Open Access Article
Open Access ArticleA mini DNA–RNA hybrid origami nanobrick†
Lifeng
Zhou
 *a,
Arun Richard
Chandrasekaran
*a,
Arun Richard
Chandrasekaran
 a,
Mengwen
Yan
a,
Mengwen
Yan
 b,
Vibhav A.
Valsangkar
b,
Vibhav A.
Valsangkar
 ab,
Jeremy I.
Feldblyum
ab,
Jeremy I.
Feldblyum
 b,
Jia
Sheng
b,
Jia
Sheng
 ab and
Ken
Halvorsen
ab and
Ken
Halvorsen
 *a
*a
aThe RNA Institute, University at Albany, State University of New York, Albany, NY,, USA. E-mail: lzhou2@albany.edu
bDepartment of Chemistry, University at Albany, State University of New York, Albany, NY, USA
First published on 7th June 2021
Abstract
DNA origami is typically used to fold a long single-stranded DNA scaffold into nanostructures with complex geometries using many short DNA staple strands. Integration of RNA into nucleic acid nanostructures is also possible, but has been less studied. In this research, we designed and characterized a hybrid RNA-scaffolded origami nanostructure with dimensions of ∼12 nm. We used 12 DNA staple strands to fold a 401 nt RNA scaffold into a ten-helix bundle with a honeycomb cross section. We verified the construction of the nanostructure using gel electrophoresis and atomic force microscopy. The DNA–RNA hybrid origami showed higher resistance to ribonuclease compared to a DNA–RNA duplex control. Our work shows potential use in folding long RNA, such as messenger RNA, into origami nanostructures that can be delivered into targeted cells as medicine or a vaccine.
Aside from being genetic information carriers, DNA and RNA can also be used as materials to construct nanostructures with pre-defined geometries.1,2 These nucleic acid nanostructures have potential for broad applications in drug delivery, biosensing, and biomaterials. For instance, nucleic acid nanostructures have been used to deliver doxorubicin, siRNA and antibodies for the treatment of cancers.3 In biosensing, dynamic DNA nanoswitches have been programmed to detect viral RNA such as SARS-CoV-2, Zika and Dengue.4 DNA nanostructures can also assist in the assembly of nanoparticles to create metamaterials such as sparsely packed colloidal crystals.5
The invention of DNA origami boosted the design and development of DNA nanostructures in the past decade.1,6 Though great progress has also been achieved in the design RNA nanostructures,7,8 DNA–RNA hybrid nanostructures have been less explored. Using branched DNA motifs, researchers have created hybrid DNA–RNA nanotubes,9 polygons10 and two-dimensional arrays.11 Hybrid DNA–RNA structures have also been constructed using the origami strategy where a set of DNA staple strands are used to fold an RNA scaffold into pre-designed geometries such as two-dimensional ribbons,12 planar tiles,12,13 and a six-helix bundle.13 The fabrication of these nanostructures has shown comparative yield as that of DNA origami nanostructures.12 The assembly and size of DNA or RNA origami structures is largely dependent on the scaffold used in these methods. This limitation has necessitated the creation of scaffold strands of different lengths for DNA origami beyond the routinely used 7249-nucleotide (nt) M13 viral genome.14 A similar need exists in the construction of size-defined DNA–RNA hybrid origami structures; so far the only RNA scaffold lengths reported are 1071 nt12 and 717 nt.13 Here, to further investigate the self-assembly quality and stability of DNA–RNA hybrid origami, we constructed a three-dimensional brick by folding a 401 nt RNA scaffold with DNA staple strands with maximum side length of ∼12 nm.
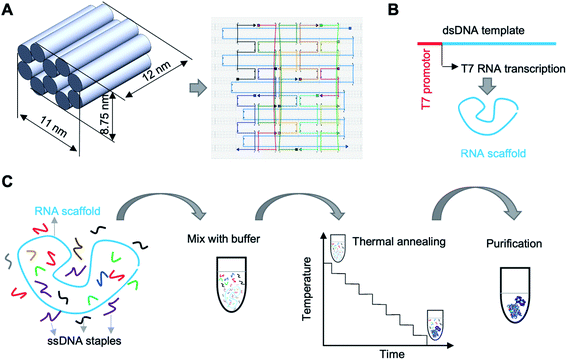
To demonstrate the approach, we designed an origami nanobrick with dimensions 12 (L) × 11 (W) × 8.75 (H) nm, and obtained the routing of the scaffold and staple strands using caDNAno15 (Fig. 1A). The nanobrick consists of ten helices arranged in a honeycomb lattice structure. Previous reports of DNA–RNA hybrid origami used 10.67 or 11 bp per turn in the design of planar tiles,12,13 while short tubular structures were designed with a helical pitch of 10.5 bp per turn.13 Since our structure also contains tightly bundled helices, we used 10.5 bp per turn in our design. The RNA scaffold we used in this work is a 3′ untranslated region of a messenger RNA.16 We created the RNA scaffold (Table S1†) by in vitro transcription of a double stranded (ds) DNA template using T7 RNA polymerase (Fig. 1B and S1†). We used 12 DNA staple oligos (Table S2†) to fold the RNA scaffold into the designed nanobrick. The entire fabrication and purification process is shown in Fig. 1C.
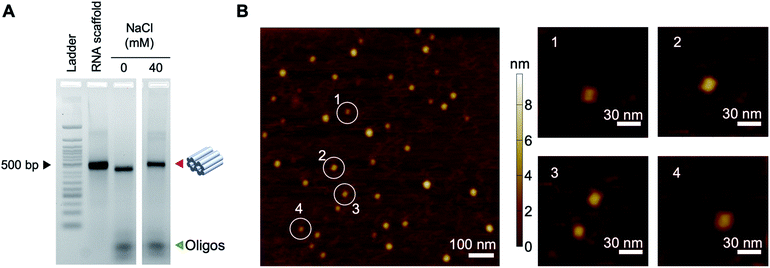
To assemble the origami structure, we mixed 50 nM RNA scaffold with 500 nM DNA staples (10-fold excess) in a solution containing 0.5× Tris-EDTA buffer (5 mM Tris, 1 mM EDTA, pH 7.5) and tested assembly in different concentrations of NaCl (0–60 mM) (Fig. S2†). Since RNA can be easily fragmented by magnesium at high temperatures, here we used NaCl for construction. The mixture was annealed from 65 °C to 20 °C over 12 hours (Table S3†). We observed similar assembly yields for the origami in 10–60 mM NaCl and chose the 40 mM condition for further experiments. The assembled structures were separated from excess DNA oligos by agarose gel electrophoresis (Fig. 2A). To further characterize the assembly, we extracted the origami structures from the gel band and verified the structures by dynamic light scattering (DLS) and atomic force microscopy (AFM). As determined by DLS, the size distribution profile of the origami nanobricks showed that ∼70% of the structures were ∼10 nm in dimension, in agreement with our design (Fig. S3†). Similarly, AFM height images of DNA–RNA nanostructures deposited and then dried on mica substrates (Fig. 2B and S4†) showed particles of geometry consistent with the folded nanostructures described schematically in Fig. 1A.
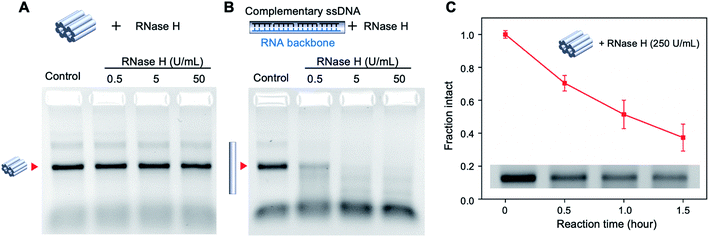
DNA nanostructures have shown remarkable resistance to the digestion by nucleases because of their compact structure and assembly.17 While a majority of DNA nanostructures have been tested against DNA nucleases such as DNase I,18–20 the nuclease resistance of DNA–RNA hybrid structures is less studied. We tested the stability of the mini origami nanobricks against ribonuclease H (RNase H), an enzyme that digests the RNA strand in the DNA–RNA hybrid duplex.21 According to the literature, the concentration of RNase in human blood is about 0.1 μg ml−1.22 We incubated the DNA–RNA hybrid nanobrick with different concentrations of RNase H at 37 °C for one hour and analyzed the structures using agarose gel electrophoresis (Fig. 3A). We quantified the band intensity corresponding to the origami structure to obtain the fraction of intact structure in different concentrations of RNase H (Fig. S5†). In addition, we designed a DNA–RNA duplex as a control structure using the same RNA scaffold and 10 DNA staple strands (Fig. 3B and Table S4†). The mini origami structure was intact even at 50 U ml−1 (∼1.25 μg ml−1) of RNase H while the duplex control of the same size was ∼20% digested with only 0.5 U ml−1 and completely digested in 5 U ml−1 of RNase H (Fig. 3A and B, S3†). This result is consistent with DNA–DNA origami, where origami structures with multi-helical bundles were more nuclease resistant compared to individual duplexes.2 We then tested the nuclease resistance of the origami nanobricks at a higher amount of RNase H (250 U ml−1) and at different reaction times (Fig. 3C and S6†). Even at this high nuclease concentration, the origami structure was still 40% intact after 1.5 h. These results further demonstrate the strong RNase H resistance ability of DNA–RNA origami nanostructures. Toward biological application, we also evaluated the stability of the origami nanobrick in cell culture medium by incubating the origami structure in DMEM containing 5% FBS at 37 °C (Fig. S7†). We observed that the origami structures were completely degraded in 5% DMEM and showed a trend of degradation over time in 1% DMEM.
In this paper, we presented the design, construction, and verification of a mini DNA–RNA hybrid origami nanobrick with ∼12 nm dimensions. The RNA scaffold we used for construction is a segment of messenger RNA that can be produced in short time by in vitro transcription. Such RNA scaffolds readily obtained from in vitro transcription or isolation of natural RNA molecules stands in contrast to the difficulty in preparing long, single DNA strands for DNA–DNA origami. Further, the DNA–RNA hybrid origami nanobrick shows enhanced resistance to RNase H digestion, rendering them useful in biological applications that require high biostability. This nuclease resistance can be further increased by modifying the component strands of the structures.23,24 The ability to produce self-folded RNA origami in vivo25 and the co-transcriptional folding of an RNA-scaffolded origami structure in vitro26 make these structure more useful in in vivo applications compared to all-DNA origami structures. The use of RNA in origami nanostructures also opens up the possibility of introducing additional functionalities such as aptamer-based ligand binding to nanostructures,27 RNA-protein interaction for cargo encapsulation,28 enzyme-mediated assembly and disassembly,9 and RNA–RNA kissing interactions for higher order assembly.29 Further, hybrid DNA–RNA nanostructures have found use in RNA interference30 and anticancer immunotherapy,31 while DNA nanostructures have used functional RNA molecules for in situ viral assembly on origami nanostructures.32 In addition, messenger RNA has shown great potential as vaccines that can significantly reduce the incidence of infectious diseases, such as COVID19,33 making RNA-scaffolded nanostructures useful in drug delivery. We envision that messenger RNA can also be packaged and delivered by DNA–RNA hybrid origami nanostructures with high efficiency in the near future.
Author contributions
L. Z. and K. H. conceived the project; L. Z. and K. H. designed experiments; V. V., J. S., M. Y., and J. I. F. obtained and analyzed the AFM images of the nanostructures; L. Z. and A. R. C. performed the nuclease digestion experiment, L. Z., A. R. C., and K. H. analyzed the data and wrote the manuscript, all authors provided comments and took part in editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Research reported in this publication was supported by the NIH through NIGMS under award R35GM124720 to K. H. and J. S. is supported by the NIH grant: R15GM124627. J. I. F. and M. Y. gratefully acknowledge support from the Donors of the American Chemical Society Petroleum Research Fund (#59835-DNI10) and start-up funds provided by The University at Albany, State University of New York. Support for atomic force microscopy is graciously provided by the Donors of the American Chemical Society Petroleum Research Fund grant #59835-DNI10. The authors also thank Wesley P. Wong and Darren Yang for assistance and training with the AFM.References
- P. W. K. Rothemund, Nature, 2006, 440, 297–302 Search PubMed.
- C. E. Castro, F. Kilchherr, D.-N. Kim, E. L. Shiao, T. Wauer, P. Wortmann, M. Bathe and H. Dietz, Nat. Methods, 2011, 8, 221–229 Search PubMed.
- J. Yan, C. Hu, X. Liu, J. Zhong, G. Sun and D. He, Curr. Pharm. Des., 2015, 21, 3181–3190 Search PubMed.
- L. Zhou, A. R. Chandrasekaran, J. A. Punnoose, G. Bonenfant, S. Charles, O. Levchenko, P. Badu, C. Cavaliere, C. T. Pager and K. Halvorsen, Sci. Adv., 2020, eabc6246 Search PubMed.
- P. Wang, S. Gaitanaros, S. Lee, M. Bathe, W. M. Shih and Y. Ke, J. Am. Chem. Soc., 2016, 138, 7733–7740 Search PubMed.
- P. Wang, T. A. Meyer, V. Pan, P. K. Dutta and Y. Ke, Chem, 2017, 2, 359–382 Search PubMed.
- P. Guo, Nat. Nanotechnol., 2010, 5, 833–842 Search PubMed.
- D. Jasinski, F. Haque, D. W. Binzel and P. Guo, ACS Nano, 2017, 11, 1142–1164 Search PubMed.
- S. Agarwal and E. Franco, J. Am. Chem. Soc., 2019, 141, 7831–7841 Search PubMed.
- A. Monferrer, D. Zhang, A. J. Lushnikov and T. Hermann, Nat. Commun., 2019, 10, 608 Search PubMed.
- S. H. Ko, M. Su, C. Zhang, A. E. Ribbe, W. Jiang and C. Mao, Nat. Chem., 2010, 2, 1050–1055 Search PubMed.
- P. Wang, S. H. Ko, C. Tian, C. Hao and C. Mao, Chem. Commun., 2013, 49, 5462–5464 Search PubMed.
- M. Endo, S. Yamamoto, K. Tatsumi, T. Emura, K. Hidaka and H. Sugiyama, Chem. Commun., 2013, 49, 2879–2881 Search PubMed.
- A. R. Chandrasekaran, M. Pushpanathan and K. Halvorsen, Mater. Lett., 2016, 170, 221–224 Search PubMed.
- S. M. Douglas, A. H. Marblestone, S. Teerapittayanon, A. Vazquez, G. M. Church and W. M. Shih, Nucleic Acids Res., 2009, 37, 5001–5006 Search PubMed.
- P. Flora, S. W. Wong-Deyrup, E. T. Martin, R. J. Palumbo, M. Nasrallah, A. Oligney, P. Blatt, D. Patel, G. Fuchs and P. Rangan, Cell Rep., 2018, 25, 3828–3843 Search PubMed.
- A. R. Chandrasekaran, Nat. Rev. Chem., 2021, 1–15 Search PubMed.
- J. Hahn, S. F. Wickham, W. M. Shih and S. D. Perrault, ACS Nano, 2014, 8, 8765–8775 Search PubMed.
- T. Gerling, M. Kube, B. Kick and H. Dietz, Sci. Adv., 2018, 4, eaau1157 Search PubMed.
- A. R. Chandrasekaran, J. Vilcapoma, P. Dey, S. W. Wong-Deyrup, B. K. Dey and K. Halvorsen, J. Am. Chem. Soc., 2020, 142, 6814–6821 Search PubMed.
- T. Tadokoro and S. Kanaya, FEBS J., 2009, 276, 1482–1493 Search PubMed.
- J. H. Connolly, R. M. Herriott and S. Gupta, Br. J. Exp. Pathol., 1962, 43, 402–408 Search PubMed.
- G. Zhu, L. Mei, H. D. Vishwasrao, O. Jacobson, Z. Wang, Y. Liu, B. C. Yung, X. Fu, A. Jin, G. Niu, Q. Wang, F. Zhang, H. Shroff and X. Chen, Nat. Commun., 2017, 8, 1482 Search PubMed.
- A. R. Chandrasekaran, J. Mathivanan, P. Ebrahimi, J. Vilcapoma, A. A. Chen, K. Halvorsen and J. Sheng, Nanoscale, 2020, 12, 21583–21590 Search PubMed.
- M. Li, M. Zheng, S. Wu, C. Tian, D. Liu, Y. Weizmann, W. Jiang, G. Wang and C. Mao, Nat. Commun., 2018, 9, 2196 Search PubMed.
- E. Torelli, J. Kozyra, B. Shirt-Ediss, L. Piantanida, K. Voïtchovsky and N. Krasnogor, ACS Synth. Biol., 2020, 9, 1682–1692 Search PubMed.
- S. Chen and T. Hermann, Nanoscale, 2020, 12, 3302–3307 Search PubMed.
- H. Ohno, T. Kobayashi, R. Kabata, K. Endo, T. Iwasa, S. H. Yoshimura, K. Takeyasu, T. Inoue and H. Saito, Nat. Nanotechnol., 2011, 6, 116–120 Search PubMed.
- D. Liu, C. W. Geary, G. Chen, Y. Shao, M. Li, C. Mao, E. S. Andersen, J. A. Piccirilli, P. W. K. Rothemund and Y. Weizmann, Nat. Chem., 2020, 12, 249–259 Search PubMed.
- K. A. Afonin, M. Viard, I. Kagiampakis, C. L. Case, M. A. Dobrovolskaia, J. Hofmann, A. Vrzak, M. Kireeva, W. K. Kasprzak, V. N. KewalRamani and B. A. Shapiro, ACS Nano, 2015, 9, 251–259 Search PubMed.
- X. Qi, X. Liu, L. Matiski, R. Rodriguez Del Villar, T. Yip, F. Zhang, S. Sokalingam, S. Jiang, L. Liu, H. Yan and Y. Chang, ACS Nano, 2020, 14, 4727–4740 Search PubMed.
- K. Zhou, Y. Ke and Q. Wang, J. Am. Chem. Soc., 2018, 140, 8074–8077 Search PubMed.
- N.-N. Zhang, X.-F. Li, Y.-Q. Deng, H. Zhao, Y.-J. Huang, G. Yang, W.-J. Huang, P. Gao, C. Zhou, R.-R. Zhang, Y. Guo, S.-H. Sun, H. Fan, S.-L. Zu, Q. Chen, Q. He, T.-S. Cao, X.-Y. Huang, H.-Y. Qiu, J.-H. Nie, Y. Jiang, H.-Y. Yan, Q. Ye, X. Zhong, X.-L. Xue, Z.-Y. Zha, D. Zhou, X. Yang, Y.-C. Wang, B. Ying and C.-F. Qin, Cell, 2020, 182, 1271–1283 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Methods and additional results. See DOI: 10.1039/d1na00026h |
| This journal is © The Royal Society of Chemistry 2021 |