Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Detection of RNA viruses from influenza and HIV to Ebola and SARS-CoV-2: a review
Rostislav
Bukasov
 a,
Dina
Dossym
a and
Olena
Filchakova
*b
a,
Dina
Dossym
a and
Olena
Filchakova
*b
aChemistry Department, SSH, Nazarbayev University, Nur-Sultan, 010000, Kazakhstan
bBiology Department, SSH, Nazarbayev University, Nur-Sultan, 010000, Kazakhstan. E-mail: olena.filchakova@nu.edu.kz
First published on 7th December 2020
Abstract
RNA-based viruses likely make up the highest pandemic threat among all known pathogens in about the last 100 years, since the Spanish Flu of 1918 with 50 M deaths up to COVID-19. Nowadays, an efficient and affordable testing strategy for such viruses have become the paramount target for the fields of virology and bioanalytical chemistry. The detection of the viruses (influenza, hepatitis, HIV, Zika, SARS, Ebola, SARS-CoV-2, etc.) and human antibodies to these viruses is described and tabulated in terms of the reported methods of detection, time to results, accuracy and specificity, if they are reported. The review is focused, but not limited to publications in the last decade. Finally, the limits of detection for each representative publication are tabulated by detection methods and discussed. These methods include PCR, lateral flow immunoassays, LAMP-based methods, ELISA, electrochemical methods (e.g., amperometry, voltammetry), fluorescence spectroscopy, AFM, SPR and SERS spectroscopy, silver staining and CRISPR-Cas based methods, bio-barcode detection, and resonance light scattering. The review is likely to be interesting for various scientists, and particularly helpful with information for establishing interdisciplinary research.
1. Introduction
Viruses are small, nanometer-scale carriers of genetic material. Unable to replicate by themselves, they live inside host cells and use their hosts for replication and assembly. Hence, they are intracellular parasites. In order to exist, viruses must be able to have multiple hosts. Viruses of bacteria, fungi, plants, and animals exist. Furthermore, a single virus can have hosts belonging to different species (e.g., rhinolophid bats and humans). Such viruses can jump from one host species to another. Viruses are not “interested” in killing their host instantaneously because the host's death would lead to the termination of viral spread. Therefore, the majority of viruses do not cause lethality. Some viruses, however, are infectious agents capable of causing illnesses with lethal outcomes. According to the CDC, the flu caused 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–61
000–61![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths in the USA each year since 2010.
000 deaths in the USA each year since 2010.
Depending on the type of nucleic acid they carry, viruses are classified into DNA-containing and RNA-containing viruses. DNA-containing viruses can be single-stranded or double-stranded; they are typically more benign than RNA-viruses and mutagenize to a lesser degree. RNA-viruses can be positive-strand, negative-strand or ambisense. Positive-strand RNA viruses contain genomic RNA, which is identical to viral mRNA, and can be translated by a host cell. Negative-strand viruses contain RNA in their genome, which is complimentary to mRNA and is used as a template for transcription by RNA-dependent RNA polymerase. Ambisense viruses contain genomic RNA, part of which behaves as positive-strand, while another part possesses negative-strand characteristics. For their replication, RNA-containing viruses rely on polymerases without stringent proofreading abilities. Contrarily, DNA-containing viruses rely on DNA-polymerase, which has proofreading properties. Therefore, the genome of RNA-containing viruses is subject to mutations at a much higher rate. This makes RNA-containing viruses rapidly evolving viruses that outnumber DNA-containing viruses. In addition, RNA-containing viruses are more pathogenic as compared to DNA-containing viruses. Examples of RNA-containing viruses that cause diseases include SARS-coronaviruses, influenza, hepatitis A, C, D, E, dengue, Ebola, HIV and other viruses.
Viruses are a diverse group of infectious agents, with more than 100 families characterized to date. Within the current review, we will focus on the laboratory diagnostics of RNA-containing viruses, which are causative agents of diseases with a high mortality rate or with a tendency for pandemic spread.
A multitude of different diagnostics tools exists. They include the detection of viruses, nucleic acids, and antibodies against viruses. The criteria that diagnostics tests should satisfy include a low limit of detection, high sensitivity, high specificity, high accuracy, and rapid speed of diagnosis. The limit of detection refers to the lowest concentration of analyte detected by a particular test. Depending on the test, the limit of detection can span a few RNA species in a reaction (for assays that detect nucleic acids) or a few ng mL−1 of analyte (for assays that detect antibodies). The sensitivity refers to the ability of a particular test to detect a virus when the virus is present in a sample, and is expressed in % (100% – % of false negative results). Specificity refers to the ability of a particular test to show a negative result when the virus is absent from a sample, and is expressed in % (100% – % of false positive results). Accuracy denotes the percentage of times at which the performed test results are correct. Thus, a high accuracy indicates low percentages of false-positive and false-negative results. The time it takes for a particular test to produce results depends on the test. Antibodies-detecting tests take less time to produce results than nucleic-acid-based tests (such as quantitative real-time PCR). The present review is focused on comparing different viral diagnostic tests through a quantitative perspective.
Cheng et al.'s review in Analytical and Bioanalytical Chemistry gave a detailed picture of viral diagnostics in 2009,1 where the detection was described using a method-to-method approach. Since the publication of that review, new lethal viruses have appeared (Ebola and SARS-coronaviruses), and many tests were developed and validated for their detection and diagnostics. The present review is focused on the quantitative analytical parameters in viral diagnostics as well, but it covers the subject from virus to virus. We selectively covered viruses that are lethal to humans, which contain RNA, and which have at least some pandemic potential. Most attention is directed towards the COVID-19 pandemic virus. The papers described in this review typically reported the limit of detection or some other analytical parameters describing the test accuracy (selectivity, sensitivity) (Fig. 1). Moreover, the orthogonal approach of viral detection classified by detection method is briefly shown as a table at the end of the review.
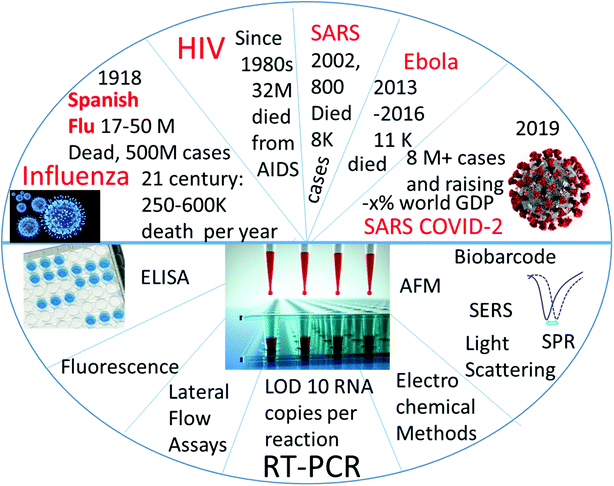 | ||
| Fig. 1 Graphic Content of the review: detection of RNA-containing pandemic-prone viruses by virus (top half) and by method of detection (bottom half). | ||
2. Detection of influenza viruses
Influenza is a respiratory disease of viral origin. Two types of viruses, influenza A and influenza B, are causative agents2 of the disease. Both viruses are negative-strand single-strand RNA viruses. The viral genome is segmented and contains 8 segments. They code for RNA-dependent RNA polymerase (needed to convert negative-strand into positive-strand RNA), haemagglutinin (HA, glycoprotein, required for viral entry), neuraminidase (NA, glycoprotein, needed for viral release), nucleoprotein (NP), matrix protein, membrane protein, nuclear export protein (NEP), and nonstructural proteins (NS). HA and NA proteins have high antigenic variability, contributing to the great diversity of viral subtypes. Influenza is characterized by seasonal epidemics, with an ability to transform into pandemics. The occurrence of pandemics is due to the zoonotic origin of the influenza A virus. It is of particular significance that the influenza A virus can spread among both animals and birds. The pandemics of 1918 and of 2009 (Spanish flu and swine flu, respectively) were caused by the H1N1 viral strain. The pandemic of 1918 resulted in more than 40 million deaths worldwide. The 2009 swine flu pandemic was due to the H1N1 influenza A virus, and caused an estimated 201![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 respiratory deaths, together with 83
200 respiratory deaths, together with 83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 cardiovascular deaths; only 18
300 cardiovascular deaths; only 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 deaths were laboratory-confirmed.3 The WHO estimates that 290
500 deaths were laboratory-confirmed.3 The WHO estimates that 290![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 650
000 to 650![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths occur annually worldwide due to influenza-associated respiratory diseases.4 The CDC estimates that influenza has resulted in between 9 million and 45 million illnesses, between 140
000 deaths occur annually worldwide due to influenza-associated respiratory diseases.4 The CDC estimates that influenza has resulted in between 9 million and 45 million illnesses, between 140![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 810
000 and 810![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 hospitalizations, and between 12
000 hospitalizations, and between 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 61
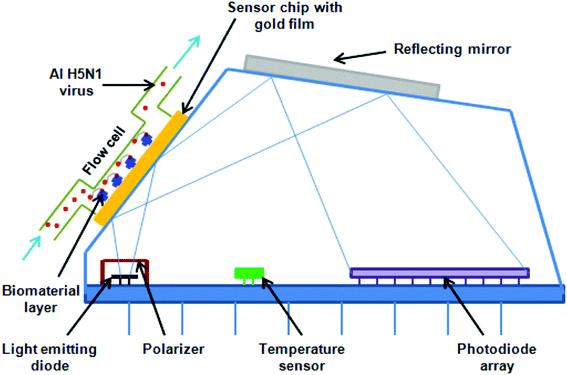
000 and 61![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths annually, since 2010 in the USA.5 The representative test for the detection of the H5N1 influenza virus is shown in Fig. 2, while different methods are presented in Table 1.
000 deaths annually, since 2010 in the USA.5 The representative test for the detection of the H5N1 influenza virus is shown in Fig. 2, while different methods are presented in Table 1.
| # | Method | Target/analyte | Volume | Accuracy | Time for test | Ref. |
|---|---|---|---|---|---|---|
| 1 | Commercial antigen detection tests and RT-qPCR | Swine-origin influenza virus (S-OIV) and seasonal influenza A (H1N1) isolate: antigens and M genes | N/A | N/A | N/A | 6 |
| 2 | – Rapid antigen test | H1N1 influenza A antigen | 200 μL of respiratory sample | Rapid antigen: 93.6% | N/A | 7 |
| – Direct immunofluorescence (DFA) | DFA: 94.5% | |||||
| – R-Mix culture | R-Mix culture: 100% | |||||
| – Respiratory Virus Panel (RVP) | RVP: 100% | |||||
| 3 | SPR aptasensor | Avian influenza virus H5N1 virions | N/A | N/A | 1.5 hours | 8 |
| 4 | ELISA | Anti-influenza A virus nucleoprotein antibodies | 15 μL of porcine sera | 99.3% | N/A | 9 |
| Sensitivity – 96.6% | ||||||
| 5 | A double-antigen sandwich ELISA | Antibodies to influenza A viruses | 50 μL of serum sample | 97.3% | N/A | 10 |
| Sensitivity – 98% | ||||||
| 6 | RT-LAMP on an integrated centrifugal disc | Influenza A (subtypes H1, H3, H5, H7, and H9) and influenza B RNA | 25 μL | N/A | 45 min | 11 |
| 7 | A nanocomposite of AuNPs and polyols with a dual response | Influenza A virions | N/A | N/A | N/A | 12 |
| 8 | Aptamer-based field-effect transistor | H5N1 avian influenza virus hemagglutinin (HA) protein | 3 μL | N/A | 5 hours | 13 |
| 9 | Magnetic particle spectroscopy | Influenza A virus subtype H1N1 nucleoprotein | 100 μL | N/A | 10 s | 14 |
3. Detection of RNA-containing viruses of health concern
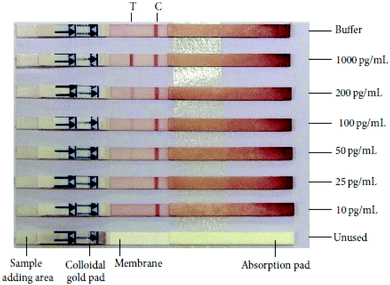
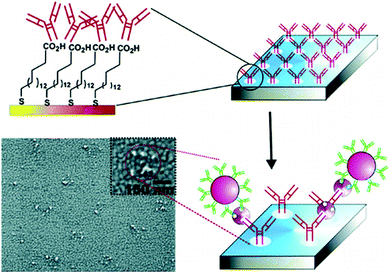
HIV/AIDS is prevalent mostly in East and Southern Africa, with a prevalence of 7.4% in the region in 2014, which is over 3 times higher than the prevalence in Western and Central Africa. Eastern Europe and Central Asia are the only regions with a rising HIV prevalence, at 0.8% in 2014. The global prevalence was 0.8% in 2014. The morbidity rate is currently at 1.1 million per year (2015).15 A variety of methods exist for HIV diagnostics. Two such methods are presented in Fig. 2 and 3. Both methods detect the p24 antigen by colloidal gold immunochromatography, and by ELISA with the use of AFM, respectively. In 2014, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people were infected with Ebola, with 4922 fatalities. The mortality rate for Ebola can reach 90%.16
000 people were infected with Ebola, with 4922 fatalities. The mortality rate for Ebola can reach 90%.16
In 2016, more than 85 countries and territories had the Zika virus infection transmitted by mosquitos. Brazil had the greatest impact, with more than 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cases of the Zika virus disease.17 The Zika virus had a mortality rate of 8.3% in Brazil.18
000 cases of the Zika virus disease.17 The Zika virus had a mortality rate of 8.3% in Brazil.18
Cases of viral hemorrhagic fever were seen in Zimbabwe, Uganda, the Democratic Republic of Congo, Kenya, and Angola, with a fatality rate of up to 15%.19
The number of dengue cases reported to the WHO has increased over 15 fold over the last two decades, from 505![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 430 cases in 2000 to over 2
430 cases in 2000 to over 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400
400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 138 in 2010 and 3
138 in 2010 and 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 312
312![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 040 in 2015. Deaths increased from 960 in 2000 to more than 4032 in 2015.20
040 in 2015. Deaths increased from 960 in 2000 to more than 4032 in 2015.20
There are an estimated 1.4 million cases per year of hepatitis A, with 0.5% of mortalities due to viral hepatitis.21 Globally, an estimated 71 million people have chronic hepatitis C virus infection. The WHO estimated that approximately 399![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people died from hepatitis C in 2016, mostly from cirrhosis and hepatocellular carcinoma (primary liver cancer).22 Hepatitis D virus (HDV) globally affects nearly 5% of people who are chronically infected with the hepatitis B virus (HBV). The superinfection of HDV on chronic hepatitis B accelerates the progression to a more severe disease in all ages and in 70–90% of persons.23 Every year, there are an estimated 20 million HEV infections worldwide. The WHO estimates that hepatitis E caused approximately 44
000 people died from hepatitis C in 2016, mostly from cirrhosis and hepatocellular carcinoma (primary liver cancer).22 Hepatitis D virus (HDV) globally affects nearly 5% of people who are chronically infected with the hepatitis B virus (HBV). The superinfection of HDV on chronic hepatitis B accelerates the progression to a more severe disease in all ages and in 70–90% of persons.23 Every year, there are an estimated 20 million HEV infections worldwide. The WHO estimates that hepatitis E caused approximately 44![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths in 2015 (accounting for 3.3% of the mortality due to viral hepatitis).24 The summary of different methods to detect the outlined viruses of health concern is presented in Table 2.
000 deaths in 2015 (accounting for 3.3% of the mortality due to viral hepatitis).24 The summary of different methods to detect the outlined viruses of health concern is presented in Table 2.
| # | Method summary | Analyte | Sample volume | Accuracy | Time | Ref. |
|---|---|---|---|---|---|---|
| 1 | Colloidal gold immunochromatographic assay (GICA) | p24 (viral protein of HIV-1) | 75 μL r-p24 and 75 μL detector mAb | Specificity – 98.03% (1.96% – false-positive) | 20 min | 25 |
| 2 | Chemiluminescent magnetic microparticle-based immunoassay (ARCHITECT HIV Ag/Ab Combo) | HIV-1 p24 antigen | N/A | N/A | N/A | 26 |
| 3 | Fluorescent microparticle enzyme immunoassay (AxSYM HIV Ag/Ab Combo) | HIV p24 antigen | N/A | N/A | N/A | 26 |
| 4 | Enzyme-linked fluorescent assay (VIDAS HIV DUO Quick) | HIV p24 antigen | N/A | N/A | N/A | 26 |
| 5 | Enzyme-linked fluorescent assay (VIDAS HIV DUO Ultra) | HIV p24 antigen | N/A | N/A | 120 min | 26 |
| 6 | Quantitative enzyme-linked fluorescent immunoassay (VIDAS HIV p24 II) | HIV p24 antigen | N/A | N/A | N/A | 26 |
| 7 | Nanometer-scale antibody array-based analysis with AFM detection | HIV p24 antigen | 1 μL | N/A | 6 hours; 2–3 hours before measurement | 27 |
| 8 | Ultrasensitive capacitive immunosensor functionalized by anti-HIV-1 p24 mAb | HIV p24 antigen | 250 μL | N/A | 20 min | 28 |
| 9 | Boosted ELISA based on immune complex dissociation and amplified signal | HIV p24 antigen | 100 μL | N/A | >120 min | 29 |
| 10 | Nanoparticle-based biobarcode amplification assay | HIV p24 antigen | 100 μL | 100% | >120 min | 30 |
| 11 | Colorimetric lateral diffusion immunochromatography | Antibody against HIV | Finger prick blood | N/A | 3–30 min | 31 |
| 12 | Electrochemical ELISA | Antibody against HIV-1 and HIV-2 peptides | 20 μL | N/A | N/A | 32 |
| 13 | Electrochemical sensor, where antibodies bind to polypeptide epitopes | anti-HIV antibodies | 200 μL | N/A | 8 min | 33 |
| 14 | Reverse transcription loop-mediated isothermal amplification (RT-LAMP) products are visualized using a lateral flow immunoassay (LFIA): microfluidic rapid and autonomous analysis device (microRAAD) | HIV-1 RNA | 12 μL of human whole blood | N/A | 90 min | 34 |
| 15 | Luminescence assay | Ebola virus oligonucleotide (RNA) | N/A | N/A | 2 days | 35 |
| 16 | Reverse transcription loop-mediated isothermal amplification (RT-LAMP) | Ebola RNA | 1 μL of RNA | N/A | 1 hour | 36 |
| 17 | ELISA | Ebola virus nucleoprotein (NP) | 100 μL rNP | N/A | N/A | 37 |
| 18 | Fluorescence signal for quantitative detection and colorimetric signal for visual detection | Ebola virus glycoprotein | 50 μL | N/A | 20 min | 38 |
| 19 | Chemiluminescent ELISA | Antibodies against ebola virus strains Zaire and Sudan | 75 μL | Low cross-reactivity | N/A | 39 |
| 20 | Electroluminescent nanospheres and immunomagnetic separation | Ebola virions | 200 μL | N/A | 2 hours | 40 |
| 21 | Fluorescence assay on a micromagnetic platform | Ebola virions | 200 μL EBOV sample | Intra-assay CV = 4.9% | N/A | 41 |
| 22 | SERS immunoassay | Zika virions | 10 μL | N/A | N/A | 42 |
| 23 | Aptamer-based ELISA | Zika NS1 protein | 100 μL | N/A | N/A | 43 |
| 24 | Field effect biosensing | Zika viral antigen ZIKV NS1 | 75 μL | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilution: CV = 19.89% 10 dilution: CV = 19.89% |
N/A | 44 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution: CV = 9.17% 100 dilution: CV = 9.17% |
||||||
| 25 | Motion-based immunological detection | Zika virions | 10 μL of ZIKV | Correlation: 89.11% with the light microscopy; 100% with CDC Zika MAC-ELISA; 80% with Aptima Zika virus assay | >40 min for assay | 45 |
| 26 | Determination of isothermally Amplified Zika Virus RNA using a Universal DNA-Hairpin Probe | Zika Virus RNA | 50 μL | N/A | 1 hour | 46 |
| 27 | Advanced strand exchange amplification (ASEA) | Zika Virus RNA | N/A | N/A | 25 min | 47 |
| 28 | ELISA | anti-ZIKA IgM antibody | N/A | 87.5% positive agreement between CDC and InBios MAC-ELISAs | N/A | 48 |
| 29 | Single particle interferometric reflectance imaging sensor (SP-IRIS) cartridge | HFV virions (model – Ebola virions) | 100 μL | N/A | 20 min | 49 |
| 30 | Real-time reverse transcription-PCR | HFV RNA | 2 μL of RNA | No amplification of HIV-1, hepatitis B and C, herpes simplex type 1, cytomegalovirus, and Modoc viruses | >1 hour | 50 |
| 31 | qRT-PCR | HFV RNA | 5 μL of viral RNA | CV < 5%, no cross-reactivity | >1 hour | 51 |
| 32 | RT-LAMP | Crimean-Congo hemorrhagic fever (CCHF) RNA | 2 μL of the target RNA | 100% agreement between RT-LAMP and the nested PCR | 60 min | 52 |
| 33 | Fiber-optic biosensor with chemiluminescence | Crimean-Congo hemorrhagic fever (CCHF) IgG antibodies | 200 μL | N/A | 90 min | 53 |
| 34 | Loop-mediated isothermal amplification, LAMP | Dengue virus RNA | 2 μL of prepared RNA | The area under the ROC curve (AUC) = 0.95 | <1 hour | 54 |
| 35 | Biosensor with isothermal nucleic acid sequence-based amplification (NASBA) | Dengue virus RNA, serotypes 1, 2, 3, and 4 | 2 μL of amplicon (amplified dengue virus RNA) | Serotype 3 displayed low cross reactivity with biosensors designed for the detection of serotypes 1 and 4 | 15 min | 55 |
| 36 | Tandem toehold-mediated displacement reactions (tTMDR) with fluorescence | Dengue virus RNA | 100 μL | N/A | 35 min | 56 |
| 37 | Magnetic paper-based ELISA | Dengue immunoglobulin M (IgM) antibodies | 5 μL | N/A | N/A | 57 |
| 38 | Long-range surface plasmon polariton (LRSPP) gold (Au) waveguides | Dengue IgM antibody | ∼10 μL plasma | N/A | N/A | 58 |
| 39 | Stacking flow immunoassay | Dengue-specific immunoglobulin antibody | 100 μL | N/A | N/A | 59 |
| 40 | Separative extended gate field-effect transistor (SEGFET) as an immunosensor | Dengue virus nonstructural protein 1 (NS1) | N/A | N/A | <1 hour | 60 |
| 41 | Lateral flow immunoassay (LFIA) | Dengue NS1 protein | 10 mL | N/A | <1 hour | 61 |
| 42 | Magnetic separation and fluorescence detection | Dengue-2 virus virions | 20 mL | High specificity in the presence of yellow fever virus | 30–60 min | 62 |
| 43 | Electrochemical membrane-based nanobiosensor | Dengue-2 virus virions | 5 μL | RSD = 5.9% | N/A | 63 |
| 44 | RT-PCR | Hepatitis A virus (HAV) RNA | 5 μL of RNA extracts | N/A | >1 hour | 64 |
| 45 | qRT-PCR | HAV RNA | 5 μL of RNA extract | Regression coefficient of 0.9999 | >1 hour | 65 |
| 46 | Indirect competitive electrochemical immunosensor | HAV antigen | 100 μL | RSD < 3% | N/A | 66 |
| 47 | Electrochemical immunosensor | HAV antigen | 1.0 mL min−1 | RSD = 3.1–5.7% | 5 min | 67 |
| 48 | Solid-phase radioimmunoassay, HAVAB®-M | anti-HAV antibodies | 100 μL | RSD = 22% | N/A | 68 |
| 49 | Immunochromatographic assay (ICA) | anti-HAV IgM antibodies | 5 μL | Specificity = 100% | <20 min | 69 |
| 50 | Resonance light scattering (RLS) sensor | HAV virions | N/A | RSD = 1.3% | N/A | 70 |
| 51 | Multifunctional molecularly imprinted fluorescence sensor | HAV virions | 200 μL | RSD < 2.7% | 20 min | 71 |
| 52 | A reduced graphene oxide-assisted hybridization chain reaction + fluorescence | Hepatitis C virus (HCV) RNA | N/A | RSD = 3–6.4% | >8 hours | 72 |
| 53 | Capture of RNA with probes and paramagnetic particle separation | HCV RNA | 300 μL of serum | 93% sensitivity and 100% specificity | >1 hour | 73 |
| 54 | Electrochemical immunosensor | HCV antigen | 1.0 mL min−1 | RSD = 2.3–5.3% | 5 min | 67 |
| 55 | Chemiluminescent magnetic particle-based immunoassay | HCV core Antigen | A few hundred μL | 99% specificity; 97.4% sensitivity | 200 assays per hour | 74 |
| 56 | Sandwich electrochemical immunosensor | HCV core antigen | ≈10 μL | RSD = 3.1% | 30 min | 75 |
| 57 | Nano-gold immunological amplification and silver staining (NIASS) | anti-HCV antibodies | 10 μL | N/A | <40 min | 76 |
| 58 | Antibody-induced DNA strand displacement and rolling circle amplification (RCA) | anti-HCV antibodies | 1 μL | “High specificity” | 30 min | 77 |
| 59 | Immunogold electron microscopy | HCV virions | 3 μL | N/A | >3 hours | 78 |
| 60 | Dual-targeting real-time RT-PCR | Hepatitis D Virus (HDV) RNA | 140 μL viral sample | N/A | >1 hour | 79 |
| 61 | Real-time PCR | HDV RNA | 200 μL specimen | N/A | >20 min | 80 |
| 62 | Electrochemical immunosensor | HDV antigen | 1.0 mL min−1 | RSD = 3.4–6.8% | 5 min | 67 |
| 63 | IgM capture enzyme immunoassay (EIA) | HDV antibodies IgM anti-HD | 100 μL | No cross-reactivity with other antibodies | 2 days | 81 |
| 64 | Real-time RT-PCR | Hepatitis E virus (HEV) subtype 3b RNA | 5 μL of extracted RNA | N/A | >1 hour | 82 |
| 65 | Array-based nano-amplification and silver stain enhancement | HEV RNA | 100 μL | N/A | 20–30 min | 83 |
| 66 | Electrochemical immunosensor | HEV antigen | 1.0 mL min−1 | RSD = 3.4–6.9% | 5 min | 67 |
| 67 | Enzyme immunoassay (EIA) | Antibody to the hepatitis E virus (anti-HEV) | 250 μL | N/A | N/A | 84 |
4. Detection of coronaviruses
Coronaviruses are single-stranded, plus-strand, enveloped RNA-containing viruses, with a relatively large genome averaging at 30 kb pairs. Human coronaviruses (HCoV) 229E, NL63, OC43, and HKU1 are known endemic human coronaviruses that cause mild respiratory infection with such symptoms as rhinorrhea and mild cough. Two other coronaviruses – Severe-Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS) coronaviruses – are more virulent, and lead to a severe respiratory disease with symptoms similar to influenza. The SARS coronavirus caused a global epidemic in 2002–2003,85–87 killing 774 people out of the 8096 infected, and thus having a 9.56% mortality rate. MERS-CoV caused an epidemic in the Middle East that started in 2012.88 By January 2020, MERS-CoV had killed 866 people out of the 2519 that were sick, and so it has a 34.4% death rate.89Diagnostic tests that use Polymerase Chain Reaction (PCR) allow for the detection of viral nucleic acid. With DNA-containing viruses, the PCR technique is more straightforward, as it depends on the amplification step of isolated viral DNA. The detection of RNA-containing viruses requires an additional step of converting RNA into DNA by reverse transcription. PCR utilizes synthetic single-stranded DNA primers and probes, and depends on the hybridization. Because primers and/or probes might bind nonspecifically to a region different from the intended one, PCR tests can result in false-positive outcomes. On the other hand, false-negative results might result from improperly collected material. The focus of the present review is to compare the key parameters of different test assays, such as specificity and selectivity, to get a quantitative outlook on the test systems.
There are different modalities of PCR, which are useful in viral detection. Among them are quantitative real-time PCR (qPCR), which allows for rapid detection with the identification of relative amounts of the nucleic acid. The quantitative real time RT-PCR (RT-qPCR) method is an efficient method of diagnostics, which allows for the rapid detection of viral nucleic acid. Many RT-qPCR methods are developed for the detection of coronaviruses. The range of methods for the detection of SARS and MERS coronaviruses is summarized in Table 3.
| # | Method summary | Analyte | Sample volume | Accuracy | Time | Ref. |
|---|---|---|---|---|---|---|
| 1 | RT-PCR and indirect immunofluorescence serologic testing | SARS-CoV RNA | 2 mL nasopharyngeal aspirates and 2 g feces | RT-PCR: 60% for positive cases; 99.4% for negative cases | RT-PCR: N/A | 90 |
| Serologic testing: 92% for positive cases; 92% for negative cases | Serologic testing: >1 hour | |||||
| 2 | Real time RT-PCR (2 assays: for upE and (ORF)1b) | SARS-CoV RNA: upstream of the E gene (upE) or within open reading frame (ORF)1b | 5 μL of extracted RNA | 100% specificity for both for upE and (ORF)1b | 36+ hours | 91 |
| 3 | Chemiluminescence immunosorbent assay with nanoarray RNA aptamer | SARS-CoV nucleocapsid protein (SARS-CoV N protein) | N/A | C-terminal domain or dimer form N protein is specifically recognized by the aptamer | N/A | 92 |
| 4 | Biosensor assay based on an optical QDs-based RNA aptamer | SARS-CoV nucleocapsid protein (SARS-CoV N protein) | N/A | QDs-conjugated RNA aptamer is selective against the SARS-CoV N protein | 1 hour | 93 |
| 5 | Asymmetric five-primer reverse transcription loop-mediated isothermal amplification (RT-LAMP) assay | MERS-CoV RNA (3 genetic loci: ORF1a, ORF1b and E) | 4 μL RNA | N/A | 30–50 minutes | 94 |
| 6 | ELISA | MERS-CoV nucleocapsid protein (NP) | 50 μL of sample | 100% specificity | N/A | 95 |
| 7 | ELISA and plaque-reduction neutralization test (PRNT) | Antibodies against MERS-CoV | N/A | No serotypic discrimination between the MERS-CoV strains | N/A | 96 |
5. Detection of SARS-CoV-2 (COVID-19 pandemic virus)
On December 31, 2019, the WHO Chinese Country Office was notified about pneumonia cases of an unknown nature, epidemiologically linked to the seafood market in Wuhan, Hubei province.97 At the same time, the Chinese CDC conducted an investigation in the field.98 The viral nature of the disease was identified, 3 viral genomes from the bronchoalveolar lavage of three disease-affected individuals were sequenced, and the sequences were submitted to GISAID (accession ID: EPI_ISL_402119; EPI_ISL_402120; EPI_ISL_402121). According to the sequencing data, the novel virus belongs to the Coronaviridae family, Orthocoronaviridae subfamily, Betacoronavirus genus, Sarbecovirus subgenus. The viral genome contains a 5′ untranslated region (5′-UTR), replicase gene (orf1ab), Spike gene (S gene), Envelope gene (E gene), M gene, Nucleocapsid gene (N gene), and open reading frames 3, 7, 8, 10b, 13, and 14. The virus was named novel coronavirus 2019-nCoV, and the disease it causes was named novel coronavirus-infected pneumonia (NCIP) by the Chinese CDC.99 Later, the WHO renamed the virus into SARS-CoV-2, and the disease into COVID-19.100 The virus is highly contagious with easy person-to-person transmission, has a variable incubation period (from 4 to 24 days),101 and leads to the development of respiratory diseases with variable symptoms, from a mild cough to pneumonia.102 Sometimes, the infected person does not show any symptoms whatsoever.103 Soon after the disease outbreak, close monitoring of the epidemiological situation around the globe started with a real-time count of new cases in the world.104 Strict quarantine measures were implemented in the countries most affected by the disease, and on March 11th, the WHO declared a state of pandemic. Accurately monitoring the virus and its spread is not possible without reliable diagnostic tools. So, soon after the outbreak, the development of tests detecting SARS-CoV-2 itself or the immune response in the affected person was initiated.A sample for the test includes material from the upper and lower respiratory tracts, and can include aspirates, oropharyngeal and nasopharyngeal swabs, bronchoalveolar lavage, and sputum, as well as nasal and nasopharyngeal aspirate. Test results depend on the quality of the collected material, as well as on the type of material.
Multiple assays were developed to test the presence of viral nucleic acid. The tests for SARS-CoV-2 detection employ a probe to detect the sequence within the RNA-dependent RNA polymerase viral gene (RdRp), as well as the nucleocapsid gene (NP), envelope protein gene (E), and spike protein gene (S). Some tests rely on the usage of fluorescently labeled TaqMan probes with a fluorescent reporter and a quencher attached to the 5′- and 3′-ends of the probe, respectively. Examples of such probes include probes with 6-carboxyfluorescein (FAM) or HEX dye at their 5′-end, and with a blackberry quencher (BBQ) or blackhole quencher (BHQ) at the probe's 3′-end.105 The 5′-exonuclease ability of DNA-polymerase removes the fluorescently-labeled 5′-end of the hybridized probe, which leads to probe degradation and unquenching of the fluorescent reporter. There are 14 probes provided by the WHO, and one described by Zhu et al.98 Most of them use TaqMan probes, with one exception from Japan that relies on nested PCR instead. Information about the limit of detection and the accuracy of the real-time reverse-transcription tests is gathered in Table 4.
| # | Method summary | Analyte | Sample volume | Accuracy | Time | Ref. |
|---|---|---|---|---|---|---|
| 1 | Real time RT-PCR Commercial kit from Altona diagnostics, Hamburg, Germany | E-gene RNA and S-gene RNA | 30 μL | N/A | >1 hour | 111 and 112 |
| 2 | ePlex-based (DNA hybridization and electrochemical detection) SARS CoV-2 | cDNA coding for nucleocapsid (N) | 200 μL of nasopharyngeal swab | Detection of positive – 94.4%; 95% CI – 74.2–99% | >1 hour | 111 and 113 |
| Detection of negative – 100%; 95% CI – 92.4–100% | ||||||
| 3 | Real-time RT-PCR | RNA coding for RdRp gene | N/A | N/A | >1 hour | 114 |
| 4 | Real-time RT-PCR | SARS-CoV-2 RNA coding for nucleocapsid gene (N1 probe) | 20 μL | N/A | >1 hour | 111 and 114 |
| 5 | Real-time RT-PCR | SARS-CoV-2 RNA coding for nucleocapsid gene (N2 probe) | 20 μL | N/A | >1 hour | 114 |
| 6 | Real-time RT-PCR (E-gene assay, and RdRp gene assay) | SARS-CoV-2 RNA coding for envelope (E) gene and RNA-dependent RNA polymerase (RdRp) gene | 25 μL | No reactivity on human coronaviruses | >1 hour | 105 |
| 7 | Real-time RT-PCR | Co-V2 viral RNA; probe against nucleocapsid gene (N and N2 assays) | N/A | N/A | >1 hour | 115 |
| 8 | Real-time RT-PCR | SARS-CoV-2 RNA | 5 μL RNA | No cross-reactivity with other human-pathogenic coronaviruses and respiratory pathogens | >1 hour | 116 |
| 9 | RT-PCR: Cepheid Xpert Xpress and Roche cobas assays | SARS-CoV-2 RNA | N/A | Both systems have agreement of 99% | 45 min (Cepheid); 90 min (Roche) | 117 |
| 10 | RT-LAMP (reverse transcription loop-mediated isothermal amplification assay) | Viral RNA coding orf1ab gene and S gene | 25 μL | Sensitivity – 100% (95% CI 92.3–100%) | 26.28 ± 4.48 min | 107 |
| Specificity – 100% (95% CI 93.7–100%) | ||||||
| 11 | Colorimetric LAMP | SARS-CoV-2 RNA | 3 μL RNA | 100% agreement with RT-PCR | 30 min | 118 |
| 12 | RT-LAMP | Viral RNA coding for conserved region within nucleocapsid gene | N/A | Sensitivity – 100% | 30 min (colorimetric visualization) | 108 |
| Specificity – 98.7% | ||||||
| 13 | RT-LAMP | Viral RNA within RdRp gene | 25 μL | 100% consistency with RT-qPCR on positive samples | 50 min real-time monitoring | 109 |
| 14 | DETECTR (SARS-CoV-2 DNA endonuclease-targeted CRISPR trans reporter) – CRISPR-Cas12-based assay | Viral RNA coding for nucleoprotein and envelope genes | N/A | 95% for positive samples, 100% for negative samples | 30–40 min (sample-to-result) | 110 |
| 15 | ELISA | SARS-CoV-2 neutralizing, spike- and nucleocapsid-specific antibodies | N/A | 87–100% specificity | 2 days | 119 |
| 16 | COVID-19 IgG/IgM rapid test Cassette | SARS-CoV-2-specific IgM and IgG | 5 μL serum | Specificity 100% for IgM and 99.2% for IgG | 15 min | 120 |
| Sensitivity 69% for IgM and 93.1% for IgG | ||||||
| 17 | Colloidal gold Antibodies test | SARS-CoV-2-specific IgM and IgG | 10 μL of serum | Specificity: IgM, 50.0%; IgG, 87.5% | N/A | 121 |
| 18 | Single molecule array (Simoa) immunoassay | SARS-CoV-2 nucleocapsid protein (N-protein) | 20 μL of whole blood | 100% specificity and 97.4% sensitivity | N/A | 122 |
| 19 | Pulse-controlled amplification (PCA) | SARS-CoV-2 RNA (E gene) | 45 μL | 100% agreement with RT-qPCR | 20 min | 123 |
| 20 | LAMP | SARS-CoV-2 RNA | N/A | N/A | <30 min | 124 |
| 21 | Reverse-transcription recombinase-aided amplification (RT-RAA) | SARS-CoV-2 RNA | 140 μL nasopharyngeal swabs and sputum | 100% agreement with RT-PCR | 5–15 min | 125 |
Alongside real-time RT-PCR, isothermal hybridization is a method of choice for viral RNA detection in a shorter period of time, as compared to real-time RT-PCR. Originally invented by Notomi,106 the so-called LAMP (loop-mediated isothermal amplification) coupled to reverse transcription is also used for SARS-CoV-2 detection,107–109 and allows for the rapid detection of viral RNA within a time frame of less than 1 hour. The method relies on the strand-displacing ability of the Bst polymerase, and needs 4 to 6 primers, which increases the target selectivity of this method, as compared to regular PCR with only 2 primers.106 The method can be run on a regular thermostat at 65 °C. The sensitivity of the RT-LAMP method is slightly lower (∼10-fold) than that of real-time RT-PCR108,109 for SARS-CoV-2 detection.
The most novel method of viral detection, and possibly the most intriguing one, uses CRISPR-Cas 12 endonuclease and isothermal amplification.110 This method, named DETECTR, is very rapid, allowing for the detection of the viral RNA within minutes. It is a very promising point-of-care test that does not require expensive equipment and can be used in developing countries.
6. Summary of detection methods
Nucleic acid detection methods including PCR and LAMP
Comparing the different methods of viral detection, PCR-based methods can detect a few copies of RNA per reaction, which places them on the high-sensitivity spectrum, corresponding to a fg mL−1 concentration range. PCR detection sensitivity is comparable between different viruses, with a range of detection from a few copies per reaction to a few hundreds of copies per reaction. PCR-based methods require trained personnel and expensive equipment, such as the PCR thermocycler. They are also time-consuming and can take several hours. In order to facilitate PCR-based techniques without compromising selectivity and specificity, alternatives to RT-qPCR exist. These include the LAMP technique. LAMP has a limit of detection comparable to that of RT-PCR, spanning from 3 copies of RNA per reaction to 100 copies of RNA per reaction, as evidenced from 5 sources presented in Table 5.| PCR-based nucleic acid detection | ||||
|---|---|---|---|---|
| Analyte | Ref. | Analytical parameters | Ref. | Analytical parameters |
| Viral hemorrhagic fever (HF) viruses: Ebola, Marburg, Lassa, Crimean-Congo HF, Rift Valley fever, dengue, yellow fever viruses | 50, 2002 | LOD = 1545 to 2835 viral genome equivalents per mL of serum (8.6 to 16 RNA copies per assay) | 51, 2014 | LOD = between 45 and 150 cRNA/rxn |
| Hepatitis A | 64, 2009 | LOD = 1 PFU/1.5 L | 65, 2010 | LOD = 10 PFU/1.5 L of bottled water, 100 PFU/1.5 L of tap water |
| Hepatitis C | 73, 2000 | LOD = 33 cRNA per mL; (∼1.74 × 10−4 pg mL−1) | ||
| Hepatitis D | 79, 2018 | LOD = 575 IU mL−1 | 80, 2013 | LOD = 7500 HDV cRNA per mL; 190 cRNA/rxn (∼0.28 pg mL−1) |
| Hepatitis E | 82, 2013 | LOD = 25 UI mL−1 | ||
| Seasonal influenza A | 6, 2009 | LOD = log10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6.5–7.1 of M gene copies 6.5–7.1 of M gene copies |
||
| Swine-origin influenza A | 6, 2009 | LOD = log10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6.5–7.3 of M gene copies 6.5–7.3 of M gene copies |
||
| SARS-CoV | 90, 2004 | LOD = 10 cRNA/rxn; Ac = 60% for positive cases, 99.4% for negative cases | 91, 2012 | LOD = 3.4 cRNA/rxn for upstream of the E gene (upE) (∼1.11 × 10−2 pg mL−1) and 64 cRNA/rxn for within open reading frame (ORF)1b, Sp = 100% |
| SARS-CoV-2 | 116, 2020 | LOD = 11.2 cRNA/rxn (∼3.67 × 10−2 pg mL−1) | 111, 2020 | LOD = 24 cRNA/rxn (∼1.97 × 10−2 pg mL−1) |
| 105, 2020 | LOD = 3.8 cRNA/rxn (∼2.49 × 10−3 pg mL−1) – RdRp-gene assay; 5.2 cRNA/rxn (∼3.41 × 10−3 pg mL−1) – E-gene assay | 115, 2020 | LOD = 25 and 250 cRNA/rxn | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Lateral flow immunoassays | ||||
| HIV-1 p24 antigen | 25, 2016 | LOD = 25 pg mL−1 | ||
| Ab against HIV | 31, 2006 | LOD = sub pmol L−1 range | ||
| HIV-1 RNA | 34, 2019 | LOD = 3 × 105 HIV-1 viral particles, or 2.3 × 107 virus copies per mL of whole blood | ||
| Dengue NS1 protein | 61, 2020 | LOD = 5 ng mL−1 | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| ELISA | ||||
| HIV p24 antigen | 29, 2003 | LOD = 0.5 pg mL−1 | ||
| Ab to HIV-1, HIV-2 | 32, 2013 | LOD = 1 ng mL−1 (6.7 pM) | ||
| Ebola virus nucleoprotein (NP) | 37, 2001 | LOD = 30 ng of purified recombinant NP (rNP) (∼3.00 × 105 pg mL−1) | ||
| Zika NS1 protein | 43, 2017 | LOD = 0.1 ng mL−1 | ||
| IgM-dengue antibodies | 57, 2017 | LOD = 0.04 μg mL−1 | ||
| MERS-CoV nucleocapsid protein (NP) | 95, 2015 | LOD = 10 TCID50/0.1 mL | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| AFM | ||||
| HIV p24 antigen | 27, 2004 | LOD = 25 fg mL−1 | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Electrochemical detection | ||||
| HIV p24 antigen | 28, 2010 | LOD = 7.9 × 10−8 pg mL−1 | ||
| Anti-HIV antibodies | 33, 2012 | LOD = 1–10 nM (∼1.50 × 105 pg mL−1) | ||
| Ebola virions | 40, 2017 | LOD = 5.2 pg mL−1 | ||
| Zika viral antigen ZIKV NS1 | 44, 2018 | LOD = 450 pM | ||
| Zika Virus RNA | 46, 2019 | LOD = 1.11 fg μL−1 (∼0.3 fM) | ||
| Dengue virus nonstructural protein 1 (NS1) | 60, 2014 | LOD = 0.25 μg mL−1 | ||
| Dengue-2 virions | 63, 2012 | LOD = 1 PFU mL−1 | ||
| H5N1 avian influenza virus hemagglutinin (HA) protein | 13, 2020 | LOD = 5.9 pM | ||
| Hepatitis A antigen | 66, 2017 | LOD = 26 × 10−5 IU/mL | 67, 2010 | LOD = 0.5 ng mL−1 |
| Hepatitis C antigen | 67, 2010 | LOD = 0.8 ng mL−1 | 75, 2017 | LOD = 3 fg mL−1 |
| Hepatitis D antigen | 67, 2010 | LOD = 0.5 ng mL−1 | ||
| Hepatitis E antigen | 67, 2010 | LOD = 1 ng mL−1 | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Chemiluminescence detection | ||||
| HIV p24 antigen | 26, 2011 | LOD = 18–25 pg mL−1 or 1.24 IU mL−1 | ||
| Hepatitis C Virus (HCV) core antigen | 74, 2006 | LOD = viral concentration equivalent to the lowest titer of 2415 cRNA/mL | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Fluorescence detection | ||||
| HIV p24 antigen | 26, 2011 | LOD = 22–77.4 pg mL−1 or 1.94–2.25 IU mL−1 | 26, 2011 | LOD = 13 pg mL−1 or 0.43 IU mL−1 |
| 26, 2011 | LOD = 11.5–25 pg mL−1 or 0.66 IU mL−1 | 26, 2011 | LOD = 11.2 pg mL−1 or 0.73–1.15 IU mL−1 | |
| Ebola RNA | 35, 2016 | LOD = femtomolar level | ||
| Ebola glycoprotein | 38, 2017 | LOD = 0.18 ng mL−1 | ||
| Ebola virions | 41, 2018 | LOD = 2.6 pg mL−1 | ||
| Dengue virus RNA | 56, 2018 | LOD = 6 cRNA per sample (∼3.62 × 10−4 pg mL−1) | ||
| Dengue-2 virions | 62, 2008 | LOD = 10 PFU mL−1 | ||
| HAV virions | 71, 2019 | LOD = 3.4 pmol L−1 | ||
| HCV RNA | 72, 2019 | LOD = 10 fM | ||
| anti-HCV Ab | 77, 2019 | LOD = 0.998 pM | ||
| SARS-CoV nucleocapsid protein (SARS-CoV N protein) | 92, 2009 | LOD = 2 pg mL−1 | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| LAMP-based nucleic acid detection | ||||
| Ebola | 36, 2017 | LOD = 100 cRNA (∼1.04 pg mL−1) | ||
| Crimean-Congo hemorrhagic fever | 52, 2013 | LOD = 0.1 fg of viral RNA (equivalent to 50 viral particles; ∼0.05 pg mL−1) | ||
| Dengue | 54, 2020 | LOD = 102 PFU per 200 μL of whole blood | ||
| Influenza A | 11, 2020 | LOD of subtypes | ||
| H1: 50 copies | ||||
| H3: 20–50 copies | ||||
| H5: 50 copies | ||||
| H7: 20–50 copies | ||||
| H9: 50–100 copies | ||||
| Influenza B | 11, 2020 | LOD = 50 copies | ||
| MERS-CoV | 94, 2015 | LOD = 0.02 to 0.2 PFU (5 to 50 PFU mL−1) | ||
| SARS-CoV-2 | 118, 2020 | LOD = 120 cRNA/rxn (or 4.8 copies per μL); ∼7.87 × 10−2 pg mL−1 | 107, 2020 | LOD = 20 cRNA/rxn – ORF1ab gene (∼1.31 × 10−2 pg mL−1) and 200 cRNA/rxn – S gene (∼0.131 pg mL−1) |
| 109, 2020 | LOD = 3 cRNA/rxn (∼1.97 × 10−3 pg mL−1) | |||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| CRISPR-Cas based | ||||
| SARS-CoV-2 | 110 and 128, 2020 | LOD = 10 cRNA/rxn (∼0.164 pg mL−1) | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Biobarcode detection | ||||
| HIV p24 antigen | 30, 2007 | LOD = 0.1 pg mL; Ac = 100% | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Motion-based detection | ||||
| Zika virions | 45, 2018 | LOD = 1 particle/μL | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| NASBA-based nucleic acid detection | ||||
| Zika | 46, 2019 | LOD = 1.11 fg μL−1 (∼0.3 fM) | ||
| Dengue | 55, 2002 | LOD = 10 PFU mL−1 | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Advanced strand exchange amplification-based nucleic acid detection | ||||
| Zika | 47, 2018 | LOD = 1.0 × 10−15 M; (∼33 pg mL−1) | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Interferometric reflectance imaging | ||||
| HFV virions | 49, 2017 | LOD = 10 viruses per spot | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Surface plasmon polariton waveguides | ||||
| Dengue-specific immunoglobulin M (IgM) antibody | 58, 2014 | LOD = ∼22 pg mm−2 | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| SERS (Surface Enhanced Raman Spectroscopy) | ||||
| Zika virions | 42, 2018 | LOD = 10 ng mL−1 | ||
| FCV virions | 129, 2005 | LOD = 106 viruses per mL | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Radioimmunoassay | ||||
| Anti-HAV Ab | 68, 1993 | LOD = 10 mIU mL−1 | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Resonance light scattering | ||||
| HAV virions | 70, 2017 | LOD = 8.6 pmol L−1 | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Silver staining | ||||
| HCV antibodies | 76, 2005 | LOD = 3 ng mL−1 | ||
| HEV RNA | 83, 2006 | LOD = 100 fM; (∼237.6 pg mL−1) | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Electron microscopy | ||||
| HCV virions | 78, 2006 | LOD = 107 virions per mL | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| SPR | ||||
| AIV H5N1 virions | 8, 2012 | LOD = 0.128 HAU | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Confocal laser scanning microscopy | ||||
| SARS-CoV nucleocapsid protein | 93, 2011 | LOD = 0.1 pg mL−1 | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Magnetic particle spectroscopy | ||||
| H1N1 nucleoprotein molecule | 14, 2020 | LOD = 4.4 pmoles | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Nanocomposite-based optical and mechanical detection | ||||
| Influenza A virions | 12, 2020 | LOD = 5 × 107 PFU mL−1 | ||
LAMP-based diagnostic tests allow for the rapid detection of the analyte, without a need for expensive equipment. Results can be obtained rapidly (within an hour), which makes these tests a suitable platform to be used for the development of point-of-care diagnostics tests. Alongside LAMP, the NASBA-based nucleic acid detection,46,55 as well as the advanced strand exchange amplification-based nucleic acid detection, are used and demonstrate a low limit of detection. A standard PCR test can also be incorporated into the assay, with other methods of separation such as laser-irradiated DNA extraction, paramagnetic particle separation, and others. Today, stationary PCR-based methods requiring expensive and complicated equipment are accompanied by bench scale PCR detection with portable devices and automated procedure, where only one step of sample loading is necessary, such as with the “Cepheid Xpert Xpress” and “Roche cobas” assays.117
Immunoassay-based methods
The average limit of detection for ELISA-based methods is within the pg mL−1 range, but has been demonstrated to be as low as a fraction of pg mL−1.29 ELISA-based methods are routinely used and have demonstrated their applicability for an extended period. Despite this, they require trained personnel and are time-consuming. Therefore, alternative methods are being developed to make diagnostics tests relatively cheap and affordable without compromising their sensitivity and selectivity. One of these methods is lateral flow immunoassay. This method has shown a limit of detection at the pg mL−1 level, which is comparable to fine ELISA-based methods, but lateral flow immunoassays are faster. For example, a standard ELISA can take anywhere from 2 hours29 to 2 days,119 as studies show. However, lateral flow immunoassays take from 20 minutes47 to 90 minutes,34 on average. This makes lateral flow immunoassays a good alternative to ELISA, especially because they can be performed at a small scale. The selectivity of immunoassay-based methods allows for the distinguishing of viruses with 100% specificity,95 but not for making distinctions between different strains of the same virus.96 Like PCR-based methods, immunoassay-based methods are now commercially available in bench scale, portable, fully automated devices, suitable for point-of-care testing.120 However, these are currently qualitative, and their sensitivity is unknown. Immunoassay-based methods can detect a viral antigen or antibodies to the virus in the sample. The latter is more widespread for diagnostic testing, but can only be conclusive after the onset of symptoms (on average, a week after infection).Electrochemical detection methods
Electrochemical detection methods provide sensitive detection of a wide range of analytes: virions,40 viral antigen,44 antibodies to the virus,33 and viral nucleic acids.46 The main advantage of these detection methods is that they are relatively inexpensive and not limited by a diffraction limit (like optical methods). The lowest limit of detection was reported by Teeparuksapun et al.28 to be at a subattogram per milliliter concentration. Other studies show a pg mL−1 to fg mL−1 limit of detection range, which is comparable to PCR and immunoassays. The specificity of these methods is also high.33 Notably, these detection methods also provide reproducible results with RSD < 5%, such as the voltammetry method of Tang et al.,67 which allows the multiplex detection of five analytes (viral antigens). Multiplex analyte detection has been reported in many research papers, which gives electrochemical methods an advantage over PCR and immunoassays. Electrochemical methods can be tuned to perform wash-free analysis33 as well. Electrochemical detection methods usually use one of three approaches: voltammetry, amperometry, and detection of the change in capacitance. All three can be used in fast analysis,28,33,67 as the reported time of experiments ranged between 5 and 20 minutes. This is much faster than the majority of PCR tests and immunoassays. However, these electrochemical methods require complicated equipment (for example, the use of gold nanoparticles with TEM and AFM imaging), highly trained operators, solution media, and are not suitable for point-of-care testing. Currently, the potential use of these methods in diagnostic testing is improbable because they require the fabrication of electrochemical sensors, often on a nanoscale level.Fluorescence
Fluorescence spectroscopy is a well-established technique of sensitive detection. Its limit of detection is in the pg mL−1 range, on average, as can be seen in Table 5. Fluorescence signal detection can be incorporated in other procedures, such as immunoassays for the detection of antibodies or antigens.26 Immunoassays with fluorescence signals have already been commercialized, and are even available in portable devices for home and point-of-care testing.26 Fluorescence is reported as a reproducible method with RSD < 5%.41 Other fluorescence-based methods use nanospheres, micromagnetic platforms, or upconversion materials for creating luminescence, which is further absorbed by nanoparticles conjugated to the target analyte. This method allows for more sensitive detection at the femtomolar level.35 However, it is complicated and requires the fabrication of assay components, as well as highly trained personnel. Fluorescence signal acquisition is fast (around 20 minutes, on average), but the whole assay would take much longer to complete if the assay components need to be fabricated for fluorescence-based detection. Fluorescence-based immunoassays are faster (the whole assay takes around 2 hours), but their limit of detection is higher.26 Fluorescence is subject to challenges such as photobleaching, autofluorescence, and dissociation of organic dyes used in live cells.Other methods
There are other promising methods of detection: for example, silver staining, surface plasmon resonance (SPR), surface-enhanced Raman spectroscopy (SERS), confocal laser scanning microscopy, resonance light scattering, and radioimmunoassay. As can be seen in Table 5, silver staining and confocal laser scanning microscopy have the lowest limits of detection, in the pg to ng mL−1 and pg mL−1 ranges, respectively. They are also suitable for the detection of different analytes, such as viral nucleic acids, antigens, and antibodies to the virus.Radioimmunoassay is another highly sensitive and inexpensive procedure, as can be seen in Table 5. The selectivity of this method is also very high, and the experimental procedure is similar to that of a standard ELISA. However, special precautions must be taken during the experiment because radiolabeled reagents are used. Resonance light scattering is another valuable technique, whose convenience and sensitivity make it a potential diagnostic tool in health care.
SPR is mainly used to track the binding dynamics of biologically important molecules. It can also be used for the quantitative detection of analytes, as was demonstrated in ref. 8. This method has an advantage over other surface spectroscopy techniques because it does not need a vacuum, and is able to produce a linear dependence of resonant energy on the analyte concentration. SPR spectroscopy has potential for multiplexing, particularly when it is integrated with multichannel microfluidic devices (Fig. 4).126
SERS is a technique that allows for producing an enhancement in the order of millions and billions over standard Raman spectroscopy, which makes it useful for sensitive detection (in ng mL−1 range) of various analytes based on the characteristic Stokes shifts. This method requires a skilled operator and the use of a Raman spectrometer, which is now available as a portable, bench-size or even handheld device.127 In comparison with fluorescence, Raman has great potential for multiplex detection of various analytes. Overall, this makes SERS a potential diagnostic test method. All abovementioned methods are listed in Table 5.
7. Post-COVID-19 trends and future perspectives
Comparing different methods, nucleic acid detection following amplification (real-time RT-PCR and LAMP) has higher sensitivity than ELISA, with a fg mL−1vs. pg mL−1 average detection limit, respectively. The average limit of detection for real-time RT-PCR is 5 fg mL−1 (n = 7), and 49 fg mL−1 (n = 6) for the LAMP method. Having a detection limit higher than that for the ELISA method, the PCR-based tests are not free of drawbacks. Their disadvantages include the need for expensive equipment, trained personnel, and time. It takes several hours from the time of sample collection for a real-time RT-PCR test to produce results. The LAMP technique overcomes this limitation and enables a shorter (less than 1 hour) time until results, without the need for expensive equipment.The limit of detection of electrochemical methods is comparable to the one for amplification-based methods of nucleic acid detection. The highest limit of detection is demonstrated by electrochemical methods with a capacitive immunosensor28 for the detection of the HIV p24 antigen, which has a limit of detection of 7.9 × 10−5 fg mL−1, and is accomplished in 20 minutes. The electrochemical methods are diverse in their principle, with some relying on capacitance measurements,28 while others employ voltammetry46 or amperometry.60 They are also less restrictive in terms of the detected analyte, and are designed for the detection of viral particles, whether RNA46 or protein28, as well as antibodies (Fig. 5). The electrochemical methods are rapid and diverse. Some of them could be developed into low-cost point-of-care tests.
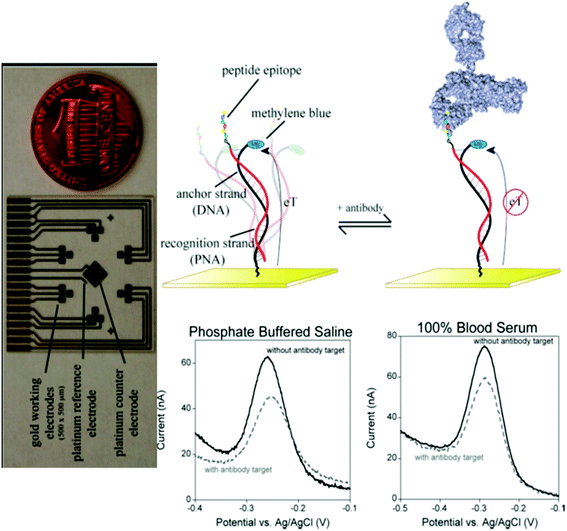 | ||
Fig. 5 E-DNA antibody sensor. The sensor (top) comprises an electrode-bound, redox-reporter-modified DNA strand, termed the “anchor strand,” that forms a duplex with a complementary “recognition strand” (here composed of PNA) to which the relevant recognition element is covalently attached. In the absence of antibody binding (top middle), the flexibility of the surface attachment chemistry supports relatively efficient electron transfer between the redox reporter and the electrode surface. Binding to the relevant target antibody (top right) decreases electron transfer, presumably by reducing the efficiency with which the reporter collides with the electrode. Binding can thus be measured as a decrease in the peak current as observed via square wave voltammetry (bottom). As shown, sensors in this class are highly selective and perform equally well in buffered saline (bottom middle), undiluted blood serum (bottom right), or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 diluted whole blood. The electrochemical E-DNA antibody sensor readily supports multiplexed detection. Here, (left, perpendicular orientation) a microfabricated chip containing eighteen 500 × 500 μm sensors, arranged in six three-pixel clusters, was employed. Each cluster is directed against a different antibody. Thus, the device supports the simultaneous, triplicate measurement of six different targets. Copyright 2012 American Chemical Society. Reprinted with permission from ref. 33, R. J. White, H. M. Kallewaard, W. Hsieh, A. S. Patterson, J. B. Kasehagen, K. J. Cash, et al. Wash-free, electrochemical platform for the quantitative, multiplexed detection of specific antibodies. Anal Chem., 2012, 84(2), 1098–1103. 4 diluted whole blood. The electrochemical E-DNA antibody sensor readily supports multiplexed detection. Here, (left, perpendicular orientation) a microfabricated chip containing eighteen 500 × 500 μm sensors, arranged in six three-pixel clusters, was employed. Each cluster is directed against a different antibody. Thus, the device supports the simultaneous, triplicate measurement of six different targets. Copyright 2012 American Chemical Society. Reprinted with permission from ref. 33, R. J. White, H. M. Kallewaard, W. Hsieh, A. S. Patterson, J. B. Kasehagen, K. J. Cash, et al. Wash-free, electrochemical platform for the quantitative, multiplexed detection of specific antibodies. Anal Chem., 2012, 84(2), 1098–1103. | ||
Amplification-based methods for the detection of nucleic acids, as well as ELISA-based methods for the detection of immune response in the form of antibodies, are two methods that are routinely used and demonstrate high sensitivity and specificity. However, they require time and proper equipment. The future of viral diagnostics lies in the point-of-care methods that can produce results within minutes, and do not require special equipment. The recent development of rapid diagnostics of SARS-CoV-2 in saliva130 demonstrates the possibility of such a method. Other possible methods that can be used in point-of-care testing include portable antibodies/antigen test kits, fluorescence immunoassay devices, as well as portable RT-PCR and RT-LAMP devices.
Comparing the time it takes for different tests to produce results, lateral flow immunoassays offer a faster time to results, as compared to other methods (usually several minutes). Electrochemical detection is another rapid diagnostics method, which takes less than 1 hour to produce results. Real-time RT-PCR takes the most time out of all test methods.
Enormous interest in virus detection on the wake of the COVID-19 pandemic is likely to reshape this area, directing efforts towards creating detection methods that have a fast time to results, high simplicity, high throughput, and are relatively low-cost. However, the specificity and selectivity of those tests should remain at least on par with well-established methods, such as PCR and ELISA.
Express testing for COVID-19 is necessary to contain the infection, and is particularly important for testing in airports, places of public gathering, transportation hubs and malls. During the pandemic, fast point-of-care tests on the scale of millions per day are required to minimize the mass quarantining of people, and to avert the harsh effects of lockdowns and travel bans on the economy and the wellbeing of people. Tests that take several days, such as the PCR test of the nasopharyngeal swabs, can result in the spread of infections since tested people will not know about their infection status until 5–7 days have passed. Today, there is a focus on developing express COVID-19 detection methods, especially using portable devices. There are several potential methods that provide test results within minutes or hours. One of them is the express RT-PCR test kit. These kits were developed by Cepheid (“Cepheid Xpert Xpress”), Roche (“Roche cobas”)117 and Abbott (“ID NOW”).131 These are already used in hospitals and ERs. According to published research, their accuracy matches that of the regular RT-PCR.117 This fact is a benefit of portable PCR devices, because with a sensitivity and specificity equal to that of stationary PCR, and with a significantly shorter time to results, they can be used for point-of-care testing. Some of these tests, such as “ID NOW”, are approved by the FDA. Another express COVID-19 testing method is RT-LAMP. RT-LAMP uses DNA polymerase with the capability of separating double-stranded DNA, which eliminates the need for cycling temperature and simplifies the procedure. Portable and rapid testing devices using this method have also been developed, such as “Talis One” by Talis Biomedical.131 This device provides results in 30 minutes, a time comparable to that of portable RT-PCR devices. Published research on the detection of RNA from SARS-COV-2 by RT-LAMP shows 100% agreement between the RT-LAMP and RT-PCR methods.118 Both RT-LAMP and RT-PCR can be used for the detection of viral RNA in the saliva from a person. Currently, the main method of detection is the same as with nasopharyngeal swabs – RT-PCR. Saliva was reported to be a suitable sample for COVID-19 testing by the University of Illinois. Researchers at the Yale School of Public Health and Hokkaido University even commented that PCR testing of saliva provides more accurate results than testing of nasopharyngeal swabs. Collecting saliva does not need medical staff and is more comfortable for a patient. This allows for more frequent testing without putting pressure on medical staff. Several procedures have been developed to pretreat saliva before PCR because saliva is more of a chemically complex matrix than nasopharyngeal swabs. Some of these procedures were proven to be successful, such as the protocols from the University of Illinois and Fluidigm's “Advanta Dx SARS-CoV-2 RT-PCR”.130 If the current saliva testing methods obtain governmental permission, they can replace the testing of nasopharyngeal swabs. The detection of viral antigens in blood or nasopharyngeal swabs is also a promising method. Benefits include it being relatively cheap, fast, and portable. One such example is the “Sofia SARS Antigen Fluorescent Immunoassay” by Quidel, which was approved for use in healthcare organizations. Detection of antibodies to the virus is also available in portable devices as a rapid test (e.g., “COVID-19 IgG/IgM Rapid Test Cassette” by Zhejiang Orient Gene Biotech Co., Ltd.). The main benefits are the ease of use and speed (results are available in 10 minutes, and the only requirement is the addition of a drop of blood and the provided buffer on a test slide).120 Antibody detection, however, is efficient only after several days have passed since the onset of symptoms. Both antibody and antigen tests are easier to use and cheaper, but they are less sensitive than RT-PCR. They can be used after the onset of symptoms to avoid a false negative during early testing. There are other rapid tests, such as pulse-controlled amplification (20 minutes),123 reverse-transcription recombinase-aided amplification (5–15 minutes),125 and other methods. However, these approaches are all new research studies requiring proficient scientists, and are confined to the laboratory, so they are not the main focus of today's trends.
Abbreviations
| AFM | Atomic-force microscopy |
| Ab | Antibodies |
| CDC | Center for Disease Control |
| CMV | Cytomegalovirus |
| CoV | Coronavirus |
| CRISPR | Clustered regularly interspaced short palindromic repeats |
| cRNA | RNA copies |
| DNA | Deoxyribonucleic acid |
| ELD50 | 50% egg lethal dose |
| ELISA | Enzyme-linked immunosorbent assay |
| HA | Haemagglutinin |
| HAU | Hemagglutination unit |
| HAV | Hepatitis A virus |
| HCV | Hepatitis C virus |
| HDV | Hepatitis D virus |
| HEV | Hepatitis E virus |
| HFV | Hemorrhagic fever virus |
| HIV | Human immunodeficiency virus |
| LAMP | Loop-mediated isothermal amplification |
| LOD | Limit of detection |
| MERS | Middle east respiratory syndrome |
| NA | Neuraminidase |
| NP | Nucleoprotein |
| NS | Nonstructural |
| PCR | Polymerase chain reaction |
| PFU | Plaque-forming unit |
| RNA | Ribonucleic acid |
| RSD | Relative standard deviation |
| RT-PCR | Reverse transcription polymerase chain reaction |
| Rxn | Reaction |
| SARS | Severe acute respiratory syndrome |
| SERS | Surface-enhanced Raman spectroscopy |
| SPR | Surface-plasmon resonance |
| TCID50 | 50% tissue culture infective dose |
| WHO | World Health Organization |
Funding statement
This research is supported by the Nazarbayev University (NU) Faculty Development Competitive Research Grant no. 090118FD5352 (Kazakhstan), and the NU Faculty Development Competitive Research Grant no. 110119FD4517 (Kazakhstan).Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to acknowledge Yunona Bukasova for proofreading the paper.References
- X. Cheng, G. Chen and W. R. Rodriguez, Micro- and nanotechnology for viral detection, Anal. Bioanal. Chem., 2009, 393(2), 487–501 CrossRef CAS.
- F. Krammer, G. J. D. Smith, R. A. M. Fouchier, M. Peiris, K. Kedzierska and P. C. Doherty, et al., Influenza, Nat. Rev. Dis. Primers, 2018, 4(1), 3 CrossRef.
- F. S. Dawood, A. D. Iuliano, C. Reed, M. I. Meltzer, D. K. Shay and P. Y. Cheng, et al., Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study, Lancet Infect. Dis., 2012, 12(9), 687–695 CrossRef.
- WHO, Burden of disease, WHO, 2020, https://www.who.int/influenza/surveillance_monitoring/bod/en/ Search PubMed.
- CDC, Disease Burden of Influenza CDC, 2020, https://www.cdc.gov/flu/about/burden/index.htmL Search PubMed.
- K. H. Chan, S. T. Lai, L. L. Poon, Y. Guan, K. Y. Yuen and J. S. Peiris, Analytical sensitivity of rapid influenza antigen detection tests for swine-origin influenza virus (H1N1), J. Clin. Virol., 2009, 45(3), 205–207 CrossRef CAS.
- C. C. Ginocchio, F. Zhang, R. Manji, S. Arora, M. Bornfreund and L. Falk, et al., Evaluation of multiple test methods for the detection of the novel 2009 influenza A (H1N1) during the New York City outbreak, J. Clin. Virol., 2009, 45(3), 191–195 CrossRef CAS.
- H. Bai, R. Wang, B. Hargis, H. Lu and Y. Li, A SPR aptasensor for detection of avian influenza virus H5N1, Sensors, 2012, 12(9), 12506–12518 CrossRef CAS.
- J. R. Ciacci-Zanella, A. L. Vincent, J. R. Prickett, S. M. Zimmerman and J. J. Zimmerman, Detection of anti-influenza A nucleoprotein antibodies in pigs using a commercial influenza epitope-blocking enzyme-linked immunosorbent assay developed for avian species, J. Vet. Diagn. Invest., 2010, 22(1), 3–9 CrossRef.
- K. Watcharatanyatip, S. Boonmoh, K. Chaichoun, T. Songserm, M. Woratanti and T. Dharakul, Multispecies detection of antibodies to influenza A viruses by a double-antigen sandwich ELISA, J. Virol. Methods, 2010, 163(2), 238–243 CrossRef CAS.
- Y. Yao, X. Chen, X. Zhang, Q. Liu, J. Zhu and W. Zhao, et al., Rapid Detection of Influenza Virus Subtypes Based on an Integrated Centrifugal Disc, ACS Sens., 2020, 5(5), 1354–1362 CrossRef CAS.
- R. Randriantsilefisoa, C. Nie, B. Parshad, Y. Pan, S. Bhatia and R. Haag, Double trouble for viruses: a hydrogel nanocomposite catches the influenza virus while shrinking and changing color, Chem. Commun., 2020, 56(24), 3547–3550 RSC.
- J. Kwon, Y. Lee, T. Lee and J.-H. Ahn, Aptamer-Based Field-Effect Transistor for Detection of Avian Influenza Virus in Chicken Serum, Anal. Chem., 2020, 92(7), 5524–5531 CrossRef CAS.
- K. Wu, J. Liu, R. Saha, D. Su, V. D. Krishna and M. C. J. Cheeran, et al., Magnetic Particle Spectroscopy for Detection of Influenza A Virus Subtype H1N1, ACS Appl. Mater. Interfaces, 2020, 12(12), 13686–13697 CrossRef CAS.
- K. Danforth, R. Granich, D. Wiedeman, S. Baxi and N. Padian, in Global Mortality and Morbidity of HIV/AIDS, ed. K. K. Holmes, S. Bertozzi, B. R. Bloom and P. Jha, Washington, DC, 3rd edn, Major Infectious Diseases, 2017 Search PubMed.
- BBC, Ebola outbreak: Cases pass 10,000, WHO reports CDC, 2014, https://www.bbc.com/news/world-africa-29769782 Search PubMed.
- Medicine BCo, Zika Virus, 2020, https://www.bcm.edu/departments/molecular-virology-and-microbiology/emerging-infections-and-biodefense/specific-agents/zika Search PubMed.
- R. Noor and T. Ahmed, Zika virus: Epidemiological study and its association with public health risk, J. Infect. Public Health, 2018, 11(5), 611–616 CrossRef.
- C. N. Fhogartaigh and E. Aarons, Viral haemorrhagic fever, Clin. Med., 2015, 15(1), 61–66 CrossRef.
- WHO, Dengue and severe dengue 2020, www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue, accessed May 26, 2020 Search PubMed.
- WHO, Hepatitis A 2020, https://www.who.int/news-room/fact-sheets/detail/hepatitis-a, accessed May 26, 2020 Search PubMed.
- WHO, Hepatitis C 2020, https://www.who.int/news-room/fact-sheets/detail/hepatitis-c, accessed May 26, 2020 Search PubMed.
- WHO, Hepatitis D 2020, www.who.int/news-room/fact-sheets/detail/hepatitis-d, accessed May 26, 2020 Search PubMed.
- WHO, Hepatitis E 2020, www.who.int/news-room/fact-sheets/detail/hepatitis-e, accessed May 26, 2020 Search PubMed.
- Y. Ma, C. Ni, E. E. Dzakah, H. Wang, K. Kang and S. Tang, et al., Development of Monoclonal Antibodies against HIV-1 p24 Protein and Its Application in Colloidal Gold Immunochromatographic Assay for HIV-1 Detection, BioMed Res. Int., 2016, 2016, 6743904 Search PubMed.
- M. Miedouge, M. Greze, A. Bailly and J. Izopet, Analytical sensitivity of four HIV combined antigen/antibody assays using the p24 WHO standard, J. Clin. Virol., 2011, 50(1), 57–60 CrossRef CAS.
- K.-B. Lee, E.-Y. Kim, C. A. Mirkin and S. M. Wolinsky, The Use of Nanoarrays for Highly Sensitive and Selective Detection of Human Immunodeficiency Virus Type 1 in Plasma, Nano Lett., 2004, 4(10), 1869–1872 CrossRef CAS.
- K. Teeparuksapun, M. Hedstrom, E. Y. Wong, S. Tang, I. K. Hewlett and B. Mattiasson, Ultrasensitive detection of HIV-1 p24 antigen using nanofunctionalized surfaces in a capacitive immunosensor, Anal. Chem., 2010, 82(20), 8406–8411 CrossRef CAS.
- R. Sutthent, N. Gaudart, K. Chokpaibulkit, N. Tanliang, C. Kanoksinsombath and P. Chaisilwatana, p24 Antigen detection assay modified with a booster step for diagnosis and monitoring of human immunodeficiency virus type 1 infection, J. Clin. Microbiol., 2003, 41(3), 1016–1022 CrossRef CAS.
- S. Tang, J. Zhao, J. J. Storhoff, P. J. Norris, R. F. Little and R. Yarchoan, et al., Nanoparticle-Based biobarcode amplification assay (BCA) for sensitive and early detection of human immunodeficiency type 1 capsid (p24) antigen, J. Acquired Immune Defic. Syndr., 2007, 46(2), 231–237 CrossRef.
- J. L. Greenwald, G. R. Burstein, J. Pincus and B. Branson, A rapid review of rapid HIV antibody tests, Curr. Infect. Dis. Rep., 2006, 8(2), 125–131 CrossRef.
- A. Bhimji, A. A. Zaragoza, L. S. Live and S. O. Kelley, Electrochemical enzyme-linked immunosorbent assay featuring proximal reagent generation: detection of human immunodeficiency virus antibodies in clinical samples, Anal. Chem., 2013, 85(14), 6813–6819 CrossRef CAS.
- R. J. White, H. M. Kallewaard, W. Hsieh, A. S. Patterson, J. B. Kasehagen and K. J. Cash, et al., Wash-free, electrochemical platform for the quantitative, multiplexed detection of specific antibodies, Anal. Chem., 2012, 84(2), 1098–1103 CrossRef CAS.
- E. A. Phillips, T. J. Moehling, K. F. K. Ejendal, O. S. Hoilett, K. M. Byers and L. A. Basing, et al., Microfluidic rapid and autonomous analytical device (microRAAD) to detect HIV from whole blood samples, Lab Chip, 2019, 19(20), 3375–3386 RSC.
- M. K. Tsang, W. Ye, G. Wang, J. Li, M. Yang and J. Hao, Ultrasensitive Detection of Ebola Virus Oligonucleotide Based on Upconversion Nanoprobe/Nanoporous Membrane System, ACS Nano, 2016, 10(1), 598–605 CrossRef CAS.
- P. Lam, R. A. Keri and N. F. Steinmetz, A Bioengineered Positive Control for Rapid Detection of the Ebola Virus by Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP), ACS Biomater. Sci. Eng., 2017, 3(3), 452–459 CrossRef CAS.
- M. Niikura, T. Ikegami, M. Saijo, I. Kurane, M. E. Miranda and S. Morikawa, Detection of Ebola viral antigen by enzyme-linked immunosorbent assay using a novel monoclonal antibody to nucleoprotein, J. Clin. Microbiol., 2001, 39(9), 3267–3271 CrossRef CAS.
- J. Hu, Y. Z. Jiang, L. L. Wu, Z. Wu, Y. Bi and G. Wong, et al., Dual-Signal Readout Nanospheres for Rapid Point-of-Care Detection of Ebola Virus Glycoprotein, Anal. Chem., 2017, 89(24), 13105–13111 CrossRef CAS.
- A. Petrosova, T. Konry, S. Cosnier, I. Trakht, J. Lutwama and E. Rwaguma, et al., Development of a highly sensitive, field operable biosensor for serological studies of Ebola virus in central Africa, Sens. Actuators, B, 2007, 122(2), 578–586 CrossRef CAS.
- Z. Wu, J. Hu, T. Zeng, Z. L. Zhang, J. Chen and G. Wong, et al., Ultrasensitive Ebola Virus Detection Based on Electroluminescent Nanospheres and Immunomagnetic Separation, Anal. Chem., 2017, 89(3), 2039–2048 CrossRef CAS.
- S. L. Hong, Y. N. Zhang, Y. H. Liu, M. Tang, D. W. Pang and G. Wong, et al., Cellular-Beacon-Mediated Counting for the Ultrasensitive Detection of Ebola Virus on an Integrated Micromagnetic Platform, Anal. Chem., 2018, 90(12), 7310–7317 CrossRef CAS.
- S. A. Camacho, R. G. Sobral-Filho, P. H. B. Aoki, C. J. L. Constantino and A. G. Brolo, Zika Immunoassay Based on Surface-Enhanced Raman Scattering Nanoprobes, ACS Sens., 2018, 3(3), 587–594 CrossRef CAS.
- K. H. Lee and H. Zeng, Aptamer-Based ELISA Assay for Highly Specific and Sensitive Detection of Zika NS1 Protein, Anal. Chem., 2017, 89(23), 12743–12748 CrossRef CAS.
- S. Afsahi, M. B. Lerner, J. M. Goldstein, J. Lee, X. Tang and D. A. Bagarozzi Jr, et al., Novel graphene-based biosensor for early detection of Zika virus infection, Biosens. Bioelectron., 2018, 100, 85–88 CrossRef CAS.
- M. S. Draz, N. K. Lakshminaraasimulu, S. Krishnakumar, D. Battalapalli, A. Vasan and M. K. Kanakasabapathy, et al., Motion-Based Immunological Detection of Zika Virus Using Pt-Nanomotors and a Cellphone, ACS Nano, 2018, 12(6), 5709–5718 CrossRef CAS.
- C. A. Lynch III, M. V. Foguel, A. J. Reed, A. M. Balcarcel, P. Calvo-Marzal and Y. V. Gerasimova, et al., Selective Determination of Isothermally Amplified Zika Virus RNA Using a Universal DNA-Hairpin Probe in Less than 1 Hour, Anal. Chem., 2019, 91(21), 13458–13464 CrossRef.
- C. Ma, H. Jing, P. Zhang, L. Han, M. Zhang and F. Wang, et al., An ultrafast one-step assay for the visual detection of RNA virus, Chem. Commun., 2018, 54(25), 3118–3121 RSC.
- D. Granger, H. Hilgart, L. Misner, J. Christensen, S. Bistodeau and J. Palm, et al., Serologic Testing for Zika Virus: Comparison of Three Zika Virus IgM-Screening Enzyme-Linked Immunosorbent Assays and Initial Laboratory Experiences, J. Clin. Microbiol., 2017, 55(7), 2127–2136 CrossRef CAS.
- S. M. Scherr, D. S. Freedman, K. N. Agans, A. Rosca, E. Carter and M. Kuroda, et al., Disposable cartridge platform for rapid detection of viral hemorrhagic fever viruses, Lab Chip, 2017, 17(5), 917–925 RSC.
- C. Drosten, S. Gottig, S. Schilling, M. Asper, M. Panning and H. Schmitz, et al., Rapid detection and quantification of RNA of Ebola and Marburg viruses, Lassa virus, Crimean-Congo hemorrhagic fever virus, Rift Valley fever virus, dengue virus, and yellow fever virus by real-time reverse transcription-PCR, J. Clin. Microbiol., 2002, 40(7), 2323–2330 CrossRef CAS.
- Z. Pang, A. Li, J. Li, J. Qu, C. He and S. Zhang, et al., Comprehensive multiplex one-step real-time TaqMan qRT-PCR assays for detection and quantification of hemorrhagic fever viruses, PLoS One, 2014, 9(4), e95635 CrossRef.
- H. A. Osman, K. H. Eltom, N. O. Musa, N. M. Bilal, M. I. Elbashir and I. E. Aradaib, Development and evaluation of loop-mediated isothermal amplification assay for detection of Crimean Congo hemorrhagic fever virus in Sudan, J. Virol. Methods, 2013, 190(1–2), 4–10 CrossRef CAS.
- F. Algaar, E. Eltzov, M. M. Vdovenko, I. Y. Sakharov, L. Fajs and M. Weidmann, et al., Fiber-optic immunosensor for detection of Crimean-Congo hemorrhagic fever IgG antibodies in patients, Anal. Chem., 2015, 87(16), 8394–8398 CrossRef CAS.
- H. J. Yoo, C. Baek, M. H. Lee and J. Min, Integrated microsystems for the in situ genetic detection of dengue virus in whole blood using direct sample preparation and isothermal amplification, Analyst, 2020, 145(6), 2405–2411 RSC.
- A. J. Baeumner, N. A. Schlesinger, N. S. Slutzki, J. Romano, E. M. Lee and R. A. Montagna, Biosensor for dengue virus detection: sensitive, rapid, and serotype specific, Anal. Chem., 2002, 74(6), 1442–1448 CrossRef CAS.
- M. Gao, D. Daniel, H. Zou, S. Jiang, S. Lin and C. Huang, et al., Rapid detection of a dengue virus RNA sequence with single molecule sensitivity using tandem toehold-mediated displacement reactions, Chem. Commun., 2018, 54(8), 968–971 RSC.
- G. A. Ortega, S. Pérez-Rodríguez and E. Reguera, Magnetic paper-based ELISA for IgM-dengue detection, RSC Adv., 2017, 7(9), 4921–4932 RSC.
- W. R. Wong, O. Krupin, S. D. Sekaran, F. R. Mahamd Adikan and P. Berini, Serological diagnosis of dengue infection in blood plasma using long-range surface plasmon waveguides, Anal. Chem., 2014, 86(3), 1735–1743 CrossRef CAS.
- Y. Zhang, J. Bai and J. Y. Ying, A stacking flow immunoassay for the detection of dengue-specific immunoglobulins in salivary fluid, Lab Chip, 2015, 15(6), 1465–1471 RSC.
- N. C. S. Vieira, A. Figueiredo, J. F. dos Santos, S. M. Aoki, F. E. G. Guimarães and V. Zucolotto, Label-free electrical recognition of a dengue virus protein using the SEGFET simplified measurement system, Anal. Methods, 2014, 6(22), 8882–8885 RSC.
- T. Axelrod, E. Eltzov and R. S. Marks, Capture-Layer Lateral Flow Immunoassay: A New Platform Validated in the Detection and Quantification of Dengue NS1, ACS Omega, 2020, 5(18), 10433–10440 CrossRef CAS.
- W. S. Chang, H. Shang, R. M. Perera, S. M. Lok, D. Sedlak and R. J. Kuhn, et al., Rapid detection of dengue virus in serum using magnetic separation and fluorescence detection, Analyst, 2008, 133(2), 233–240 RSC.
- M. S. Cheng, J. S. Ho, C. H. Tan, J. P. Wong, L. C. Ng and C. S. Toh, Development of an electrochemical membrane-based nanobiosensor for ultrasensitive detection of dengue virus, Anal. Chim. Acta, 2012, 725, 74–80 CrossRef CAS.
- S. Perelle, L. Cavellini, C. Burger, S. Blaise-Boisseau, C. Hennechart-Collette and G. Merle, et al., Use of a robotic RNA purification protocol based on the NucliSens easyMAG for real-time RT-PCR detection of hepatitis A virus in bottled water, J. Virol. Methods, 2009, 157(1), 80–83 CrossRef CAS.
- S. Blaise-Boisseau, C. Hennechart-Collette, L. Guillier and S. Perelle, Duplex real-time qRT-PCR for the detection of hepatitis A virus in water and raspberries using the MS2 bacteriophage as a process control, J. Virol. Methods, 2010, 166(1–2), 48–53 CrossRef CAS.
- J. Mandli, A. Attar, M. M. Ennaji and A. Amine, Indirect competitive electrochemical immunosensor for hepatitis A virus antigen detection, J. Electroanal. Chem., 2017, 799, 213–221 CrossRef CAS.
- D. Tang, J. Tang, B. Su, J. Ren and G. Chen, Simultaneous determination of five-type hepatitis virus antigens in 5 min using an integrated automatic electrochemical immunosensor array, Biosens. Bioelectron., 2010, 25(7), 1658–1662 CrossRef CAS.
- W. J. Miller, W. Clark, W. Hurni, B. Kuter, T. Schofield and D. Nalin, Sensitive assays for hepatitis A antibodies, J. Med. Virol., 1993, 41(3), 201–204 CrossRef CAS.
- H. J. Lee, H. S. Jeong, B. K. Cho, M. J. Ji, J. H. Kim and A. N. Lee, et al., Evaluation of an immunochromatographic assay for the detection of anti-hepatitis A virus IgM, Virol. J., 2010, 7, 164 CrossRef.
- B. Yang, H. Gong, C. Chen, X. Chen and C. Cai, A virus resonance light scattering sensor based on mussel-inspired molecularly imprinted polymers for high sensitive and high selective detection of Hepatitis A Virus, Biosens. Bioelectron., 2017, 87, 679–685 CrossRef CAS.
- L. Luo, F. Zhang, C. Chen and C. Cai, Visual Simultaneous Detection of Hepatitis A and B Viruses Based on a Multifunctional Molecularly Imprinted Fluorescence Sensor, Anal. Chem., 2019, 91(24), 15748–15756 CrossRef CAS.
- J. Fan, L. Yuan, Q. Liu, C. Tong, W. Wang and F. Xiao, et al., An ultrasensitive and simple assay for the Hepatitis C virus using a reduced graphene oxide-assisted hybridization chain reaction, Analyst, 2019, 144(13), 3972–3979 RSC.
- H. Miyachi, A. Masukawa, T. Ohshima, T. Hirose, C. Impraim and Y. Ando, Automated specific capture of hepatitis C virus RNA with probes and paramagnetic particle separation, J. Clin. Microbiol., 2000, 38(1), 18–21 CAS.
- T. P. Leary, R. A. Gutierrez, A. S. Muerhoff, L. G. Birkenmeyer, S. M. Desai and G. J. Dawson, A chemiluminescent, magnetic particle-based immunoassay for the detection of hepatitis C virus core antigen in human serum or plasma, J. Med. Virol., 2006, 78(11), 1436–1440 CrossRef CAS.
- A. Valipour and M. Roushani, Using silver nanoparticle and thiol graphene quantum dots nanocomposite as a substratum to load antibody for detection of hepatitis C virus core antigen: Electrochemical oxidation of riboflavin was used as redox probe, Biosens. Bioelectron., 2017, 89(Pt 2), 946–951 CrossRef CAS.
- L. Duan, Y. Wang, S. S. Li, Z. Wan and J. Zhai, Rapid and simultaneous detection of human hepatitis B virus and hepatitis C virus antibodies based on a protein chip assay using nano-gold immunological amplification and silver staining method, BMC Infect. Dis., 2005, 5, 53 CrossRef.
- T. Gao, W. Chai, L. Shi, H. Shi, A. Sheng and J. Yang, et al., A new colorimetric assay method for the detection of anti-hepatitis C virus antibody with high sensitivity, Analyst, 2019, 144(21), 6365–6370 RSC.
- M. Kaito, S. Ishida, H. Tanaka, S. Horiike, N. Fujita and Y. Adachi, et al., Morphology of hepatitis C and hepatitis B virus particles as detected by immunogold electron microscopy, Med. Mol. Morphol., 2006, 39(2), 63–71 CrossRef.
- Y. Wang, J. S. Glenn, M. A. Winters, L. P. Shen, I. Choong and Y. L. Shi, et al., A new dual-targeting real-time RT-PCR assay for hepatitis D virus RNA detection, Diagn. Microbiol. Infect. Dis., 2018, 92(2), 112–117 CrossRef CAS.
- M. Kodani, A. Martin, T. Mixson-Hayden, J. Drobeniuc, R. R. Gish and S. Kamili, One-step real-time PCR assay for detection and quantitation of hepatitis D virus RNA, J. Virol. Methods, 2013, 193(2), 531–535 CrossRef CAS.
- A. G. Shattock, M. Morris, K. Kinane and C. Fagan, The serology of delta hepatitis and the detection of IgM anti-HD by EIA using serum derived delta antigen, J. Virol. Methods, 1989, 23(3), 233–240 CrossRef CAS.
- C. Mokhtari, E. Marchadier, S. Haim-Boukobza, A. Jeblaoui, S. Tesse and J. Savary, et al., Comparison of real-time RT-PCR assays for hepatitis E virus RNA detection, J. Clin. Virol., 2013, 58(1), 36–40 CrossRef CAS.
- H. H. Liu, X. Cao, Y. Yang, M. G. Liu and Y. F. Wang, Array-based nano-amplification technique was applied in detection of hepatitis E virus, J. Biochem. Mol. Biol., 2006, 39(3), 247–252 CAS.
- E. E. Mast, M. J. Alter, P. V. Holland and R. H. Purcell, Evaluation of assays for antibody to hepatitis E virus by a serum panel. Hepatitis E Virus Antibody Serum Panel Evaluation Group, Hepatology, 1998, 27(3), 857–861 CrossRef CAS.
- N. S. Zhong, B. J. Zheng, Y. M. Li, L. L. M. Poon, Z. H. Xie and K. H. Chan, et al., Epidemiology and cause of severe acute respiratory syndrome (SARS) in Guangdong, People’s Republic of China, in February, 2003, Lancet, 2003, 362(9393), 1353–1358 CrossRef CAS.
- T. G. Ksiazek, D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret and S. Emery, et al., A novel coronavirus associated with severe acute respiratory syndrome, N. Engl. J. Med., 2003, 348(20), 1953–1966 CrossRef CAS.
- C. Drosten, S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt and S. Becker, et al., Identification of a novel coronavirus in patients with severe acute respiratory syndrome, N. Engl. J. Med., 2003, 348(20), 1967–1976 CrossRef CAS.
- A. M. Zaki, S. van Boheemen, T. M. Bestebroer, A. D. Osterhaus and R. A. Fouchier, Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia, N. Engl. J. Med., 2012, 367(19), 1814–1820 CrossRef CAS.
- WHO, MERS situation update, January 2020, http://www.emro.who.int/health-topics/mers-cov/mers-outbreaks.html Search PubMed.
- K. H. Chan, L. L. Poon, V. C. Cheng, Y. Guan, I. F. Hung and J. Kong, et al., Detection of SARS coronavirus in patients with suspected SARS, Emerging Infect. Dis., 2004, 10(2), 294–299 CrossRef.
- V. M. Corman, I. Eckerle, T. Bleicker, A. Zaki, O. Landt and M. Eschbach-Bludau, et al., Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction, Eurosurveillance, 2012, 17(39), 20285 CrossRef.
- D. G. Ahn, I. J. Jeon, J. D. Kim, M. S. Song, S. R. Han and S. W. Lee, et al., RNA aptamer-based sensitive detection of SARS coronavirus nucleocapsid protein, Analyst, 2009, 134(9), 1896–1901 RSC.
- C. Roh and S. K. Jo, Quantitative and sensitive detection of SARS coronavirus nucleocapsid protein using quantum dots-conjugated RNA aptamer on chip, J. Chem. Technol. Biotechnol., 2011, 86(12), 1475–1479 CrossRef CAS.
- S. Bhadra, Y. S. Jiang, M. R. Kumar, R. F. Johnson, L. E. Hensley and A. D. Ellington, Real-time sequence-validated loop-mediated isothermal amplification assays for detection of Middle East respiratory syndrome coronavirus (MERS-CoV), PLoS One, 2015, 10(4), e0123126 CrossRef.
- Y. Chen, K. H. Chan, Y. Kang, H. Chen, H. K. Luk and R. W. Poon, et al., A sensitive and specific antigen detection assay for Middle East respiratory syndrome coronavirus, Emerging Microbes Infect., 2015, 4(4), e26 CAS.
- A. Liljander, B. Meyer, J. Jores, M. A. Muller, E. Lattwein and I. Njeru, et al., MERS-CoV Antibodies in Humans, Africa, 2013-2014, Emerging Infect. Dis., 2016, 22(6), 1086–1089 CrossRef CAS.
- WHO, Novel Coronavirus – China, WHO, 2020, https://www.who.int/csr/don/12-january-2020-novel-coronavirus-china/en/ Search PubMed.
- N. Zhu, D. Zhang, W. Wang, X. Li, B. Yang and J. Song, et al., A Novel Coronavirus from Patients with Pneumonia in China, 2019, N. Engl. J. Med., 2020, 382(8), 727–733 CrossRef CAS.
- T. Wenjie, Z. Xiang, M. Xuejun, W. Wenling, N. Peihua, X. Wenbo, et al.,Notes from the Field: A Novel Coronavirus Genome Identified in a Cluster of Pneumonia Cases — Wuhan, China 2019−2020 China CDC Weekly, 2020 Search PubMed.
- WHO, Naming the coronavirus disease (COVID-19) and the virus that causes it, WHO, 2020, https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it Search PubMed.
- W. J. Guan, Z. Y. Ni, Y. Hu, W. H. Liang, C. Q. Ou and J. X. He, et al., Clinical Characteristics of Coronavirus Disease 2019 in China, N. Engl. J. Med., 2020, 382(18), 1708–1720 CrossRef CAS.
- W. Wang, Y. Xu, R. Gao, R. Lu, K. Han and G. Wu, et al., Detection of SARS-CoV-2 in Different Types of Clinical Specimens, JAMA, J. Am. Med. Assoc., 2020, 323(18), 1843–1844 CAS.
- C. Rothe, M. Schunk, P. Sothmann, G. Bretzel, G. Froeschl and C. Wallrauch, et al., Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany, N. Engl. J. Med., 2020, 382(10), 970–971 CrossRef.
- Worldometer, COVID-19 Coronavirus Pandemic, 2020, https://www.worldometers.info/coronavirus/ Search PubMed.
- V. M. Corman, O. Landt, M. Kaiser, R. Molenkamp, A. Meijer and D. K. Chu, et al., Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR, Eurosurveillance, 2020, 25(3), 2000045 CrossRef.
- T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe and N. Amino, et al., Loop-mediated isothermal amplification of DNA, Nucleic Acids Res., 2000, 28(12), E63 CrossRef CAS.
- C. Yan, J. Cui, L. Huang, B. Du, L. Chen and G. Xue, et al., Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay, Clin. Microbiol. Infect., 2020, 26(6), 773–779 CrossRef CAS.
- Y. H. Baek, J. Um, K. J. C. Antigua, J. H. Park, Y. Kim and S. Oh, et al., Development of a reverse transcription-loop-mediated isothermal amplification as a rapid early-detection method for novel SARS-CoV-2, Emerging Microbes Infect., 2020, 9(1), 998–1007 CrossRef CAS.
- R. Lu, X. Wu, Z. Wan, Y. Li, L. Zuo and J. Qin, et al., Development of a Novel Reverse Transcription Loop-Mediated Isothermal Amplification Method for Rapid Detection of SARS-CoV-2, Virol. Sin., 2020, 35, 344–347 CrossRef CAS.
- J. P. Broughton, X. Deng, G. Yu, C. L. Fasching, V. Servellita and J. Singh, et al., CRISPR-Cas12-based detection of SARS-CoV-2, Nat. Biotechnol., 2020, 38, 870–874 CrossRef CAS.
- K. Uhteg, J. Jarrett, M. Richards, C. Howard, E. Morehead and M. Geahr, et al., Comparing the analytical performance of three SARS-CoV-2 molecular diagnostic assays, J. Clin. Virol., 2020, 127, 104384 CrossRef CAS.
- A. Diagnostics, Instructions for use RealStar SARS-CoV-2 RT-PCR Kit 1.0, 2020 Search PubMed.
- ePLEX, ePLEX SARS-CoV2 assay manual, 2020 Search PubMed.
- WHO, WHO in-house molecular assays, 2020, https://www.who.int/docs/defaultsource/coronaviruse/whoinhouseassays.pdf?sfvrsn=de3a76aa_2 Search PubMed.
- K. Okamaoto, K. Shirato, N. Nao, S. Saito, T. Kageyama and H. Hasegawa, et al., An assessment of real-time RT-PCR kits for SARS-CoV-2 detection, Jpn. J. Infect. Dis., 2020, 73(5), 366–368 CrossRef CAS.
- J. F. Chan, C. C. Yip, K. K. To, T. H. Tang, S. C. Wong and K. H. Leung, et al., Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated In Vitro and with Clinical Specimens, J. Clin. Microbiol., 2020, 58(5), e00310-20 CrossRef.
- A. Moran, K. G. Beavis, S. M. Matushek, C. Ciaglia, N. Francois and V. Tesic, et al., The Detection of SARS-CoV-2 using the Cepheid Xpert Xpress SARS-CoV-2 and Roche cobas SARS-CoV-2 Assays, J. Clin. Microbiol., 2020, 58(8), e00772-20 CrossRef.
- Y. Zhang, N. Odiwuor, J. Xiong, L. Sun, R. O. Nyaruaba, H. Wei, et al., Rapid Molecular Detection of SARS-CoV-2 (COVID-19) Virus RNA Using Colorimetric LAMP, medRxiv, 2020, 2020.02.26.20028373 Search PubMed.
- N. M. A. Okba, M. A. Muller, W. Li, C. Wang, C. H. GeurtsvanKessel, V. M. Corman, et al., SARS-CoV-2 specific antibody responses in COVID-19 patients, medRxiv, 2020, 2020.03.18.20038059 Search PubMed.
- T. Hoffman, K. Nissen, J. Krambrich, B. Ronnberg, D. Akaberi and M. Esmaeilzadeh, et al., Evaluation of a COVID-19 IgM and IgG rapid test; an efficient tool for assessment of past exposure to SARS-CoV-2, Infect. Ecol. Epidemiology, 2020, 10(1), 1754538 CrossRef.
- Y. Gao, Y. Yuan, T. T. Li, W. X. Wang, Y. X. Li, A. Li, et al., Evaluation the auxiliary diagnosis value of antibodies assays for detection of novel coronavirus (SARS-Cov-2) causing an outbreak of pneumonia (COVID-19), medRxiv, 2020, 2020.03.26.20042044 Search PubMed.
- D. Shan, J. M. Johnson, S. C. Fernandes, M. Mendes, H. Suib, M. Holdridge, et al., SARS-Coronavirus-2 nucleocapsid protein measured in blood using a Simoa ultra-sensitive immunoassay differentiates COVID-19 infection with high clinical sensitivity, medRxiv, 2020, 2020.08.14.20175356 Search PubMed.
- K. Zwirglmaier, M. Weyh, C. Krueger, R. Ehmann, K. Mueller, R. Woelfel, et al., Rapid detection of SARS-CoV-2 by pulse-controlled amplification (PCA), medRxiv, 2020, 2020.07.29.20154104 Search PubMed.
- W. S. Yoo, H. S. Han, J. G. Kim, K. Kang, H.-S. Jeon and J.-Y. Moon, et al., Development of a tablet PC-based portable device for colorimetric determination of assays including COVID-19 and other pathogenic microorganisms, RSC Adv., 2020, 10(54), 32946–32952 RSC.
- G. Xue, S. Li, W. Zhang, B. Du, J. Cui and C. Yan, et al., Reverse-Transcription Recombinase-Aided Amplification Assay for Rapid Detection of the 2019 Novel Coronavirus (SARS-CoV-2), Anal. Chem., 2020, 92(14), 9699–9705 CrossRef CAS.
- J. Liu, M. A. Eddings, A. R. Miles, R. Bukasov, B. K. Gale and J. S. Shumaker-Parry, In Situ Microarray Fabrication and Analysis Using a Microfluidic Flow Cell Array Integrated with Surface Plasmon Resonance Microscopy, Anal. Chem., 2009, 81(11), 4296–4301 CrossRef CAS.
- R. Pilot, R. Signorini, C. Durante, L. Orian, M. Bhamidipati and L. Fabris, A Review on Surface-Enhanced Raman Scattering, Biosensors, 2019, 9(2), 57 CrossRef CAS.
- H. C. Metsky, C. A. Freije, T.-S. F. Kosoko-Thoroddsen, P. C. Sabeti and C. Myhrvold, CRISPR-based surveillance for COVID-19 using genomically-comprehensive machine learning design, bioRxiv, 2020, 2020.02.26.967026 Search PubMed.
- J. D. Driskell, K. M. Kwarta, R. J. Lipert, M. D. Porter, J. D. Neill and J. F. Ridpath, Low-Level Detection of Viral Pathogens by a Surface-Enhanced Raman Scattering Based Immunoassay, Anal. Chem., 2005, 77(19), 6147–6154 CrossRef CAS.
- M. Peplow, Saliva tests show promise for widespread COVID-19 surveillance at universities and workplaces C. E. N., 2020, https://cen.acs.org/analytical-chemistry/diagnostics/Saliva-tests-show-promise-widespread/98/web/2020/08 Search PubMed.
- M. Peplow, Rapid COVID-19 testing breaks free from the lab C. E. N., 2020, https://cen.acs.org/analytical-chemistry/diagnostics/Rapid-COVID-19-testing-breaks/98/web/2020/08 Search PubMed.
| This journal is © The Royal Society of Chemistry 2021 |