Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Nitric oxide imaging in cancer enabled by steric relaxation of a photoacoustic probe platform†
Christopher J.
Reinhardt
 ,
Ruiwen
Xu
,
Ruiwen
Xu
 and
Jefferson
Chan
and
Jefferson
Chan
 *
*
Department of Chemistry, The Beckman Institute for Advanced Science and Technology, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA. E-mail: jeffchan@illinois.edu
First published on 7th January 2020
Abstract
Nitric oxide (NO) is a key signaling molecule involved in a variety of physiological and pathological processes. Over the past few decades it has become clear that the microenvironment and concentration of NO are critically linked to its bioactivity. Direct visualization of NO in vivo remains difficult due to a lack of sensitive analytical tools with deep tissue compatibility. Herein, we report the optimization of an activatable photoacoustic probe for NO by planarizing the boron-azadipyrromethene (aza-BODIPY) dye platform via steric relaxation. The lead compound, SR-APNO-3, exhibits maximal absorption at 790 and 704 nm before and after N-nitrosation with NO, respectively, and a 4.4-fold increase in the theoretical maximal ratiometric response compared to the non-sterically relaxed parent compound, APNO-5. This circa 30 nm red-shift enabled direct visualization with the laser system commonly employed in commercially available photoacoustic tomographers and a 1.1-fold increase in sensitivity within an intramuscular lipopolysaccharide-induced inflammation model. Moreover, this optimization facilitated the detection of endogenously produced NO in an allograft murine breast cancer model, where steady-state concentrations are several orders of magnitude less than during the immune response.
Introduction
Reactive oxygen and nitrogen species represent a major class of biological signaling molecules. Nitric oxide (NO) was initially identified in the 1980s for its role in the regulation of vascular tone.1–4 Since then, NO has been implicated in a wide range of pharmacologically important processes, such as inflammation,5 neuronal signaling,6 and cancer,7–9 through distinct chemical mechanisms.10 A general and underappreciated conclusion from these works is that there are intrinsic connections between the concentration, cellular environment, and resulting biological effects of NO. For example, high concentrations of NO are antitumoural, while low concentrations promote angiogenesis and tumour growth.8,11–15 For this reason, it is critical to develop new methods that facilitate non-invasive detection of NO within complex biological systems. This remains an unsolved problem due to the lack of imaging technologies with the requisite sensitivity, spatiotemporal resolution, and/or depth penetration along with the capabilities of performing molecular imaging.16Photoacoustic (PA) imaging is a non-invasive modality that has the promise to expand bioimaging beyond purely optical limits (millimeter depths). Mechanistically, pulsed laser excitations are used to induce heating–cooling cycles (∼2 kJ) with corresponding thermoelastic expansion events, which propagate through the tissue as ultrasound. These ultrasound signals are then detected by piezoelectric detectors and reconstructed to yield high-resolution images. By detecting sound, rather than emitted fluorescence, PA imaging depths are limited only by the excitation light and provide high resolution (empirically ∼1/200 of imaging depth)17 within deep tissue (centimeter depths).18
To facilitate PA-based molecular imaging, a range of nanoparticle- and small-molecule-based contrast agents and molecular probes have been developed.18–21 We reported the first activatable PA probe for NO, APNO-5, and applied it for the detection of NO in an LPS-induced murine model of inflammation.16 It is important to note that while APNO-5 was able to detect NO concentrations relevant to the immune response (millimolar range), it was ineffective for NO detection in cancer (103-fold lower concentrations). For this reason, it is essential to enhance the sensitivity of the probe platform for imaging within deeper tissue and at lower NO concentrations.15
To date, only a handful of approaches have been successfully employed for increasing the sensitivity and ratiometric properties of small-molecule PA probes. Specifically, increasing the electron density within the aza-BODIPY22 and conformational restriction of the pendant phenyl rings23 have led to improved designs for ratiometric, activatable PA probes compared to non-ratiometric24 or poorly ratiometric probes.25 While successful in the selected example, increasing electron density may not be trivial, depending on the application. On the other hand, conformational restriction yields reliable red-shifts (∼30 nm per fusion) but increases the molecular weight (26 Da per fusion) and cLogP of the dye platform (increase from the parent aza-BODIPY cLogP = 7.8 to conformationally restricted aza-BODIPY cLogP = 8.2–8.6). Together, these properties decrease the probability of passive uptake into the cell and increase propensity for aggregate formation and peak broadening.
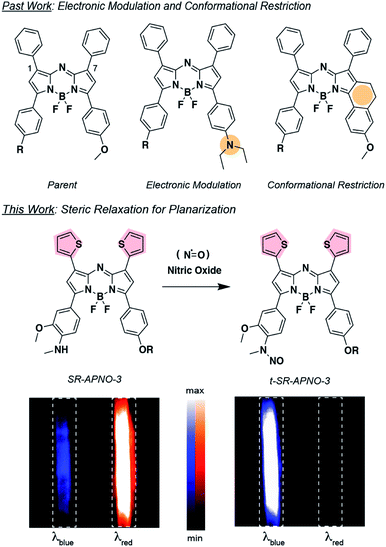
We decided to investigate the photophysical effects of planarizing the pendant aromatic rings on the aza-BODIPY core by relaxing steric clash. We generally refer to this approach as “steric relaxation for planarization” with the corresponding PA platform being named SR-aza-BODIPY for sterically relaxed aza-BODIPY (Fig. 1). Specifically, we proposed to replace the phenyls with thiophenes because the average diameter is ∼75% the size. Similar thiophene-modified aza-BODIPYs have been previously prepared and analyzed using X-ray crystallography and UV-vis spectroscopy.26–29 These studies confirmed a substantial planarization of the dye backbone and red-shifting. However, their applicability to activity-based sensors and PA imaging remain unexplored. Moreover, we hypothesized that incorporation of sulphur atoms would induce the heavy-atom effect, causing a reduction in fluorescence quantum yield and increase in PA signal. Herein, we report the optimization of APNO-5 via the systematic replacement of the 1- and 7-position phenyl rings with thiophenes and the application of the optimized compound for imaging cancer-derived NO in live-animal models.
Results and discussion
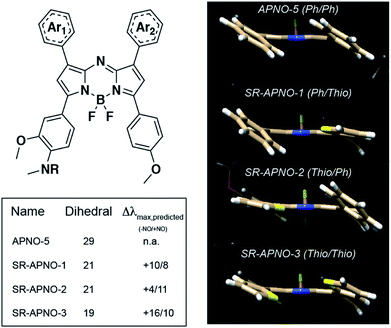
To investigate the feasibility of this approach, we utilized density functional theory (DFT) and time-dependent DFT (TD-DFT) to study the effect of thiophene substitution on the dihedral angle and predicted absorption of the dyes (calculated in implicit methanol solvent). The method was validated by comparing the dihedral angle between the planes of the aromatic rings and the aza-BODIPY core to previous experimental data in which the computational and experimental data were in good agreement (∼28° and ∼18°, respectively, Fig. S1a–d†).26,27 Replacement of the phenyls with thiophenes resulted in a decreased dihedral angle with a corresponding ∼25 nm red-shift per thiophene. Importantly, we observed an inverse linear relationship between the calculated dihedral angle and experimental absorption maximum, suggesting the dihedral angle is predictive (R2 = 0.60, Fig. S1e†).With the validated computational workflow, the probes and N-nitrosated products were built in silico with the polyethylene solubilizing group truncated to a methyl for computational analyses. The geometries were optimized, and the wavelength of maximal absorbance was calculated for each compound. In all cases, we observed a near complete decrease (8°, 80% of the disubstituted analogue's effect) in dihedral angle after replacing a single phenyl and a 14 nm red-shift per thiophene (Fig. 2). Additionally, inverse correlations were observed between the dihedral angle and wavelength of maximal absorbance for both the initial probe (R2 = 0.64) and the N-nitrosated product (R2 = 0.59). These trends were consistent with the results from gas phase optimized geometries with only the universal force field (R2 = 0.60, 0.90) and B3LYP (R2 = 0.69, 0.90), suggesting these should be sufficient for predicting the effect of steric interactions on the wavelength for the aza-BODIPY platform (Fig. S1e†).
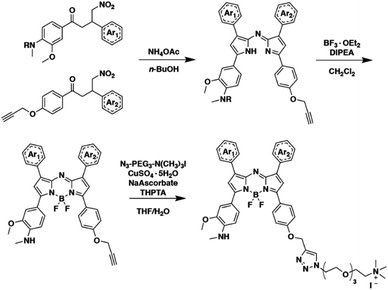
Encouraged by the computational results, we prepared both singly substituted constitutional isomers, SR-APNO-1 and SR-APNO-2, and the disubstituted analogue, SR-APNO-3. First, the desired functionality is installed through the Claisen–Schmidt condensation of the appropriate acetophenone and aromatic aldehyde to afford a chalcone. Next, the chalcone is subjected to Michael-type addition of the nitromethane anion followed by Paal–Knorr cyclization and boron chelation. Finally, the appended alkyne handle was utilized for installation of a solubilizing moiety through copper-catalysed 1,3-dipolar cycloaddition (Schemes 1 and S1-5†).16
Photophysical characterization indicated that all probes and N-nitrosated products displayed maximal absorbance at greater than 680 nm, maintained high extinction coefficients and low quantum yields, and exhibited pH-independent absorbance and fluorescence properties (Table 1, Fig. 3c and S2-3†). The oxidative nitrosation products were confirmed using high resolution mass spectrometry and clean conversion was observed by liquid chromatography mass spectrometry (Fig. S4-6†).
| Compound | −Nitric oxide | +Nitric oxide | ||||
|---|---|---|---|---|---|---|
| λ max | ε × 104 | ϕ | λ max | ε × 104 | ϕ | |
| a λ (nm) and ε (M−1 cm−1) are reported in methanol and ϕ is reported in ethanolic 20 mM potassium phosphate buffer (pH 7.4, 50% v/v). Data is reported as the average of experimental replicates (n = 3) with less than 15% error. | ||||||
| APNO-5 | 759 | 3.38 | 0.0003 | 670 | 2.91 | 0.23 |
| SR-APNO-1 | 775 | 3.18 | 0.004 | 685 | 2.25 | 0.26 |
| SR-APNO-2 | 770 | 1.75 | 0.015 | 688 | 1.56 | 0.17 |
| SR-APNO-3 | 790 | 7.24 | 0.002 | 704 | 5.31 | 0.10 |
As anticipated, each probe underwent rapid NO-mediated oxidation to afford the blue-shifted product. Incorporation of two sulphurs into the chromophore resulted in a decrease in N-nitrosated product's fluorescence quantum yield, presumably due to the heavy atom effect, and the planarization resulted in an increase in molar absorptivity due to more efficient orbital overlap. Striking similarities between the two constitutional isomers confirmed that the planarization is not dependent on the location relative to the trigger (consistent with our computational predictions, Table 1 and Fig. S1-2†). This is in contrast to the conformational restriction approach in which the location of restriction effects both the photophysical and PA properties.23 The SR-APNO series, along with previous reports,26–29 indicate that planarization via steric relaxation is similarly efficient for red-shifting the absorbance (15 versus 0–30 nm and 30–67 versus 0–52 nm after one and two modifications, respectively).23,30,31
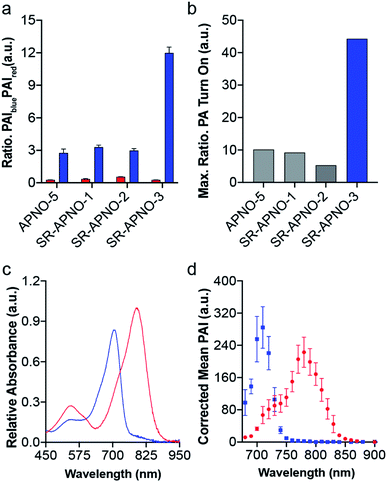
To evaluate the effect on the PA properties, the maximal ratiometric responses were measured for each compound following N-nitrosation (Fig. 3 and S2†). Interestingly, neither SR-APNO-1 nor SR-APNO-2 resulted in any increase in sensitivity. On the other hand, SR-APNO-3 was 4.4× more sensitive than the parent molecule with a 44-fold maximal ratiometric PA turn on. It is important to note that the increased sensitivity comes predominantly from more efficient detection of the N-nitrosated product rather than changes in the initial probe (Fig. 3b).
With the ideal probe in hand, SR-APNO-3's applicability for NO imaging was evaluated. First, SR-APNO-3 and t-SR-APNO-3 were compared using UV-vis spectroscopy and PA tomography, where maximal PA ratiometric responses were observed at 690 and 790 nm (Fig. 3c and d). Changing the percent ethanol content of the buffer resulted in different ratiometric responses to NO; however, N-nitrosated product was observed at all tested concentrations (0–80% ethanol, Fig. S7a and b†). The spectral changes were consistent with the formation of J-aggregates under aqueous conditions, which has been observed previously with the aza-BODIPY dye platform.16,32 Comparison between reactions performed in the presence and absence of ethanol suggest that the differences in ratiometric response occur due to differences in reaction depending on aggregation state and lower sensitivity due to beak broadening (Fig. S7c and d†). Next, the selectivity of SR-APNO-3 was confirmed against a panel of reactive metals, carbonyl, oxygen, sulphur, and nitrogen species. The panel of potential cross-reactive biomolecules was selected due to the possibility of binding to the thiophenes or oxidation of the trigger, which could lead to false positives. Importantly, no changes were observed via fluorescence or UV-vis (Fig. S8†). Finally, the photostability of SR-APNO-3 and the corresponding N-nitrosated product were measured using an OPO laser for irradiation. All of the SR-aza-BODIPYs were photostable (>90% over 60 s) and the N-nitrosated products showed reasonable stability towards de-nitrosylation (greater than 50% over 30 s, typical imaging is ∼10 s per wavelength, Fig. S9†). t-SR-APNO-3 proved to be the most photostable product. This finding is consistent with the proposed de-nitrosylation mechanism which includes photo-induced electron transfer from the chromophore into the N–NO followed by NO release.33,34 More broadly, this suggests that tuning intersystem crossing rates can be used to optimize both N-nitrosation based NO probes (e.g., APNO-5)16 and photo-activatable NO donors (e.g., photoNOD-1).35
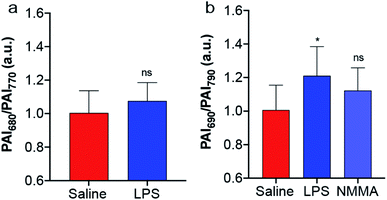
The biocompatibility of SR-APNO-3 was assessed in RAW 264.7 macrophages and 4T1 murine breast carcinoma cells, where good viability was observed (Fig. S10†). The cellular performance was then investigated in 4T1 cells to confirm both uptake and sufficient reaction kinetics for intracellular detection. Cells were stained with SR-APNO-3, washed to remove excess probe, and then incubated with DEA-NONOate. The cells were collected via centrifugation and the cell pellet was imaged using PA tomography. Quantification revealed a dose-dependent, ratiometric response to NO (Fig. S11†). Next, SR-APNO-3 was applied for imaging LPS-induced inflammation in BALB/c mice. To more accurately simulate the deeper tissue involved in cancer imaging, inflammation was induced through intramuscular administration of LPS rather than the subcutaneous model reported in our initial report.16 Under these conditions, APNO-5 was no longer able to detect NO. On the other hand, a statistically significant increase was observed using SR-APNO-3 after 3 h (Fig. 4 and S12†). These results are consistent with the in vitro characterization, where SR-APNO-3 was shown to be more sensitive than APNO-5. Mice were then co-administered SR-APNO-3 and L-NG-monomethyl-arginine (L-NMMA), a pan-selective nitric oxide synthase inhibitor,36 to confirm that the signal enhancement was a result of NO. Under these conditions, we observed a 12% inhibition in the ratiometric response (Fig. 4b and S12†). This suggests that SR-APNO-3 could serve as a tool for the evaluation of anti-inflammatory small molecules within live mouse models.
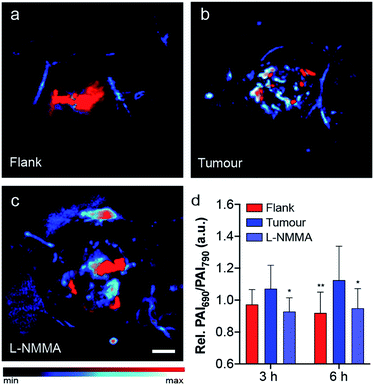
As previously mentioned, NO plays a key role in cancer biology, where high concentrations are initially biosynthesized by macrophages to elicit antitumoural effects. After this brief window of inflammation, emerging evidence suggests that the tumour microenvironment decreases macrophage motility, thereby trapping the macrophages within the tumour. These immune cells, often referred to as tumour-associated macrophages, produce lower levels of NO which promote tumour progression, angiogenesis and premetastatic potential.37 After local administration of SR-APNO-3 we observed a clear difference in the ratiometric PA response between the flank and tumour indicating that the probe was nitrosated. After 6 h, a 1.22-fold increase in the relative ratiometric PA response was observed between the flank and tumour (Fig. 5a and b and S13†). To confirm these results, we deemed it necessary to inhibit NO biosynthesis within the tumour, where co-administration of L-NMMA and SR-APNO-3 resulted in an 81% decrease in the relative ratiometric PA response (Fig. 5 and S13†). This represents the first example of where the PA imaging of cancer-derived NO has been validated using an inhibitor of nitric oxide synthase. To confirm that the signal change was not due to changes in any endogenous absorbers (e.g., haemoglobin or deoxyhaemoglobin), we performed an intratumoural injection of L-NMMA without SR-APNO-3. Under these conditions, we observed no change in the relative ratiometric PA response (Fig. S14†). Together, these imaging studies demonstrate that SR-APNO-3 can be applied to facilitate deep tissue imaging of cancer NO, where concentrations are approximated to be ∼103-fold lower than in the immune response. We propose that SR-APNO-3 provides a new, previously inaccessible window of imaging NO within cancer using a single contrast agent. Ongoing efforts in our group are focused on utilizing the increased sensitivity of SR-APNO-3 to study the role of NO within cancer.
Conclusion
In this work, we report the first systematic photophysical optimization of the aza-BODIPY dye platform that interrogates the effect of relieving steric interactions for dye planarization and PA signal optimization. A computational model was validated for predicting the effect of steric relaxation on the dihedral angle, which was confirmed experimentally. Replacement of a phenyl with a thiophene resulted in a circa 15 nm shift in the absorbance maxima, regardless of where the replacement was in comparison to the trigger. This contrasts with the conformational restriction approach, in which the location was essential for obtaining desirable PA properties. Replacing both aromatic rings with thiophenes had an additive effect and yielded the optimal compound, SR-APNO-3, which was 4.4-fold more sensitive than the parent molecule, APNO-5. This increase in sensitivity allowed for higher ratiometric responses in an intramuscular inflammation model and the first successful detection of cancer-derived NO with a small-molecule PA contrast agent. This small molecule approach for PA probe optimization is in contrast to recent nanoparticle-based examples which focus on for enhancing the sensitivity of NO-responsive PA by co-administering inert or analyte-selective probes for internal calibration or added sensitivity.38,39 More broadly, this work provides a new approach for the optimization of activatable PA probes and bridging the gap in sensitivity between optical and photoacoustic imaging methods.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Alfred P. Sloan fellowship (FG-2017-8964 to J. C.). C. J. R. thanks the Chemistry-Biology Interface Training Grant (T32 GM070421) and the Seemon Pines Graduate Fellowship for support. R. X. acknowledges the Summer Undergraduate Research Fellowship. Major funding for the 500 MHz Bruker CryoProbe was provided by the Roy J. Carver Charitable Trust (Muscatine, Iowa; Grant No. 15-4521) to the School of Chemical Sciences NMR Lab. The Q-Tof Ultima mass spectrometer was purchased in part with a grant from the National Science Foundation, Division of Biological Infrastructure (DBI-0100085). We acknowledge Professor Elvira de Mejia (Food Science and Human Nutrition, UIUC) for providing RAW 264.7 macrophage cells. Molecular graphics and analyses performed with UCSF Chimera, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco, with support from NIH P41-GM103311.Notes and references
- W. P. Arnold, C. K. Mittal, S. Katsuki and F. Murad, Proc. Natl. Acad. Sci. U. S. A., 1977, 74, 3203–3207 CrossRef CAS PubMed.
- R. Furchgott and J. V. Zawadski, Nature, 1980, 288, 373–376 CrossRef CAS PubMed.
- L. J. Ignarro, R. E. Byrns, G. M. Buga and K. S. Wood, Circ. Res., 1987, 61, 866–879 CrossRef CAS PubMed.
- R. M. J. Palmer, A. G. Ferrige and S. Moncada, Nature, 1987, 327, 524–526 CrossRef CAS PubMed.
- D. A. Wink, H. B. Hines, R. Y. S. Cheng, C. H. Switzer, W. Flores-Santana, M. P. Vitek, L. A. Ridnour and C. A. Colton, J. Leukocyte Biol., 2011, 89, 873–891 CrossRef CAS PubMed.
- S. Snyder, Science, 1992, 257, 494–496 CrossRef CAS PubMed.
- D. Fukumura, S. Kashiwagi and R. K. Jain, Nat. Rev. Cancer, 2006, 6, 521–534 CrossRef CAS PubMed.
- F. Vanini, K. Kashfi and N. Nath, Redox Biol., 2015, 6, 334–343 CrossRef CAS PubMed.
- J. L. Heinecke, L. A. Ridnour, R. Y. S. Cheng, C. H. Switzer, M. M. Lizardo, C. Khanna, S. A. Glynn, S. P. Hussain, H. A. Young, S. Ambs and D. A. Wink, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 6323–6328 CrossRef CAS PubMed.
- T. A. Heinrich, R. S. Da Silva, K. M. Miranda, C. H. Switzer, D. A. Wink and J. M. Fukuto, Br. J. Pharmacol., 2013, 169, 1417–1429 CrossRef CAS PubMed.
- L. A. Ridnour, D. D. Thomas, C. Switzer, W. Flores-Santana, J. S. Isenberg, S. Ambs, D. D. Roberts and D. A. Wink, Nitric Oxide, 2008, 19, 73–76 CrossRef CAS PubMed.
- J. R. Hickok, S. Sahni, H. Shen, A. Arvind, C. Antoniou, L. W. M. Fung and D. D. Thomas, Free Radicals Biol. Med., 2011, 51, 1558–1566 CrossRef CAS PubMed.
- J. R. Hickok, D. Vasudevan, K. Jablonski and D. D. Thomasn, Redox Biol., 2013, 1, 203–209 CrossRef CAS PubMed.
- D. D. Thomas, M. G. Espey, L. A. Ridnour, L. J. Hofseth, D. Mancardi, C. C. Harris and D. A. Wink, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 8894–8899 CrossRef CAS PubMed.
- D. D. Thomas, L. A. Ridnour, J. S. Isenberg, W. Flores-Santana, C. H. Switzer, S. Donzelli, P. Hussain, C. Vecoli, N. Paolocci, S. Ambs, C. A. Colton, C. C. Harris, D. D. Roberts and D. A. Wink, Free Radicals Biol. Med., 2008, 45, 18–31 CrossRef CAS PubMed.
- C. J. Reinhardt, E. Y. Zhou, M. D. Jorgensen, G. Partipilo and J. Chan, J. Am. Chem. Soc., 2018, 140, 1011–1018 CrossRef CAS PubMed.
- L. V. Wang and S. Hu, Science, 2012, 335, 1458–1462 CrossRef CAS PubMed.
- C. J. Reinhardt and J. Chan, Biochemistry, 2018, 57, 194–199 CrossRef CAS PubMed.
- Q. Miao and K. Pu, Bioconjugate Chem., 2016, 27, 2808–2823 CrossRef CAS PubMed.
- Y. Jiang and K. Pu, Small, 2017, 13, 1–19 Search PubMed.
- L. Zeng, G. Ma, J. Lin and P. Huang, Small, 2018, 14, 1–18 Search PubMed.
- H. J. Knox, T. W. Kim, Z. Zhu and J. Chan, ACS Chem. Biol., 2018, 13, 1838–1843 CrossRef CAS PubMed.
- E. Y. Zhou, H. J. Knox, C. Liu, W. Zhao and J. Chan, J. Am. Chem. Soc., 2019, 141, 17601–17609 CrossRef CAS PubMed.
- H. J. Knox, J. Hedhli, T. W. Kim, K. Khalili, L. W. Dobrucki and J. Chan, Nat. Commun., 2017, 8, 1794 CrossRef PubMed.
- H. Li, P. Zhang, L. P. Smaga, R. A. Hoffman and J. Chan, J. Am. Chem. Soc., 2015, 137, 15628–15631 CrossRef CAS.
- X. Zhang, H. Yu and Y. Xiao, J. Org. Chem., 2012, 77, 669–673 CrossRef CAS PubMed.
- Q. Bellier, F. Dalier, E. Jeanneau, O. Maury and C. Andraud, New J. Chem., 2012, 36, 768 RSC.
- S. Liu, Z. Shi, W. Xu, H. Yang, N. Xi, X. Liu, Q. Zhao and W. Huang, Dyes Pigm., 2014, 103, 145–153 CrossRef CAS.
- X. D. Jiang, J. Zhao, Q. Li, C. L. Sun, J. Guan, G. T. Sun and L. J. Xiao, Dyes Pigm., 2016, 125, 136–141 CrossRef CAS.
- W. Zhao and E. M. Carreira, Angew. Chem., Int. Ed., 2005, 44, 1677–1679 CrossRef CAS PubMed.
- W. Zhao and E. M. Carreira, Chem.–Eur. J., 2006, 12, 7254–7263 CrossRef CAS PubMed.
- Z. Chen, Y. Liu, W. Wagner, V. Stepanenko, X. Ren, S. Ogi and F. Würthner, Angew. Chem., Int. Ed., 2017, 56, 5729–5733 CrossRef CAS PubMed.
- S. Namiki, T. Arai and K. Fujimori, J. Am. Chem. Soc., 1997, 119, 3840–3841 CrossRef CAS.
- N. Ieda, Y. Hotta, N. Miyata, K. Kimura and H. Nakagawa, J. Am. Chem. Soc., 2014, 136, 7085–7091 CrossRef CAS.
- E. Y. Zhou, H. J. Knox, C. J. Reinhardt, G. Partipilo, M. J. Nilges and J. Chan, J. Am. Chem. Soc., 2018, 140, 11686–11697 CrossRef CAS.
- J. Víteček, A. Lojek, G. Valacchi and L. Kubala, Mediators Inflammation, 2012, 2012, 318087 Search PubMed.
- A. Mantovani, F. Marchesi, A. Malesci, L. Laghi and P. Allavena, Nat. Rev. Clin. Oncol., 2017, 14, 399–416 CrossRef CAS PubMed.
- Y. Wu, L. Sun, F. Zeng and S. Wu, Photoacoustics, 2019, 13, 6–17 CrossRef PubMed.
- L. Teng, G. Song, Y. Liu, X. Han, Z. Li, Y. Wang, S. Huan, X.-B. Zhang and W. Tan, J. Am. Chem. Soc., 2019, 141, 13572–13581 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9sc05600a |
| This journal is © The Royal Society of Chemistry 2020 |