Open Access Article
Open Access ArticleWall teichoic acids govern cationic gold nanoparticle interaction with Gram-positive bacterial cell walls†
Emily R.
Caudill
 a,
Rodrigo Tapia
Hernandez‡
a,
Rodrigo Tapia
Hernandez‡
 b,
Kyle P.
Johnson§
c,
James T.
O'Rourke¶
b,
Kyle P.
Johnson§
c,
James T.
O'Rourke¶
 a,
Lingchao
Zhu||
d,
Christy L.
Haynes
a,
Lingchao
Zhu||
d,
Christy L.
Haynes
 c,
Z. Vivian
Feng
c,
Z. Vivian
Feng
 *b and
Joel A.
Pedersen
*b and
Joel A.
Pedersen
 *aefg
*aefg
aDepartment of Chemistry, University of Wisconsin-Madison, 1101 University Avenue, Madison, WI 53706, USA. E-mail: joelpedersen@wisc.edu
bChemistry Department, Augsburg University, Minneapolis, MN 55454, USA. E-mail: feng@augsburg.edu
cDepartment of Chemistry, University of Minnesota, Minneapolis, MN 55455, USA
dDepartment of Chemistry, University of Pennsylvania, 231 S 34th St, Philadelphia, PA 19104, USA
eEnvironmental Chemistry and Technology Program, University of Wisconsin-Madison, 660 North Part Street, Madison, WI 53706, USA
fDepartment of Soil Science, University of Wisconsin-Madison, 1525 Observatory Drive, Madison, WI 53706, USA
gDepartment of Civil & Environmental Engineering, University of Wisconsin-Madison, 1415 Engineering Drive, Madison, WI 53706, USA
First published on 23rd March 2020
Abstract
Molecular-level understanding of nanomaterial interactions with bacterial cell surfaces can facilitate design of antimicrobial and antifouling surfaces and inform assessment of potential consequences of nanomaterial release into the environment. Here, we investigate the interaction of cationic nanoparticles with the main surface components of Gram-positive bacteria: peptidoglycan and teichoic acids. We employed intact cells and isolated cell walls from wild type Bacillus subtilis and two mutant strains differing in wall teichoic acid composition to investigate interaction with gold nanoparticles functionalized with cationic, branched polyethylenimine. We quantified nanoparticle association with intact cells by flow cytometry and determined sites of interaction by solid-state 31P- and 13C-NMR spectroscopy. We find that wall teichoic acid structure and composition were important determinants for the extent of interaction with cationic gold nanoparticles. The nanoparticles interacted more with wall teichoic acids from the wild type and mutant lacking glucose in its wall teichoic acids than those from the mutant having wall teichoic acids lacking alanine and exhibiting more restricted molecular motion. Our experimental evidence supports the interpretation that electrostatic forces contributed to nanoparticle–cell interactions and that the accessibility of negatively charged moieties in teichoic acid chains influences the degree of interaction. The approaches employed in this study can be applied to engineered nanomaterials differing in core composition, shape, or surface functional groups as well as to other types of bacteria to elucidate the influence of nanoparticle and cell surface properties on interactions with Gram-positive bacteria.
Introduction
A molecular-level understanding of nanomaterial interactions with bacterial cell surfaces is needed to effectively design nanomaterial-enabled products intended to modulate bacterial populations (e.g., antimicrobials, antifouling surfaces) and for assessing the potential effects of engineered nanomaterials released into the environment. The increase in engineered nanomaterial production and their incorporation into commercial products makes their introduction into the environment inevitable during some portion of their life cycle.1 Bacteria are vital to many biogeochemical processes and may play a role in the introduction of nanomaterials into some food webs. Molecular-level insight into the interactions of bacteria with nanomaterials may facilitate their design to either target harmful bacteria more effectively or reduce their negative impacts on beneficial microbiota.The distinct cell envelope architectures of Gram-negative and Gram-positive bacteria2 lead to the expectation that their interactions with charged nanoparticles differ, as indeed has been shown in earlier studies.3–7 The Gram-negative cell envelope is composed of two phospholipid membranes with a thin layer of peptidoglycan sandwiched between them; the outer leaflet of the outer membrane is composed primarily of lipopolysaccharides. In contrast, Gram-positive bacterial cells are bounded by a single cell membrane, which includes lipoteichoic acids anchored into it, and a thick cell wall composed of peptidoglycan with covalently attached wall teichoic acids (WTAs). Previous investigation of electrostatically driven association of nanoparticles with the Gram-negative bacterial outer membrane employed intact and lipopolysaccharide-depleted bacteria and supported lipid bilayers incorporating lipopolysaccharides to demonstrate that lipopolysaccharide structure influenced the extent and distance from the outer membrane lipid bilayer of nanoparticle binding.8 Experimental modeling Gram-positive cell surfaces to probe the nanomaterial–bacterium interface requires a different approach be taken. The prominent Gram-positive bacterial cell wall surface structures lack lipid-like character, precluding the use of lipid bilayers to study interactions with nanoparticles. Also, given that teichoic acids can account for up to 50% of the mass of the cell wall9 and assist in maintaining cation homeostasis for the cell, understanding their role in interacting with nanoparticles is vital, particularly for nanoparticles functionalized with cationic groups.10
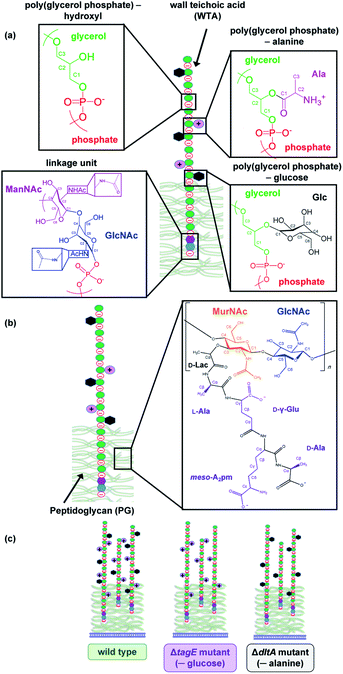
For Bacillus subtilis SB491, a Gram-positive bacterium found in soil and in human and ruminant gastrointestinal tracts, the hydroxyl groups of the glycerol units of the wall teichoic acid poly(glycerolphosphate) backbone can be substituted with glucose (Glc) or alanine (Ala) residues (Fig. 1). Wall teichoic acids are covalently bound to cell wall peptidoglycan via a phosphodiester bond formed between the phosphate group bound to the N-acetylglucosamine (GlcNAc) residue in the WTA disaccharide linkage unit and the C6 hydroxyl of N-acetylmuramic acid (MurNAc) residue in peptidoglycan. Each WTA molecule contains between 45 to 60 glycerolphosphate units.11 Peptidoglycan is composed of alternating N-acetylglucosamine (GlcNAc) and MurNAc residues, connected via a MurNAc D-lactyl group to a tetrapeptide (L-alanyl-D-γ-glutamyl-meso-diaminopimelyl-D-alanine). The D-Ala of the tetrapeptide is bound to the meso-diaminopimelyl of a pentapeptide (L-alanyl-D-γ-glutamyl-meso-diaminopimelyl-D-alanine-D-alanine), which in turn is bound to another MurNAc residue. Approximately every ninth MurNAc unit contains an attached WTA polymer. For Bacillus subtilis, the crosslinking of peptidoglycan strands results in a meshwork having a reported effective pore size of 4.24 or 5 nm.12,13
Bacteria synthesize WTAs within the cytoplasm and translocate them through the cell membrane. Prior to translocation, Glc is added to the C2 hydroxyl of the WTA glycerol via the wall teichoic acid glycosyltransferase Tag E (encoded by the teichoic acid glycerol (tagE) gene).14 After translocation, a proposed D-alanyl carrier protein ligase (encoded by the D-alanyl-lipoteichoic acid A (dltA) gene) attaches D-Ala to the WTAs also to the C2 hydroxyl of the WTA glycerol.10,15 In the lipoteichoic acids of B. subtilis, approximately 9% of the glycerolphosphate moieties become substituted with D-Ala and 64% with Glc.15 The degree of substitutions of wall teichoic acids have not been reported to our knowledge. Deletion of the tagE gene results in elaboration of WTA lacking Glc attached to the poly(glycerolphosphate) backbone.14 Deletion of genes in the dlt operon of B. subtilis results in the production of teichoic acids lacking D-Ala and a concomitant increase in methicillin susceptibility.16 Similarly, the absence of D-alanylation in Gram-positive Lactococcus lactis resulted in decreased resistance to cationic antimicrobials nisin and lysozyme.17 These results reinforce the notion that the lack of D-Ala in teichoic acids can alter interaction of the cell surface with external factors, such as antimicrobial agents and potentially nanomaterials.
In the present study, we investigated the interactions of WTA from the Gram-positive bacterium Bacillus subtilis with gold nanoparticles functionalized with cationic branched polyethylenimine (bPEI-AuNPs). The bPEI-AuNPs were chosen for the colloidal stability conferred by their strong positive charge (vide infra) as well as for the availability of bPEI for use as an experimental control. We focused on the cell surface components responsible for the interaction of cationic nanoparticles with bacterial cell surfaces because prior investigation demonstrated negligible interaction of wild type B. subtilis cells with anionic gold nanoparticles.3 We employed three strains of B. subtilis possessing the same peptidoglycan structure but differing in WTA structure due to genetic modifications: wild type (SB 491), the tagE knockout strain ΔtagE with non-glycosylated WTA, and the dltA knockout strain ΔdltA with WTA lacking D-Ala. Experiments employed intact cells or bacterial exoskeletons (sacculi) composed of peptidoglycan and covalently bound WTAs. We note that the sacculi did not include lipoteichoic acids which are anchored in the cell membrane or can be noncovalently associated with the peptidoglycan matrix.11 Stable isotope-labelled sacculi were used to facilitate investigation of nanoparticle interaction with WTA and peptidoglycan via solid-state nuclear magnetic resonance (NMR) spectroscopy. Solid-state NMR has been previously applied to characterize WTA of different bacterial species, propose divalent cation binding sites on bacterial sacculi, investigate the interaction of cationic polymers with sacculi, and determine the influence of antibiotics on cell wall composition.18–20 To our knowledge, the present work represents the first solid-state NMR investigation of nanomaterial interaction with bacterial cell wall components. Flow cytometry performed on intact bacterial cells showed that WTA composition impacts cationic gold nanoparticle association with bacterial cells. Solid-state 31P- and 13C-NMR experiments performed on hydrated sacculi allowed identification of the chemical groups in peptidoglycan and WTA involved in binding.
Results and discussion
Bacterial cell surface charge
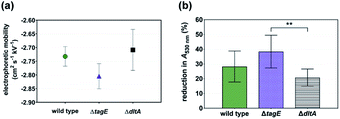
We investigated the interaction of cationic bPEI-AuNPs with the surfaces of wild type and WTA-mutant Gram-positive Bacillus subtilis cells. We hypothesized that bPEI-AuNP association with the cell surfaces is governed predominantly by electrostatic interactions. We evaluated the impact of the genetic mutations on overall cell surface charge by two approaches: whole-cell electrophoretic mobility (ue) measurement and the binding of the cationic protein cytochrome c. The permeability of bacterial surface structures (e.g., WTA) to ions and water (i.e., the “softness” of the surface) precludes determination of cell surface ζ-potential from electrophoretic mobility measurements.21 We therefore compared electrophoretic mobilities of the bacterial cells directly22,23 recognizing that in addition to the potential at the polyelectrolyte–solution interface, ue depends on the thickness, charge density, permeability, and homogeneity of the soft layer.24 All three strains exhibited negative electrophoretic mobility (Fig. 2a), largely attributable to the phosphate groups in the WTA on the cell surfaces. The electrophoretic mobilities of the three strains were statistically indistinguishable.We measured the binding capacity of the cationic protein cytochrome c (+8 at pH 7) for B. subtilis cells of each strain as a proxy for their relative amounts of anionic surface charge. The absorbance of cytochrome c solutions at 530 nm was used to determine the amount of the protein removed from solution upon exposure to bacterial suspensions to provide a measure of the amount of negative charge on the cell surfaces.16Fig. 2b shows that the reduction in A530 was larger for ΔtagE strain than for the ΔdltA strain indicating that the former possessed a larger number of negatively charged surface sites. The extent of cytochrome c binding to the wild type and ΔdltA strains was statistically indistinguishable, suggesting comparable anionic surface site densities. This and the equivalent ue for these strains contrasts with earlier reports of increased negative surface charge in dltA-gene deleted strains.11,16 Our results are, however, consistent with those reported for a different Gram-positive bacterium, Lactococcus lactis, for which changes in the extent of D-alanization did not impact electrophoretic mobility.17
WTA composition
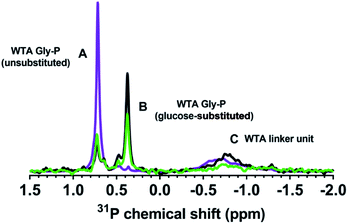
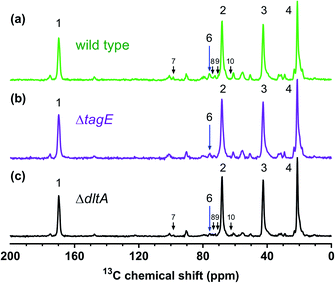
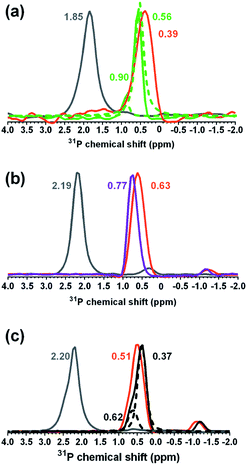
To verify that the B. subtilis mutants expressed the expected phenotypes, we examined the Ala and Glc contents of WTA isolated from the three strains. We hydrolyzed Ala from WTA and derivatized it with Marfey's reagent to allow detection via absorbance at 820 nm.25,26D-Ala was clearly present in WTA from both wild type and ΔtagE strains, but was significantly diminished in the ΔdltA sample (Fig. S1†). The clear differences in the chromatograms indicate that knockout of the D-alanyl-lipoteichoic acid A (ΔdltA) gene resulted in the expected phenotype.We conducted solution 31P-NMR experiments to obtain evidence for the Glc content of isolated WTA from each B. subtilis strain. Fig. 3 compares the 31P-NMR spectra from the strains collected using a pulse sequence to directly observe P nuclei. The 31P-NMR spectra of isolated WTA exhibit three main peaks centered at approximately 0.73 (peak A), 0.38 (peak B), and −0.80 (peak C) ppm. Peaks A and B are both assigned to the phosphorus in the WTA poly(glycerolphosphate) backbone (Fig. 1) based on previous reports for wall teichoic18 and lipoteichoic acids.27 Peak C is assigned to phosphorus in the linker unit as previously reported.28–30 A distinctive feature of the ΔtagE mutant (WTA glycosyltransferase Tag E knockout) WTA spectrum is the absence of peak B. Because peak A arises from the unsubstituted glycerol,51 we deduce that peak B corresponds to glycerolphosphate substituted with Glc. The proximity in space of an electron-rich Glc residue to the phosphorus nuclei likely causes the observed upfield position of peak B relative to peak A.31 The absence of peak B in the ΔtagE spectrum provides clear evidence that the tagE gene deletion resulted in the intended phenotypic changes.
Peak positions in 31P-NMR spectra of WTA from the ΔdltA strain closely resemble those in the spectrum from the wild type. We attribute the lack of a unique 31P feature corresponding to the presence of D-Ala on WTA to hydrolysis of the ester bond and loss of this substituent during the WTA isolation process.32,33 Quantitative comparison of the wild type and ΔdltA WTA spectra leads to three important conclusions. First, the ΔdltA mutant likely has more linker units and therefore a larger number of WTA chains than does the wild type. Peak C is more prominent in the spectrum from the ΔdltA mutant than in that from the wild type strain. The ratio of the integrated areas of peaks A + B (WTA backbone phosphates) to that of peak C (phosphorus in the linker unit) was higher for the wild type than for the ΔdltA strain (p < 0.05, n = 4; Table 1). (The peak ratio for the ΔtagE mutant did not differ from the other two strains (p > 0.05).) The broadness of peak C likely reflects a range of chemical environments for the phosphorus in the linker units. Second, the ΔdltA mutant has shorter WTA chains based on the first conclusion and the observation that the integrated peak areas of A + B, which correspond to the total glycerophosphates in WTA chains do not differ significantly in wild type and ΔdltA mutant. Third, the extent of Glc substitution (which gives rise to peak B) is higher for the ΔdltA strain than the wild type as indicated by comparison of the integrated areas of peak A and B (A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B in Table 1). In summary, the 31P-NMR data are consistent with ΔdltA mutant having more numerous and shorter WTA chains with more Glc substitution than wild type.
B in Table 1). In summary, the 31P-NMR data are consistent with ΔdltA mutant having more numerous and shorter WTA chains with more Glc substitution than wild type.
The properties of the bacterial cells and their WTA described above confirmed that the mutations led to the expected changes in WTA structure. However, the trends in net cell surface charge ran counter to the straightforward explanation that ΔtagE differed only in lacking Glc and ΔdltA differed only in lacking D-Ala. Rather, the results suggest that cell surface characteristics of the ΔtagE and ΔdltA mutants differ beyond the mere absence of Glc and D-Ala, respectively. Comparison of signal intensities in the 31P-NMR spectra suggests that WTA from the ΔtagE strain contained a larger amount of phosphate relative to that from the other two strains. This was corroborated by colorimetric phosphorous analysis (Fig. S2†).34 Therefore, we hypothesized that the small variations in surface charge among B. subtilis mutants arose from differences in the length or number of WTA chains. Wall teichoic acid contributes substantially to the surface charge of Gram-positive bacteria. Phosphorus analysis alone cannot discriminate whether the higher P content of the WTA from the ΔtagE strain was due to a larger number of or longer WTA chains. Nonetheless, more negative surface charge of the ΔtagE relative to the ΔdltA strain (Fig. 2b) may be related to the larger amount of glycerophosphate in the former. Comparison of the phosphate content of wild type and ΔtagE strains suggests that factors in addition to WTA glycerophosphate (e.g., lipoteichoic acids, not included in the total phosphate determination here) may contribute to overall negative surface charge of the cells.
Solid-state 31P-NMR analysis of sacculi
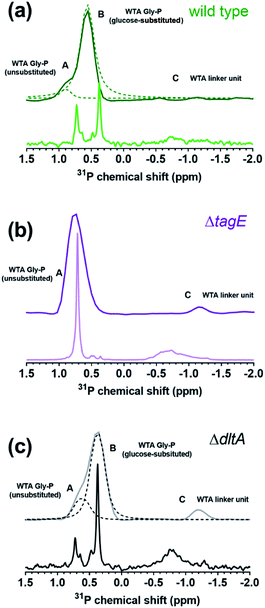
We acquired solid-state 31P-NMR spectra of isolated sacculi, which contain only peptidoglycan and WTA, from each strain and compared them with the solution 31P-NMR spectra for the corresponding isolated WTAs (Fig. 4a–c). The solution and solid-state NMR spectra resembled one another for each strain, although the peaks in the solid-state spectra exhibited the broadening expected from the restricted molecular motion of WTA covalently bound to and embedded within the peptidoglycan meshwork of the sacculi. Larger molecules have shorter transverse, T2, relaxation times, resulting in broader peaks.35 Peak shifts are presented in Tables S1 and S2,† and additional observations are provided in the ESI.†The positions of peaks A and B differ slightly in the deconvolved solid-state spectra for wild type (Fig. 4a) and ΔdltA (Fig. 4c) relative to the solution NMR spectra; this is attributable to either the presence of shoulders on the peaks in the solution spectra or differences in chemical environment of the phosphorus nuclei (i.e., the presence of peptidoglycan in the sacculi samples). Overall, the close resemblance of 31P-NMR spectra of isolated WTA and cell wall sacculi suggests that the phosphorus signals in the sacculi samples arise exclusively from WTA, confirming the purity of the isolated sacculi with respect to other P-containing species. Though unlikely given the thorough washing steps, any phospholipids remaining in the sacculi or WTA samples would produce signals in the same spectral region (between 1 and −1 ppm).36,37 Specifically, the 31P chemical shifts produced by phosphatidylethanolamine and phosphatidylglycerol lipids (the dominant phospholipids in Gram-positive bacterial membranes) occur at approximately −0.25 ppm and 0.24 ppm, respectively.38 The absence of such peaks confirms the purity of both sacculi and isolated WTA.
Solid-state 13C-NMR analysis of sacculi
We further probed the sacculi by solid-state 13C-NMR (Fig. 5). Labeled structures are in Fig. 1 and enlarged in Fig. S3.† Peaks in these spectra reflect the C atoms in the peptidoglycan amino acid and amino sugar residues and in the WTA (amino) sugar residues and glycerol. The four large peaks dominating the spectra reflect the most abundant cell wall components, which are primarily associated with the GlcNAc and MurNAc amino sugar residues in peptidoglycan. Peak assignments were based on previous reports39,40 and are provided in Table S3.† The carbons of the WTA glycerol and Glc (when present) produces maller peaks. The Glc carbon peaks were often difficult to discern; we consider the assignments of these peaks tentative. The other smaller peaks reflect the remaining C atoms in the amino sugar residues in peptidoglycan and the WTA linker unit, and the peptide and D-lactyl group in peptidoglycan.The 13C spectra for the three strains closely resemble one another with respect to peak positions. The peaks attributed to Glc residues were quite small and are indicated in Fig. 5 by arrows in the spectra of the sacculi from the wild type and ΔdltA strains; these peaks are displayed more clearly in Fig. S4.† Wall teichoic acid from the wild type and ΔdltA strains contains Glc, while the WTA of ΔtagE is unsubstituted (see also Fig. 4). Given the nature of cross polarization experiments, rigorous, quantitative comparison of peak areas among the three strains is not possible. Cross polarization transfers magnetization from a nearby network of abundant ½-spin nuclei (1H in our case) to the observed nucleus (13C) during the contact period. Thus, peak intensity and area depend on both the abundance of the observed nuclei and the number of protons on nearby carbon nuclei (within 3–4 Å). The solution 31P-NMR results indicated that the ΔdltA strain contains more Glc than wild type. We further note that the ΔtagE strain exhibited the largest 13C chemical shift at C1 and C3 and the smallest at C2 of the glycerolphosphate (Table S3†).
To provide a measure of the rigidity of the peptidoglycan meshwork and the covalently bound WTA molecules, we quantified longitudinal relaxation time-constants, T1, for 13C in sacculi from the three B. subtilis strains using inversion-recovery experiments (Fig. S6 and S7†). Our analysis focused on nuclei for which we can assume dipole–dipole interaction dominates relaxation rather than chemical shift anisotropy. We therefore omitted data from carbonyl carbons from the analysis. Relaxation times were measured between −8 and 30 °C to determine the molecular regime (small or large) based on the increase in molecular motion with temperature. With the exception of Cγ in the Glu residues of ΔtagE sacculi, T1 decreased as temperature increased for all carbon nuclei analyzed (fitted slopes showed non-zero linear dependence; Fig. S5 and Table S4†). For residues exhibiting a decrease in T1 with increasing temperature, larger T1 values indicate hampered motion and higher structural rigidity and smaller T1 values reflect more rapid relaxation due to higher molecular motion.18,41
We determined T1 values for the 13C nuclei in peptidoglycan and WTA indicated in Fig. 6, as well as for Cγ of Glu and Cβ of meso-A2pm in peptidoglycan (Fig. S5†). We focused on peaks attributable solely to peptidoglycan or to WTA. At 30 °C, the temperature closest to the sample temperature in the solid-state NMR experiments, T1 values for peptidoglycan amino sugar residues and the WTA glycerolphosphate of ΔdltA were larger than those of the wild type and ΔtagE strains (Fig. 6). In particular, T1 of C2 (the location of Glc substitution in the wild type and ΔdltA strains) was larger in ΔdltA than for wild type and ΔtagE, reflecting more restricted molecular motion for the ΔdltA strain. Differences among T1 values for the three strains were also apparent at 18 and 12 °C, but not at lower temperatures (p > 0.05; Fig. S6†). The T1 values indicate that the molecular motion of peptidoglycan strands from the ΔdltA strain are more restricted than those from the wild type and ΔtagE strains, which exhibit similar molecular motion. Likewise, the WTA from the ΔdltA strain exhibits more restricted molecular motion than does that from the other two strains. The glycerolphosphate carbons bearing –OH groups (viz. C1 and C3) exhibit somewhat higher molecular motion in the ΔtagE strain than in the other two strains. For resonances associated with amino acid residues in peptidoglycan, T1 values did not differ among the three strains at any temperature (p > 0.05).
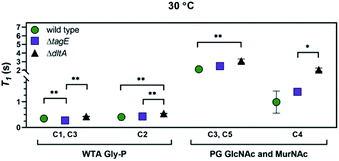 | ||
| Fig. 6 T 1 relaxation time constants at 30 °C for the indicated 13C resonances of sacculi from wild type, ΔtagE, and ΔdltA strains of B. subtilis: GlcNAc C3 and MurNAc C5 of peptidoglycan, GlcNAc and MurNAc C4 of peptidoglycan, glycerolphosphate C1 and C3 of wall teichoic acids, and glycerolphosphate C2 of wall teichoic acids. Non-linear fit statistical significance: *, p < 0.05; **, p < 0.01. Error bars represent standard error in non-linear fitting from T1 relaxation time equation. See Fig S6† for T1 relaxation times for other temperatures. Abbreviations: GlcNAc, N-acetylglucosamine; MurNAc, N-acetylmuramic acid. | ||
The lower molecular motion of WTA in ΔdltA sacculi relative to those of the other two strains is consistent with the more abundant Glc substitution of its glycerolphosphate (Table 1), the larger number of WTA molecules bound to peptidoglycan (Fig. 3), and less protrusion into solution due to fewer glycerolphosphate repeat units per WTA molecule (Fig. S7†). As noted above, the ΔdltA strain appears to produce a larger number of shorter WTA molecules than the other two strains. Shorter WTA strands dissipate energy less efficiently, leading to longer relaxation times. Both solution 31P-NMR and phosphorus analysis indicate that the ΔtagE strain contains the most glycerophosphate groups, although this strain lacks Glc side chains, and the distance that WTA molecules protrude beyond the peptidoglycan meshwork is unknown. Overall, we hypothesize that differences in molecular motion result from the difference in type and number of substituents on the WTA glycerol, and the extent to which the WTA molecules protrude from the peptidoglycan layer. Specifically, the glycerol substituent Glc is larger than –OH; high amounts of Glc substitution and close proximity of Glc residues to other Glc residues would slow the molecular motion of WTA. Furthermore, more hindered molecular motion is expected for shorter WTA as they extend into solution from the peptidoglycan matrix to a smaller degree.
Nanoparticle properties
To investigate the differential interactions between the B. subtilis strains and cationic nanoparticles, we exposed the bacteria to bPEI-AuNPs with core diameters of 10.9 ± 1.8 nm as indicated by their localized surface plasmon resonance peak,42 in good agreement with the diameter reported by the manufacturer based on analysis of transmission electron microscopy images (12.1 ± 0.8 nm). We characterized the hydrodynamic and electrokinetic properties of the particles in 0.025 M NaCl buffered to pH 7.4 with 0.002 M HEPES, the solution we used to study the interaction of the bPEI-AuNPs with intact cells. Under these solution conditions, the number mean hydrodynamic diameter was 37.7 ± 0.3 nm, and the ζ-potential was +23 ± 5.9 mV. The bPEI ligands are covalently attached to the nanoparticle surfaces. Nevertheless, we determined that a 0.93 nM solution of bPEI-AuNPs contained 792 ± 72 nM bPEI, using the reported average molecular mass for bPEI of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da.
000 Da.
Nanoparticle–cell surface association
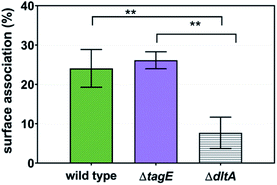
We employed flow cytometry to quantify the number of B. subtilis cells of each strain that had bPEI-AuNPs associated with them.3,8 We previously demonstrated that flow cytometry can be used to quantify the number of bacterial cells having AuNPs associated with cell surfaces and that the measurements correlate well with qualitative TEM observations.3,43 Cells were stained with a membrane-permeant nucleic acid stain to allow intact cells to be sorted from cellular debris. The strong localized surface plasmon resonance scattering signal from the AuNPs allowed cells with surface-associated AuNPs to be discriminated from those lacking associated AuNPs. The detected AuNP signals in flow cytometry did not necessarily originate from individual AuNPs; previous TEM analyses have revealed the likelihood of NP aggregation on bacterial surfaces.3,6,7Fig. 7 shows the proportion of cells from each strain with bPEI-AuNPs associated with their surfaces (of 104 bacterial cells). Fewer ΔdltA cells had AuNPs associated with them than did the wild type and ΔtagE strains (p < 0.01). No difference was observed between the wild type and ΔtagE strains.To gain further insight into the chemical basis for the difference in bPEI-AuNP association with cells from the three strains, we employed solid-state 31P- and 13C-NMR. Solid-state 31P-NMR spectra were collected from sacculi from the three B. subtilis strains before and after exposure to 34 nM bPEI-AuNPs and to 29.1 μM free bPEI polymer, the estimated amount of free polymer in a 34 nM AuNP solution (Fig. 8). For the wild type and ΔtagE strains, the peaks associated with backbone phosphates shifted upfield upon nanoparticle interaction (Table S2†). For the ΔdltA strain, the dominant peak in the spectrum (peak B) appears to shift downfield upon nanoparticle addition. For all three strains, the peaks associated with backbone phosphates shifted substantially downfield upon exposure to free bPEI polymer.
Upfield peak shifts typically result from shielding due to addition of electron density to the observed nuclei. Downfield peak shifts arise from deshielding, due to removal of electron density. The downfield peak shifts observed for all strains upon exposure to bPEI polymer (grey traces in Fig. 8) reflect removal of electron density from the P nuclei, likely due to formation of hydrogen bonds between primary amines of bPEI and WTA phosphate oxygen.44 In contrast, addition of bPEI-AuNPs produced peak shifts in the opposite direction for the wild type and ΔtagE strains. This change is clearly not the result of free bPEI polymer. These results imply that wild type and ΔtagE phosphorus nuclei experienced an increase in electron density in the presence of bPEI-AuNPs, which we attribute to the proximity of the AuNPs which contain delocalized electrons.45
Exposure to either the bPEI-AuNPs or the free bPEI caused broadening of the 31P peaks in spectra acquired from all three strains (Tables S5–S7†). This broadening may be due to heterogeneity in the interaction of the WTA phosphorus atoms with both bPEI-AuNPs and free bPEI polymer.41 Peak broadening may also arise from reduced mobility and slower relaxation times due to the presence of bPEI-AuNPs. Restricted WTA motion could result from the interactions with bPEI-AuNPs or free bPEI polymer. In a similar manner, peak broadening in 1H-NMR spectra of a potential cancer therapeutic molecule was attributed to its reduced mobility in proximity to the metal core of poly(ethylene glycol)-AuNPs.46
We further investigated changes in the solid-state 13C-NMR spectra induced by interaction of sacculi from each strain with bPEI-AuNPs (Fig. 9) and free bPEI polymer (Fig. S8†). Carbon nuclei in wild type and ΔtagE strains clearly experienced more changes in their chemical environments when exposed to bPEI-AuNPs and free bPEI than did the ΔdltA strain as indicated by changes in chemical shift, peak width, and normalized intensity. Changes to these parameters indicate an alteration in the chemical environment of observed nuclei and suggest possible interaction sites. Previous studies have suggested that nanoparticles can affect the signal intensities of interacting biomolecules to a much larger extent than chemical shifts.47
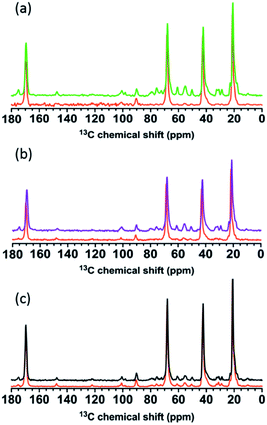 | ||
| Fig. 9 Solid-state 13C-NMR spectra of (a) wild type, (b) ΔtagE, and (c) ΔdltA before (green, purple, and black, respectively) and after (orange) exposure to 34 nM bPEI-AuNPs. | ||
Spectral changes upon introduction of bPEI-AuNPs were similar for the wild type and ΔtagE strains, with the exception of those associated with the WTA Glc in the wild type. For both strains, exposure to bPEI-AuNPs resulted in substantial attenuation of the intensities of peaks associated primarily with carbons in the amino sugar residues, glycerolphosphate, and Glc (wild type only) of WTA (Tables S5 and S6†). The glycerolphosphate peaks decrease substantially in intensity, and many of the peaks corresponding to the linker unit GlcNAc and ManNAc residues broaden to the point of disappearing into the baseline (Fig. S9†). Other peaks exhibiting decreased intensity, albeit less pronounced than those of the WTA groups, are associated with carbons in the amino acid residues of peptidoglycan (meso-A2pm, L- and D-Ala, Glu) and in the D-Lac connecting MurNAc to L-Ala (Fig. S9†). The negative charges of D-γ-Glu (and meso-A2pm if not crosslinked) carboxylate groups likely enhanced interaction with bPEI polymer relative to carbons in the glycan sugar residues. Peaks associated with amino acid and amino sugar residues in peptidoglycan decreased somewhat in intensity and shifted slightly downfield (<0.35 ppm), but otherwise remain largely unaltered. Exposure to free bPEI polymer produced spectral changes that resembled those observed in the presence of bPEI-AuNPs, although peak intensity also decreased for some carbon nuclei in the GlcNAc and MurNAc residues of peptidoglycan (Fig. S10†). The spectral changes indicate that WTAs are the primary binding sites for wild type and ΔtagE sacculi for both AuNPs and free bPEI polymer, and the chemical environment of some of the carbon residues in the peptide of peptidoglycan are also affected. More peaks decrease in the presence of free bPEI polymer relative to bPEI-AuNPs, suggesting that the peptidoglycan meshwork is less accessible to bPEI bound to the NPs relative to free bPEI.
Peak intensities in the spectra from ΔdltA sacculi remained largely unaltered upon exposure to bPEI-AuNPs or free bPEI polymer (Table S8†). Specifically, peaks corresponding to WTA glycerolphosphate, GlcNAc, and ManNAc were minimally impacted by the presence of nanoparticles. Small decreases in peak intensity and narrowing of peak shoulders were apparent for a few resonances, including the D-Lac group between MurNAc and L-Ala, the WTA linker GlcNAc and ManNAc C5 and C3, and glycerolphosphate C1, 2, and 3 (Fig. S11†). Similar, rather minor changes are observed in the presence of free bPEI.
Both flow cytometry analysis of whole bacteria cells and solid-state NMR analysis of bacterial sacculi revealed low association of bPEI-AuNPs with the surface of the ΔdltA mutant relative to the other two strains. The flow cytometry results for the ΔdltA and ΔtagE strains are consistent with expectations based on cell surface charge as reflected by cytochrome c binding (Fig. 2b). The less negatively charged strain, ΔdltA, had much lower proportion of cells associated with AuNPs, suggesting that electrostatic forces contributed to the interactions between whole cells and bPEI-AuNPs. Foxley et al. previously reported on the importance of Coulombic interactions between the cationic primary amines of bPEI polymer and the anionic phosphate groups in WTA in methicillin-resistant Staphylococcus aureus with slightly different WTA structure from B. subtilis.44 Similar interactions between the bPEI coating the AuNPs and the WTA glycerophosphate in the three B. subtilis strains is expected. Upon addition of free bPEI polymer, we observed downfield shifts of the dominant peaks in 31P-NMR spectra for each strain which corresponded to the most abundant form of glycerol substitution (glucose-substituted (peak B) for wild type and ΔdltA, and unsubstituted (peak A) for ΔtagE). Similar downfield peak shifts were previously reported for a strain of B. subtilis with poly(ribitol phosphate) WTA molecules upon exposure to bPEI in solution 31P-NMR experiments.44 In contrast, we observe small upfield peak shifts in 31P-NMR for wild type and ΔtagE strains upon exposure to bPEI-AuNPs. Comparison of flow cytometry results from the wild type and ΔdltA strains suggests the electrostatic charge at the cell surface is not the sole factor influencing bPEI-AuNP association with the bacteria. The surface potentials/charges of wild type and ΔdltA were not distinguishable by the methods employed; nonetheless, a significantly higher proportion of wild type than ΔdltA cells were associated with bPEI. We hypothesize that the accessibility of WTA at the cell surface affects association between the bPEI-AuNPs and the bacterial cells.
The T1 measurements show that specific carbons in ΔdltA sacculi exhibit more restricted molecular motion than in those of the wild type, ΔtagE, or both. This more constrained molecular motion may be due to a larger proportion of the WTA being embedded within the peptidoglycan meshwork in the ΔdltA mutant than in the wild type (supported by the evidence for a larger number of shorter WTA strands in the ΔdltA mutant than in the wild type), making the WTA physically unable to interact with the bPEI-coated AuNPs. The lower flexibility of the ΔdltA WTA may also reflect the presence of more Glc substituents than are attached to the WTA of the wild type. The ΔdltA WTA that is available to interact with AuNPs may be less flexible and unable to form as many contacts with the nanoparticles as the WTA from the wild type. Interestingly, although both wild type and ΔdltA WTA contain Glc, T1 values are higher for a larger number of ΔdltA WTA carbons than in the wild type. This further suggests wild type has longer WTA than ΔdltA, supporting the interpretation that the accessibility of the WTA to the AuNPs influences the extent of NP association. Overall, the T1 results are consistent with observations that wild type and ΔtagE strains are similar in their interaction with bPEI-AuNPs, while the ΔdltA strain interacts with the nanoparticles to a much smaller extent as shown by flow cytometry. Peaks in the 31P-NMR spectra from wild type and ΔtagE sacculi exhibit small upfield peak shifts in the presence of AuNPs, indicating enhanced electron density around the phosphorus nuclei, while a small downfield shift in the main peak is apparent for ΔdltA. Many changes are apparent in the 13C-NMR spectra for sacculi from wild type and ΔtagE strains upon interaction with bPEI-AuNPs, while minimal changes are observed for sacculi from the ΔdltA strain. Although the ΔdltA WTA may be shorter or more buried within the peptidoglycan matrix relative to that in wild type B. subtilis, free polymer can more readily penetrate the cell wall than bPEI-AuNPs. This hypothesis is consistent with the pronounced downfield peak shift seen in 31P-NMR for ΔdltA exposed to free bPEI, while the downfield shift upon exposure to bPEI-AuNPs is quite small.
Conclusions
We have examined the interaction of cationic bPEI-AuNPs with intact bacteria and their exoskeletons (sacculi) using wild type and two mutant strains of Bacillus subtilis that differ in the composition of their wall teichoic acids. Through a combination of flow cytometry and solution and solid-state NMR, we have established a relationship between bacterial cell wall composition and the association of cationic AuNPs. Flow cytometry data reveal significantly more bPEI-AuNP associated to the wild type and ΔtagE strains, the latter of which lacks Glc in its WTA, than to the ΔdltA strain, the WTA of which lacks Ala and exhibits the most restricted molecular motion. Solid-state 31P-NMR spectra suggest AuNPs come into closer proximity to the wall teichoic acid molecules of wild type and ΔtagE, compared to ΔdltA, as evidenced by an upfield shift when nanoparticles are present. Solid-state 13C-NMR spectra reveal that bPEI-AuNPs induced spectral changes primarily to the resonances corresponding to the linker unit between WTA and peptidoglycan, to glycerol in the WTA backbone for wild type and ΔtagE, and to the Glc carbons in wild type. For the ΔdltA strain, exposure to bPEI-AuNPs impacted the chemical environment of WTA and peptidoglycan carbon nuclei to only a small extent. The ΔdltA strain GlcNAc and MurNAc of peptidoglycan and the WTA glycerol generally exhibited more restricted molecular motion relative to wild type and ΔtagE as ascertained from temperature-dependent T1 relaxation experiments. The reduced molecular motion of the WTA in the ΔdltA strain likely reflected WTA extending to a lesser extent into solution relative to the other two strains and resulted in corresponding diminished interaction with the cationic AuNPs. Similar effects may be seen in cell wall interaction with cationic antimicrobial peptides. The nanoparticle cores used here are inherently larger than the effective pore size of peptidoglycan meshwork and are not expected to penetrate into the cell wall.48Our experimental evidence supports the interpretation that electrostatic forces contribute importantly to interactions of Gram-positive bacterial surfaces with cationic bPEI-AuNPs and that the accessibility of negatively charged moieties in teichoic acid chains influence the degree of interaction. We note that wall teichoic acid structural variations did not significantly impact cell viability (Fig. S12†) indicating that cell wall mutations may alter NP binding, but these binding differences do not necessarily result in changes to bacterial viability.
We found that major components of B. subtilis walls that interacted with nanoparticles include the poly(glycerolphosphate) backbone in WTA, the amino sugars in the disaccharide linker unit, and the peptides in peptidoglycan. Although our findings were based on one type of cationic nanoparticle and one Gram-negative bacterial species, the results demonstrate the importance of the structure and composition of wall teichoic acid in modulating interaction with nanoparticles. The approaches taken in the present study can be extended to additional systems, including engineered nanomaterials differing in core composition, shape, or surface functional groups to elucidate the influence of these properties on interactions with Gram-positive bacteria. Knowledge of such relationships can aid the design of nanomaterials to intentionally target cells to limit microbial growth or to minimize undesired impacts on bacteria.
Materials and methods
Chemicals and nanoparticles
The chemicals and nanoparticles used, suppliers, and purities are described in the ESI.†Nanoparticle characterization
To verify the nanoparticle properties reported by the manufacturer, the stock particle solution was diluted 100× in 0.025 M NaCl buffered to pH 7.4 with 0.002 M HEPES, and UV-Vis extinction spectra were acquired (Beckman DU 7500) to determine AuNP core diameter and concentration.42 Particle hydrodynamic diameter and ζ-potential were derived from dynamic and electrophoretic light scattering measurements (Brookhaven ZetaPALS). All measurements were made in triplicate. The particle suspension was stored at 4 °C, and the colloidal stability of the suspension was determined prior to all experiments. The structure of cationic branched polyethylenimine (bPEI) is shown in Fig. S13.†Bacterial culture and characterization
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 15 min). The supernatants containing unbound cytochrome c were then removed and oxidized with 20 μL of 30% H2O2. The difference between absorbance at 530 nm of supernatants after exposure to cells and of the negative control provide an indication of the relative surface charge of the cell surfaces.
000g, 15 min). The supernatants containing unbound cytochrome c were then removed and oxidized with 20 μL of 30% H2O2. The difference between absorbance at 530 nm of supernatants after exposure to cells and of the negative control provide an indication of the relative surface charge of the cell surfaces.
Wall teichoic acid isolation and characterization
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 10 min), and washed with 2% NaCl [wt/vol] buffered to pH 6.5 with 0.05 M MES, followed by 0.05 M MES at pH 6.5. The samples were incubated with proteinase K (0.02 M Tris–HCl, 0.5% [wt/vol] SDS, 20 μL mL−1 of proteinase K, pH 8.0) at 50 °C for ∼4 h and washed again with 2% NaCl [wt/vol] buffered to pH 6.5 with 0.05 M MES and at least twice with ultrapure water. Finally, the samples were suspended in 1 mL of 0.1 M NaOH and gently shaken at room temperature for ∼16 h. Cell wall debris was removed by centrifugation (14
000g, 10 min), and washed with 2% NaCl [wt/vol] buffered to pH 6.5 with 0.05 M MES, followed by 0.05 M MES at pH 6.5. The samples were incubated with proteinase K (0.02 M Tris–HCl, 0.5% [wt/vol] SDS, 20 μL mL−1 of proteinase K, pH 8.0) at 50 °C for ∼4 h and washed again with 2% NaCl [wt/vol] buffered to pH 6.5 with 0.05 M MES and at least twice with ultrapure water. Finally, the samples were suspended in 1 mL of 0.1 M NaOH and gently shaken at room temperature for ∼16 h. Cell wall debris was removed by centrifugation (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 10 min), and the supernatant containing isolated WTA was collected for further analysis.
000g, 10 min), and the supernatant containing isolated WTA was collected for further analysis.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of 10% ascorbic acid and 0.42% ammonium molybdate in 1 M H2SO4 was used to complex inorganic phosphate from WTA during incubation for 20 min at 45 °C. The absorbances of reaction mixtures were measured at 820 nm to quantify the amount of phosphate against a calibration curve constructed from potassium phosphate standards.
1 mixture of 10% ascorbic acid and 0.42% ammonium molybdate in 1 M H2SO4 was used to complex inorganic phosphate from WTA during incubation for 20 min at 45 °C. The absorbances of reaction mixtures were measured at 820 nm to quantify the amount of phosphate against a calibration curve constructed from potassium phosphate standards.
Isolation and characterization of bacterial sacculi
Following thorough sonication and vortexing, samples were sedimented for 30 min at 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g in a tabletop centrifuge at room temperature (Eppendorf, 5417R). After removing the supernatant, samples were packed into solid-state NMR inserts (Bruker, Kel-F). Inserts were placed into 4 mm diameter rotors (Bruker, Kel-F) and spun at 10
000g in a tabletop centrifuge at room temperature (Eppendorf, 5417R). After removing the supernatant, samples were packed into solid-state NMR inserts (Bruker, Kel-F). Inserts were placed into 4 mm diameter rotors (Bruker, Kel-F) and spun at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Hz during data collection. We collected 17
000 Hz during data collection. We collected 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 920 scans for each 31P spectrum and 512 scans for each 13C spectrum. The maximum distance between 13C nuclei sensed through space was roughly 20 Å.57 As a control experiment, 34 nM bPEI-AuNPs were analyzed without sacculi by 13C-NMR. No signal was produced from these samples over the spectral acquisition time. We referenced 13C-NMR spectra to an external adamantine sample (chemical shifts of 38.5 and 29.5 ppm).58 For 31P-NMR spectra, hexamethylphosphoramide was used as an internal reference (chemical shift at 29.97 ppm). All spectra were processed in MestReNova, including baseline correction and curve smoothing. 13C-NMR spectra were normalized by scaling the peak at ∼20 ppm to 100 with respect to intensity.
920 scans for each 31P spectrum and 512 scans for each 13C spectrum. The maximum distance between 13C nuclei sensed through space was roughly 20 Å.57 As a control experiment, 34 nM bPEI-AuNPs were analyzed without sacculi by 13C-NMR. No signal was produced from these samples over the spectral acquisition time. We referenced 13C-NMR spectra to an external adamantine sample (chemical shifts of 38.5 and 29.5 ppm).58 For 31P-NMR spectra, hexamethylphosphoramide was used as an internal reference (chemical shift at 29.97 ppm). All spectra were processed in MestReNova, including baseline correction and curve smoothing. 13C-NMR spectra were normalized by scaling the peak at ∼20 ppm to 100 with respect to intensity.
Sample temperatures were calibrated following a published method.59 The chemical shifts of K79Br were measured over a range of temperature settings, and the actual sample temperatures (Tactual) were calculated based on the expected relationship between Tactual and the measured 79Br chemical shift: (K). The reference chemical shift was obtained at ambient temperature of 22 °C at a magic angle spinning frequency of 3 kHz, decoupler off (PLW2 = PLW12 = 0 W), repetition rate d1 = 5 s. Parameters were then changed to the experimental values of a spinning frequency of 8 kHz, 1H decoupler strength = 104 kHz (pw90 = 2.4 μs at 200 W), aq = 30 ms, d1 = 15 s. Additional temperature points were then set, and the actual sample temperatures were obtained from the chemical shift difference from the reference.59 One measurement was taken at each temperature and interscan delay time.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Science Foundation under the Center for Sustainable Nanotechnology, CHE-1503408. The CSN is part of the Centers for Chemical Innovation Program. We thank Charlie Fry, Director of the University of Wisconsin-Madison Magnetic Resonance Facility, for help with the T1 relaxation measurements, Letitia Yao at the University of Minnesota NMR facility for help with the solution 31P-NMR experiments, and Michael Liou for help with statistical analysis. The National Magnetic Resonance Facility at Madison is supported by NIH grants P41 GM103399 (NIGMS) and P41GM66326 (NIGMS). Additional equipment was purchased with funds from the University of Wisconsin, NIH (RR02781, RR08438), NSF (DMB-8415048, OIA-9977486, BIR-9214394), and USDA. The Bruker AVANCE III 500 NMR spectrometer was supported by a UW2020 grant and a generous gift from Paul J. and Margaret M. Bender. The solution NMR conducted at University of Minnesota was supported by the Office of the Vice President of Research, College of Science and Engineering, and the Department of Chemistry at the University of Minnesota.Notes and references
- A. Helland, M. Scheringer, M. Siegrist, H. G. Kastenholz, A. Wiek and R. W. Scholz, Environ. Sci. Technol., 2008, 42, 640–646 CrossRef CAS PubMed.
- T. J. Silhavy, D. Kahne and S. Walker, Cold Spring Harbor Perspect. Biol., 2010, 2, 1–16 Search PubMed.
- Z. V. Feng, I. L. Gunsolus, T. A. Qiu, K. R. Hurley, L. H. Nyberg, H. Frew, K. P. Johnson, A. M. Vartanian, L. M. Jacob, S. E. Lohse, M. D. Torelli, R. J. Hamers, C. J. Murphy, C. L. Haynes, J. Murphy and C. L. Haynes, Chem. Sci., 2015, 6, 5186–5196 RSC.
- J. Beranová, G. Seydlová, H. Kozak, O. Benada, R. Fišer, A. Artemenko, I. Konopásek and A. Kromka, FEMS Microbiol. Lett., 2014, 351, 179–186 CrossRef PubMed.
- Y. Liu, J. Tan, A. Thomas, D. Ou-Yang and V. R. Muzykantov, Ther. Delivery, 2012, 3, 181–194 CrossRef CAS PubMed.
- S. C. Hayden, G. Zhao, K. Saha, R. L. Phillips, X. Li, O. R. Miranda, V. M. Rotello, M. A. El-Sayed, I. Schmidt-Krey and U. H. F. Bunz, J. Am. Chem. Soc., 2012, 134, 6920–6923 CrossRef CAS PubMed.
- W. Pajerski, D. Ochonska, M. Brzychczy-Wloch, P. Indyka, M. Jarosz, M. Golda-Cepa, Z. Sojka and A. Kotarba, J. Nanopart. Res., 2019, 21, 1–12 CrossRef CAS.
- K. H. Jacobson, I. L. Gunsolus, T. R. Kuech, J. M. Troiano, E. S. Melby, S. E. Lohse, D. Hu, W. B. Chrisler, C. J. Murphy, G. Orr, F. M. Geiger, C. L. Haynes and J. A. Pedersen, Environ. Sci. Technol., 2015, 49, 10642–10650 CrossRef CAS PubMed.
- W. Vollmer, D. Blanot and M. a de Pedro, FEMS Microbiol. Rev., 2008, 32, 149–167 CrossRef CAS PubMed.
- S. Brown, J. P. Santa Maria and S. Walker, Annu. Rev. Microbiol., 2013, 67, 313–336 CrossRef CAS PubMed.
- F. C. Neuhaus and J. Baddiley, Microbiol. Mol. Biol. Rev., 2003, 67, 686–723 CrossRef CAS PubMed.
- P. Demchick and A. L. Koch, J. Bacteriol., 1996, 178, 768–773 CrossRef CAS PubMed.
- R. C. Hughes, P. F. Thurman and E. Stokes, Z. für Immunitatsforsch. Exp. Klin. Immunol., 1975, 49, 26–135 Search PubMed.
- S. E. Allison, M. A. D'Elia, S. Arar, M. A. Monteiro and E. D. Brown, J. Biol. Chem., 2011, 286, 23708–23716 CrossRef CAS PubMed.
- M. Perego, P. Glaser, A. Minutello, M. A. Strauch, K. Leopold and W. Fischer, J. Biol. Chem., 1995, 270, 15598–15606 CrossRef CAS PubMed.
- J. Wecke, K. Madela and W. Fischer, Microbiology, 1997, 143, 2953–2960 CrossRef CAS.
- E. Giaouris, R. Briandet, M. Meyrand, P. Courtin and M. P. Chapot-Chartier, Appl. Environ. Microbiol., 2008, 74, 4764–4767 CrossRef CAS PubMed.
- T. Kern, M. Giffard, S. Hediger, A. Amoroso, C. Giustini, N. K. Bui, B. Joris, C. Bougault, W. Vollmer and J.-P. Simorre, J. Am. Chem. Soc., 2010, 132, 10911–10919 CrossRef CAS PubMed.
- J. A. H. Romaniuk and L. Cegelski, Biochemistry, 2018, 57, 3966–3975 CrossRef CAS PubMed.
- R. Nygaard, J. A. H. Romaniuk, D. M. Rice and L. Cegelski, Biophys. J., 2015, 108, 1380–1389 CrossRef CAS PubMed.
- K. Makino and H. Ohshima, Sci. Technol. Adv. Mater., 2011, 12, 1–13 CrossRef PubMed.
- M. Dittrich and S. Sibler, J. Colloid Interface Sci., 2005, 286, 487–495 CrossRef CAS PubMed.
- H. Y. Buse, J. M. Hoelle, C. Muhlen and D. A. Lytle, FEMS Microbiol. Lett., 2018, 365, 1–7 CrossRef PubMed.
- S. M. Louie, T. Phenrat, M. J. Small, R. D. Tilton and G. V. Lowry, Langmuir, 2012, 28, 10334–10347 CrossRef CAS PubMed.
- J. G. Adamson, T. Hoang, A. Crivici and G. A. Lajoie, Anal. Biochem., 1992, 202, 210–214 CrossRef CAS PubMed.
- M. Kovács, A. Halfmann, I. Fedtke, M. Heintz, A. Peschel, W. Vollmer, R. Hakenbeck and R. Brückner, J. Bacteriol., 2006, 188, 5797–5805 CrossRef PubMed.
- M. Batley, J. W. Redmond and A. J. Wicken, Biochim. Biophys. Acta, Biomembr., 1987, 901, 127–137 CrossRef CAS.
- N. K. Bui, A. Eberhardt, D. Vollmer, T. Kern, C. Bougault, A. Tomasz, J. P. Simorre and W. Vollmer, Anal. Biochem., 2012, 421, 657–666 CrossRef CAS PubMed.
- C. Schäffer, N. Müller, P. K. Mandal, R. Christian, S. Zayni and P. Messner, Glycoconjugate J., 2000, 17, 681–690 CrossRef PubMed.
- C. Schäffer, H. Kählig, R. Christian, G. Schulz, S. Zayni and P. Messner, Microbiology, 1999, 145, 1575–1583 CrossRef PubMed.
- L. D. Quin and A. J. Williams, Practical Interpretation of P-31 NMR Spectra and Computer Assisted Structure Verification, Advanced Chemistry Development, Toronto, Canada, 2004 Search PubMed.
- T. C. Meredith, J. G. Swoboda and S. Walker, J. Bacteriol., 2008, 190, 3046–3056 CrossRef CAS PubMed.
- D. Mirelman, B. D. Beck and D. R. D. Shaw, Biochem. Biophys. Res. Commun., 1970, 39, 712–717 CrossRef CAS PubMed.
- B. N. Ames, B. Garry and L. a Herzenberg, J. Gen. Microbiol., 1960, 22, 369–378 CrossRef CAS PubMed.
- N. Satoh and K. Kimura, J. Am. Chem. Soc., 1990, 112, 4688–4692 CrossRef CAS.
- S. Clejan, T. A. Krulwich, K. R. Mondrus and D. Seto-Young, J. Bacteriol., 1986, 168, 334–340 CrossRef CAS PubMed.
- S. Thong, B. Ercan, F. Torta, Z. Y. Fong, H. Y. Alvina Wong, M. R. Wenk and S. S. Chng, Elife, 2016, 5, 1–19 CrossRef PubMed.
- K. L. Lanier, J. D. Moore, D. Smith, S. Li, B. Davis and W. A. Shaw, Quantitative Phospholipid Analysis of Soy Lecithin and Krill Lecithin by 31P NMR, 2008 Search PubMed.
- T. Kern, Approches innovantes en RMN biomoléculaire: Cinétiques moléculaires par RMN rapide et paroi bactérienne par RMN du solide, Université Joseph-Fourier – Grenoble I, 2009 Search PubMed.
- J. Lee and R. I. Hollingsworth, Carbohydr. Res., 1997, 303, 103–112 CrossRef.
- C. V. Rice and J. R. Wickham, J. Am. Chem. Soc., 2005, 127, 856–857 CrossRef CAS PubMed.
- W. Haiss, N. T. K. Thanh, J. Aveyard and D. G. Fernig, Anal. Chem., 2007, 79, 4215–4221 CrossRef CAS PubMed.
- J. T. Buchman, A. Rahnamoun, K. M. Landy, X. Zhang, A. M. Vartanian, L. M. Jacob, C. J. Murphy, R. Hernandez and C. L. Haynes, Environ. Sci.: Nano, 2018, 5, 279–288 RSC.
- M. A. Foxley, A. W. Friedline, J. M. Jensen, S. L. Nimmo, E. M. Scull, J. B. King, S. Strange, M. T. Xiao, B. E. Smith, K. J. Thomas, D. T. Glatzhofer, R. H. Cichewicz and C. V. Rice, J. Antibiot., 2016, 69, 871–878 CrossRef CAS PubMed.
- M. A. Bakar, M. Sugiuchi, M. Iwasaki, Y. Shichibu and K. Konishi, Nat. Commun., 2017, 8, 2–8 CrossRef PubMed.
- S. C. Coelho, M. Rangel, M. C. Pereira, M. A. N. Coelho and G. Ivanova, Phys. Chem. Chem. Phys., 2015, 17, 18971–18979 RSC.
- M. Assfalg, L. Ragona, K. Pagano, M. D'Onofrio, S. Zanzoni, S. Tomaselli and H. Molinari, Biochim. Biophys. Acta, Proteins Proteomics, 2016, 1864, 102–114 CrossRef CAS PubMed.
- S. O. Meroueh, K. Z. Bencze, D. Hesek, M. Lee, J. F. Fisher, T. L. Stemmler and S. Mobashery, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 4404–4409 CrossRef CAS PubMed.
- W. W. Wilson, M. M. Wade, S. C. Holman and F. R. Champlin, J. Microbiol. Methods, 2001, 43, 153–164 CrossRef CAS PubMed.
- T. Ishikawa, B. L. Zhu and H. Maeda, Toxicol. Ind. Health, 2006, 22, 337–341 CrossRef CAS PubMed.
- R. E. Martinez, O. S. Pokrovsky, J. Schott and E. H. Oelkers, J. Colloid Interface Sci., 2008, 323, 317–325 CrossRef CAS PubMed.
- A. J. de Kerchove and M. Elimelech, Langmuir, 2005, 21, 6462–6472 CrossRef CAS PubMed.
- V. Vadyvaloo, S. Arous, A. Gravesen, Y. Héchard, R. Chauhan-Haubrock, J. W. Hastings and M. Rautenbach, Microbiology, 2004, 150, 3025–3033 CrossRef CAS PubMed.
- S. Pricelius, C. Held, S. Sollner, S. Deller, M. Murkovic, R. Ullrich, M. Hofrichter, A. Cavaco-Paulo, P. Macheroux and G. M. Guebitz, Enzyme Microb. Technol., 2007, 40, 1732–1738 CrossRef CAS.
- B. N. Ames and D. T. Dubin, J. Biol. Chem., 1960, 235, 769–775 CAS.
- P. S. Chen, T. Y. Toribara and H. Warner, Anal. Chem., 1956, 28, 1756–1758 CrossRef CAS.
- K. Gawrisch and B. W. Koenig, Curr. Top. Membr., 2002, 52, 163–190 CAS.
- C. R. Morcombe and K. W. Zilm, J. Magn. Reson., 2003, 162, 479–486 CrossRef CAS PubMed.
- K. R. Thurber and R. Tycko, J. Magn. Reson., 2009, 196, 84–87 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9sc05436g |
| ‡ Current address: Department of Chemistry, University of Illinois at Urbana-Champaign, 505 South Matthews, Urbana, IL 61801, USA. |
| § Current address: 3M Center, Saint Paul, MN 55144, USA. |
| ¶ Current address: Carteret Community College, 3505 Arendell Street, Morehead City, NC 28557, USA. |
| || Current address: Department of Chemistry, University of California-Riverside, 501 Big Springs Road, Riverside, CA 92521, USA. |
| This journal is © The Royal Society of Chemistry 2020 |