Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A rapid variant-tolerant reverse transcription loop-mediated isothermal amplification assay for the point of care detection of HIV-1†
Yingxue
Li‡
abc,
Xin
Chen‡
d,
Yongjuan
Zhao‡
a,
Zhenzhou
Wan
e,
Yi
Zeng
a,
Yingying
Ma
a,
Lianqun
Zhou
 bc,
Gaolian
Xu
bc,
Gaolian
Xu
 f,
Julien
Reboud
f,
Julien
Reboud
 g,
Jonathan M.
Cooper
g,
Jonathan M.
Cooper
 *g and
Chiyu
Zhang
*g and
Chiyu
Zhang
 *a
*a
aShanghai Clinical Research Center for Infectious Disease (HIV/AIDS), Shanghai Public Health Clinical Center, Fudan University, Shanghai 201508, China. E-mail: zhangcy1999@shphc.org.cn
bCAS key Laboratory of Bio-medical Diagnostics, Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Sciences, Suzhou 215163, China
cSchool of Biomedical Engineering, University of Science and Technology of China, Hefei 260026, China
dDepartment of Pathogenic Biology, School of Basic Medical Sciences, Gannan Medical University, Ganzhou 341000, China
eMedical Laboratory of Taizhou Fourth People's Hospital, Taizhou 225300, China
fNano Biomedical Research Centre, School of Biomedical Engineering, Shanghai Jiao Tong University, Shanghai 200030, China
gDivision of Biomedical Engineering, University of Glasgow, G12 8LT Glasgow, UK. E-mail: jon.cooper@glasgow.ac.uk
First published on 2nd July 2021
Abstract
Human immunodeficiency virus (HIV) continues to be a major burden on public health globally with on-going increases in the number of new infections each year. Rapid and sensitive point-of-care tests allow timely interventions and are essential to control the spread of the disease. However the highly variable nature of the virus, resulting in the evolution of many subtypes and inter-subtype recombinants, poses important challenges for its diagnosis. Here we describe a variant-tolerant reverse-transcription RT-LAMP amplification of the virus's INT gene, providing a simple to use, rapid (<30 min) in vitro point-of-care diagnostic test with a limit of detection <18 copies/reaction. The assay was first validated in clinical studies of patient samples, using both established RT-LAMP and RT-qPCR assays for reference, with results showing that this new variant-tolerant HIV-1 RT-LAMP diagnostic test is highly sensitive without compromising its high specificity for HIV-1 subtypes. The diagnostic test was subsequently configured within an easy-to-read paper microfluidic lateral flow test and was validated clinically using patient samples, demonstrating its future potential for use in timely, effective, low cost HIV diagnostics in global regions where healthcare resources may be limited.
Introduction
Human immunodeficiency virus type 1 (HIV-1) continues to pose a significant global public burden1 with current care pathways indicating that all infected people should receive antiretroviral therapy (ART). The Joint United Nations Program on HIV/AIDS (UNAIDS) proposed the global project termed “90–90–90” to end the global HIV/AIDS epidemic,2 stressing the importance of diagnostic tests and aiming for 90% of all people living with HIV-1 to know their HIV-1 status, for 90% of all people with diagnosed HIV-1 infection to receive sustained ART, as well as for 90% of all people on treatment to achieve virological suppression. The establishment of a fast and efficient HIV-1 testing method is essential to meet these goals and to implement timely public health interventions to control the spread of the disease.Real-time quantitative polymerase chain reaction (qPCR) remains the most widely used method for quantifying viral load and for the early detection of HIV-1 infections.3,4 However, the requirements of relatively sophisticated equipment and expertise in sample processing and results analysis imply that these tests need to be carried out in centralized laboratories. Furthermore, PCR reactions need a relatively long time (often 1.5 h), thus greatly limiting the application of the technique in resource-limited settings such as sub-Saharan Africa, South Asia and Southeast Asia, where many HIV-1 infected people live.5 To enable decentralized, rapid and convenient detection, such as HIV-1 point-of-care-testing immunodiagnostic lateral flow tests are now available although these may have a reduced efficacy because of the “window period”, as the lag between the infection and detection of seroconversion in infected individuals. Nucleic acid amplification testing (NAAT) can reduce this diagnostic window period for HIV-1 to about 10–15 days and is thus often used for donor blood screening. The development of HIV-1 point-of-care nucleic acid amplification tests is important for the timely diagnosis of HIV-1 infection with high clinical sensitivity. Unlike qPCR, isothermal amplification is performed at a constant temperature, decreasing the requirement for complex equipment.6 Tests, including nucleic acid sequence amplification (NASBA), loop-mediated isothermal amplification (LAMP),7,8 rolling circle amplification (RCA), and recombinase polymerase amplification (RPA)9 have been developed. However, many of these isothermal assays, particularly those based on RCA and RPA strategies are unable to detect viruses that mutate rapidly, since any mismatch with the primers can lead to a significant decrease in their amplification efficacy.9 HIV-1 is an example of one such virus, which has high mutation and recombination rates due to the lack of error-prone repair of viral reverse transcriptase, and a high degree of genetic heterogeneity.5,10
HIV-1 contains four groups (M, N, O and P) and the M group is responsible for the vast majority of global HIV infections. The HIV-1 M group is further classified into 11 subtype/sub-subtypes, over 102 circulating recombinant forms (CRFs), as well as a large number of unique recombinant forms (URFs) (https://www.hiv.lanl.gov/content/sequence/HIV/CRFs/CRFs.html). The prevalence of HIV-1 CRFs has increased dramatically, currently accounting for ∼20% of all known HIV infections.11,12 In some regions of East and South-East Asia, ∼80% of HIV-1 circulating strains are derived from CRFs and diverse URFs.13–15
We have recently developed a mismatch-resistant RT-LAMP method, which involves a new amplification mechanism using a small amount of high-fidelity DNA polymerase, where the 3′-5′ exonuclease activity removes potential mismatched bases at the 3′end of the primer, thereby improving its specificity for a range of infectious diseases including those caused by dengue and SARS-CoV-2 viruses.8,16–18 In this study, we now apply this mechanism to the challenge of point-of-care diagnostic tests of diverse genotypes (subtypes/CRFs/URFs) of HIV-1 M group, and demonstrated how this same strategy can be adapted to develop a variant-resistant RT-LAMP method for detecting various genotypes of the HIV-1 M group, especially URFs.
Our results show that the assay is highly sensitive without compromising its high specificity for HIV-1 M group strains, and has a high degree of consistency with the widely used RT-qPCR assay. By typing and analyzing positive HIV-1 samples, we found that our new RT-LAMP method has a higher detection rate of all involved genotypes of the HIV-1 M group when compared to the conventional RT-LAMP system, enabling us to detect some positive samples that were missed with the conventional RT-LAMP. Following the successful clinical evaluation of the assay in vitro, we implemented the test into a lateral flow device for testing patient samples. We propose that the method can be used for easy-to-read diagnosis of the HIV-1 M group, without the need for complex instrumentation, demonstrating its future potential to be used in this format in resource-limited settings as a point-of-care test.
Experimental
Clinical samples
To evaluate the performance of the novel RT-LAMP for HIV-1 detection, two batches of clinical samples were used. The first batch of 82 plasma samples, including 60 positive and 22 negative for HIV-1, was collected in Dehong Prefecture, Yunnan province of China. These samples were transported to Gannan Medical University for amplification and sequencing of the HIV-1 pol gene. The second batch of 45 plasma samples including 33 positive and 12 negative for HIV-1 was collected in Ganzhou city, Jiangxi province of China. HIV-1 infection was firstly screened using ELISA in the involved hospitals, and then confirmed by western blot at the loc at the local Centre for Disease Control. The use of the clinical samples was approved by the Ethics Committee of Gannan Medical University (2019206). Written informed consent was obtained from all participants.HIV-1 genotyping and viral load measurement
Viral RNA was extracted from 200 μL of plasma with a High Pure Viral RNA Kit (Roche Molecular Systems, Inc.), according to the manufacturer's instruction. Among the first batch of 60 HIV-1 positive samples, 44 were successfully amplified by nested-RT-PCR, sequenced by Sanger sequencing and typed by phylogenetic analysis (Chen et al. Unpublished data). Sixteen failed to be amplified.HIV-1 positive samples in the second batch were subjected to viral load measurement. HIV-1 viral load was measured using COBAS AmpliPrep/COBAS TaqMan HIV-1 Test v2.0 (Roche Molecular System Inc. Shanghai) at the time before storage.
To evaluate the performance of the novel RT-LAMP for HIV-1 detection, the RNA of two batches of clinical samples was re-extracted using a High Pure Viral RNA Kit (Roche Molecular Systems, Inc.) at Gannan Medical University, and transported to the Shanghai Public Health Clinical Center in dry ice for evaluating the performance of the novel RT-LAMP strip assay.
RNA extraction and viruses
To evaluate the specificity of the novel RT-LAMP assay. Nine standard virus strains, including influenza A and B viruses (VR-333 and VR-789), parainfluenza viruses 3 (VR-93), HCoV-229E (VR-740), HCoV-OC43 (VR-1558), VZV (VR-1367), EBV (VR-1492), CMV (VR-538), HHV-6 (VR-1480) and eight clinical samples confirmed positive for other viruses, including HPV52, HPV58, HPV66, HPV81, HBV, adenovirus, HSV-1 and HSV-2, were used as control viruses to test the assay specificity. Viral RNA was extracted from 140 μL of plasma using the QIAamp Viral RNA Mini Kit (Qiagen, Germany) according to the manufacturer's instructions, and eluted in 50 μL of nuclease-free water for storage at −80 °C until use.Construction of the RNA standard and mutants
To prepare the RNA standard, a HIV-1 plasmid containing a partial fragment of HIV-1 integrase gene was synthesized by BioSune Biotechnology Co., Ltd (reference sequence GenBank number is AF033819.3). A series of mutants of the HIV-1 plasmid that form mismatches with F3, FIP, and LB primers were constructed using a rapid mutagenesis system (TransGen, Beijing, China) and subjected to the proof-of-concept experiments of variant-tolerant RT-LAMP. The different mutants were constructed using site-directed mutagenesis with mutagenic primers (ESI Table S1†) and confirmed by Sanger sequencing. The fragment of HIV-1 integrase gene was amplified from the plasmid with primers containing the T7 promoter. The RNA standard and mutant RNA were obtained by in vitro transcription using a HiScribe T7 high-yield RNA synthesis kit (NEB, Beijing, China). After digestion with DNase I at 37 °C for 15 min, RNA was quantified using a Qubit® 4.0 Fluorometer (Thermo Fisher Scientific, USA). The copy number of each mutant RNA was calculated using the following formula: RNA copies per ml = [RNA concentration (ng mL−1)/(nt transcript length × 340)] × 6.022 × 1023.Reaction system of the novel RT-LAMP assay
0.15 unit of Q5 high-fidelity DNA polymerase (New England Biolabs, USA) was added per 25 μL of the conventional LAMP reaction mixture to establish the reaction system of the variant-tolerant RT-LAMP assay. This amount was optimised previously to obtain an efficient proof-reading function to remove mismatched bases, without competing significantly with other DNA polymerases (e.g. Taq polymerase and Bst 2.0 DNA polymerase) for primer extension.8,19,20 The optimized reaction mix of the novel assay thus included 2.5 μL of 10× isothermal amplification buffer, 4 mM MgSO4, 1.4 mM dNTPs, 8 units of Bst 2.0 DNA polymerase, 7.5 units of WarmStart RT, 0.15 unit of Q5 High-Fidelity DNA Polymerase, 0.2 μM each of primers AceIN-F3, AceIN-B3a, and AceIN-B3b, 0.8 μM each of primers AceIN-FIPf, AceIN-FIPe, AceIN-LF, and AceIN-LB, and 1.6 μM of AceIN-BIP. Three μL of RNA was added to each reaction mixture. The primer information was previously described and is provided as ESI Table S1.†16,21 The RT-LAMP reaction was performed at 62 °C for 60 min with 0.4 mM SYTO 9 (Life technologies, Carlsbad, CA, United States) as the fluorescent dye for real-time monitoring using the Light Cycler 96 real-time PCR System (Roche Diagnostics, Mannheim, Germany) or Bio-rad CFX 96 Real-Time System (Bio-Rad, CA, USA).Sensitivity and limit of detection (LOD)
Ten-fold serial dilutions of the RNA standard, from 104 to 100 copies per μL, were used as the standard to determine the sensitivity of the novel HIV-1 RT-LAMP assay. For the LOD test, ten-fold serial dilutions of the RNA standard, from 104 to 103 copies per μL, and five-fold dilutions of the RNA standard (1875 copies, 375 copies, 75 copies, 15 copies and 3 copies) were also used in the 25 μL reaction mixture. Each dilution was tested in a set of 10 replicates. The LOD was defined as a 95% probability of obtaining a positive result, using probit regression analysis with SPSS 17.0 software.22RT-qPCR assay of the first batch of clinical samples
The RT-qPCR reaction was carried out in a Light Cycler 96 Real-Time PCR System (Roche Diagnostics, Germany) using the 2×StarScript II Probe One-Step qRT-PCR Buffer (GenStar BioSolutions Co., Ltd, Beijing, China). The reaction contains 10 μL of 2×StarScript II Probe One-Step qRT-PCR Buffer, 2 μL of StarScript II One-Step Enzyme Mix, 200 nM HIV-1 probe (5′-FAM-ACAGTGCAGGGGAAAG-MGB-3′), 200 nM HIV-1 forward primer (5′-CAATTTTAAAAGAAAAGGGGGGATTG-3′), and 200 nM reverse primer (5′-TAAACCCGAAAATTTTGAATT-3′). The cycling condition was as follows: RT at 42 °C for 20 min, enzyme activation at 94 °C for 2 min followed by 45 cycles of denaturation at 94 °C for 15s, and annealing and extension at 60 °C for 30s. The fluorescence signal was collected at the annealing and extension step of each cycle. The threshold value was set to 0.05 automatically. The concordance rate between both assays was calculated by the formula: (number of consistent results by both methods/total number) × 100%.Lateral flow strip test of the novel HIV-1 detection assay
In order to apply the novel HIV-1 LAMP assay to POCT diagnosis, we combined this method with the lateral flow strip established by Xu et al. to verify the sensitivity of the novel RT-LAMP on strip and the accuracy of detecting clinical samples.21 We firstly used ten-fold serial dilutions of the RNA standard, from 105 to 100 copies per μL RNA standards, to determine the sensitivity of the novel HIV-1 RT-LAMP assay, and then used a lateral flow strip to show the reaction products. For the strip detection, the 5′-ends of loop primers LF and LB were labelled with biotin and FITC, respectively. The anti-FITC antibodies and immobilized streptavidin was labelled in the test and control lines of the detection strips, respectively. The results can be observed by the naked eye.The second batch of 45 clinical samples was also subjected to the novel RT-LAMP assay, which was performed as described above. The products were measured using a lateral flow strip.
Results
Principle and validation of the variant-tolerant RT-LAMP assay
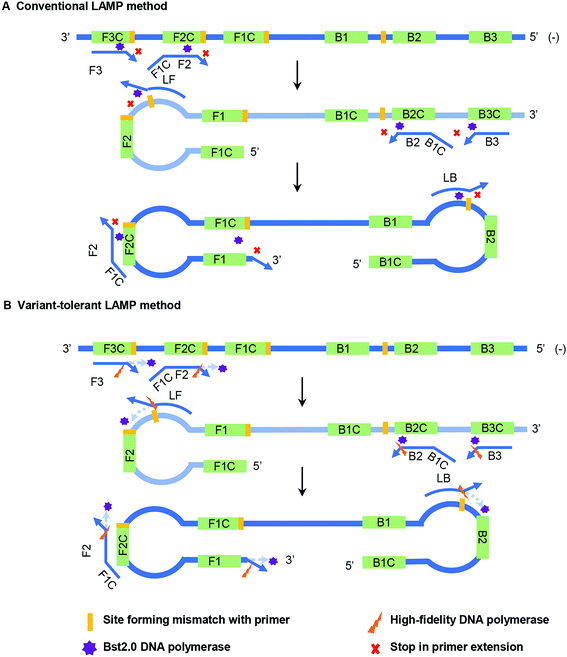
Bst DNA polymerase lacks a proofreading function, and therefore the appearance of 3′ mismatch in the primers prevents their elongation by Bst DNA polymerase in the conventional LAMP reaction (Fig. 1A). Furthermore, the existence of the newly synthesized forward and reverse strands guarantee the subsequent self-priming cycle in the LAMP reaction. A 3′ mismatch in FIP or BIP will prevent or dramatically reduce the amount of initial self-priming DNA products (newly synthesized forward or reverse strands) (Fig. 1A). The main difference between the variant-tolerant and conventional LAMP assay is that the former contains a small amount of additional high-fidelity DNA polymerase. High-fidelity DNA polymerase has a proofreading function by recognizing and removing a 3′ mismatched base in a newly synthesized DNA strand to initiate DNA elongation. The role of the proofreading polymerase in the variant-tolerant LAMP is to recognize and remove 3′-mismatched bases of the primers and/or newly synthesized DNA strands, which guarantees the effective elongation of 3′ mismatched primers and the self-priming of newly synthesized dumb-bell form DNA with mismatch in its 3′-end by Bst DNA polymerase (Fig. 1B).To assess the influence of various variants leading to mismatches with primers on RT-LAMP amplification and, at the same time, to validate the effectiveness of the novel variant-tolerant RT-LAMP assay, we constructed a series of mutant RNAs of HIV-1 that can form two different types of mismatches with the 3′-ends of F1, FIP, F3 and LB, as well as the penultimate, third and fifth bases at the 3′-end of FIP (Table 1 and ESI Fig. S2†). We firstly used a conventional RT-LAMP assay to detect 3 × 104 copies of these mutant RNAs with a wild type template as a control. Relative to the wild type template (Time threshold, Tt): 13.0 ± 0.2 min, the mutants generated slower RT-LAMP amplification with Tt values of 13.3–20.2 min (Table 1). In particular, some mismatches formed at the 3′-end of FIP prevented amplification (e.g. FIP-Mu-C and FIP-Mu-A) by conventional RT-LAMP, but had less impact on the amplification using our variant-tolerant RT-LAMP. The mismatches in different primers led to a marked difference in the inhibition of RT-LAMP amplification. The mutants forming mismatches with the 3′-end of the F1 (corresponding to the 5′-end of the FIP) and F3 primers had Tt values of 13.3–16.1 min (about 0.3–3.0 min slower than the wild type), while the mutants forming mismatches with the 3′-end of the FIP and LB primers had no Tt values (no amplification) and Tt values of 18.8–20.2 min (about 5.8–7.1 min slower than the wild type), respectively. The results indicated that the mismatches occurring in the 3′-end of the internal primer (i.e. FIP and BIP) and loop primer (i.e. LF and LB) led to greater inhibition than those occurring at the 3′-end of the outer primer (F3 and B3) and F1 (corresponding to the 5′-end of internal primers FIP). When compared to the mismatches occurring at the 3′-end of FIP that had no Tt values, the mismatches occurring at the penultimate, third and fifth bases at the 3′-end of FIP had Tt values of 14.5–18.2 min (about 1.5–5.2 min slower than the wild type, Table 1). These results indicated that mismatches occurring within a primer showed relatively less inhibition in amplification than those occurring at the 3′-end of the primer.
| Template | The novel variant-tolerant RT-LAMP | The conventional RT-LAMP | T t Diff (Novel-Conv.) | P values | ||||
|---|---|---|---|---|---|---|---|---|
| T t Mean (min) | SD | T t Diff (Mut-WT) | T t Mean (min) | SD | T t Diff (Mut-WT) | |||
| SD, standard deviation; NA, not applicable; None, none of three replicates generates amplification; Diff, difference; Mut, mutants; WT, wild type; Tt, time threshold; Conv., conventional RT-LAMP assay; Novel, the variant-tolerant RT-LAMP assay; P values: from the t-test; min, minute.a Only one of three replicates generated an amplification curve with a Tt value of 42.5 minutes, and the only one Tt value was shown.b Based on the Tt value of the unique one amplification in three replicates. | ||||||||
| Wild | 12.2 | 0.8 | NA | 13.0 | 0.2 | NA | −0.8 | 0.17 |
| F1-Mu-A | 12.1 | 0.5 | −0.1 | 13.3 | 0.5 | 0.3 | −1.2 | 0.02 |
| F1-Mu-C | 12.0 | 0.8 | −0.2 | 14.9 | 2.0 | 1.9 | −2.9 | 0.06 |
| F3-Mu-A | 14.9 | 0.9 | 2.7 | 16.1 | 0.9 | 3.1 | −1.2 | 0.09 |
| F3-Mu-G | 13.4 | 0.6 | 1.2 | 14.7 | 0.5 | 1.7 | −1.3 | 0.02 |
| LB-Mu-A | 13.2 | 1.2 | 1 | 18.8 | 3.5 | 5.8 | −5.6 | 0.05 |
| LB-Mu-C | 15.4 | 1.3 | 3.2 | 20.2 | 2.7 | 7.2 | −4.8 | 0.04 |
| FIP-Mu-A | 19.7 | 2.3 | 7.5 | 42.5a | NA | 29.5b | 22.8b | NA |
| FIP-Mu-C | 20.0 | 0.6 | 7.8 | None | NA | NA | NA | NA |
| FIP-Mu-2G | 13.8 | 0.9 | 1.6 | 16.0 | 1.0 | 3 | −2.2 | 0.03 |
| FIP-Mu-2C | 14.3 | 1.0 | 2.1 | 18.2 | 1.7 | 5.2 | −3.9 | 0.02 |
| FIP-Mu-3A | 15.1 | 1.8 | 2.9 | 16.6 | 0.2 | 3.6 | −1.5 | 0.14 |
| FIP-Mu-3C | 13.4 | 1.1 | 1.2 | 15.8 | 0.8 | 2.8 | −2.4 | 0.02 |
| FIP-Mu-5T | 14.0 | 0.2 | 1.8 | 14.5 | 0.3 | 1.5 | −0.5 | 0.04 |
| FIP-Mu-5G | 14.0 | 0.3 | 1.8 | 14.9 | 0.2 | 1.9 | −0.9 | 0.01 |
When our new variant-tolerant RT-LAMP assay was used, slightly larger Tt values (12.0–20.0 min) were obtained for all of the mutant RNAs than for the wild-type RNA (12.2 min) (Table 1). Importantly, however, the variant-tolerant RT-LAMP assay decreased the Tt values, regardless of any mutant RNAs (0.5–5.6 min, P values: 0.01–0.14; wild-type RNA, 0.8 min, P value: 0.17) compared to the conventional RT-LAMP (Table 1). In detail, the mutant RNAs FIP-Mu-C and FIP-Mu-A were amplified with Tt values of 19.7–20.0 min in the variant-tolerant RT-LAMP assay, but failed in the conventional RT-LAMP assay. These results indicated that the variant-tolerant RT-LAMP assay significantly improved the amplification of various mutant RNA templates when compared to the conventional RT-LAMP assay and showed excellent tolerance to a range of mismatches occurring in LAMP primers.
Sequence analysis of HIV-1 LAMP primer-binding regions
The LAMP primers to detect HIV-1 were previously designed in the INT gene region of the genome (ESI Fig. S1A†). To further evaluate the relevance of the primer-binding regions, we retrieved all available INT gene sequences of various subtypes and recombination forms of the HIV-1 M group and performed sequence alignments using MEGA 7.0 software. A total of 661 CRF01_AE, 555 CRF07_BC, 322 B, 98 CRF08_BC, 25 CRFC, 9 CRF59_01B, 9 CRF69_BC, 9 CRF85_BC, 8 CRF55_01B, 7 CRF65_cpx, 7 CRF57_BC, 5 G, 4 CRF61_BC, 4 CRF68_01B, 3 CRF78_cpx, 3 CRF79_0107, 3 CRF80_0107, 3 CRF86_BC, 3 CRF87_cpx, 3 CRF88_BC, 3 CRF96_cpx, and 2 CRF67_01B HIV-1 sequences were obtained (ESI Fig. S1B and S1C†). The vast majority of sequences are conserved for all subtypes, and only a few variants in each subtype formed mismatches with the LAMP primers (ESI Fig. S1B and S1C†).Specificity and sensitivity of the novel variant-tolerant HIV-1 RT-LAMP assay
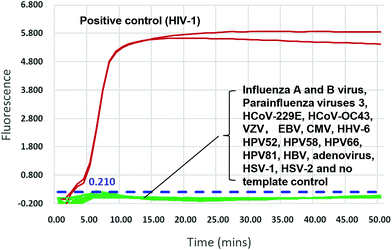
We validated the specificity of our assay against 17 other human viruses after 50 min amplification. No amplification signal was observed for these viruses (Fig. 2 and ESI Fig. S3†), indicating specificity to HIV-1. Ten-fold serial dilutions of wild-type HIV-1 RNA standard, from 104 to 100 copies per μL, were used to determine the sensitivity of the RT-LAMP assay. RNA inputs of 3 × 104 to 3 × 100 copies per 25 μL reaction mixture generated S-shaped amplification curves for all three replicates, whereas the input of 3 copies yielded only one amplification curve in three replicates in two independent experiments and no amplification in another experiment (Fig. 3). Importantly, almost all amplification curves appeared within 15 min and entered a plateau phase within 20 min (Fig. 3), indicating that our novel variant-tolerant RT-LAMP assay is fast.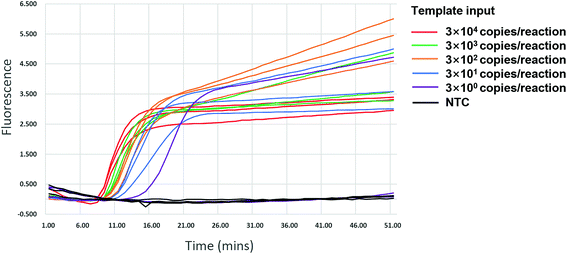 | ||
| Fig. 3 Sensitivity of the novel HIV-1 RT-LAMP assay. Positive amplification was observed in all replicates of more than 30 copies input. NTC: no template control. | ||
Further experiments showed that when the template input was more than 75 copies of HIV-1 RNA, all 10 reactions (100%) displayed positive amplification. When the template input was 15 and 3 copies, nine and five of the 10 replicates showed positive, respectively (Table 2). The LOD of the new RT-LAMP assay (defined by a probit regression with 95% confidence, see Methods) is thus below 18 copies per reaction.
| Dilution | Standard (copies/reaction) | Positive/total tested |
|---|---|---|
| 1× | 1875 | 10/10 |
| 5× | 375 | 10/10 |
| 5× | 75 | 10/10 |
| 5× | 15 | 9/10 |
| 5× | 3 | 5/10 |
Clinical evaluation of the novel variant-tolerant HIV-1 RT-LAMP assay
To evaluate the clinical application of the novel RT-LAMP assay, comparisons with a RT-qPCR assay and conventional RT-LAMP assay were performed, using 82 clinical samples previously collected from both HIV-1 infected individuals and healthy people. The novel RT-LAMP, the RT-qPCR assay, and the conventional RT-LAMP assays detected 39 (47.6%), 45 (54.9%), and 21 (25.6%) HIV-1 positive from 60 previously confirmed positive samples, respectively (Table 3). Only 21 (25.6%) of these samples were detected as HIV-1 positive by all three assays (ESI Fig. S3†). The concordance rates of the novel RT-LAMP assay were 92.7% with RT-qPCR assay, substantially higher than 78% with the conventional RT-LAMP assay (Table 3). All six positive samples that failed to be detected by the novel RT-LAMP assay showed high Ct values of >38 in the RT-qPCR analysis, implying a very low copy number.| Methods | RT-qPCR assay | The conventional RT-LAMP | Total | |||
|---|---|---|---|---|---|---|
| Pos., positive; Neg., negative. | ||||||
| Items | Pos. | Neg. | Pos. | Neg. | ||
| The novel RT-LAMP assay | Pos. | 39 | 0 | 21 | 18 | 39 |
| Neg. | 6 | 37 | 0 | 43 | 43 | |
| Total | 45 | 37 | 21 | 61 | 82 | |
The novel RT-LAMP assay detected 18 more positive samples than the conventional RT-LAMP assay. In order to investigate whether the failure by the conventional RT-LAMP assay was caused by the occurrence of mismatches between viral RNA and the primers, we further amplified 18 samples that were positive using the novel RT-LAMP assay but negative by the conventional ones, using a nested RT-PCR. Thirteen of them were amplified and sequenced. Seven were identified as HIV-1 M group URFs, two as CRF01_AE, one each as CRF08_BC, CRF57_BC, CRF96_cpx and B (ESI Fig. S4†). Further sequence analysis identified 6 mutant bases in the F2 primer-binding region, 5 mutant bases in the B3 primer-binding region, and 1 to 3 mutations in the binding regions of other primers (ESI Fig. S4†). Importantly, the vast majority of these mutations have locations corresponding to the 5′-end or in the middle of the primers.
We further evaluated the ability of the new method to detect different subtypes and recombinants of the HIV-1 M group. Forty four of 60 positive samples were previously genotyped, including 4 B, 2 C, 6 CRF01_AE, 3 CRF08_BC, 2 CRF57_BC, 2 CRF87_cpx, 1 CRF07_BC, 1 CRF62_BC, 1 CRF62_BC, 1 CRF65_cpx, and 21 URFs (Unpublished data). Twenty-nine of them (65.9%) were detected as positive by the novel RT-LAMP assay. The novel RT-LAMP assay showed better performance for CRF01_AE (66.7% vs. 33.3%), CRF08_BC (66.7% vs. 33.3%), CRF57_BC (100% vs. 50%), CRF96_cpx (66.7% vs. 33.3%) and URF (66.7% vs. 33.3%) than the conventional assay, and identical performance for CRF07_BC, B, C, CRF87_cpx, CRF62_BC and CR F65_cpx (ESI Table S2†).
Point-of-care detection using lateral flow strips
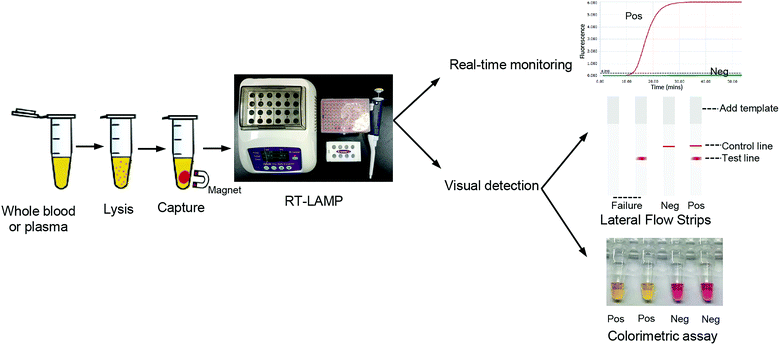
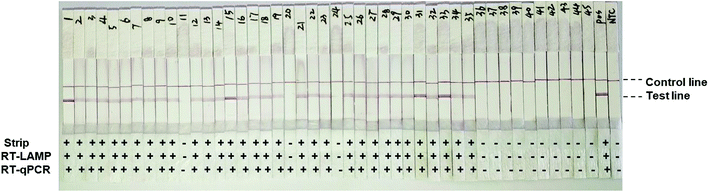
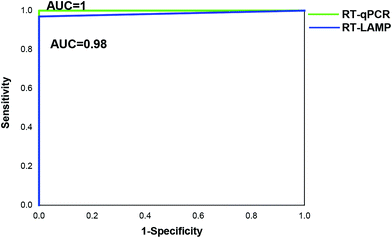
To develop point-of-care detection of the HIV-1 M group, we developed visual detection by combining the variant-tolerant RT-LAMP assay with a lateral flow strip and/or colorimetric reaction (Fig. 4). We first extracted viral RNA from plasma using the magnetic bead-based RNA extraction method, which can avoid multiple centrifugation steps using a centrifuge. The results demonstrated the efficacy of the magnetic bead-based method in RNA extraction from plasma (ESI Fig. S5†). To apply the novel HIV-1 LAMP assay to point-of-care diagnosis, we combined our new variant-tolerant RT-LAMP method with the lateral flow strip detection previously established by Xu et al.,23 which can avoid the difficulties in assessing color changes under variable lighting conditions in field-based work for example. We first verified the sensitivity of the variant-tolerant RT-LAMP assay using ten-fold serial dilutions of the wild-type HIV-1 RNA standard, from 105 to 100 copies per μL. Specific bands at the test line location were observed for all reactions with 3 × 105 to 3 × 100 copies input, indicating that the lateral flow strip can detect as low as 3 copies of template input per reaction (ESI Fig. S6†), which is consistent with the real-time monitoring with fluorescent dye SYTO 9 in the real-time analysis (Fig. 3).We further evaluated the performance of the point-of-care diagnostics using the second batch of 45 clinical samples. Among the 45 samples, the strips showed positive for 32 samples, which was consistent with the results by real-time qPCR monitoring (Table 4 and Fig. 5). The sensitivity and specificity of the assay was 97% and 100%, respectively. Only one sample was detected as negative by the novel RT-LAMP assay but positive by the RT-qPCR assay. ROC curve analysis showed that the clinical performance of the novel RT-LAMP assay was close to that of RT-qPCR for this study (Fig. 6). In our novel RT-LAMP assay, 81.3% of positive samples had Tt values less than 20 min, and only one sample with a viral load of 4050 copies per mL had a Tt value of more than 30 min. One failure and one slow amplification by RT-LAMP might be due to very low viral loads caused by the freezing/thawing cycle and/or degradation of nucleic acids during transportation. We further analyzed the relationship between the viral load of the 32 HIV-1 positive samples and the Tt values of the novel RT-LAMP, and found that Tt values by the novel RT-LAMP assay did not linearly correlate with the viral load (ESI Fig. S7†).
| The novel RT-LAMP assay | Total | |||
|---|---|---|---|---|
| Items | Pos. | Neg. | ||
| Pos., positive; Neg., negative. | ||||
| RT-qPCR assay | Pos. | 32 | 1 | 33 |
| Neg. | 0 | 12 | 12 | |
| Total | 32 | 13 | 45 | |
Visual detection using the colorimetric version of the LAMP assay is another useful alternative for point-of-care diagnostics. We also developed the detection of HIV-1 M groups using the colorimetric version of the variant-tolerant RT-LAMP assay. The positive result is easily judged by the colorimetric change from red to orange or yellow. The results showed that the sensitivity of the colorimetric version assay for the HIV-1 M group was 4 copies per reaction, consistent with that of the real-time monitoring system (ESI Fig. S8†).
Discussion
Despite the introduction of efficient ART, HIV/AIDS continues to cause a worldwide health crisis.2,24,25 Easy and rapid detection of HIV-1 infection is required for new global strategies to curb the impact of the virus. A number of detection methods have already been established, based upon either immunodiagnostics or NAAT.24 The immunodiagnostics methods that target the detection of HIV-1 specific antibodies often fail to detect HIV-1 infection within the window period (about two-four weeks before seroconversion), enabling further transmission and delaying treatment. Although NAAT can qualitatively detect the presence of HIV-1 as early as 7 days after infection, greatly shortening the “window period” of HIV infection,26,27 current tests either require centralized facilities (sophisticated equipment and skilled personnel) or are generally unable to detect rapidly evolving virus variants,13,28 posing a significant challenge for HIV-1 nucleic acid testing.10 HIV-1 is one of the most variable human viruses and has very high heterogeneity. Currently, Africa and Southeast Asia are the worst-hit regions by HIV/AIDS, and many subtypes and CRFs, as well as a large number of URFs of the HIV-1 M group are circulating in these areas.14,29 Fast, simple and sensitive POCT diagnosis of HIV-1 infection will benefit the prevention and control of HIV/AIDS in these low-resource regions.In our previous work, we added a small amount of high-fidelity DNA polymerase to the conventional RT-LAMP mix to increase its ability to detect highly variable viruses, establishing a fast and sensitive variant-tolerant RT-LAMP technology.16 We now build upon this initial work and demonstrate a new variant-tolerant RT-LAMP technique that has high sensitivity for detecting various subtypes, CRFs and URFs of the HIV-1 M group, with a LOD of 18 copies per reaction and no cross-reactivity with other common human viruses. The assay was evaluated using 82 clinical samples from HIV-1 infected individuals and control populations and showed a high degree of consistency with the widely used RT-qPCR assay (92.7%). However, the conventional RT-LAMP assay showed low consistency with the RT-qPCR assay and the new RT-LAMP assay, especially for HIV-1 positive samples. By typing and analyzing the positive HIV-1 samples, it was found that the new RT-LAMP method has a higher detection rate of various subtypes, CRFs and URFs of the HIV-1 M group than that of the conventional RT-LAMP system, explaining the low agreement between the two techniques (47.6%). Our new RT-LAMP system enabled us to detect 18 HIV-1 positive samples that were missed by the conventional RT-LAMP assay, giving an 85.7% (18/21) improvement in the positive detection rate and confirming that the new format is more suitable for detecting high-variation viruses. Finally, we also showed that the novel HIV-1 RT-LAMP assay can be conveniently implemented as an easy-to-read visual detection on lateral flow strips or via colorimetric change. Whilst colorimetric detection enables to perform the assay in a closed-tube format, limiting the risk of contamination, the interpretation of the color under varying lighting conditions, as often encountered in field situations, can be challenging. The lateral flow strips are a commonly used format in rapid tests and facilitate interpretation. Using new cartridge formats,30 the amplicons can be transferred to the strip for detection in a closed system as well. In a separate study using 45 clinical samples, we obtained a high sensitivity and accuracy for the proposed POCT format.
Conclusions
In conclusion, we have developed a novel variant-tolerant RT-LAMP assay for detecting various genotypes and recombinants of the HIV-1 M group using the addition of tiny amounts of high-fidelity DNA polymerase to the conventional HIV-1 RT-LAMP mixture. The new LAMP method can tolerate significant mismatches between primers and viral genomic RNA, and shows higher amplification efficiency than the conventional RT-LAMP method. Combined with lateral flow strips or colorimetric version for easy visual detection, the method's short response time and simple instrument requirements show great potential for its application in resource-limited regions.Author contributions
YL, XC and Y Zh contributed equally to this work. Conceptualisation, CZ, YL and JC; methodology, YL, XC, Y Zh, JR, JC and YM; software, YL; validation, ZW, LZ, YL and GX; formal analysis, YL, Y Zh, JR, JC and XC; investigation, YL, XC, Y Zh, Y Ze and YM; resources, XC; writing – original draft preparation, YL; writing – review and editing, JR, JC, and CZ; project administration, CZ. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Shanghai Science & Technology Innovation Action Program [20MC1920100], the National Science and Technology Major Project of China [2019YFC1200603], Shenzhen Health System Research Project [SZGW201810003] and the UK Global Challenge Research Fund, through funding from the Engineering and Physical Sciences Research Council (EP/R01437X/1, co-funded by the UK National Institute for Health Research) and the Scottish Funding Council [SFC/AN/18/2020].References
- UNAIDS, Indicators for monitoring the 2016 Political Declaration on Ending AIDS—Global AIDS Monitoring 2021. Available from: https://www.unaids.org/sites/default/files/media_asset/global-aids-monitoring_en.pdf.
- UNAIDS: 90-90-90: An Ambitious Treatment Target to Help End the AIDS Epidemic 2017. Available from: https://www.unaids.org/sites/default/files/media_asset/90-90-90_en.pdf.
- J. Zhao, L. Chang and L. Wang, Eur. J. Clin. Microbiol. Infect. Dis., 2019, 38, 829–842 CrossRef CAS PubMed.
- S. Yang and R. E. Rothman, Lancet Infect. Dis., 2004, 4, 337–348 CrossRef CAS.
- J. Hemelaar, Trends Mol. Med., 2012, 18, 182–192 CrossRef PubMed.
- H. D. de Paz, P. Brotons and C. Muñoz-Almagro, Expert Rev. Mol. Diagn., 2014, 14, 827–843 CrossRef CAS.
- T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe, N. Amino and T. Hase, Nucleic Acids Res., 2000, 28, e63 CrossRef CAS PubMed.
- Y. Zhou, Z. Wan, S. Yang, Y. Li, M. Li, B. Wang, Y. Hu, X. Xia, X. Jin, N. Yu and C. Zhang, Front. Microbiol., 2019, 10, 1056 CrossRef PubMed.
- L. Yan, J. Zhou, Y. Zheng, A. S. Gamson, B. T. Roembke, S. Nakayama and H. O. Sintim, Mol. Biosyst., 2014, 10, 970–1003 RSC.
- N. R. Faria, N. Vidal, J. Lourenco, J. Raghwani, K. C. E. Sigaloff, A. J. Tatem, D. A. M. van de Vijver, A. C. Pineda-Pena, R. Rose, C. L. Wallis, S. Ahuka-Mundeke, J. J. Muyembe-Tamfum, J. Muwonga, M. A. Suchard, T. F. Rinke de Wit, R. L. Hamers, N. Ndembi, G. Baele, M. Peeters, O. G. Pybus, P. Lemey and S. Dellicour, PLoS Pathog., 2019, 15, e1007976 CrossRef CAS PubMed.
- H. Yuan, Z. Liu, X. Wu, M. Wu, Q. Fang, D. C. Tully and T. Zhang, Int. J. Infect. Dis., 2019, 90, 125–131 CrossRef.
- L. Zhang, Y. J. Wang, B. X. Wang, J. W. Yan, Y. N. Wan and J. Wang, Int. J. STD AIDS, 2015, 26, 291–305 CrossRef PubMed.
- J. Hemelaar, R. Elangovan, J. Yun, L. Dickson-Tetteh, I. Fleminger, S. Kirtley, B. Williams, E. Gouws-Williams, P. D. Ghys, A. l. G. Abimiku, S. Agwale, C. Archibald, B. Avidor, M. G. Barbás, F. Barre-Sinoussi, B. Barugahare, E. H. Belabbes, S. Bertagnolio, D. Birx, A. F. Bobkov, J. Brandful, H. Bredell, C. A. Brennan, J. Brooks, M. Bruckova, L. Buonaguro, F. Buonaguro, S. Buttò, A. Buve, M. Campbell, J. Carr, A. Carrera, M. G. Carrillo, C. Celum, B. Chaplin, M. Charles, D. Chatzidimitriou, Z. Chen, K. Chijiwa, D. Cooper, P. Cunningham, A. Dagnra, C. F. de Gascun, J. Del Amo, E. Delgado, U. Dietrich, D. Dwyer, D. Ellenberger, B. Ensoli, M. Essex, F. Gao, H. Fleury, P. N. Fonjungo, V. Foulongne, D. A. Gadkari, F. García, R. Garsia, G. M. Gershy-Damet, J. R. Glynn, R. Goodall, Z. Grossman, M. Lindenmeyer-Guimarães, B. Hahn, R. L. Hamers, O. Hamouda, R. Handema, X. He, J. Herbeck, D. D. Ho, A. Holguin, M. Hosseinipour, G. Hunt, M. Ito, M. A. Bel Hadj Kacem, E. Kahle, P. K. Kaleebu, M. Kalish, A. Kamarulzaman, C. Kang, P. Kanki, E. Karamov, J.-C. Karasi, K. Kayitenkore, T. Kelleher, D. Kitayaporn, L. G. Kostrikis, C. Kucherer, C. Lara, T. Leitner, K. Liitsola, J. Lingappa, M. Linka, I. Lorenzana de Rivera, V. Lukashov, S. Maayan, L. Mayr, F. McCutchan, N. Meda, E. Menu, F. Mhalu, D. Mloka, J. L. Mokili, B. Montes, O. Mor, M. Morgado, F. Mosha, A. Moussi, J. Mullins, R. Najera, M. Nasr, N. Ndembi, J. R. Neilson, V. R. Nerurkar, F. Neuhann, C. Nolte, V. Novitsky, P. Nyambi, M. Ofner, F. J. Paladin, A. Papa, J. Pape, N. Parkin, C. Parry, M. Peeters, A. Pelletier, L. Pérez-Álvarez, D. Pillay, A. Pinto, T. D. Quang, C. Rademeyer, F. Raikanikoda, M. A. Rayfield, J.-M. Reynes, T. Rinke de Wit, K. E. Robbins, M. Rolland, C. Rousseau, J. Salazar-Gonzales, H. Salem, M. Salminen, H. Salomon, P. Sandstrom, M. L. Santiago, A. D. Sarr, B. Schroeder, M. Segondy, P. Selhorst, S. Sempala, J. Servais, A. Shaik, Y. Shao, A. Slim, M. A. Soares, E. Songok, D. Stewart, J. Stokes, S. Subbarao, R. Sutthent, J. Takehisa, A. Tanuri, K. K. Tee, K. Thapa, M. Thomson, T. Tran, W. Urassa, H. Ushijima, P. van de Perre, G. van der Groen, K. van Laethem, J. van Oosterhout, A. van Sighem, E. van Wijngaerden, A.-M. Vandamme, J. Vercauteren, N. Vidal, L. Wallace, C. Williamson, D. Wolday, J. Xu, C. Yang, L. Zhang and R. Zhang, Lancet Infect. Dis., 2019, 19, 143–155 CrossRef PubMed.
- W. Pang, C. Zhang, L. Duo, Y. H. Zhou, Z. H. Yao, F. L. Liu, H. Li, Y. Q. Tu and Y. T. Zheng, AIDS, 2012, 26, 1121–1129 CrossRef PubMed.
- R. Yang, X. Xia, S. Kusagawa, C. Zhang, K. Ben and Y. Takebe, AIDS, 2002, 16, 1401–1407 CrossRef PubMed.
- R. H. Wang, H. Zhang, Y. Zhang, X. N. Li, X. X. Shen, J. J. Qi, G. H. Fan, X. Y. Xiang, Z. F. Zhan, Z. W. Chen and X. J. Ma, Virol. J., 2019, 16, 86 CrossRef PubMed.
- R. Lu, X. Wu, Z. Wan, Y. Li, L. Zuo, J. Qin, X. Jin and C. Zhang, Virol. Sin., 2020, 35, 344–347 CrossRef CAS PubMed.
- R. Lu, X. Wu, Z. Wan, Y. Li, X. Jin and C. Zhang, Int. J. Mol. Sci., 2020, 21, 2826 CrossRef CAS PubMed.
- W. Hao, L. Fan, Q. Chen, X. Chen, S. Zhang, K. Lan, J. Lu and C. Zhang, PLoS One, 2015, 10, e0123468 CrossRef PubMed.
- Y. Li, Z. Wan, Y. Hu, Y. Zhou, Q. Chen and C. Zhang, BioTechniques, 2019, 66, 225–230 CrossRef CAS PubMed.
- K. E. Ocwieja, S. Sherrill-Mix, C. Liu, J. Song, H. Bau and F. D. Bushman, PLoS One, 2015, e0117852 CrossRef.
- D. J. Anderson, Clin. Chem., 1989, 35, 2152–2153 CrossRef CAS.
- G. Xu, Q. You, S. Pickerill, H. Zhong, H. Wang, J. Shi, Y. Luo, P. You, H. Kong, F. Lu and L. Hu, J. Med. Virol., 2010, 82, 1143–1149 CrossRef CAS PubMed.
- J. Guarner, Semin. Diagn. Pathol., 2017, 34, 318–324 CrossRef PubMed.
- C. J. L. Murray, K. F. Ortblad, C. Guinovart, S. S. Lim, T. M. Wolock, D. A. Roberts, E. A. Dansereau and N. Graetz, Lancet, 2014, 384, 1–127 CrossRef.
- K. A. Sollis, P. W. Smit, S. Fiscus, N. Ford, M. Vitoria, S. Essajee, D. Barnett, B. Cheng, S. M. Crowe, T. Denny, A. Landay, W. Stevens, V. Habiyambere, J. Perrins and R. W. Peeling, PLoS One, 2014, 9, e85869 CrossRef PubMed.
- S. A. Glynn, S. H. Kleinman, D. J. Wright and M. P. Busch, Transfusion, 2002, 42, 966–972 CrossRef PubMed.
- Y. Liu, L. Jia, B. Su, H. Li, Z. Li, J. Han, Y. Zhang, T. Zhang, T. Li, H. Wu, J. Li and L. Li, AIDS Res. Hum. Retroviruses, 2020, 36, 440–449 CrossRef CAS.
- M. Rolland and K. Modjarrad, AIDS, 2015, 29, 1417–1419 CrossRef PubMed.
- J. Reboud, G. Xu, A. Garrett, M. Adriko, Z. Yang, E. M. Tukahebwa, C. Rowell and J. M. Cooper, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 4834–4842 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1an00598g. The associated data is available from the University of Glasgow repository at the following DOI http://dx.doi.org/10.5525/gla.researchdata.1168 |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |