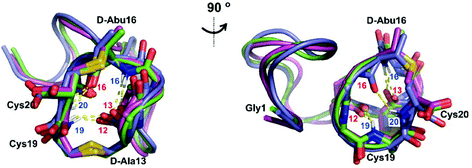Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Correction: Structural determinants of macrocyclization in substrate-controlled lanthipeptide biosynthetic pathways
Silvia C.
Bobeica
 a,
Lingyang
Zhu
a,
Lingyang
Zhu
 b,
Jeella Z.
Acedo
b,
Jeella Z.
Acedo
 a,
Weixin
Tang
a,
Weixin
Tang
 a and
Wilfred A.
van der Donk
a and
Wilfred A.
van der Donk
 *a
*a
aDepartment of Chemistry and Howard Hughes Medical Institute, University of Illinois at Urbana-Champaign, 600 South Mathews Avenue, Urbana, Illinois 61801, USA. E-mail: vddonk@illinois.edu; Fax: +1-217-244-8533; Tel: +1-217-244-5360
bSchool of Chemical Sciences NMR Laboratory, University of Illinois at Urbana-Champaign, 505 South Mathews Avenue, Urbana, Illinois 61801, USA
First published on 30th September 2020
Abstract
Correction for ‘Structural determinants of macrocyclization in substrate-controlled lanthipeptide biosynthetic pathways’ by Silvia C. Bobeica et al., Chem. Sci., 2020, DOI: 10.1039/d0sc01651a.
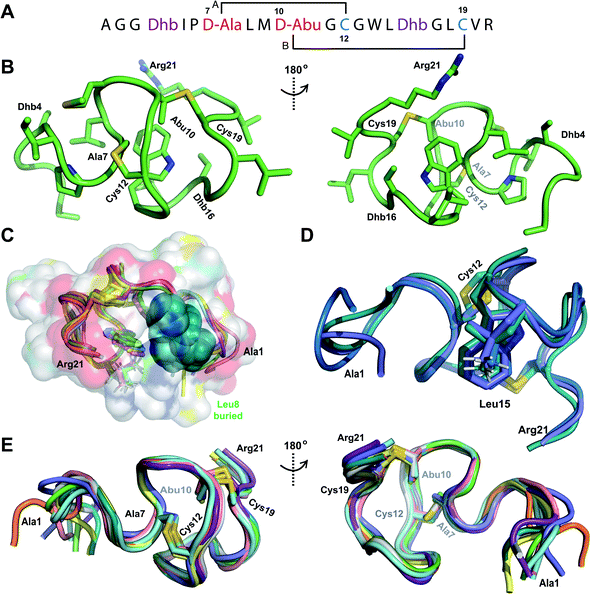
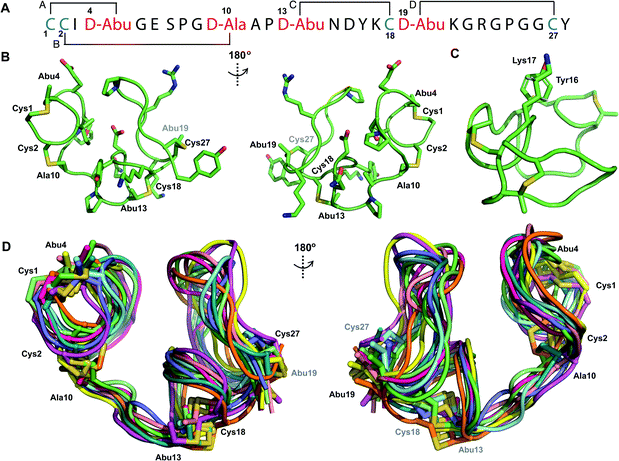
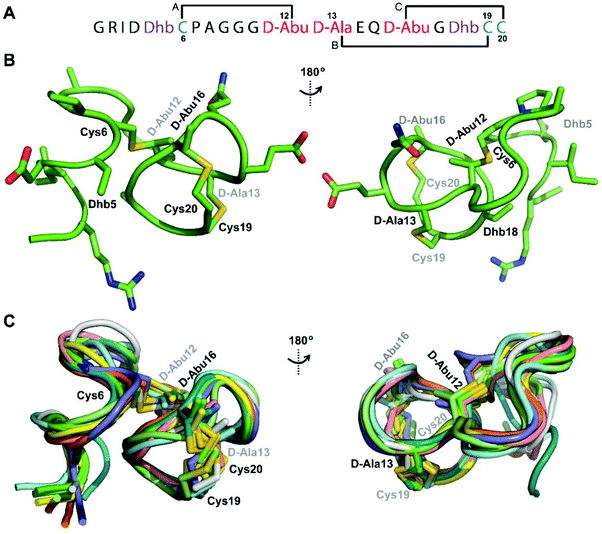
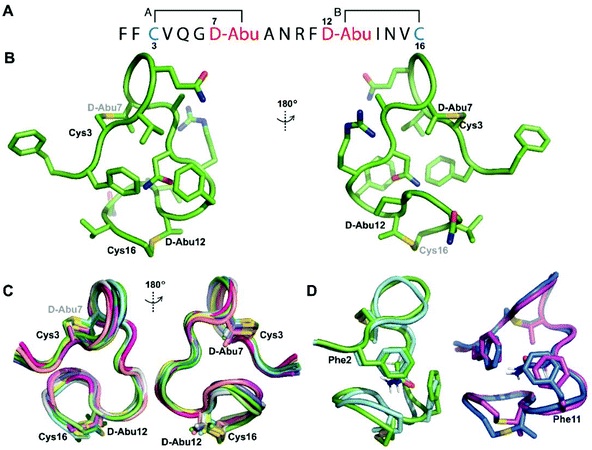
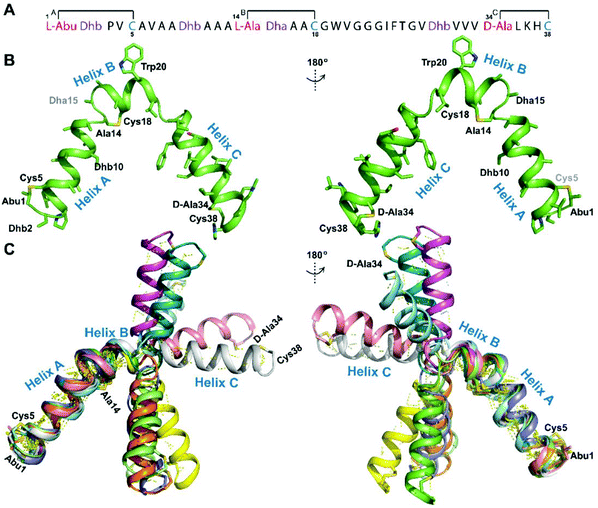
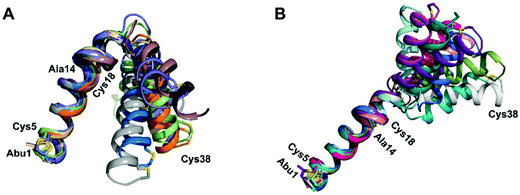
We recently reported the three-dimensional structures of seven lanthipeptides determined by NMR spectroscopy (Chem. Sci., DOI: 10.1039/d0sc01651a). After publication we became aware that the custom written definition of the non-proteinogenic bis-amino acid methyllanthionine (MeLan) for use by the software package Xplor does not sufficiently define the stereochemistry at C3. As a result, a subset of the seven structures contained a single MeLan with incorrect stereochemistry at C3. We rewrote the definition as shown in the updated ESI, redid all calculations using the original NMR data using the new definition, and manually inspected all 20 lowest energy structures to make sure they all had the correct stereochemistry of MeLan. All of the interactions observed in the original publication were still observed and none of the original conclusions were affected. We replaced the Protein Data Bank (PDB) files for prochlorosins 1.1 and 2.1 and cytolysin L using the same PDB identification numbers as in the original publication (PDB 6VHJ, 6VJQ, and 6VGT, respectively). For prochlorosins 2.10 and 2.11, we deposited the new structures with a new PDB ID (PDB 7JVF and 7JU9, respectively) and rendered the old pdb files obsolete. All BMRB codes remained the same except that BMRB 30789 is now associated with PDB 7JU9. Several of the figures in the original paper are affected in subtle ways and we provide new figures with this correction (Fig. 2–6, 12, and 13).
We apologize for this error and thank Prof. Gonzalo Jiménez-Osés (CIC bioGUNE, Spain) for alerting us to the incorrect stereochemistry in a subset of the final structures.
The Royal Society of Chemistry apologises for these errors and any consequent inconvenience to authors and readers.
| This journal is © The Royal Society of Chemistry 2020 |