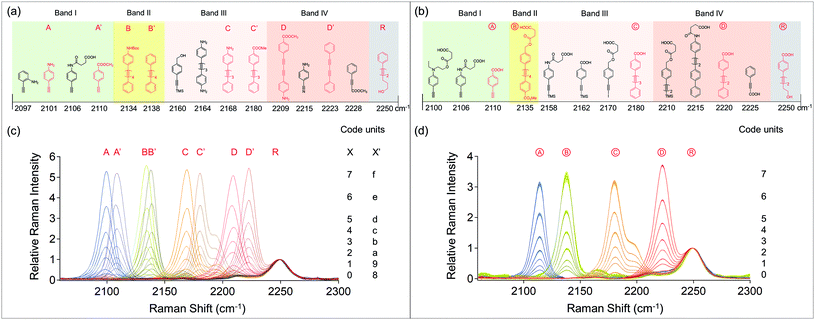
We generated a library of alkyne compounds with distinct Raman peaks. The Raman shifts of these compounds are located from 2000–2300 cm−1 and clearly separated into 4 bands. In each band, the peaks are at least ∼4 cm−1 away from those of the adjacent compounds. They are categorized into two groups. 15 of the compounds can be used for the strategy of simple mixing in solutions and then spotting onto surfaces, and the other 13 compounds have carboxyl groups which can be covalently attached to aminolated resin beads. The Relative Raman Intensities (RRI) of these compounds (Fig. S1†) provide a base for calculating and designing the amount ratios to mix compounds and generate Raman codes.As a demonstration, the compounds A, A′, B, B′, C, C′, D, D′ and R were selected to be spotted onto a surface. We tuned and obtained signals of octal code units for each of these compounds. Each of them was mixed with the reference compound R at the designed ratios (see the ESI†), and then spotted onto a quartz surface. Fig. 2c shows the 8 spectra of the obtained surface spots for each compound, after being normalized by the intensity of the reference peak at 2250 cm−1. The results showed that 8 distinguishable intensities were clearly obtained without cross talking.
The compounds Ⓐ, Ⓑ, Ⓒ, Ⓓ, and Ⓡ were selected to covalently attach to resin beads. Each of them was mixed with the reference compound Ⓡ at the designed ratios (see the ESI†), and then reacted with aminolated beads by the amide coupling reaction.18Fig. 2d shows the 8 spectra of the obtained bead samples for each compound. For each bead sample, 5 randomly picked beads from the same batch were measured independently. They were normalized by the reference peak and then averaged to a single curve with the standard deviation shown as shades above and below the curve. The results evidenced that 8 distinguishable intensities were clearly obtained without cross talking.
For such optical multiplex barcode systems described above, it can be inferred that the coding capacity is determined by three major factors: the number of Raman spectral bands being used, the number of compounds in each band being used, and the number of distinguishable intensities being used for each compound. Multiple compound candidates in the same band can be selected. As some compounds in the same band have close Raman peaks (4–10 cm−1 separation), they are not used simultaneously in order to avoid cross talking between signals. If they are employed in parallel, the code units will be multiplied in each band. For example, Fig. 2a shows the discovered compounds for the spotting strategy. Assuming 8 distinguishable intensities are available for each compound in Band I, the code units for this band will be 8 × 3. Then the capacity is calculated by the general formula (8 × NI) × (8 × NII) × (8 × NIII) × (8 × NIV), where Ni stands for the number of available compounds in each band. Based on this calculation formula, the total coding capacity is (8 × 4) × (8 × 2) × (8 × 4) × (8 × 4) = 524![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 288, when all of the discovered compounds are used for coding with the spotting strategy. The total capacity may expand further to a higher level if more coding compounds are found, or if the Raman intensity for each reporter is tuned more precisely to 10, 16, or even 25.
288, when all of the discovered compounds are used for coding with the spotting strategy. The total capacity may expand further to a higher level if more coding compounds are found, or if the Raman intensity for each reporter is tuned more precisely to 10, 16, or even 25.
We applied such a super-capacity information-carrying system to write, read and store data. While molecules related to life sciences, e.g. nucleic acids, peptides and carbohydrate polymers, provide ideas to store massive and stable genetic information efficiently, the synthetic counterparts as popularly studied information carriers, have to create a sequence-based macromolecule to encode each unique piece of information.23–27 In addition, the decoding methods are typically complicated. For instance, nucleic acids need to be sequenced using PCR and Sanger techniques, and peptides or polymers have to be decoded by mass spectrometry.24–28 In comparison, the Raman barcode systems allow us to carry massive information and decode information in situ and non-invasively for potential applications in different fields, e.g. data storage and anti-counterfeiting.29–33
To demonstrate these capabilities, we encoded the ASCII (American Standard Code for Information Interchange) system using the combinations of A (2100 cm−1) in Band I, and D (2209 cm−1)/D′ (2223 cm−1) in Band IV. ASCII is an 8-digit binary system where every single code represents a unique character, so that a computer can use it to store text and numbers that human beings can understand. To make the respective 128 Raman barcodes, the signal compounds A, D or D′ were thoroughly mixed with the reference compound R at the designed ratios in the solvent N-methylpyrrolidone (NMP), and then spotted onto a clean quartz slide. Table S1 and Fig. S3 in the ESI† list all the 128 ASCII–Raman codes and their octal codes, including the first 32 non-printing characters of actions and the remaining 96 printing characters.
Fig. 3a shows the overlaid spectra of 128 spots of solutions on a 75 × 25 mm quartz slide (Fig. S3a†), identifying 128 different codes, including 64 codes of AmDj (m = 0–7, j = 0–7), and 64 codes of AmD′j (m = 0–7, j = 0–7). While different compounds D and D′ were used to combine with A, their spectra have slight differences in peak positions. The overlaid spectra clearly present the variations in Raman shifts and intensities, therefore providing evidence for accurate identification of codes.
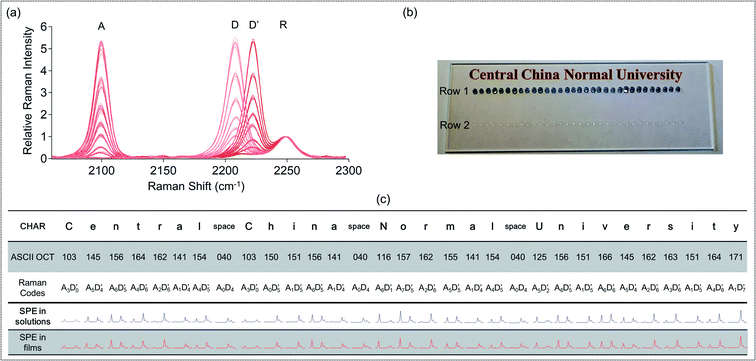
As ASCII can be used to express alphanumeric text, we demonstrated an application of this Raman–ASCII system to write a few English words. Fig. 3b and c show how the affiliation of this paper's authors, Central China Normal University, was written by this code system. All the letters and spaces between the words have their specific ASCII codes and were designed as unique Raman codes. These designed codes were fabricated using the simple strategy of mixing and spotting. The second row of Fig. 3c shows the measured spectra of the respective Raman codes in solutions. The third row of Fig. 3c shows the measured spectra of the respective Raman codes in the solid polymethylmethacrylate (PMMA) films on a quartz slide, as it is inconvenient to store the codes in a wet form. The decoding of these spectra was done by comparing them with the 128 standard ASCII codes in Fig. 3a. The decoded results were consistent with the intended designed codes.
In order to demonstrate the coding capability of our Raman barcode system, we chose Unicode as another example. The Unicode system contains significantly larger diversities than ASCII, and allows the representation and transportation of languages and symbols through many different platforms, devices and applications without confusion. The Unicode standard defines values for characters that have been split into 17 different sections. The most commonly used characters are included in the first section, known as the Basic Multilingual Plane (BMP). It defines 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 536 characters. Each character is written as a Unicode, e.g. U+xxxx, including a prefix of U+ and combinations of 4 hexadecimal code units. The expected capacity of our designed Raman–Unicode system will match these 65
536 characters. Each character is written as a Unicode, e.g. U+xxxx, including a prefix of U+ and combinations of 4 hexadecimal code units. The expected capacity of our designed Raman–Unicode system will match these 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 536 codes.
536 codes.
We used combinations of the compounds A (2100 cm−1)/A′ (2110 cm−1) in Band I, B (2134 cm−1)/B′ (2138 cm−1) in Band II, C (2168 cm−1)/C′ (2180 cm−1) in Band III, and D (2209 cm−1)/D′ (2223 cm−1) in Band IV. Since 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 536 is a large number, experimentally we did not make all the 65
536 is a large number, experimentally we did not make all the 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 536 samples. We simplified the test by measuring the code units at each frequency, which has been shown in Fig. 2c. As discussed previously, two compounds are used in parallel instead of simultaneously within the same bands, in order to avoid cross talking between codes of X and X′, thus allowing hexadecimal code units (8 + 8) in each band. Then, the total capacity of this specific Raman–Unicode is 16 × 16 × 16 × 16 = 65
536 samples. We simplified the test by measuring the code units at each frequency, which has been shown in Fig. 2c. As discussed previously, two compounds are used in parallel instead of simultaneously within the same bands, in order to avoid cross talking between codes of X and X′, thus allowing hexadecimal code units (8 + 8) in each band. Then, the total capacity of this specific Raman–Unicode is 16 × 16 × 16 × 16 = 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 536. This exactly matches the diversity of the Unicode BMP.
536. This exactly matches the diversity of the Unicode BMP.
We used this Raman–Unicode system to write a few Chinese words. Fig. 4 shows how the authors' affiliation in Chinese,  , was written by this code system. All the characters have their unique Raman codes listed in the third row. These designed codes were fabricated using the mixing and spotting strategy described previously. The 4th row of Fig. 4 shows the measured spectra of the respective Raman codes in solutions. Since it was not realistic to make all the 65
, was written by this code system. All the characters have their unique Raman codes listed in the third row. These designed codes were fabricated using the mixing and spotting strategy described previously. The 4th row of Fig. 4 shows the measured spectra of the respective Raman codes in solutions. Since it was not realistic to make all the 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 536 standard codes manually in the lab, we decoded the experimental spectra of “
536 standard codes manually in the lab, we decoded the experimental spectra of “ ” by comparing it with the coding unit graph in Fig. 2a. The decoded results were consistent with the intended designed codes.
” by comparing it with the coding unit graph in Fig. 2a. The decoded results were consistent with the intended designed codes.
Besides creating large-capacity 2-D Raman codes on surfaces, the codes can be fabricated by covalently attaching the Raman compounds to 3-D carriers as well. As the octal code units on resin beads were verified in the previous discussion, we tested a design of 64 codes using the code units of Ⓐ and Ⓓ. They were mixed together with Ⓡ at designed ratios, and then reacted with aminolated beads in a single step. The corresponding Raman spectra of the products were measured and plotted in Fig. 5a. Referring to the standard octal codes in Fig. 2d, the 64 spectra were easily decoded as 64 different 2-digit codes respectively, corresponding to ⒶmⒹn (m = 0–7, n = 0–7). We continued to test combinations of Ⓐ, Ⓒ and Ⓓ, by fixing the intensity of Ⓐ at Code #3 and Ⓓ at Code #4, and tuned the intensities of Ⓒ to Code #0–7. The orange spectra in Fig. 5b confirm the distinction of these 8 spectra, corresponding to 8 different 3-digit codes Ⓐ3ⒸnⒹ4 (n = 0–7). To continue the experiment and expand the combinations, we fixed the intensity of Ⓐ at Code #3, Ⓓ at Code #4, and Ⓒ at Code #5, and tuned the intensities of Ⓑ to Code #0–7. The green spectra in Fig. 5c confirm the distinction of these 8 spectra, referring to 8 different 4-digit codes Ⓐ3ⒷnⒸ5Ⓓ4 (n = 0–7). These encoded beads were stored at the room light and temperature for 5 months and the coded information was fully maintained (see the ESI†).
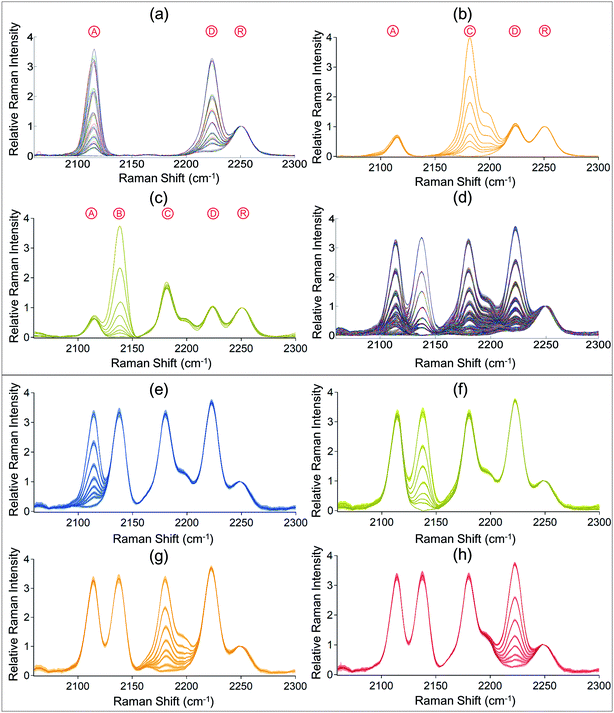 | ||
| Fig. 5 Verification of the coding capacity based on the experimental and simulated spectra of barcodes on beads. (a) 64 spectra of beads with covalently bound Ⓐ and Ⓓ at 64 different dosages, corresponding to ⒶmⒹn (m = 0–7, n = 0–7). (b) 8 spectra of beads with covalently bound Ⓐ, Ⓒ and Ⓓ at 8 different dosages, corresponding to Ⓐ3ⒸnⒹ4 (n = 0–7). (c) 8 spectra of beads with covalently bound Ⓐ, Ⓑ, Ⓒ and Ⓓ at 8 different dosages, corresponding to Ⓐ3ⒷnⒸ5Ⓓ4 (n = 0–7). (d) Plot of all the 4096 spectra (8 × 8 × 8 × 8) by combining and adding the spectra in Fig. 2d one by one. They represent the 4096 codes, ⒶmⒷnⒸiⒹj (m = 0–7, n = 0–7, i = 0–7, and j = 0–7). (e–h) The selected spectra from (b), representing ⒶmⒷ7Ⓒ7Ⓓ7 (m = 0–7), Ⓐ7ⒷnⒸ7Ⓓ7 (n = 0–7), Ⓐ7Ⓑ7ⒸiⒹ7 (i = 0–7) and Ⓐ7Ⓑ7Ⓒ7Ⓓj (j = 0–7), respectively. | ||
Based on the design of combining Ⓐ, Ⓑ, Ⓒ and Ⓓ, we employed one compound in each band and 8 Raman intensities of each compound in 4 spectral bands. Then the diversity of barcodes is calculated by 8 × 8 × 8 × 8 = 4096. As 4096 is a large number, experimentally we did not make all the 4096 samples. We simulated 4096 (84) spectra by sequentially adding up spectra of these 4 individual series of code units. Compared to the individual series of octal code units in Fig. 2d, Fig. 5d presents the simulations with similar trends. However, because these simulations contain massive information of all the 4096 spectra, it is not straightforward to determine whether each spectrum in the figure is reliably different from its nearby spectra. The cross talking between them would be serious when the adjacent peaks have high intensities, causing substantial interference from their signals at shoulder areas. Based on this ratiocination, from the 4096 spectra, we dug out the spectra of ⒶmⒷ7Ⓒ7Ⓓ7 (m = 0–7), Ⓐ7ⒷnⒸ7Ⓓ7 (n = 0–7), Ⓐ7Ⓑ7ⒸiⒹ7 (i = 0–7), and Ⓐ7Ⓑ7Ⓒ7Ⓓj (j = 0–7), respectively standing for Code #0–7 for one of the 4 peaks while the other 3 peaks were fixed at the maximum intensities (Code #7). If these 8 codes are distinguishable, all the remaining codes are identifiable at all the other situations when the adjacent peaks have lower intensities. Fig. 5e–h present the simulation results. With standard deviations of 5 independent measurements included, we did not observe cross talking between the nearby codes. These findings further confirm our expectation with high accountability that a capacity of 4096 for the encoded resin bead system is practical and feasible.
While a capacity of 4096 was demonstrated, it is further expandable by employing more coding compounds inside each band. As Fig. 2b shows the currently discovered carboxylated compounds that can be employed to covalently attach to beads, based on the calculation formula described previously, the total coding capacity will be (8 × 3) × (8 × 1) × (8 × 4) × (8 × 4) = 196![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 608.
608.
Such a super-capacity information-carrying system can solve many problems in the field of life sciences. Specifically, there is a great need for increasing the throughput of discovering or screening molecular/cellular targets, by simultaneously detecting multiple entities to significantly decrease the turn-around times.1–5,19 We demonstrate here a simple application of the encoded resin beads as information carriers allowing multiplex screening of a specific peptide target binding to cancer cells in a One-Bead-One-Compound (OBOC) library.
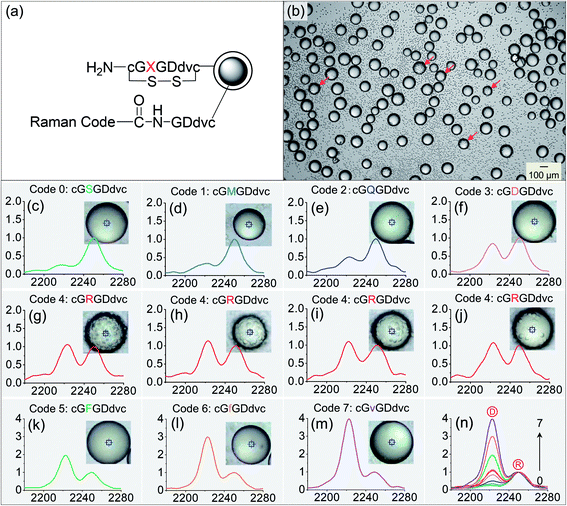
Since traditional peptide decoding procedures in OBOC libraries such as Edman degradation or mass spectrometry methods usually rely on liquid media, the decoding speed is restricted by tedious elution and separation steps after screening.20 We developed a new method to in situ decode peptide sequences rapidly using Raman barcodes. With a bi-layer strategy21 shown in Fig. 6a, 8 Raman codes were written on TentaGel beads to express the diversity of 8 different amino acids in the 8-mer cyclic peptide sequences -cGXGDdvc-, where X is the amino acid that varies. Among them, Code #4 stands for the peptide of -cGRGDdvc-, containing the well-known RGD sequence that strongly binds to the highly expressed αvβ3 integrin on the cell line U-87MG.22 Code #0, 1, 2, 3, 5, 6, and 7 represent other different peptides at the X position (-cGXGDdvc-, X ≠ R).
The mixed beads of 8 different peptides were incubated with U-87MG cells. The spectra of all the 8 codes were first obtained and saved for subsequent decoding use (Fig. 6n). Fig. 6b shows a bright-field image of the binding results. On approximately one eighth of the beads, there appeared positive binding. The closer views of the positive beads are shown in the insets of Fig. 6g–j. They present different binding strengths, e.g. very strong, strong, mild and weak binding, respectively. After identifying positive and negative beads, we measured their Raman spectra, and interestingly, found that no matter what the binding strength was, the decoded outcome was consistently Code #4 (-cGRGDdvc-). We also randomly picked a number of negative beads, and identified their codes (Fig. 6c–f and k–m). All of them corresponded to other codes, e.g. Code #0, 1, 2, 3, 5, 6, and 7. This confirms our expectation that the non-specific sequences were -cGXGDdvc- (X ≠ R).
The experiment above is an example of applying the information-carrying system to rapidly identify a cell-binding peptide. Based on the findings that the Raman codes on resin beads were compatible with synthetic reactions and biological recognitions, this strategy may be combined with the “split and pool” technique to boost the development of OBOC library screening. It should be feasible to encode thousands to millions of compounds within one library. In the subsequent screening process, positive beads were identified under a confocal microscope. Without the tedious process of isolation, recovery and elution steps used for the conventional decoding techniques, each single positive bead can be decoded in situ within 1 min to determine the exact structures of the positive compounds. This information-carrying system provides a novel solution to the essential challenges of analyzing sufficient molecular or cellular targets simultaneously, including creating coding systems with super capacity, stability, and efficiency, and obtaining the decoded information with accuracy and simplicity.
Conclusions
Optical multiplex barcode systems aim to carry massive and stable information that can be decoded with ease and accountability. In order to take advantage of the narrow bandwidths and the stable signals of molecular vibrational spectra, and overcome the difficulties of tuning the signals into distinguishable intensities for enhancing the coding capacity, we obtained a library of multiple alkyne compounds in 4 spectral bands. In each band, every single compound maintains a specific Raman shift and allows for a design of octal code units using the strategy of mixing with a reference compound at designated dosages. The edited codes can be written on 2-D surfaces and 3-D beads through spotting and covalent bonding, respectively. The practical capacity of 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 536 (164) and 4096 (84) for these two systems was easily achieved by selecting two and one compounds from each band and using them in combination, respectively. This design can be further extended to achieve a super capacity of 524
536 (164) and 4096 (84) for these two systems was easily achieved by selecting two and one compounds from each band and using them in combination, respectively. This design can be further extended to achieve a super capacity of 524![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 288 and 196
288 and 196![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 608, respectively, when all the discovered compounds are used for coding.
608, respectively, when all the discovered compounds are used for coding.
We demonstrated an application of encoding ASCII and Unicode for data storage, by writing the Raman octal and hexadecimal code units onto quartz surfaces, respectively. The example of writing the authors' affiliation in English (Central China Normal University) and in Chinese ( ) implies that these super-capacity information-carrying systems have great potential of storing massive data in a novel method. In addition, by encoding information onto resin beads and effectively decoding a specific peptide binding to cancer cells in situ, the super-capacity information-carrying system opens new prospects to simultaneously track multiple molecular/cellular targets in large quantities and meet the current significant demand in increasing the throughput of discovery and screening in life sciences. The future direction would be the automation of the encoding reactions involving high-throughput fabrication of practical super-capacity information carriers, and the automation of the decoding process allowing large measurement statistics spanning the total library of all standard codes and computer-based decoding from standard algorithms.
) implies that these super-capacity information-carrying systems have great potential of storing massive data in a novel method. In addition, by encoding information onto resin beads and effectively decoding a specific peptide binding to cancer cells in situ, the super-capacity information-carrying system opens new prospects to simultaneously track multiple molecular/cellular targets in large quantities and meet the current significant demand in increasing the throughput of discovery and screening in life sciences. The future direction would be the automation of the encoding reactions involving high-throughput fabrication of practical super-capacity information carriers, and the automation of the decoding process allowing large measurement statistics spanning the total library of all standard codes and computer-based decoding from standard algorithms.
Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (21605054), the Central China Normal University (CCNU) new PI start-up fund (210-31102) and self-determined research funds of CCNU (234-20205016002 and 234-20205170355) provided by the colleges' basic research and operation of the Ministry of Education (MOE) to the central universities. TG acknowledges support from the Program of Introducing Talents of Discipline to the University of China (111 program, B17019).Notes and references
- D. C. Pregibon, M. Toner and P. S. Doyle, Science, 2007, 315, 1393–1396 CrossRef PubMed.
- R. Wilson, A. R. Cossins and D. G. Spiller, Angew. Chem., Int. Ed., 2006, 45, 6104–6117 CrossRef.
- Y. Leng, K. Sun, X. Chen and W. Li, Chem. Soc. Rev., 2015, 44, 5552–5595 RSC.
- S. Shikha, T. Salafi, J. Cheng and Y. Zhang, Chem. Soc. Rev., 2017, 46, 7054–7093 RSC.
- G. Lin, M. A. B. Baker, M. Hong and D. Jin, Chem, 2018, 4, 1–25 Search PubMed.
- M. Han, X. Gao, J. Z. Su and S. Nie, Nat. Biotechnol., 2001, 19, 631–635 CrossRef.
- B. Houser, Arch. Physiol. Biochem., 2012, 118, 192–196 CrossRef.
- S. Lu, D. S. Zhang, D. Wei, Y. Lin, S. Zhang, H. He and H. Xu, Chem. Mater., 2017, 29, 10398–10408 CrossRef.
- H. H. Gorris and O. S. Wolfbeis, Angew. Chem., Int. Ed., 2013, 52, 3584–3600 CrossRef.
- Z. Wang, S. Zong, L. Wu, D. Zhu and Y. Cui, Chem. Rev., 2017, 117, 7910–7963 CrossRef.
- Z. Wang, S. Zong, W. Li, C. Wang and Y. Cui, J. Am. Chem. Soc., 2012, 134, 2993–3000 CrossRef.
- S. Jeong, H. Kang, M. G. Cha, S. G. Lee, J. Kim, H. Chang and D. H. Jeong, Chem. Commun., 2019, 55, 2700–2703 RSC.
- H. Yamakoshi, K. Dodo, M. Okada, J. Ando, A. Palonpon, K. Fujita, S. Kawata and M. Sodeoka, J. Am. Chem. Soc., 2011, 133, 6102–6105 CrossRef.
- Y. Zeng, J. Ren, A. Shen and J. Hu, J. Am. Chem. Soc., 2018, 140, 10649–10652 CrossRef.
- F. Hu, C. Zeng, R. Long, Y. Miao, L. Wei, Q. Xu and W. Min, Nat. Methods, 2018, 15, 194–200 CrossRef.
- C. Zong, M. Xu, L. J. Xu, T. Wei, X. Ma, X. S. Zheng and B. Ren, Chem. Rev., 2018, 118, 4946–4980 CrossRef.
- B. Mir-Simon, I. Reche-Perez, L. Guerrini, N. Pazos-Perez and R. A. Alvarez-Puebla, Chem. Mater., 2015, 27, 950–958 CrossRef.
- Y. Tang, Y. Thillier, R. Liu, X. Li, K. S. Lam and T. Gao, Anal. Chem., 2017, 89, 7000–7008 CrossRef CAS.
- V. V. Komnatnyy, T. E. Nielsen and K. Qvortrup, Chem. Commun., 2018, 54, 6759–6771 RSC.
- S. H. Joo, Q. Xiao, Y. Ling, B. Gopishetty and D. Pei, J. Am. Chem. Soc., 2006, 128, 13000–13009 CrossRef CAS.
- R. Liu, J. Marik and K. S. Lam, J. Am. Chem. Soc., 2002, 124, 7678–7680 CrossRef CAS PubMed.
- Y. Wang, W. Xiao, Y. Zhang, L. Meza, H. Tseng, Y. Takada and K. S. Lam, Mol. Cancer Ther., 2016, 15, 232–240 CrossRef CAS.
- N. Goldman, P. Bertone, S. Chen, C. Dessimoz, E. M. LeProust, B. Sipos and E. Birney, Nature, 2013, 494, 77–80 CrossRef CAS.
- L. Ceze, J. Nivala and K. Strauss, Nat. Rev. Genet., 2019, 20, 456–466 CrossRef CAS.
- M. G. Rutten, F. W. Vaandrager, J. A. Elemans and R. J. Nolte, Nat. Rev. Chem., 2018, 2, 365–381 CrossRef.
- J. F. Lutz, Macromolecules, 2015, 48, 4759–4767 CrossRef CAS.
- J. A. Amalian, G. Cavallo, A. Al Ouahabi, J. F. Lutz and L. Charles, Anal. Chem., 2019, 91, 7266–7272 CrossRef CAS.
- B. J. Cafferty, A. S. Ten, M. J. Fink, S. Morey, D. J. Preston, M. Mrksich and G. M. Whitesides, ACS Cent. Sci., 2019, 5, 911–916 CAS.
- K. Degardin, Y. Roggo, F. Been and P. Margot, Anal. Chim. Acta, 2011, 705, 334–341 CrossRef CAS.
- J. D. Rodriguez, B. J. Westenberger, L. F. Buhse and J. F. Kauffman, Anal. Chem., 2011, 83, 4061–4067 CrossRef CAS.
- K. Kwok and L. S. Taylor, Vib. Spectrosc., 2012, 61, 176–182 CrossRef CAS.
- P. de Peinder, M. J. Vredenbregt, T. Visser and D. de Kaste, J. Pharm. Biomed. Anal., 2008, 47, 688–694 CrossRef CAS.
- L. S. Lawson and J. D. Rodriguez, Anal. Chem., 2016, 88, 4706–4713 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental section including materials, synthesis, preparation, and characterization procedures. See DOI: 10.1039/c9sc05133c |
| This journal is © The Royal Society of Chemistry 2020 |