Open Access Article
Open Access ArticlePhotocontrol of CRISPR/Cas9 function by site-specific chemical modification of guide RNA†
Yang
Wang
,
Yan
Liu
,
Fan
Xie
,
Jiao
Lin
and
Liang
Xu
 *
*
MOE Key Laboratory of Bioinorganic and Synthetic Chemistry, School of Chemistry, Sun Yat-Sen University, Guangzhou, 510275, China. E-mail: xuliang33@mail.sysu.edu.cn
First published on 25th September 2020
Abstract
The function of CRISPR/Cas9 can be conditionally controlled by the rational engineering of guide RNA (gRNA) to target the gene of choice for precise manipulation of the genome. Particularly, chemically modified gRNA that can be activated by using specific stimuli provides a unique tool to expand the versatility of conditional control. Herein, unlike previous engineering of gRNA that generally focused on the RNA part only but neglected RNA–protein interactions, we aimed at the interactive sites between 2′-OH of ribose in the seed region of gRNA and the Cas9 protein and identified that chemical modifications at specific sites could be utilized to regulate the Cas9 activity. By introducing a photolabile group at these specific sites, we achieved optical control of Cas9 activity without disrupting the Watson–Crick base pairing. We further examined our design through CRISPR-mediated gene activation and nuclease cleavage in living cells and successfully manipulated the gene expression by using light irradiation. Our site-specific modification strategy exhibited a highly efficient and dynamic optical response and presented a new perspective for manipulating gRNA based on the RNA–protein interaction rather than the structure of RNA itself. In addition, these specific sites could also be potentially utilized for modification of other stimuli-responsive groups, which would further enrich the toolbox for conditional control of CRISPR/Cas9 function.
Introduction
The RNA-guided CRISPR/Cas9 system, a powerful genome-editing toolbox adapted from the prokaryotic acquired immune system,1 has been gaining increasing attention for both biological research and therapeutic applications in the past few years.2–5 The genomic locus-targeting ability of CRISPR/Cas9 relies on the base pairing between the spacer region of guide RNA (gRNA) and the genomic DNA.6 The nuclease-dead Cas9 (dCas9) that removes the nuclease activity but retains the DNA targeting ability has been repurposed to further expand the applicable scenarios of the CRISPR/Cas9 system by employing different protein effectors,7 such as gene activation and repression,8,9 genomic loci imaging,10,11 nucleobase editing12,13 and epigenetic modifications.14,15 Compared with the constantly active RNA–Cas9 complex, development of conditionally controlled CRISPR function is of particular interest as its activity can be restricted in a desired spatial and temporal manner to achieve the precise manipulation of the genome with reduced side effects.16 Conditional control can be achieved through rational engineering of either (d)Cas9 or gRNA. The activity of (d)Cas9 protein can be induced by small molecules,17–20 activated by light,21–24 or controlled by tissue-specific promotors25 or miRNAs,26 but this approach encounters an inevitable limitation that all the targeted genes are subject to the same regulatory scope. In contrast, engineering of gRNA provides another approach to conditionally control CRISPR/Cas9 function that can manipulate specific genes via structural changes of gRNA induced by small molecules,27–30 protein binding,31 nuclease cleavage,28 RNA expressions,32,33 or miRNA/antisense RNA processing.34,35Besides engineering of genetically expressed gRNA, chemical modifications of gRNA also provide unique tools to expand the versatility of conditional control. By introducing different kinds of chemically reactive groups, modified gRNA can further enrich the toolbox for orthogonal activation of CRISPR/Cas9 function by using different stimuli. Several recent reports have shed some new light on this approach. These studies include utilization of photocleavable antisense DNA to regulate the activity of gRNA,36 photoactivation of directly caged gRNA,37–40 and ligand-induced deprotection of gRNA.41,42 The general principle for these designs relies on significant structural alteration of gRNA either by blocking the base pairing36–39 or by changing the overall chemical identity through unspecific multiple modifications.40–42 The former approach needs an individual sequence design for insertion of multiple modified nucleotides with relative difficulties for sample preparation; the latter one utilizes chemical reactive agents to achieve unspecific and heavy modifications of gRNA that may potentially encounter incomplete deprotection and result in reduced activation efficiency. To this end, an ideal strategy should utilize a single but universal site of gRNA that can be simply modified to switch on the activity of CRISPR/Cas9 function with an efficient response.
As a part of the CRISPR/Cas9 complex, gRNA is not an independently structured nucleic acid but tightly interacts with the Cas9 protein. Previous studies on engineering of gRNA generally focused on the RNA part, but neglected the interactive role of the Cas9 protein. In our work, taking a unique perspective, we aimed at the interaction between the spacer region of gRNA and Cas9 protein, and identified a site-specific chemical modification in the guide sequence that can achieve photocontrol of CRISPR/Cas9 function without disrupting the Watson–Crick base pairing.
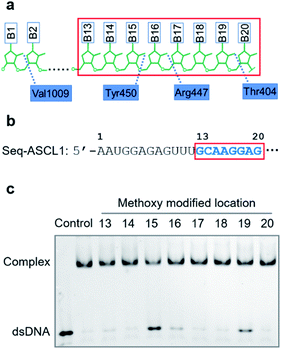
Results
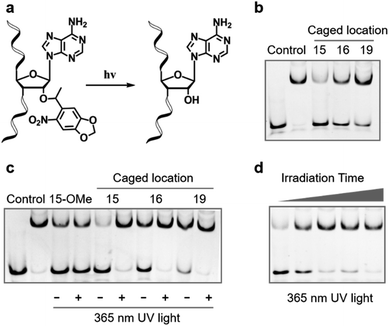
Compared with other gRNA regions, engineering of the variable guide sequence can precisely manipulate the gene of choice without affecting the binding ability between gRNA and Cas9. Analysis of the crystal structure of the Cas9/gRNA complex43 revealed that 2′-OH groups of ribose in the guide region are well recognized by residues of Cas9 (Fig. 1a). Recent investigations using the deoxynucleotide substitution44 or the truncation of spacer RNA45 indicated that the region near the 5′-end is not essential for the activity of the Cas9 complex. Several studies employing chemical substitutions suggested that these hydroxy groups in the seed region are relatively more sensitive.46–48 Based on these reports, firstly, we evaluated the role of 2′-OH in the seed region to identify whether there would be a critical site for regulating the function of the Cas9 complex. We selected a guide sequence to target the ASCL1 gene in human cells49 (Seq-ASCL1) and replaced the hydroxy group with the methoxy group starting from the 13th to 20th nucleotide one by one as shown in Fig. 1b. Given that the function of the Cas9 complex must rely on the binding of duplex DNA, we measured the binding behaviour between the modified gRNA and catalytically dead Cas9 (dCas9). The dCas9 protein was expressed and purified as described in the ESI;† the methoxy modified crRNA was chemically synthesized through solid phase synthesis and annealed with the tracrRNA prepared by in vitro T7 transcription to form the complete gRNA. The 5′-FAM-labeled duplex DNA with the Seq-ASCL1 targeting sequence was analysed by the gel shift assay to characterize the formation of the Cas9-RNA-DNA ternary complex. Initial screening of the binding ability indicated that the 2′-methoxy substitution of ribose in the 15th, 16th, and 19th nucleotides presented a more significant impact to block the complex formation than other positions (Fig. 1c). Intriguingly, these three positions were exactly in accordance with the protein recognition sites (Fig. 1a), demonstrating that disrupting the protein–RNA interaction in the guide sequence is an effective approach to regulate the Cas9 activity. Compared with the unmodified gRNA, concentration-dependent binding analysis (Fig. S1†) suggested that methoxy modification in the 15th nucleotide greatly reduced the ability for complex formation, whereas substitution in the 16th and 19th positions exhibited a weaker impact.Having confirmed that 2′-OH of ribose in the 15th, 16th, and 19th nucleotides was important for maintaining the activity of Cas9, we then aimed to test whether chemical caging of these 2′-OH groups could be utilized as a switch to control the CRISPR/Cas9 function. Herein, we selected a well-established photolabile group, 6-nitropiperonyloxymethylene,50 as a proof of principle, and introduced this caging group into ribose to mask the function of 2′-OH (Fig. 2a). The caged nucleoside was synthesized as described in the ESI,† and incorporated into the crRNA by solid phase synthesis. Based on the above observation, we placed the caging nucleotide at the 15th, 16th, and 19th positions and verified the blocking ability of each site through the binding analysis. Overall, the introduction of the caging group can significantly reduce the formation of the ternary complex (Fig. 2b). Notably, besides disrupting the role of 2′-OH, the steric effect of the caging group induced a more severe impact than the methoxy group. Concentration-dependent analysis of binding behaviour suggested that the caging of the 15th position caused the strongest inhibition, which greatly reduced the binding ability with very weak formation of the ternary complex even in the micromolar range of dCas9/gRNA (Fig. S2†). To exclude the possibility that the modification of 2′-OH would disrupt the formation of a stable RNA/DNA hybrid, we measured the melting temperatures of the 20-nt RNA/DNA guide duplex. As shown in Fig. S3,† both the single methoxy and the single caged modification could hardly destabilize the RNA/DNA duplex. Therefore, this negligible alteration in the RNA/DNA binding was unlikely to significantly affect the CRISPR/Cas9 activity.
Next, we examined whether the caged gRNA can be photoactivated to recover the activity. Taking the unmodified and the methoxy substituted gRNA as control, photo-irradiation could not affect the complex formation (Fig. 2c). In contrast, irradiation at the caged gRNA with the photolabile group in the 15th, 16th, and 19th nucleotides could completely recover the binding ability of Cas9, respectively (Fig. 2c). Particularly, the caged nucleotide at the 15th position exhibited the most significant dynamic change in the Cas9 binding ability before and after UV irradiation. Furthermore, taking the 15th position as a representative, we analysed the time-dependent photo-activation process, and the results indicated that a 30 second irradiation could recover the full activity of gRNA (Fig. 2d), revealing a quick response via light control. Collectively, these results suggested that photoinduced decaging of 2′-OH at the specific site of gRNA could achieve prompt and efficient control of the Cas9 activity.
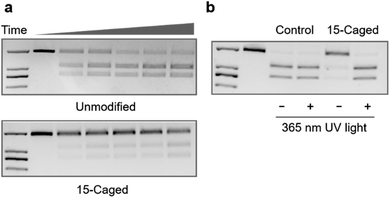
Furthermore, given that the application of CRISPR/Cas9 in gene editing requires the nuclease activity of Cas9 after the formation of the ternary complex, we also tested whether the site-specific modification on 2′-OH of gRNA could also regulate the cleavage behaviour of catalytically active Cas9. Compared with the unmodified gRNA, methoxy substitution suggested that the modification at the 15th, 16th, and 19th positions was still effective to inhibit the nuclease activity of Cas9 (Fig. S4–S7†) with the strongest effect at the 15th nucleotide (Fig. S5†), which was consistent with the binding behaviour. Similarly, modifications with the 6-nitropiperonyloxymethylene group could significantly inhibit the cleavage behaviour as shown in Fig. 3a, with the strongest effect at the 15th position (Fig. S8†). The quantitative analysis suggested that the observed cleavage activity was weakened by ∼30 fold for the caged gRNA at the 15th position (Fig. S8†), whereas caging at the other two locations exhibited a relatively weaker impact (Fig. S9 and S10†). Light irradiation on the photocaged gRNA could efficiently recover the nuclease activity of Cas9 (Fig. 3b), which indicated a highly dynamic responding process. However, it also needs to be mentioned that unlike the binding behaviour, cleavage is an irreversible process, and consequently, the highly inhibited nuclease activity could still induce significant breakage of the DNA duplex with prolonged incubation for hours (Fig. S8†). Nevertheless, the nuclease activity of Cas9 was also effectively controlled by photoactivation with the site-specifically caged 2′-OH of gRNA.
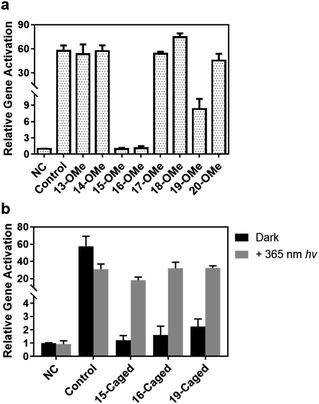
Since the Cas9 activity can be optically controlled in the test tube by the site-specifically modified gRNA without disruption of Watson–Crick base pairing, we further pursued to achieve photoactivation of CRISPR/Cas9 function in living cells. Among a variety of CRISPR-based applications in living cells, we first selected CRISPR mediated gene activation (CRISPRa) as a representative. We prepared HEK293T cells with stably expressed Cas9-VP64-p65-Rta (VPR) as the model cell line (see the ESI† for details) to determine whether CRISPRa can be conditionally controlled by irradiation of light through the modified gRNA. First, we verified whether the impact of site-specific modification of gRNA observed in test tubes would be the same with the cellular behaviour. Herein, we transfected the cell with the methoxy substituted gRNA in different positions, and took the same concentration of unmodified gRNA as control. The expression level of the target gene ASCL1 under different conditions was then determined by qPCR. As shown in Fig. 4a, compared with the unmodified one, methoxy substituted gRNA exhibited a similar enhancing effect for gene activation except those modifications in the 15th, 16th, and 19th nucleotides (∼1–2 fold for 15th and 16th, and ∼8 fold for 19th), which was consistent with binding behaviours. Notably, modifications at these specific sites were much more influential in the cellular context than those in vitro.
Based on these results, we further examined the effect of photocaged gRNA targeting the same gene for CRISPRa. After transfection, the cells were exposed to 365 nm UV light for 3 min. The irradiated and unirradiated cells were cultured under the same conditions, and the expression levels of the ASCL1 gene were compared and is shown in Fig. 4b. It is noteworthy that the 3 minute UV exposure caused a slightly negative impact on the cell viability (Fig. S11†), which might result in a reduced activation effect to some extent as observed in the control sample with the unmodified gRNA (the control columns in Fig. 4b), but the overall influence of this short-time UV exposure is not critical for transcriptional activation. Clearly, modified gRNA with photocaged 2′-OH significantly blocked gene activation with the strongest blockage at the 15th position. Upon photoirradiation, the active gRNA was released and the activation effect was greatly recovered. As shown in Fig. 4b, photo-irradiation induced the activation effect changing from ∼1.2-fold to ∼18-fold with a ∼15-fold dynamic range for the 15th position. Similar results were also observed for the 16th (∼20-fold change) and 19th (∼14-fold change) positions, suggesting an effective photoactivation process with the site-specifically caged 2′-OH of gRNA to regulate gene expression.
In order to demonstrate the universality of our site-specific caging strategy, additional gRNA sequences were selected to target different genes.51 First, we checked another different sequence (Seq2-ASCL1) to target the ASCL1 gene for photoinduced CRISPR activation in HEK293T cells and observed a similar effect of transcriptional activation changing from ∼2-fold to ∼60-fold with a ∼30-fold dynamic range for this 15th-caged gRNA by light irradiation (Fig. S12†). Besides, we also generated another sequence of light-activated gRNA to target the human gene CXCR4 for photocontrol of transcriptional activation (Fig. S13†). Notably, the CRISPR activation effect for this CXCR4 gene was less efficient than the ASCL1 gene, but the 15th-caged gRNA could still achieve an efficient photocontrol of gene activation.
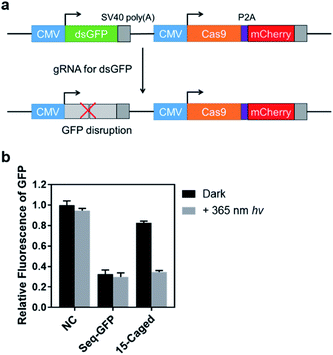
In addition to the activation of transcription, we also tested the photo-controlled nuclease cleavage of CRISPR/Cas9 in living cells by targeting the GFP gene (Fig. 5). In this experiment, we designed a construct in which expressions of destabilized GFP (dsGFP) and nuclease active Cas9 along with mCherry were packed into one plasmid under the control of two independent CMV promoters (Fig. 5a). The dsGFP expression was intervened upon transfection of the target gRNA. Therefore, the nuclease cleavage of CRISPR/Cas9 could be directly determined by the measurement of average GFP fluorescence from the mCherry positive cells through flow cytometry. As shown in Fig. 5b, the expression level of destabilized GFP was significantly disrupted upon light irradiation using the 15th-caged gRNA with the nuclease-active Cas9. Collectively, these results together with the transcriptional activation data suggested that the strategy using the site-specifically photocaged gRNA can optically control the CRISPR/Cas9 function in general.
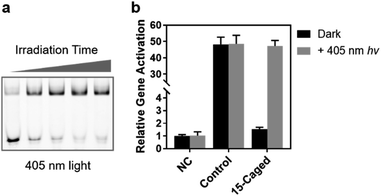
Finally, we verified the feasibility of visible light control of CRISPR function by our site-specifically caged gRNA, given that the 6-nitropiperonyloxymethylene group is also photolabile upon irradiation of 405 nm light.39 Compared with the 365 nm UV light, the time-dependent photo-irradiation process suggested that although a more powerful energy intensity was needed (16 mW cm−2 for 405 nm vs. 3.5 mW cm−2 for 365 nm), the 405 nm light could still induce the full activity of caged gRNA to form the ternary complex within an exposure of several minutes (Fig. 6a). Therefore, we further examined the photoinduced gene activation targeting the ASCL1 gene (Seq-ASCL1) in HEK293T cells through the 405 nm visible light. Intriguingly, a 3 min irradiation of 405 nm light could fully recover the activity of the 15th-caged gRNA with the transcriptional activation changing from ∼1.5-fold to ∼47-fold with about a 30-fold dynamic range (Fig. 6b). Importantly, unlike the 365 nm UV light, the 3 min 405 nm irradiation did not significantly impact the cell viability (Fig. S14†), and consequently, the activation effect was comparable to the unirradiated control sample (Fig. 6b). This visible light control could potentially provide a more ideal approach for photo-regulation of CRISPR/Cas9 function.
Discussion
Chemical modifications have enriched the versatility of conditional control of RNA-based technology, such as siRNA for silencing of mRNA52–54 and gRNA for CRISPR function.36–42 However, previously reported RNA caging strategies were generally based on structural variation of nucleic acids through disrupting base pairing or significant alteration of molecular identities, but they neglected the interactive impacts between RNA and RNA binding proteins. Our current study indicates that interruption of the interactive sites between 2′-OH of ribose in the seed region of gRNA and the Cas9 protein would significantly disturb the Cas9 activity, and these sites can be utilized to introduce photolabile groups for optical control of CRISPR function. Herein, our successful application paved a new way for developing more versatile strategies for chemical caging of RNA based on the RNA–protein interaction rather than the structure of RNA itself.Another important feature of our site-specifically modified gRNA was that structure alteration for RNA was very limited with only one photocaged 2′-OH in ribose and no disruption of Watson–Crick base pairing. Therefore, modified gRNA can still maintain the overall structural and chemical identity of nucleic acids. One can envision that this caged gRNA can still function like regular RNA to form base pairing and co-exist with other RNA regulatory elements, such as the recently reported toehold-switch,32,33 which could introduce more regulatory scope at the same time for conditional control based on RNA engineering.
Moreover, our single modified site was located in the 2′-position of ribose, independent of the nucleotide sequence, which could bring a great advantage and convenience for both design and synthesis of modified RNA. Particularly, our results showed that the 15th position of 2′-OH was the most sensitive site that could be potentially utilized to regulate any other gRNA sequences for gene manipulation. Besides, the single modification site at 2′-OH also made the synthesis of modified RNA much easier than introducing multiple modification sites. In addition, compared with previously reported multiple modifications of gRNA by using UV labile groups,37–40 our single-site modification made the optical response highly efficient with a relatively low energy dosage, which would be practically beneficial for prompt and harmless temporal control. Notably, given the fact that optical control by light with a longer wavelength is more suitable for applications in biomedical samples, we believe introduction of visible or even near-infrared (NIR) light activated moieties55 for visible or NIR light control could also be feasible through this convenient strategy.
Finally, we also wanted to point out that although the site-specific modification in the interface of gRNA and Cas9 protein can greatly inhibit the Cas9 activity, complete abolishment would be still difficult in the presence of a high concentration of Cas9 (enhanced binding ability with a high protein concentration) with a prolonged incubation time (irreversible nuclease activity with a long-time incubation), which might result in unwanted leakage before activation. However, some previous studies suggested that modifications at specific sites of gRNA exhibited highly sensitive impacts in the cellular context,46–48 implying that a weak leakage might not be robust enough to significantly alter the chromosome. Our gene regulation study also indicated that the caging effect of gRNA at specific sites was much more influential in living cells than in biochemical assays. Nevertheless, we believe the leakage, if it occurs, would be well solved by finetuning the concentration of modified gRNA and incubation time to achieve a dynamic range of conditional control.
Conclusions
In summary, we developed a strategy of photoactivable gRNA by site-specific chemical modification to regulate CRISPR-Cas9 function. Unlike previous engineering of gRNA that generally focused on the RNA part only but neglected RNA–protein interactions, we aimed at the interactive sites between 2′-OH of ribose in the seed region of gRNA and Cas9 protein and identified that modifications of 2′-OH at specific sites could be utilized to regulate the Cas9 activity. Introducing the photolabile group, 6-nitropiperonyloxymethylene, in these specific sites can achieve photocontrol of CRISPR/Cas9 function without disrupting the Watson–Crick base pairing. Taking CRISPR/Cas9 mediated gene activation and nuclease cleavage as representatives for versatile CRISPR applications, we achieved efficient and dynamic photocontrol of gene expression in living cells. Our design of site-specific photoactivable gRNA presented a new pathway for modification of RNA based on the RNA–protein interaction rather than RNA itself. More than optical control, these specific sites could be also modified with other stimuli-responsive groups,54,56,57 which would potentially further enrich the toolbox for conditional control of CRISPR-Cas9 function in the future.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the National Natural Science Foundation of China (No. 21708054 and No. 21977122) and the Innovative Research Team in University of Ministry of Education of China (IRT_17R111).Notes and references
- F. Hille, H. Richter, S. P. Wong, M. Bratovic, S. Ressel and E. Charpentier, Cell, 2018, 172, 1239–1259 CrossRef CAS
.
- P. Mali, K. M. Esvelt and G. M. Church, Nat. Methods, 2013, 10, 957–963 CrossRef CAS
.
- R. Barrangou and J. A. Doudna, Nat. Biotechnol., 2016, 34, 933–941 CrossRef CAS
.
- H. Wang, M. La Russa and L. S. Qi, Annu. Rev. Biochem., 2016, 85, 227–264 CrossRef CAS
.
- J. A. Doudna, Nature, 2020, 578, 229–236 CrossRef CAS
.
- M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna and E. Charpentier, Science, 2012, 337, 816–821 CrossRef CAS
.
- A. A. Dominguez, W. A. Lim and L. S. Qi, Nat. Rev. Mol. Cell Biol., 2016, 17, 5–15 CrossRef CAS
.
- L. S. Qi, M. H. Larson, L. A. Gilbert, J. A. Doudna, J. S. Weissman, A. P. Arkin and W. A. Lim, Cell, 2013, 152, 1173–1183 CrossRef CAS
.
- L. A. Gilbert, M. A. Horlbeck, B. Adamson, J. E. Villalta, Y. Chen, E. H. Whitehead, C. Guimaraes, B. Panning, H. L. Ploegh, M. C. Bassik, L. S. Qi, M. Kampmann and J. S. Weissman, Cell, 2014, 159, 647–661 CrossRef CAS
.
- B. Chen, L. A. Gilbert, B. A. Cimini, J. Schnitzbauer, W. Zhang, G. W. Li, J. Park, E. H. Blackburn, J. S. Weissman, L. S. Qi and B. Huang, Cell, 2013, 155, 1479–1491 CrossRef CAS
.
- S. C. Knight, R. Tjian and J. A. Doudna, Angew. Chem., Int. Ed., 2018, 57, 4329–4337 CrossRef CAS
.
- N. M. Gaudelli, A. C. Komor, H. A. Rees, M. S. Packer, A. H. Badran, D. I. Bryson and D. R. Liu, Nature, 2017, 551, 464–471 CrossRef CAS
.
- G. T. Hess, J. Tycko, D. Yao and M. C. Bassik, Mol. Cell, 2017, 68, 26–43 CrossRef CAS
.
- I. B. Hilton, A. M. D'Ippolito, C. M. Vockley, P. I. Thakore, G. E. Crawford, T. E. Reddy and C. A. Gersbach, Nat. Biotechnol., 2015, 33, 510–517 CrossRef CAS
.
- X. Xu, X. Tan, B. Tampe, T. Wilhelmi, M. S. Hulshoff, S. Saito, T. Moser, R. Kalluri, G. Hasenfuss, E. M. Zeisberg and M. Zeisberg, Nat. Commun., 2018, 9, 3509 CrossRef
.
- W. Zhou and A. Deiters, Angew. Chem., Int. Ed., 2016, 55, 5394–5399 CrossRef CAS
.
- K. M. Davis, V. Pattanayak, D. B. Thompson, J. A. Zuris and D. R. Liu, Nat. Chem. Biol., 2015, 11, 316–318 CrossRef CAS
.
- B. Zetsche, S. E. Volz and F. Zhang, Nat. Biotechnol., 2015, 33, 139–142 CrossRef CAS
.
- K. I. Liu, M. N. Ramli, C. W. Woo, Y. Wang, T. Zhao, X. Zhang, G. R. Yim, B. Y. Chong, A. Gowher, M. Z. Chua, J. Jung, J. H. Lee and M. H. Tan, Nat. Chem. Biol., 2016, 12, 980–987 CrossRef CAS
.
- T. Chen, D. Gao, R. Zhang, G. Zeng, H. Yan, E. Lim and F. S. Liang, J. Am. Chem. Soc., 2017, 139, 11337–11340 CrossRef CAS
.
- J. Hemphill, E. K. Borchardt, K. Brown, A. Asokan and A. Deiters, J. Am. Chem. Soc., 2015, 137, 5642–5645 CrossRef CAS
.
- Y. Nihongaki, F. Kawano, T. Nakajima and M. Sato, Nat. Biotechnol., 2015, 33, 755–760 CrossRef CAS
.
- L. R. Polstein and C. A. Gersbach, Nat. Chem. Biol., 2015, 11, 198–200 CrossRef CAS
.
- J. Shao, M. Wang, G. Yu, S. Zhu, Y. Yu, B. C. Heng, J. Wu and H. Ye, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E6722–E6730 CrossRef CAS
.
- J. Ablain, E. M. Durand, S. Yang, Y. Zhou and L. I. Zon, Dev. Cell, 2015, 32, 756–764 CrossRef CAS
.
- M. Hirosawa, Y. Fujita, C. J. C. Parr, K. Hayashi, S. Kashida, A. Hotta, K. Woltjen and H. Saito, Nucleic Acids Res., 2017, 45, e118 CrossRef CAS
.
- Y. Liu, Y. Zhan, Z. Chen, A. He, J. Li, H. Wu, L. Liu, C. Zhuang, J. Lin, X. Guo, Q. Zhang, W. Huang and Z. Cai, Nat. Methods, 2016, 13, 938–944 CrossRef CAS
.
- Q. R. Ferry, R. Lyutova and T. A. Fulga, Nat. Commun., 2017, 8, 14633 CrossRef
.
- W. Tang, J. H. Hu and D. R. Liu, Nat. Commun., 2017, 8, 15939 CrossRef CAS
.
- K. Kundert, J. E. Lucas, K. E. Watters, C. Fellmann, A. H. Ng, B. M. Heineike, C. M. Fitzsimmons, B. L. Oakes, J. Qu, N. Prasad, O. S. Rosenberg, D. F. Savage, H. El-Samad, J. A. Doudna and T. Kortemme, Nat. Commun., 2019, 10, 2127 CrossRef
.
- B. Maji, C. L. Moore, B. Zetsche, S. E. Volz, F. Zhang, M. D. Shoulders and A. Choudhary, Nat. Chem. Biol., 2017, 13, 9–11 CrossRef CAS
.
- M. H. Hanewich-Hollatz, Z. Chen, L. M. Hochrein, J. Huang and N. A. Pierce, ACS Cent. Sci., 2019, 5, 1241–1249 CrossRef CAS
.
- K. H. Siu and W. Chen, Nat. Chem. Biol., 2019, 15, 217–220 CrossRef CAS
.
- Y. J. Lee, A. Hoynes-O'Connor, M. C. Leong and T. S. Moon, Nucleic Acids Res., 2016, 44, 2462–2473 CrossRef CAS
.
- X. W. Wang, L. F. Hu, J. Hao, L. Q. Liao, Y. Z. Chiu, M. Shi and Y. M. Wang, Nat. Cell Biol., 2019, 21, 522–530 CrossRef
.
- P. K. Jain, V. Ramanan, A. G. Schepers, N. S. Dalvie, A. Panda, H. E. Fleming and S. N. Bhatia, Angew. Chem., Int. Ed., 2016, 55, 12440–12444 CrossRef CAS
.
- W. Zhou, W. Brown, A. Bardhan, M. Delaney, A. S. Ilk, R. R. Rauen, S. I. Kahn, M. Tsang and A. Deiters, Angew. Chem., Int. Ed., 2020, 59, 8998–9003 CrossRef CAS
.
- E. V. Moroz-Omori, D. Satyapertiwi, M.-C. Ramel, H. Høgset, I. K. Sunyovszki, Z. Liu, J. P. Wojciechowski, Y. Zhang, C. L. Grigsby, L. Brito, L. Bugeon, M. J. Dallman and M. M. Stevens, ACS Cent. Sci., 2020, 6, 695–703 CrossRef CAS
.
- Y. Liu, R. S. Zou, S. X. He, Y. Nihongaki, X. G. Li, S. Razavi, B. Wu and T. Ha, Science, 2020, 368, 1265–1269 CrossRef CAS
.
- S. Wang, L. Wei, J. Q. Wang, H. Ji, W. Xiong, J. Liu, P. Yin, T. Tian and X. Zhou, ACS Chem. Biol., 2020, 15, 1455–1463 CrossRef CAS
.
- S. R. Wang, L. Y. Wu, H. Y. Huang, W. Xiong, J. Liu, L. Wei, P. Yin, T. Tian and X. Zhou, Nat. Commun., 2020, 11, 91 CrossRef CAS
.
- M. Habibian, C. McKinlay, T. R. Blake, A. M. Kietrys, R. M. Waymouth, P. A. Wender and E. T. Kool, Chem. Sci., 2020, 11, 1011–1016 RSC
.
- H. Nishimasu, F. A. Ran, P. D. Hsu, S. Konermann, S. I. Shehata, N. Dohmae, R. Ishitani, F. Zhang and O. Nureki, Cell, 2014, 156, 935–949 CrossRef CAS
.
- H. Yin, C. Q. Song, S. Suresh, S. Y. Kwan, Q. Wu, S. Walsh, J. Ding, R. L. Bogorad, L. J. Zhu, S. A. Wolfe, V. Koteliansky, W. Xue, R. Langer and D. G. Anderson, Nat. Chem. Biol., 2018, 14, 311–316 CrossRef CAS
.
- Y. Fu, J. D. Sander, D. Reyon, V. M. Cascio and J. K. Joung, Nat. Biotechnol., 2014, 32, 279–284 CrossRef CAS
.
- H. Yin, C. Q. Song, S. Suresh, Q. Wu, S. Walsh, L. H. Rhym, E. Mintzer, M. F. Bolukbasi, L. J. Zhu, K. Kauffman, H. Mou, A. Oberholzer, J. Ding, S. Y. Kwan, R. L. Bogorad, T. Zatsepin, V. Koteliansky, S. A. Wolfe, W. Xue, R. Langer and D. G. Anderson, Nat. Biotechnol., 2017, 35, 1179–1187 CrossRef CAS
.
- D. O'Reilly, Z. J. Kartje, E. A. Ageely, E. Malek-Adamian, M. Habibian, A. Schofield, C. L. Barkau, K. J. Rohilla, L. B. DeRossett, A. T. Weigle, M. J. Damha and K. T. Gagnon, Nucleic Acids Res., 2019, 47, 546–558 Search PubMed
.
- F. O. Rueda, M. Bista, M. D. Newton, A. U. Goeppert, M. E. Cuomo, E. Gordon, F. Kroner, J. A. Read, J. D. Wrigley, D. Rueda and B. J. M. Taylor, Nat. Commun., 2017, 8, 1610 CrossRef
.
- P. Perez-Pinera, D. D. Kocak, C. M. Vockley, A. F. Adler, A. M. Kabadi, L. R. Polstein, P. I. Thakore, K. A. Glass, D. G. Ousterout, K. W. Leong, F. Guilak, G. E. Crawford, T. E. Reddy and C. A. Gersbach, Nat. Methods, 2013, 10, 973–976 CrossRef CAS
.
- N. Ankenbruck, T. Courtney, Y. Naro and A. Deiters, Angew. Chem., Int. Ed., 2018, 57, 2768–2798 CrossRef CAS
.
- D. Du and L. S. Qi, Cold Spring Harb. Protoc., 2016, 2016, pdb.prot090175 CrossRef
.
- I. A. Shestopalov, S. Sinha and J. K. Chen, Nat. Chem. Biol., 2007, 3, 650–651 CrossRef CAS
.
- F. Schafer, J. Wagner, A. Knau, S. Dimmeler and A. Heckel, Angew. Chem., Int. Ed., 2013, 52, 13558–13561 CrossRef
.
- S. Mori, K. Morihiro, T. Okuda, Y. Kasahara and S. Obika, Chem. Sci., 2018, 9, 1112–1118 RSC
.
- Q. Liu and A. Deiters, Acc. Chem. Res., 2014, 47, 45–55 CrossRef CAS
.
- A. Kadina, A. M. Kietrys and E. T. Kool, Angew. Chem., Int. Ed., 2018, 57, 3059–3063 CrossRef CAS
.
- L. Xiao, C. Gu and Y. Xiang, Angew. Chem., Int. Ed., 2019, 58, 14167–14172 CrossRef CAS
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0sc04343e |
| This journal is © The Royal Society of Chemistry 2020 |