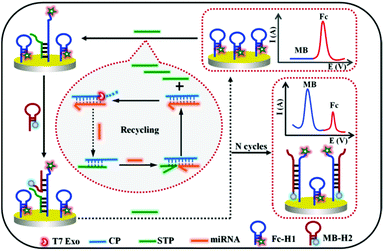
A novel ratiometric electrochemical biosensing strategy based on T7 exonuclease-assisted homogenous target recycling coupling hairpin assembly-triggered double-signal output for the multiple amplified detection of miRNA†
Qing-Yun
Zhou‡
a,
Rong-Na
Ma‡
 *a,
Chao-Long
Hu
a,
Fei
Sun
b,
Li-Ping
Jia
*a,
Chao-Long
Hu
a,
Fei
Sun
b,
Li-Ping
Jia
 a,
Wei
Zhang
a,
Lei
Shang
a,
Qing-Wang
Xue
a,
Wen-Li
Jia
a and
Huai-Sheng
Wang
a,
Wei
Zhang
a,
Lei
Shang
a,
Qing-Wang
Xue
a,
Wen-Li
Jia
a and
Huai-Sheng
Wang
 *a
*a
aSchool of Chemistry and Chemical Engineering, Liaocheng University, Liaocheng 252000, Shandong, P.R. China. E-mail: marongna@lcu.edu.cn; hswang@lcu.edu.cn
bOncology Department, Hospital of Traditional Chinese Medicine of Jinan City, Jinan 250000, Shandong, P.R. China
First published on 25th February 2021
Abstract
A novel ratiometric electrochemical biosensing strategy based on T7 exonuclease (T7 Exo)-assisted homogenous target recycling coupling hairpin assembly triggered dual-signal output was proposed for the accurate and sensitive detection of microRNA-141 (miRNA-141). Concretely, in the presence of target miRNA, abundant signal transduction probes were released via the T7 Exo-assisted homogenous target recycling amplification, which could be captured by the specially designed ferrocene-labeled hairpin probe (Fc-H1) on -electrode interface and triggered the nonenzymatic catalytic hairpin assembly (Fc-H1 + MB-H2) to realize the cascade signal amplification and dual-signal output. Through such a conformational change process, the electrochemical signal of Fc (IFc) and MB (IMB) is proportionally and substantially decreased and increased. Therefore, the signal ratio of IMB/IFc can be employed to accurately reflect the true level of original miRNA. Benefiting from the efficient integration of the T7 Exo-assisted target recycle, nonenzymatic hairpin assembly and dual-signal output mode, the proposed sensor could realize the amplified detection of miRNA-141 effectively with a wide detection range from 1 fM to 100 pM, and a detection limit of 200 aM. Furthermore, it exhibits outstanding sequence specificity to discriminate mismatched RNA, acceptable reproducibility and feasibility for real sample. This strategy effectively integrated the advantages of multiple amplification and ratiometric output modes, which could provide an accurate and efficient method in biosensing and clinical diagnosis.
Introduction
MicroRNAs (miRNAs) are a class of short (approximately 18–25 nucleotides in length), endogenous, nonprotein coding single-stranded RNA molecules, which play a critical role in gene expression and are associated with important biological processes such as cell multiplication,1,2 apoptosis,3 differentiation4 and metabolism.5 Research reported that the abnormal expression of miRNAs is closely related to serious cancer-related diseases.6,7 In particular, latest studies show that miRNAs released from tumour cells could be present in various biological fluids such as serum and plasma in stable form, designated circulating miRNAs, which can be utilized as new potential invasive biomarkers for early cancer screening and diagnosis.8–10Northern blotting,11 quantitative real-time polymerase chain reaction (qRT-PCR)12,13 and microarray14,15 are widely used standard methods for identifying and quantifying miRNAs. However, these techniques suffer from certain defects such as high cost, time- and sample-consuming, complex and tedious procedures. Hence, numerous other techniques including electrochemistry,16 surface plasmon resonance,17,18 photoelectrochemistry,19 electro-chemiluminescence20 and fluorescence21 have been extensively explored to overcome these shortcomings for the sensitive detection of miRNAs. Among all of the methodologies, the electrochemical-based technique is particularly promising with the outstanding advantages of high sensitivity, satisfactory selectivity, fast response, low cost, and handy operation.22,23 However, considering the short lengths, ultralow abundance, and high sequence similarity of miRNAs,24 and the nature of huge fluctuations as the disease detection progresses, it is still challenging to develop advanced electrochemical sensing strategies to confront the high demands of sensitivity and accuracy in practical miRNA assay.
The enzyme-assisted recycle amplification strategy, in which one target molecule triggers numerous cycles of target-dependent nuclease enzymolysis for powerful signal amplification, is meaningful to improve the target-to-mimic target conversion ratio and enhance the amplification efficiency.25–29 Unfortunately, the traditional electrochemical-based enzymatic heterogeneous hydrolysis usually occurs at the interface of electrode, which restricts the enzyme kinetics and substrate-binding efficiency compared with the homogeneous assay as the spatial hindrance effect and the loss of configuration freedom.30 Besides, the traditional single-signal output strategy usually has drawbacks of the low reproducibility and poor reliability owing to the electrode modification, intrinsic background electrochemical signals, and environmental fluctuations.31,32 The double-signal based ratiometric strategy with dual-signal ratiometric conversion modes instead of absolute single signals, which could effectively normalize the variation in experimental microenvironment, has been applied to improve the accuracy.33–38 Herein, the combination of homogenous recognition amplification and dual-signal output provides the feasibility for the construction of an accurate and efficient miRNA sensor.
In the study, a novel electrochemical biosensing strategy by coupling the advantages of enzyme-assisted homogenous recognition amplification and dual-signal ratiometric output is presented for the accurate and sensitive detection of miRNA-141 (Scheme 1). By combining the T7 exonuclease (Exo)-assisted homogenous recognition cascade recycle and the separation of recognition and transduction probe, the presence of target miRNA would realize numerous signal transduction probe (STP) releases. The released STP could be captured to the electrode surface by the ferrocene-labeled hairpin probe (Fc-H1), and further initiated the methylene blue-labeled hairpin DNA (MB-H2) assembly to trigger another cycle amplification and realize Fc and MB dual-signal output. The decrease and increase in the two labeling signals can accurately reflect the true level of the original miRNA with ratiometric signals. The proposed strategy has some notable features: first, the unique design by integrating T7 Exo-assisted target recycle and hairpin assembly could realize the multiple amplification and improve the sensitivity. Second, the strategy is generalizable owing to the separation of the molecular recognition element and signal reporter. Third, the dual-signal mode has superior anti-interference ability. Therefore, the designed strategy has immense value for highly sensitive and accurate target analysis, and clinical noninvasive liquid biopsy.
Experimental section
The descriptions of “Reagents and apparatus” and “Pretreatment of electrode” are described in the ESI.†Fabrication of the sensing platform
First, 1 μM Fc-H1 probe was obtained by incubating 10 μM Fc-H1 with freshly prepared TCEP to cleave disulfide bonds and diluting with 1× TAE per Mg buffer (20 mM Tris, 2 mM EDTA, 12.5 mM MgCl2; pH 7.4, the same below). Subsequently, 6 μL of the above solution was incubated on the pretreated electrode at 37 °C for 2.5 h. Then, it was rinsed, dried, and further treated with MCH to block the nonspecific binding sites. After being washed and dried once again, the Fc-H1-modified sensing electrode was acquired and stored at 4 °C in the Tris buffer for further use.Procedure for target miRNA assay
10 μM Capture probe (CP) and STP were first dissolved in 10 mM PBS (pH 7.4, 0.2 M NaCl), respectively. Next, a total of solution with equal volumes of CP and STP was mixed and heated to 95 °C for 3 min. The CP/STP probe stock solution was finally obtained after slowly cooling down the mixture to room temperature.For the detection of target miRNA, the T7 Exo-assisted homogeneous target recycling amplification was first realized by incubating 0.8 μM CP/STP, 20 U T7 Exo and miRNA with different concentrations in the reaction buffer (pH 7.9, 50 mM KAc, 20 mM Tris-Ac, 10 mM Mg(Ac)2, and 1 mM DTT) at 37 °C for 2 h. Subsequently, 6 μL of the resulted solution was dipped on the Fc-H1-modified sensing electrode and incubated at 37 °C for 1.5 h to fulfill the released STP hybridizes with Fc-H1; after being washed with Tris buffer, the resulting electrode was then incubated with 6 μL of 1.2 μM MB-H2 at 37 °C for 2 h to introduce MB signal and replace STP. Finally, the replaced STP further triggered another hairpin assembly to fulfill the recycling amplification and dual-signal output. The finally obtained electrode was rinsed thoroughly with washing buffer solution and dried with nitrogen for electrochemical measurements. The whole procedure is shown in Scheme 1.
Measurement method
All electrochemical experiments were performed on a conventional three-electrode system composed of a platinum wire auxiliary electrode, an Ag/AgCl reference electrode and a gold working electrode. The alternating current voltammogram (ACV) was recorded in 20 mM Tris buffer within the potential window from −0.4 V to 0.6 V, potential increment of 4 mV, frequency of 25 Hz, and amplitude of 25 mV. Electrochemical impedance spectroscopy (EIS) measurements were performed in a 5 mM [Fe(CN)6]3−/4− (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solution containing 0.1 M KCl in the frequency range from 0.1 Hz to 10 kHz to monitor changes of the electrode surface. The amplitude of the applied sine wave potential was 5 mV and the formal potential of the redox probe [Fe(CN)6]3−/4− is 220 mV.
1) solution containing 0.1 M KCl in the frequency range from 0.1 Hz to 10 kHz to monitor changes of the electrode surface. The amplitude of the applied sine wave potential was 5 mV and the formal potential of the redox probe [Fe(CN)6]3−/4− is 220 mV.
Results and discussion
Detection mechanism
The working principle of the designed dual-signal ratiometric biosensor for the multiple amplified detection of miRNA is schematically depicted in Scheme 1. To be specific, the entire design includes two key factors: (1) the CP/STP probe, which is resistant to T7 Exo digestion owing to the 5′ protruding terminuses, was tactfully constructed for both target recognition and signal transduction. The presence of target miRNA could trigger the toehold exchange-mediated strand displacement reaction, release the signal transduction probe, and generate RNA/DNA duplexes containing 5′ blunt terminus. Subsequently, the molecular recognition probe in RNA/DNA duplexes is specifically degraded by T7 Exo. Moreover, the miRNA molecule is released, and triggers more and more cycles of strand displacement to generate abundant free STP. By combining the homogeneous recognition and T7 Exo-assisted CP-STP probe separation, the first cycle of target recycling amplification was fulfilled. (2) The released STP hybridizes with the hairpin Fc-H1 on the electrode surface and the exposed sequences in the Fc-H1 probe triggers another hairpin assembly-mediated recycling amplification, realizing the STP probe recycling and the hairpin MB-H2 introducing. As a result, a large number of Fc-H1 + MB-H2 are formed and the IMB “signal-on” and IFc “signal-off” for the ratiometric electrochemical dual-signal output were realized. This strategy can realize multiple amplification since one copy of the miRNA can yield numerous Fc-H1 + MB-H2 and it can be easily extended for general gene-related molecular diagnosis.Feasibility of the sensing strategy
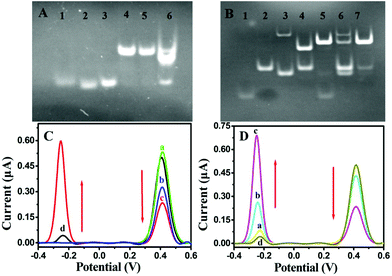
The T7 Exo-assisted homogeneous recognition and cascade extension was first confirmed by gel electrophoresis as the cascaded nucleic acid circuit was a key factor for signal amplification. As shown in Fig. 1A, miRNA, CP and STP exhibited individual distinct band, respectively (lanes 1, 2 and 3). The mixture of probe CP and STP showed a new band with significantly slow migration accompanied by the band disappearance corresponding to CP and STP, indicating that a newly cooperative complex probe CP/STP was formed (lane 4). In the absence of target miRNA, no newly formed band appeared (lane 5). On the contrary, an obvious change in the band shift was displayed in the positive control experiment in the presence of target miRNA (lane 6). These results confirmed that the T7 Exo-assisted cascade extension was successfully performed.The transductive probe STP-triggered successful formation of Fc-H1 + MB-H2 was a prerequisite for signal transduction and dual-signal output; therefore, the corresponding gel electrophoresis was also tested. As shown in Fig. 1B, the band in lane 2 and lane 4 were the Fc-H1 and the mixture of Fc-H1 and STP (lane 1), respectively, indicating the formation of Fc-H1 + STP conjugation. Compared with lane 4, the band of Fc-H1 + STP conjugation disappeared upon the introduction of MB-H2, along with a high molecular weight of product, indicating the successful formation of Fc-H1 + MB-H2 (lane 5). However, in the absence of STP, the mixture of MB-H2 and Fc-H1 showed negligible band intensity corresponding to Fc-H1 + MB-H2 (lane 6), which could be attributed to the low level of spontaneous interactions between MB-H2 and Fc-H1. The mixture of MB-H2 and Fc-H1 via the annealing treatment was also operated, which indicates that the Fc-H1 + MB-H2 (lane 7) conjugation products can be formed. Herein, these results clearly confirm that probe STP is the determining factor of the successful formation of the Fc-H1 + MB-H2 complex.
In addition, the feasibility of the STP-mediated DNA hairpin assembly and dual-signal output was further verified by a series of control experiments for the ACV measurements. As shown in Fig. 1C, an intensive oxidation peak at about +0.44 V is observed for the Fc-H1-modified gold electrode (curve a), indicating the successful modification of Fc-H1. After incubation with probe STP, the intensity of IFc decreased as a result of the conformational change in the Fc-H1 hairpin structure (curve b). On subsequent incubation with MB-H2, a new peak appeared at −0.28 V, accompanied by a significant decrease in IFc (curve c). However, as hypothesized, when only MB-H2 was incubated with Fc-H1, the intensity of Fc was almost similar to that of the Fc-H1-modified electrode and MB just showed a tiny peak (curve d). Therefore, these results demonstrated that probe STP is indispensable to mediate the electrode interface DNA hairpin assembly and dual-signal output.
Furthermore, the feasibility of the designed dual-signal biosensing strategy for target miRNA detection was also substantiated by ACV measurements (Fig. 1D). As expected, when the sensing platform MCH/Fc-H1/Au was incubated with the homogeneous solution without miRNA-141 (curve a), the ACV curve was almost similar to that of only MB-H2 incubated with Fc-H1 (curve d), indicating that the CP/STP probe hardly dissociated and released STP to induce the following STP-mediated hairpin assembly of the sensing surface without miRNA-141. In contrast, the presence of target miRNA-141 could induce a decrease in IFc but an increase in IMB (curve b), which indicates that the specific recognition triggers the STP release and subsequent hairpin assembly. In particular, the tendency was further strengthened with the presence of T7 Exo (curve c), implying that T7 Exo could trigger more cycles of strand displacement and the coupling of the hairpin assembly is efficient for signal enhancement. Therefore, all the above experimental results indicated that the design of dual-signal readout and amplification by integrating T7 Exo-assisted target recycling and nonenzymatic hairpin assembly was practicable.
EIS characterization of the developed biosensor
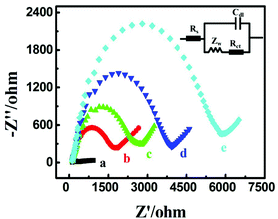
Electrochemical impedance spectroscopy was employed to investigate the electrode interface construction and probe STP-mediated hairpin assembly process as it has been proven to be a powerful tool for interfacial investigation (Fig. 2). In comparison to the bare Au electrode (curve a), when successively incubated with the negatively charged Fc-H1 (curve b) and nonconductive MCH (curve c), with the increasing of the electrostatic repelling force and insulativity, the charge transfer resistance (Rct) continuously increased, respectively, demonstrating the successful immobilization of Fc-H1. Subsequently, with the step-by-step incubation of STP (curve d) and MB-H2 (curve e), Rct sequentially increased due to the successful hairpin assembly of the negatively charged DNA phosphate backbone suppressed the diffusion of ferricyanide. Through the comparison of these experiments, the electrode modification of Fc-H1 and subsequent probe STP-mediated hairpin assembly have been successfully verified.Optimization of experimental conditions
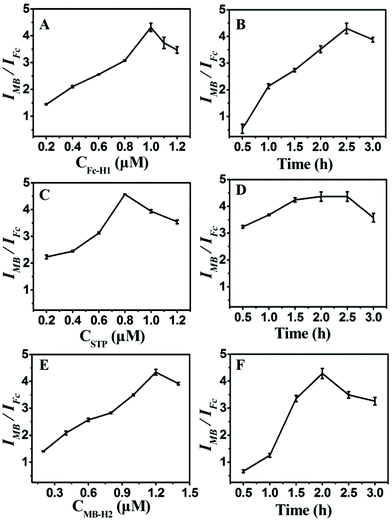
To achieve the best analytical performance of the designed strategy for the miRNA assay, several experimental parameters were examined and optimized (Fig. 3). The ratiometric of IMB to IFc was used to assess the performance of the experimental conditions. The concentration of Fc-H1 and the corresponding incubation time were first investigated since the packing density of Fc-H1 was critical for the subsequent assembly and dual-signal output. As shown in Fig. 3A, B, IMB/IFc increased with the increase in the Fc-H1 concentration and incubation time, and then tended to descend above 1 μM and 2.5 h. Thus, 1 μM Fc-H1 and 2.5 h incubation time were chosen in the experiment.The probe STP plays a role of a connector between target recognition and signal output, and an enormous amount of STP can lead to the low detection sensitivity, but low concentration would narrow the linear range of detection; therefore, the concentration of STP was also investigated. With the successive increase in the STP concentration, IMB/IFc increased and reached a maximum value at 0.8 μM (Fig. 3C). Thus, 0.8 μM STP were used for the construction of CP/STP, at which the CP/STP probe concentration is sufficient enough and even the tiny target miRNA can cause the signal conversion. The incubation time of probe STP was also investigated. As illustrated in Fig. 3D, IMB/IFc increased continuously with the increase in the incubation time, and almost reached the saturation value within 1.5 h, indicating that 1.5 h was sufficient for probe STP incubation.
Moreover, the concentration and incubation time of MB-H2 were also critical factors for signal amplification and dual-signal output. It could be seen that IMB/IFc increased continuously with the increase in the MB-H2 concentration (Fig. 3E) and incubation time (Fig. 3F) and then gradually decreased. Thus, 1.2 μM MB-H2 and 2 h incubation time were chosen as the most suitable concentration and incubation time, respectively, to fulfill the hairpin assembly.
Analytical performance corresponding to miRNA-141 biosensing
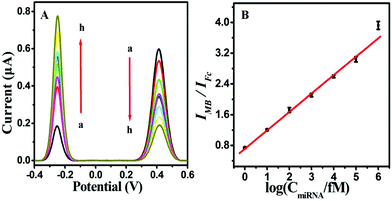
Under optimized experimental conditions, the analytical performance of the proposed biosensing strategy for the accurate and sensitive detection of miRNA-141 was studied. As hypothesized, the Fc oxidation peak current signal decreased and the MB oxidation peak current signal increased with the increase in the target miRNA concentration from 0 M to 1 nM (Fig. 4A). As a result, a desirable linear correlation between the logarithmic value of the miRNA-141 concentration and the ratio of the dual current peak intensity (IMB/IFc) was manifested in the concentration range of 1.0 fM–100 pM (Fig. 4B). The limit of detection (LOD) was calculated to be 200 aM at the signal-to-noise ratio S/N of 3. Moreover, the assay performance of the presented strategy compared with that of other previously reported detection strategies for miRNA in the detection limit and linear range was systematically summarized in Table S2 (ESI).† Notably, the linear response range is relatively wider than or comparable to previously reported miRNA sensors. Furthermore, the detection limit value was better than or comparable to the previously reported methods for miRNA. The prominent performance of the proposed biosensing strategy could be attributed to the T7 Exo-assisted cascade signal amplification, hairpin assembly amplification and dual-signal ratiometric output mode.Specificity, reproducibility and stability of the biosensor
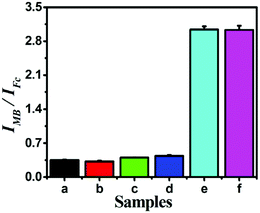
Diagnostic specificity is very important to evaluate the performance of a biosensor for accurate disease diagnosis; therefore, miRNAs containing single, double or total mismatched sites compared with target miRNA-141 were employed to examine the selectivity of the proposed strategy. As illustrated in Fig. 5, the signal output significantly increased with the introduction of target miRNA-141, while the other miRNAs had negligible response signals, indicating that only target miRNA can trigger the release of probe STP and subsequent signal transduction and amplification. This proves that the designed biosensing strategy showed excellent sequence specificity to discriminate miRNA family members. The remarkable specificity was due to the limited cleaving property of T7 Exo-based catalyzed reactions and the ingenious design to perform the ratiometric detection strategy.Moreover, reproducibility is extremely important for assessing the performance of the biosensing strategy, which was explored by a series of five repetitive measurements with 1 fM, 1 pM and 10 pM miRNA-141 (Table S3†). The relative standard deviations (RSD) are 3.77, 2.04 and 3.76%, respectively, demonstrating pleasurable reproducibility and accuracy of the proposed biosensor. Moreover, the sensor remains bioactive too and the response signal had no obvious changes after two-week storage at 4 °C, indicating the satisfied stability of the proposed Fc-H1 sensing platforms. These results illustrated that the proposed dual-signal ratiometric electrochemical biosensing strategy had satisfactory reproducibility and stability.
Real sample analysis
The feasibility of the proposed dual-signal ratiometric electrochemical biosensor for practical applications was investigated by adding numerous concentrations miRNA-141 into the 10% diluted human serum samples obtained from healthy volunteers. As shown in Table 1, the results showed acceptable recovery in the range of 97.60–102.49% and the RSDs from 2.31% to 5.62%. These results demonstrate that the proposed dual-signal ratiometric strategy possesses high potential for real sample determination.| Samples | Added (fM) | Found (fM) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| 1 | 10 | 10.95 ± 0.08 | 100.95 | 2.31 |
| 2 | 100 | 102.49 ± 0.14 | 102.49 | 5.62 |
| 3 | 1000 | 976.00± 0.09 | 97.60 | 4.56 |
Conclusions
In summary, a novel dual-signal ratiometric electrochemical biosensor based on the T7 Exo-assisted homogenous target cycle amplification coupling hairpin assembly-triggered dual-signal output has been successfully constructed for the accurate and sensitive detection of miRNA-141. Benefiting from the unique integration of the homogenous recognition amplification and anti-interference ratiometric electrochemical readout, the designed biosensor showed excellent analytical performance for miRNA-141 in a linear range of 5 orders of magnitude and the detection limit as low as aM level. Moreover, the method also exhibited preeminent specificity and acceptable accuracy, thus providing great promise in clinical application. This proposed biosensing strategy would provide favourable expansibility for the detection of biomolecules with high sensitivity, good selectivity and wide detection range, showing a promising avenue in early clinical disease diagnostics.Author contributions
Qing-Yun Zhou: methodology, investigation, data curation, writing original draft. Rong-Na Ma: conceptualization, project administration, funding acquisition, writing review & editing. Chao-Long Hu: investigation, data curation. Fei Sun: resources. Li-Ping Jia: methodology, investigation. Wei Zhang: methodology, investigation. Lei Shang: methodology, investigation. Qing-Wang Xue: writing review & editing. Wen-Li Jia: writing review & editing. Huai-Sheng Wang: supervision, project administration, funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (21505063 and 21804063), Natural Science Foundation of Shandong Province (ZR2015BQ007, ZR2017BB084, ZR2017QB010) and the Young Innovative Talents Introduction & Cultivation Program for Colleges and Universities of Shandong Province: Innovative Research Team on biomedical sensing and food safety Research.Notes and references
- J. F. Chen, E. M. Mandel, J. M. Thomson, Q. L. Wu, T. E. Callis, S. M. Hammond, F. L. Conlon and D. Z. Wang, Nat. Genet., 2006, 38, 228–233 CrossRef CAS PubMed.
- C. D. Johnson, A. E. Kerscher, G. Stefani, M. Byrom, K. Kelnar, D. Ovcharenko, M. Wilson, X. W. Wang, J. Shelton, J. Shingara, L. Chin, D. Brown and F. J. Slack, Cancer Res., 2007, 67, 7713–7722 CrossRef CAS PubMed.
- J. A. Chan, A. M. Krichevsky and K. S. Kosik, Cancer Res., 2005, 65, 6029–6033 CrossRef CAS PubMed.
- C. Z. Chen, L. Li, H. F. Lodish and D. P. Bartel, Science, 2004, 33, 83–86 CrossRef PubMed.
- B. Chen, H. Li, X. Zeng, P. Yang, X. Liu, X. Zhao and S. Liang, J. Transl. Med., 2012, 10, 228 CrossRef CAS PubMed.
- Z. Xie, L. Wroblewska, L. Prochazka, R. Weiss and Y. Benenson, Science, 2011, 333, 1307–1311 CrossRef CAS PubMed.
- Y. Zhang, Z. H. Shuai, H. Zhou, Z. M. Luo, B. Liu, Y. N. Zhang, L. Zhang, S. F. Chen, J. Chao, L. X. Weng, Q. L. Fan, C. H. Fan, W. Huang and L. H. Wang, J. Am. Chem. Soc., 2018, 140, 3988–3993 CrossRef CAS PubMed.
- J. Y. Li, W. X. Fu, Z. Y. Wang and Z. H. Dai, Chem. Sci., 2019, 10, 5616–5623 RSC.
- Z. H. Xu, H. Wang, J. Wang, W. Zhao, J. J. Xu and H. Y. Chen, Anal. Chem., 2019, 91, 12000–12005 CrossRef CAS PubMed.
- S. S. Zhang, S. L. Xu, X. Li, R. N. Ma, G. G. Cheng, Q. W. Xue and H. S. Wang, Chem. Commun., 2020, 56, 4288–4291 RSC.
- R. C. Lee and V. Ambros, Science, 2001, 294, 862–864 CrossRef CAS PubMed.
- C. Chen, R. Tan, L. Wong, R. Fekete and J. Halsey, Methods Mol. Biol., 2011, 687, 113–134 CrossRef CAS PubMed.
- J. Li, B. Yao, H. Huang, Z. Wang, C. H. Sun, Y. Fan, Q. Chang, S. L. Li, X. Wang and J. Z. Xi, Anal. Chem., 2009, 81, 5446–5451 CrossRef CAS PubMed.
- M. Castoldi, S. Schmidt, V. Benes, M. Noerholm, A. E. Kulozik, M. W. Hentze and M. U. Muckenthaler, RNA, 2006, 12, 913–920 CrossRef CAS PubMed.
- M. Schena, D. Shalon, R. W. Davis and P. O. Brown, Science, 1995, 270, 467–470 CrossRef CAS PubMed.
- P. Miao, T. Zhang, J. H. Xu and Y. G. Tang, Anal. Chem., 2018, 90, 11154–11160 CrossRef CAS PubMed.
- X. Liu, R. L. Huang, R. X. Su, W. Qi, L. B. Wang and Z. M. He, ACS Appl. Mater. Interfaces, 2014, 6, 13034–13042 CrossRef CAS PubMed.
- W. Y. Nie, Q. Wang, L. Y. Zou, Y. Zheng, X. F. Liu, X. H. Yang and K. M. Wang, Anal. Chem., 2018, 90, 12584–12591 CrossRef CAS PubMed.
- L. Y. Xia, M. J. Li, H. J. Wang, R. Yuan and Y. Q. Chai, Anal. Chem., 2020, 92, 14550–14557 CrossRef PubMed.
- Z. H. Xu, H. Wang, J. Wang, W. Zhao, J. J. Xu and H. Y. Chen, Anal. Chem., 2019, 91, 12000–12005 CrossRef CAS PubMed.
- D. X. Li, T. T. Zhang, F. Yang, R. Yuan and Y. Xiang, Anal. Chem., 2020, 92, 2074–2079 CrossRef CAS PubMed.
- S. Azzouzi, Z. Fredj, A. P. F. Turner, M. B. Ali and W. C. Mak, ACS Sens., 2019, 4, 326–334 CrossRef CAS PubMed.
- N. Zhang, X. M. Shi, H. Q. Guo, X. Z. Zhao, W. W. Zhao, J. J. Xu and H. Y. Chen, Anal. Chem., 2018, 90, 11892–11898 CrossRef CAS PubMed.
- Z. H. Xu, H. Wang, J. Wang, W. Zhao, J. J. Xu and H. Y. Chen, Anal. Chem., 2019, 91, 12000–12005 CrossRef CAS PubMed.
- T. Hou, N. N. Xu, W. X. Wang, L. Ge and F. Li, Anal. Chem., 2018, 90, 9591–9597 CrossRef CAS PubMed.
- H. F. Wang, R. N. Ma, F. Sun, L. P. Jia, W. Zhang, L. Shang, Q. W. Xue, W. L. Jia and H. S. Wang, Biosens. Bioelectron., 2018, 122, 224–230 CrossRef CAS PubMed.
- C. Wang, Q. Han, F. J. Mo, M. Chen, Z. W. Xiong and Y. Z. Fu, Anal. Chem., 2020, 92, 12145–12151 CrossRef CAS PubMed.
- S. P. Wen, Y. Su, C. X. Dai, J. R. Jia, G. C. Fan, L. P. Jiang, R. B. Song and J. J. Zhu, Anal. Chem., 2019, 91, 12298–12306 CrossRef CAS PubMed.
- Q. F. Xu, F. Ma, S. Q. Huang, B. Tang and C. Y. Zhang, Anal. Chem., 2017, 89, 7077–7083 CrossRef CAS PubMed.
- X. Y. Gao, M. X. Geng, Y. C. Li, X. L. Wang and H. Z. Yu, Anal. Chem., 2017, 89, 2464–2471 CrossRef CAS PubMed.
- H. Jin, R. J. Gui, J. B. Yu, W. Lv and Z. H. Wang, Biosens. Bioelectron., 2017, 91, 523–537 CrossRef CAS PubMed.
- C. X. Zhu, D. Liu, Y. Y. Li, X. L. Shen, L. B. Li, Y. Liu and T. Y. You, Curr. Opin. Electrochem., 2019, 17, 47–55 CrossRef CAS.
- L. Cui, M. F. Lu, Y. Li, B. Tang and C. Y. Zhang, Biosens. Bioelectron., 2018, 102, 87–93 CrossRef CAS PubMed.
- F. L. Gao, Y. Qian, L. Zhang, S. Z. Dai, Y. F. Lan, Y. Zhang, L. L. Du and D. Q. Tang, Biosens. Bioelectron., 2015, 71, 158–163 CrossRef CAS PubMed.
- R. N. Ma, L. L. Wang, M. Zhang, L. P. Jia, W. Zhang, L. Shang, W. L. Jia and H. S. Wang, Sens. Actuators, B, 2018, 257, 678–684 CrossRef CAS.
- L. L. Wang, R. N. Ma, L. S. Jiang, L. P. Jia, W. L. Jia and H. S. Wang, Biosens. Bioelectron., 2017, 92, 390–395 CrossRef CAS PubMed.
- Y. H. Yuan, B. Z. Chi, S. H. Wen, R. P. Liang, Z. M. Li and J. D. Qiu, Biosens. Bioelectron., 2018, 102, 211–216 CrossRef CAS PubMed.
- Y. Li, Y. Chang, J. Ma, Z. Wu, R. Yuan and Y. Chai, Anal. Chem., 2019, 91, 6127–6133 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Supplemental experimental section and tables. See DOI: 10.1039/d1an00204j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |