Open Access Article
Open Access ArticleDetermination of sitagliptin in human plasma using a smart electrochemical sensor based on electroactive molecularly imprinted nanoparticles†
Isma
Haq
a,
Kaseb
Alanazi
b,
Joanna
Czulak
c,
Sabrina
Di Masi
 d,
Elena
Piletska
b,
Adnan
Mujahid
d,
Elena
Piletska
b,
Adnan
Mujahid
 a,
Tajamal
Hussain
a,
Sergey A.
Piletsky
b and
Alvaro
Garcia-Cruz
a,
Tajamal
Hussain
a,
Sergey A.
Piletsky
b and
Alvaro
Garcia-Cruz
 *b
*b
aInstitute of Chemistry, University of the Punjab, Quaid-e-Azam Campus, Lahore, Pakistan
bDepartment of Chemistry, University of Leicester, University Rd, Leicester, LE1 7RH, UK. E-mail: agc14@leicester.ac.uk
cMIP Diagnostics Ltd, Colworth Science Park, Sharnbrook, Bedford, MK44 1LQ, UK
dDepartment of Biological and Environmental Sciences and Technologies, University of Salento, Via Monteroni, Lecce, IT 73100, Italy
First published on 28th May 2021
Abstract
Sitagliptin is a hypoglycaemic agent used to reduce blood sugar levels in patients with type 2 diabetes mellitus (T2DM). Real time monitoring of sitagliptin levels is crucial to prevent overdose, which might cause liver, kidney and pancreatic diseases. As an alternative solution, a sitagliptin voltammetric sensor was fabricated using artificial receptors called electroactive molecularly imprinted polymer nanoparticles (nanoMIPs). The nanoMIP tagged with a redox probe (ferrocene) combines both the recognition and reporting functions. Traditional electrochemical sensors determine the redox activity of an analyte. Thus, they are influenced by interfering molecules and the nature of the sample. These innovative nanoMIPs allow us to easily design and customise sensors, increase their sensitivity and minimise the cross reactivity in biological samples. The present technology replaces the traditional enzyme–mediator pairs used in traditional biosensors. The polymer composition was optimized “in silico” using docking and screening methods. Nanoparticles were synthesized via free radical polymerization and a solid phase method and then characterized by infrared spectroscopy (FTIR), transmission electron microscopy (TEM) and dynamic light scattering (DLS). The specific sitagliptin nanoparticles were covalently immobilized on platinum electrodes via silane and carbodiimide chemistry. The determination of sitagliptin in human plasma by a nanoMIP sensor was assessed by differential pulse voltammetry (DPV). The sensor current response was directly related to the change in nanoMIP conformation triggered by the analyte. The optimisation of the sensor response was made by adjusting (i) the silane concentration, (ii) nanoMIP concentration, and (iii) immobilization time. The sensor measurements in plasma revealed high selectivity and a sensitivity of 32.5 ± 0.6 nA pM−1 towards sitagliptin, and the limit of detection of the fabricated sensor was found to be 0.06 pM. The sensor displayed a satisfactory performance for the determination of sitagliptin in spiked human plasma, demonstrating the potential of this technology for drug monitoring and clinical diagnosis.
1. Introduction
Type 2 diabetes mellitus (T2DM) is an increasing global health problem, closely linked to the epidemic of obesity and genetic predisposition.1,2 Over the past three decades, the number of people with diabetes mellitus has more than doubled globally and mostly observed among children, adolescents and younger adults.3 T2DM is a chronic disease in human metabolism that causes an unusual increase in the blood sugar level due to insulin resistance, which is a pancreatic hormone that regulates body metabolism.2 No cure has yet been found for T2DM. The multiple pathogenetic disturbances involved in T2DM require the use of a combination of lifestyle modifications and multiple antidiabetic agents to maintain normoglycaemia.1Predominantly, sitagliptin and metformin are commonly used together to improve the blood sugar levels in adults with T2DM.2,4–6 Sitagliptin regulates the levels of insulin in the body produced after eating and reduces blood sugar levels.7,8 Additionally, metformin decreases the glucose production in the liver and the absorption of glucose by the intestines.9 It is very important to monitor sitagliptin blood levels because periodic doses can seriously disturb the immune system.10 Sitagliptin can also be associated with the development of serious hypersensitivity reactions including e.g. anaphylaxis, angioedema and serious skin reactions.11,12 To determine sitagliptin in human plasma, several analytical methods are used; the most common include chromatography,13,14 tandem mass spectrometry,15,16 HPLC,17,18 spectrofluorometry,19 and spectrophotometry.20,21 The existing methods are summarized in Table 1. These techniques present high sensitivity with low detection limits; nonetheless, these methods are time consuming due to the tedious protocols and extensive sample pre-treatment steps that limit their use for point of care (PoC) diagnostics. Thus, there is a need for an efficient, rapid and low cost method for sitagliptin determination.
| Detection method | Linear dynamic range | LOD | RSD (%) | Accuracy (%) | Recovery (%) | Ref. |
|---|---|---|---|---|---|---|
| a High performance liquid chromatography mass spectrometry (HPLC-MS), high turbulence liquid chromatography (HTLC), capillary zone electrophoresis (CZE), liquid chromatography/mass spectrometry (LC/MS), and reversed-phase liquid chromatography (RPLC). | ||||||
| HPLC-MS/MS | 9.9–1583 nM | 9.89 nM | <7.5 | 101.23 | ≥64 | 16 |
| HTLC-MS/MS | 0.02–12.26 nM | 0.025 nM | <4.2 | 99.2 | 99.5 | 17 |
| CZE | 19.1–191 μM | 0.936 μM | ≤1.50 | 100.4 | 99.81 | 22 |
| Spectrofluorometry | 99–1385 nM | 32.5 nM | <2 | 99.3 | 99.87 | 23 |
| Spectrofluorometry | 0.4–2.7 μM | 61.4 nM | 0.96 | 100.2 | 100.54 | 24 |
| LC-MS/MS | 2.5–2455 nM | 2.5 nM | 2.68 | 100.3 | 100.01 | 25 |
| LC-MS/MS | 24.5–1227 nM | 0.417 nM | <1 | >90 | 99.08 | 26 |
| LC-MS/MS | 0.25–1230 nM | 0.736 nM | <15 | 110 | 102.1 | 27 |
| HPLC | 0.19–5.73 μM | 0.134 μM | <5 | 100.07 | 101.41 | 28 |
| RP-LC | 0.5–15.83 μM | 0.099 μM | <2 | 99.6 | 100.81 | 29 |
To overcome these analytical and practical challenges, electrochemical sensors have been proposed.30 The electrochemical sensors are a growing approach offering several benefits over traditional analytical techniques such as miniaturization of devices, portability, rapid response, reproducibility and high sensitivity.31 In addition, electrochemical sensing is a straightforward and effective approach for drug determination for PoC diagnostics.32 The classical approach in electrochemistry for drug determination is the oxidation of the target analyte.33–35 Alternatively, the indirect detection of the target is possible by using an external redox probe in solution.30,36 Both electrochemical methods present disadvantages such as poor performance due to the matrix effect, deposition of by products and cross-reactivity.37 The matrix effect is the signal distortion in a reading, that occurs during the analysis of biological samples (urine, plasma, saliva, etc.), caused by the presence of cells, biomolecules, heterogeneity in ionic strength, pH, temperature, etc.38
Molecularly imprinted polymers (MIPs) were employed successfully as biomimetic receptors in sensors for range of different targets.39–41 These artificial receptors selectively bind to the target analyte and give a highly sensitive response when immobilized on electrochemical detectors.42–45 Nevertheless, there is still plenty of room for improvement of device integration and fabrication. The present technology is an improvement of the “classic” MIP technology and provides an opportunity to develop a superior alternative to existing biosensors.46
Previous MIP sensor technology incorporated ferrocene as a redox marker into rigid MIP films and hard MIP microbeads prepared by precipitation polymerization methods.47–49 The working principle of these polymers is known as the “gate effect”, which is frequently used in sensors as a result of changes in the morphology, diffusion and permeability of the polymer triggered by specific interactions with the template molecule.50,51 However, the operation mode based on the permeability of the polymeric membrane leads to the saturation of the sensors and limits their use due to the solvents, pH and buffers employed. Also, these sensors show cross-reactivity especially in biological samples, limiting their applications and performance in clinical applications.30
The specific actuation of the polymers and selective binding are obtained only at the nanoscale using electroactive molecularly imprinted polymer nanoparticles (nanoMIPs), easily prepared by solid phase synthesis.52 The operating mode of the nanoMIPs relies on polymer actuation triggered by the recognition of the analyte.53 Consequently, the analyte binds to the nanoMIPs prompting a polymer conformational change, which leads to the exposure of the ferrocene moieties and the increase of the electron transfer at the electrode surface.50,54 Thus, swelling (or shrinking) would affect the density of ferrocene moieties exposed on the surface of nanoparticles that are anchored onto the electrode, thus affecting the electron transfer rate.50 Consequently, the analyte recognition event generates a detectable signal monitored via electrochemical techniques. As a result, the sensor current response is directly related to the analyte concentration, increasing the sensitivity and minimising the cross reactivity in biological samples.54
Therefore, the present approach eliminates the necessity of mediators or enzymes employed in traditional biosensors. The present technology uses an advanced type of nanoMIP, which combines the receptor and signalling functions.50 For this reason, the nanoMIP formulation includes an electrochemically active ferrocene monomer (FcMMA).55,56 The nanoMIP actuation mechanism allows us to fabricate sensors with high selectivity and sensitivity in biological samples.50,54 Therefore, the concentration of the analyte can be measured using voltammetric techniques such as differential pulse voltammetry (DPV) by following the nanoMIP electro-activity (Scheme 1). This detection method is very convenient because it does not require monitoring the redox activity of the analyte, which might be affected by interferents present in biological samples.
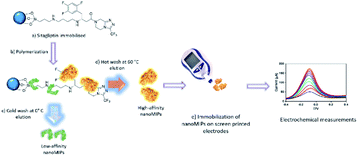 | ||
| Scheme 1 Preparation and elution of high affinity sitagliptin nanoMIPs (a) to (d) and (e) their use in the fabrication of sitagliptin sensors. | ||
Herein, a sensor for sitagliptin was devised using electroactive nanoMIPs. The specific sitagliptin nanoMIPs were synthesized via free radical polymerization using the well-established solid phase synthesis of MIP nanoparticles.52 Afterwards, the voltammetric sensors were prepared by immobilizing nanoMIPs on screen-printed platinum electrodes (SPPE) via silane and carbodiimide chemistry as shown in Scheme 1. Finally, the voltammetric sensors were successfully applied for the selective determination of sitagliptin in spiked human plasma samples. In this work, various factors, such as the nanoMIP concentration and immobilization conditions, affecting the sensor response were investigated and optimized. The technology presented here has several advantages, such as simple preparation, portability, high sensitivity and selectivity, extended shelf life and easy operation.
2. Experimental
2.1 Chemicals and reagents
Ammonium persulfate (APS), 1,2-bis(triethoxysilyl)ethane (BTSE), N-(2-aminoethyl)-3-aminopropyltrimethoxysilane (DAMO), 5-(dimethylamino)naphthalene-1-sulfonyl chloride (DNSCl), ethanol amine, ethylene glycol dimethyl bisacrylate (EGDMA), 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (EDC), ferrocenylmethyl methacrylate (FcMMA), N,N-diethyldithiocarbamic acid benzyl ester (iniferter), methacrylic acid (MAA), N,N′-methylene-bis-acrylamide (MBA), N-(3-aminopropyl)methacrylamide hydrochloride (NAPMA), N-isopropylacrylamide (NIPAM), sodium cyanoborohydride (NaBH3CN), sodium hydroxide (NaOH), N-hydroxysuccinimide (NHS), phosphate buffered saline (PBS), pentaerythritol tetrakis(3-mercaptopropionate) (PETMP), sulphuric acid (H2SO4), tetramethyl-ethylene-diamine (TEMED), 2-(trifluoromethyl)acrylic acid (TFMAA), trimethylolpropane trimethacrylate (TRIM), sitagliptin and metformin were purchased from Sigma-Aldrich, UK. Glass microspheres, SPHERIGLASS® A-Glass 2429 (106–150 μm diameter), were purchased from Potters Industries LLC. Ethylene glycol methacrylate phosphate (EGMP) was acquired from Alfa-chemistry, USA. Double-distilled ultrapure water (Millipore, UK) was used for all experiments. All solvents and chemicals were of HPLC or analytical grade and used without purification.2.2 Instrumentation
Nitrogen plasma was used to activate and clean the surface of screen printed electrodes (Emitech, K1050X RF Plasma Cleaner, 50 W, 13.56 MHz RF for 5 min). Transmission electron microscopy (TEM) images were obtained by using a JEOL JEM-1400 TEM (accelerating voltage of 100 kV) coupled with a Megaview III digital camera with iTEM software. Dynamic light scattering (DLS) measurements were performed using a Zetasizer Nano (Nano-S) from Richmond Scientific Instruments Ltd (Lancashire, UK). For these measurements, 1 mL solution of nanoparticles was ultra-sonicated for 3 min in order to disrupt potential agglomerates. The infrared spectroscopy analysis was performed using a Bruker Alpha platinum-ATR FTIR spectrometer. For the elution of nanoMIPs, solid phase extraction (SPE) tubes with a polyethylene frit (20 μm porosity, Supelco), disposable plastic syringes with a cellulose acetate syringe filter (25 mm, 0.45 μm, Whatman) and a SnakeSkin dialysis tubing (10 K MWCO, 22 mm diameter, 11 cm tubing length, 3.3 mL cm−1 Thermo Scientific) were used. The screen printed platinum electrodes (SPPE, Dropsens DRP-550, Metrohm, UK) with dimensions of 3.4 × 1.0 × 0.05 cm (length × width × height) were used for the sensor development. All the electrochemical measurements were performed by using a potentiostat/galvanostat/impedance analyzer (PalmSens4) equipped with a cable connector for screen-printed electrodes. The PSTrace software (PalmSens, Netherlands) controlled the data acquisition.2.3 Molecular modelling
Computational modelling of imprinted polymers was pioneered by our group, and it is based on the screening and selection of functional monomers using molecular mechanics.57,58 Molecular modelling was carried out in an HP Elite-Desk with two Intel Core™Duo CPU E8400 and 3 GHz processors running on a CentOS Linux 7 operating system. The software package used was Sybil™ version 7.3 (Tripos Inc.) in a Gnome 2.28.2 (Linux) environment. The sitagliptin molecule was screened against a database of common functional monomers used in MIP synthesis using the Leapfrog automated method available in the Sybil molecular modelling package.59,60 The database used consisted of 44 monomers, representing charged and neutral forms, and the screening was performed for 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 iterations.
000 iterations.
2.4 Synthesis of nanoMIPs
2.5 Electrochemical measurements
The electrochemical response of sensors was investigated by using differential pulse voltammetry (DPV) in the potential range from −0.4 to 0.4 V (vs. Ag/AgCl), scan rate of 33 mV s−1, modulation amplitude of 200 mV, modulation time at 20 ms and step potential of 50 mV. Samples (100 μL) were analyzed by drop casting on the sensor surface and incubating for 3 minutes. Then, the DPV measurements were assessed. After measuring the voltammetric sensor response, the data were normalized by calculating the current change response using the following equation: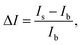 , where “ΔI” represents the current change response, “Is” indicates the current response of the sample, and “Ib”, the current response for zero concentration of sitagliptin, background signal (buffer or plasma). Thus, calibration plots were obtained by representing a normalized current change response (ΔI) against the sitagliptin concentration. The limit of detection (LOD) was calculated conventionally from calibration curves (3 × SD/slope) and each measurement was done in replicate. The limit of quantification (LOQ) was calculated as follows: (10 × SD/slope).
, where “ΔI” represents the current change response, “Is” indicates the current response of the sample, and “Ib”, the current response for zero concentration of sitagliptin, background signal (buffer or plasma). Thus, calibration plots were obtained by representing a normalized current change response (ΔI) against the sitagliptin concentration. The limit of detection (LOD) was calculated conventionally from calibration curves (3 × SD/slope) and each measurement was done in replicate. The limit of quantification (LOQ) was calculated as follows: (10 × SD/slope).
2.6 Determination of sitagliptin in human plasma samples
Sitagliptin standard solutions were prepared in PBS buffer (5 mM, pH 7.2) in a concentration range of 100 to 2000 pM. The spiked human plasma was prepared as follows: firstly, 0.5 mL of human plasma was diluted in 5 mL of 10 mM PBS (pH 7.2). After agitating for 3 min with a vortex, samples were centrifuged for 3 min at 3500 rpm. As a result, the supernatant was recovered and filtered using a syringe with a micro membrane. Finally, the human plasma was spiked in the concentration range from 100 pM to 2000 pM of sitagliptin.3. Results and discussion
3.1 Molecular modelling
Molecular modelling is particularly essential for the design of polymer compositions that involve different monomers and for enhancing the molecular recognition of a target.58 First, the sitagliptin structure was minimized (Fig. S2†) and then screened against the monomer database. The results obtained after the monomer screening resulted in five potential functional monomers, TFMAA, MBA, EGMP, MAA and NAPMA as shown in Table S2.† From that selection, the highest binding scores were found for the combination of TFMAA (−37.6 kJ mol−1) and MBA (−31.84 kJ mol−1) as shown in Fig. S2.† The combination of MAA (−28.18 kJ mol−1) and EGMP (−29.32 kJ mol−1) also showed high binding scores. The TFMAA and MAA monomers interact mainly with the primary amine group of sitagliptin. Additionally, TFMAA interacts with the amino group through polar interactions and with the carbonyl group as well by Bürgi–Dunitz interactions. Similarly, MBA contributes to the polymer interaction with the sitagliptin molecule via dipole–dipole and hydrogen bond interactions in the triazole cycle. The EGMP monomer interacts with the primary amine group predominately via hydrogen bonds. The NAPMA monomer interacts with the fluorine groups by polar interactions and with the carbonyl group by hydrogen bonds. Besides, NAPMA is used to confer thermos-responsive properties to the polymers.61,62 This feature is used to remove the nanoparticles from the solid phase as shown in Scheme 1. From these calculations, two formulations were proposed, nanoMIP-1 involving the MAA–sitagliptin–MBA complex (Fig. S3†) and the second formulation (nanoMIP-2) including the TFMAA–sitagliptin–MBA complex (Fig. S2†).3.2 Characterization of nanoparticles
The chemical structure of nanoMIPs was analyzed by ATR-FTIR spectroscopy as shown in Fig. S4.† NanoMIP-1 spectra display the characteristic polyacrylamide network structure bands at 1296 cm−1 (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N, δ), 1309 cm−1 (C–N, δ), and 1722 cm−1 (C
C–N, δ), 1309 cm−1 (C–N, δ), and 1722 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, δ). The polymer hydrocarbon bands appeared at 449, 2666, and 2783 cm−1 for (C–H, δ) and also (C–H, ν) at 956 and 1477 cm−1. In addition, the carboxylic group from MAA was observed at 2967 cm−1 (O–H, δ). However, nanoMIP-2 spectra showed distinctive bands from the alkane chain at 438 cm−1 (C–H, δ), 809, 944 and 1457 cm−1 (C–H, ν). The polyacrylamide bands appeared at 1292 cm−1 (
O, δ). The polymer hydrocarbon bands appeared at 449, 2666, and 2783 cm−1 for (C–H, δ) and also (C–H, ν) at 956 and 1477 cm−1. In addition, the carboxylic group from MAA was observed at 2967 cm−1 (O–H, δ). However, nanoMIP-2 spectra showed distinctive bands from the alkane chain at 438 cm−1 (C–H, δ), 809, 944 and 1457 cm−1 (C–H, ν). The polyacrylamide bands appeared at 1292 cm−1 (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N, δ), 1304 cm−1 (C–N, δ), and 1718 cm−1 (C
C–N, δ), 1304 cm−1 (C–N, δ), and 1718 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, δ). The carboxylic group from TFMAA was observed at 2929 cm−1 (O–H, δ). The amine groups from NAPMA monomer were observed at 3304 cm−1 (N–H, δ).
O, δ). The carboxylic group from TFMAA was observed at 2929 cm−1 (O–H, δ). The amine groups from NAPMA monomer were observed at 3304 cm−1 (N–H, δ).
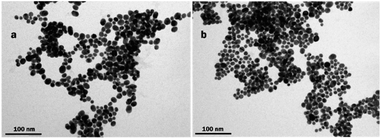
Transmission electron microscopy (TEM) analysis revealed highly aggregated spherical homogeneous nanoparticles with a diameter of 15 ± 4.5 nm for nanoMIP-1 and 21 ± 3.2 nm for nanoMIP-2 as shown in Fig. 1. The nanoparticles observed are aggregated and smaller than expected. Presumably, the TEM sample preparation, which involves water removal and ethanol evaporation, induced a collapse of the nanostructure and prompted agglomeration of particles into condensed aggregates. These particles were also analyzed in water solutions by dynamic light scattering (DLS). The hydrodynamic size measurements displayed uniform populations with a satisfactory poly-dispersity index (≤0.36) and diameter of 233 ± 5.8 nm for nanoMIP-1 and 193 ± 9.5 nm for nanoMIP-2. Seemingly, these nanoMIPs are present mainly as homogenous clusters in solution and not as individual particles. It was also noticed that the hydration of the polymer produced swelling increasing the volume of the cluster to almost 90% as previously reported for polymers containing NAPMA.45,46 The nanoparticle size analysis is summarized in Table S3.†
3.3 Optimisation of the sensor
As synthetic recognition elements, nanoMIPs have a key role in the functioning of the electrochemical sensor; thus the immobilization of this polymer sensing layer is crucial for the sensor performance. Therefore, they are key parameters affecting the covalent immobilization of the nanoMIPs, particularly the concentration of silanes (APTES), nanoMIP concentration and the incubation time. These parameters were studied and the resulting sensors were tested in a concentration range from 400 to 2000 pM of sitagliptin. The APTES concentration was studied at 2%, 6% and 10%. The optimal sensor sensitivity was found at 6% APTES as shown in Fig. S5.† It was noticed that, low concentrations of silane led to poor immobilization of nanoMIPs on the electrodes, which reduce the sensor response. Oppositely, the excess of silane produced some intramolecular interactions causing aggregation.63 Seemingly, these interactions lead to the formation of multi-layered silane structures, and as a result, an isolation coating was deposited on the working electrode, which reduced the charge transfer, and consequently, the sensor response was reduced as well.The concentration of nanoMIPs and time of immobilisation were optimized. The enhanced sensor response was demonstrated when the concentration of nanoMIPs was set at 0.5 mg mL−1 (Fig. S6A†). Presumably, sensors with low concentrations of nanoparticles showed reduced performance, and higher concentrations of nanoMIPs lead to a decrease of sensitivity. Certainly, the excess of particles induces aggregations and also a deposition of multiple polymer layers. Therefore, recognition sites are lost and actuation of the nanoMIPs is hindered, resulting in a decrease of the electron transfer, and therefore, a decrease of the sensor response was observed. The optimal time for the immobilisation of nanoparticles on SPPE that gave a linear response was 20 h. The sensor response did not change dramatically after 2 h (sensor response at 85%, R2 = 0.95) and 4 h (sensor response at 79%, R2 = 0.68) of immobilization as shown in Fig. S6B.† From these results, it can be concluded that the carbodiimide reaction and formation of the sensing layer is a fast process. Nevertheless, the linear response was improved after 20 h (sensor response at 100%, R2 = 0.99). To conclude, the FcMMA concentration was also optimized. For that, sensors were prepared with 15, 20, and 25% of the molar polymer composition. The 20% molar content showed the highest sensitivity (Fig. S7†). The optimized parameters are summarized in Table S4.†
3.4 Electrochemical measurements of the fabricated nanoMIP sensors
The voltammetric response of the nanoMIP sensors to sitagliptin was analyzed under optimized conditions. The sitagliptin was measured in a concentration range from 100 to 2000 pM in PBS buffer (5 mM, pH 7.2). The DPV response was recorded and calibration curve was plotted for both sensors, nanoMIP-1 and nanoMIP-2 as shown in Fig. S8.†The sensor response relied on the electroactivity of the nanoMIPs conferred by the ferrocene redox marker in the polymer structure. The nanoMIP sensor displayed a current increase triggered by the analyte recognition. Presumably, the recognition prompted changes in the polymer conformation promoting the electron transfer of the ferrocene moieties, which are responsible for the overall observed electrochemical processes. The nanoMIP actuating process is similar to the “induce fit” mechanism in enzymes.64,65 NanoMIP actuation is dependent on the analyte's selective recognition, which results in a polymer conformation change and swelling, causing ferrocene moieties to be exposed at the particle surface. Thus, polymer swelling is directly proportional to the analyte concentration, changing the density of ferrocene exposed on the surface of the nanoparticles. This polymer alteration stimulates electron transfer, which is responsible for the overall electrochemical processes detected in the sensor, and displayed as an increase of the current response. The specific “induce fit” mechanism in nanoMIPs was previously evidenced by measuring the increase of the diameter of nanoparticles using dynamic light scattering (DLS). In this study, actuation was detected only in presence of the analyte, while no changes were observed in presence of other molecules as described in the manuscript published earlier.50
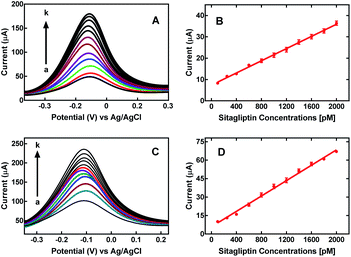
Therefore, the nanoMIPs tagged with a redox probe combines both recognition and reporting functions. The nanoMIP signal was observed at −0.1 V (vs. Ag/AgCl), corresponding to the ferrocene redox marker as shown in Fig. S8.† The signal increments were directly correlated to the sitagliptin concentration. During the recognition process, the analyte remained intact, since the sitagliptin electro-oxidation was not observed at this working potential (−0.4 to +0.4 V vs. Ag/AgCl) as shown in Fig. S9.†
Both sensors, nanoMIP-1 and nanoMIP-2, showed an increase of response directly proportional to the sitagliptin concentration (Fig. S8†). The sensors showed a satisfactory linear response; the nanoMIP-1 sensor displayed a sensitivity of 65 ± 1 nA pM−1 (R2 = 0.998) with a relative standard deviation (RSD) of 4.7%. Similarly, the nanoMIP-2 sensor sensitivity was found to be 60 ± 1 nA pM−1 (R2 = 0.996) with a RSD of 4.3%. The LOD and LOQ of sensors were found to be 0.05 pM and 0.18 pM for the nanoMIP-1 sensor and 0.06 pM and 0.20 pM for the nanoMIP-2 sensor. From these parameters, it can be concluded that both sensors exhibited similar performance for sitagliptin determination.
3.5 Cross reactivity of the fabricated nanoMIP sensors
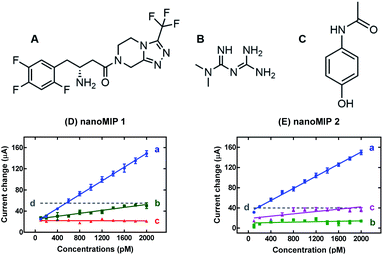
The selectivity of the sensors was tested by measuring the response towards possible interferents such as metformin and paracetamol and then compared to the sitagliptin sensor response. These results were represented as calibration plots in Fig. 3. The sensors displayed different selectivity, apparently caused by the polymer composition. Presumably, small molecules can interact non-specifically with the nanoMIPs, and these interactions may include hydrogen bonds and van der Waals depending on the polymer formulation and the properties of the interfering compounds.The response of both polymers is similar in terms of sensitivity towards sitagliptin as shown in Fig. S8.† From these results, it can be observed that nanoMIP-2 exhibited higher selectivity to sitagliptin, when compared to nanoMIP-1. Moreover, the selectivity of nanoMIP-1 was inferior in the presence of metformin or paracetamol as presented in Fig. S10 and S11.† Thus, nanoMIP-1 showed a cross reactivity of 21.5% to metformin (R2 = 0.857) and 0.5% to paracetamol (R2 = 0.011). Oppositely, the nanoMIP-2 sensor displayed a cross reactivity of 19.2% to paracetamol (R2 = 0.687) and 3.5% to metformin (R2 = 0.117). Nevertheless, the sensor response to interferents was non-specific and not linear. These results revealed that nanoMIP-2 was 29 times more sensitive to sitagliptin when compared to the metformin response, as summarized in Table S5.†
Alternatively, the interferents can be analytically removed from sensors by placing a “current cut-off” at 55 and 40 μA for the nanoMIP-1 sensor and nanoMIP-2 sensor, respectively. In this case, the concentration working range is reduced for nanoMIP-1 from 600 to 2000 pM and for the nanoMIP-2 sensor from 200 to 2000 pM as shown in Fig. 2. Therefore, the nanoMIP-2 sensor can be successfully used for the selective determination of sitagliptin without any significant interference from metformin. This strategy can be applied in the case of real scenarios. This aspect is important because sitagliptin is used mostly in binary combination with metformin for therapeutic purposes.
The cross-reactivity was mainly observed in nanoMIP-1, apparently due to the polymer structure. Seemingly, the negligible response to paracetamol was due to the non-specific interactions with the recognition sites such as dipole interactions with the polymer chain or non-specific adsorption on the sensor surface. The potential interference of metformin was apparently due to the small size of the molecule (Fig. 2), allowing it to interact non-specifically and reversibly with the nanoMIP cavity. Possible interactions included hydrogen bonds, electrostatic and dipole interactions.
Moreover, the nanoMIP-1 interaction with sitagliptin was less stable (9.4 kJ mol−1) when compared to the nanoMIP-2 interaction, as previously shown in the molecular modelling section (Fig. S3†). Apparently, the TFMAA monomer (nanoMIP-2) played an important role in the sitagliptin recognition, which cannot be replaced with the MAA monomer (nanoMIP-1), as shown in the molecular modelling results. Seemingly, the nanoMIP-2 interaction with sitagliptin is a specific key-lock system, where there is a poor interaction with metformin. This selective interaction is possibly directed by the multiple hydrogen bond system formed in the sitagliptin complex, as shown in Fig. S2b.† As a result, only nanoMIP-2 was considered for further applications.
3.6 Determination of sitagliptin in human plasma samples
The sensor was evaluated for the determination of sitagliptin in spiked human plasma samples in a concentration range from 100 to 2000 pM. Before the analysis, samples were diluted, centrifuged and filtered as described in the Experimental section. The DPV measurements were carried out under the optimal conditions and calibration curves were obtained to analyse the sensor performance. The sensitivity was found to be 14.4 ± 0.4 nA pM−1 (R2 = 0.994) and 32.5 ± 0.6 nA pM−1 (R2 = 0.996) for nanoMIP-1 and nanoMIP-2, respectively. The sensitivity and linearity was higher for nanoMIP-2 when compared to nanoMIP-1, as shown in Fig. 3. Human plasma is a complex matrix and difficult to analyse due to the presence of biological components. However, it was possible to determine sitagliptin using nanoMIP-2 with a LOD and LOQ of 0.06 pM and 0.20 pM, respectively. Therefore, the nanoMIP-2 sensor can be successfully used for sitagliptin determination in human plasma with high sensitivity in the presence of interfering substances. Potentially, the sensor can be applied for point of care (PoC) diagnostics in the clinically relevant concentration, which is reported in the range from 24.5 nM to 950 mM in human plasma.24,28,66,673.7 Performance of the nanoMIP sensor
The nanoMIP-2 sensor performance, the repeatability, reproducibility, and stability were evaluated. The repeatability was evaluated by calculating the relative standard deviation (RSD%) of three sensor measurements of a sitagliptin standard (1200 pM) in buffer under the same conditions. The repeatability of the measurements for a single sensor was found to be 6.6%. The reproducibility was also assessed by calculating the percentage of the RSD of the response of four different sensors to a sitagliptin standard (1200 pM) under identical conditions, and it was found to be 7.3%. The previous analysis proves that the sensor performance is in compliance with the industrial accuracy standards (ISO-151917), which recommends that 95% of the results should be within ±15% of the laboratory standard.The stability and shelf life of the nanoMIP-2 sensor were also assessed by evaluating the performance of the stored sensors. For this, nine sensors were prepared under similar conditions and then stored. The sensor current response to sitagliptin was tested in a concentration range from 200 to 2000 pM. Each individual sensor (1–9) was tested independently for a period ranging from 1 to 112 days. The stability of the sensors is summarized in Fig. S12;† here, the average response and standard deviation is displayed per sensor per day. According to these results, after 30 days, the sensor response decreased by only 6%, and after 60 days, the drop in response was around 15%, which is in compliance with the industrial standards. The overall decrease in the sensor response after 112 days of storage was 27%; nevertheless, the sensors remained functional. The stability of the proposed sensor device is higher compared to that of other devices as described in Table 2. To overcome the sensor drop response, the measured current can be compensated by applying the storage factor to obtain the equivalent response. Eventually, the storage conditions of the present sensors need to be improved.
| Transducer | PVC-membrane/ammonium reineckate | PVC-membrane/calix-8-arene | MIP membrane | NanoMIPa |
|---|---|---|---|---|
| a NanoMIP advantages: (a) high selectivity and sensitivity, (b) relevant to the clinical concentration range, (d) robustness and high stability, (e) compatible with industrial production, and (f) fast measurements. | ||||
| Detection method | Potentiometry | Potentiometry | Potentiometry | Voltammetry |
| Linear concentration range | 0.01–10 mM | 0.1 nM to 10 mM | 5 μM to 10 mM | 100–2000 pM |
| LOD | 2 μM | 0.11 nM | 2.6 μM | 0.06 pM |
| Sensitivity | 40.9 mV per decade | 59.9 mV per decade | 52.7 mV per decade | 32.5 nA pM−1 |
| Stability (days) | 30 | 31 | 15 | 112 |
| Sample | Tablets | Human plasma | Human plasma | Human plasma |
| Recovery (%) | 100.06 | 100.33 | 97.3 | 98.7 |
| Ref. | 68 | 69 | 70 | This work |
3.8 Comparison to the available sensor technology
Different transducers and fabrication strategies have been used to fabricate sitagliptin sensors and monitor the levels in human plasma (Table 2). Some of them include the combination of MIPs and macromolecules to increase the selectivity. Unfortunately these methods face low selectivity and a lack of adequate sensitivity.There is a clear need for improving the compatibility of the fabrication process. For that, the sensors need to be simplified and the complexity of traditional systems reduced. The “classic” MIP-based sensor technology is difficult to apply in mass manufacturing or scaling up.44,71 Consequently, these systems are not useful for PoC applications. To address these technological challenges, the present technology uses electroactive nanoMIPs. NanoMIPs confer several advantages when compared to previous MIP technology and traditional biosensors. For instance, nanoMIPs are precisely designed for corresponding targets using molecular modelling. Besides, the electro-activity of nanoMIPs permits the direct recognition of analytes and reporting of binding events at the same time, which results in higher sensitivity and low cross-reactivity even when measuring in complex biological samples.
The key advantages of the proposed technology are the following: (a) nanoMIPs can be designed computationally for a large range of targets; (b) their polymeric nature make them robust and resistant to harsh conditions, and they possess long shelf-life; (c) their synthetic nature make them easy to be prepared and fabricates at relatively low cost using high throughput industry-standard protocols; (d) the synthetic nature of the nanoMIPs makes them ideal for industrial production which does not require the use of animals (or recombinant DNA technology) when compared to the production of antibodies and enzymes. The sensor presented here displays higher performance in terms of sensitivity, selectivity and stability of the sensor as shown in Table 2. The LOD and LOQ of the nanoMIP-2 sensor were 0.10 pM and 0.34 pM in the PBS buffer and 0.12 pM and 0.42 pM for the spiked human plasma, respectively, which is way below the clinically relevant concentration of sitagliptin concentration in human plasma (24.5 nM to 950 mM). The sensing ranges of the nanoMIP-based electrodes could easily be reached by diluting the plasma sample in PBS buffer.
4. Conclusions
A sitagliptin sensor was devised using nanoMIPs immobilized on SPPE. Herein, the nanoMIP capability as an artificial biomimetic receptor for the determination of sitagliptin was demonstrated. This nanoMIP-2 sensor shows high sensitivity and selectivity towards sitagliptin due to specific recognition binding sites imprinted in nanoMIPs. The nanoMIPs that contain ferrocene as a redox probe combine both the recognition and the reporting functions. The proposed sitagliptin sensor system is compact, simple and cost effective when compared to other traditional analytical methods. The sensors exhibit a shelf life of almost 4 months (at 4 ± 0.5 °C and 30% relative humidity), remaining functional even after that time. The nanoMIP-based sensor technology has tremendous potential, which could have a positive impact in drug monitoring, forensics, in the clinical area and point-of-care diagnostics.The present technology is a step forward in developing robust sensors, compatible with industrial production. Future work will include the improvement of the fabrication process including industrial techniques such as screen printing of electrodes using conductive inks (e.g. graphene or carbon), and further optimization of the storage conditions. These sensors can be operated as a near patient test by relatively inexperienced personnel, and a design of a user-friendly interface will be considered. Thus, future work will also include efforts to adapt this device to a hospital environment.
Author contributions
I. Haq: methodology, investigation, data curation, writing – original draft preparation, writing – reviewing and editing. K. Alanazi: investigation. A. Mujahid: supervision, reviewing and editing. J. Czulak: software (molecular modelling). S. Di-Masi: reviewing and editing. T. Hussain: reviewing. E. Piletska: conceptualization, methodology, supervision. S. A. Piletsky: conceptualization, methodology, supervision. A. Garcia-Cruz: conceptualization, methodology, investigation, data curation, writing – original draft preparation, visualization, writing – reviewing and editing, project administration.Conflicts of interest
They are no conflict of interest.Acknowledgements
The authors are thankful to the Higher Education Commission (HEC) for IRSIP scholarship and the Department of Chemistry, University of Leicester UK for the research fellowship.References
- R. A. DeFronzo, E. Ferrannini, L. Groop, R. R. Henry, W. H. Herman, J. J. Holst, F. B. Hu, C. R. Kahn, I. Raz and G. I. Shulman, Nat. Rev. Dis. Primers, 2015, 1, 1–22 CrossRef PubMed.
- A. B. Olokoba, O. A. Obateru and L. B. Olokoba, Oman Med. J., 2012, 27, 269 CrossRef CAS PubMed.
- L. Chen, D. J. Magliano and P. Z. Zimmet, Nat. Rev. Endocrinol., 2012, 8, 228–236 CrossRef CAS PubMed.
- G. Derosa, A. Carbone, I. Franzetti, F. Querci, E. Fogari, L. Bianchi, A. Bonaventura, D. Romano, A. F. Cicero and P. Maffioli, Diabetes Res. Clin. Pract., 2012, 98, 51–60 CrossRef CAS PubMed.
- A. J. Scheen, Expert Opin. Drug Metab. Toxicol., 2010, 6, 1265–1276 CrossRef CAS PubMed.
- B. Seyoum, Expert Opin. Pharmacother., 2011, 12, 641–646 CrossRef CAS PubMed.
- G. L. Plosker, Drugs, 2014, 74, 223–242 CrossRef CAS PubMed.
- A. Karasik, P. Aschner, H. Katzeff, M. J. Davies and P. P. Stein, Curr. Med. Res. Opin., 2008, 24, 489–496 CrossRef CAS PubMed.
- R. Yendapally, D. Sikazwe, S. S. Kim, S. Ramsinghani, R. Fraser-Spears, A. P. Witte and B. La-Viola, Drug Dev. Res., 2020, 81(4), 390–401 CrossRef CAS.
- K. Yokota and N. Igaki, Intern. Med., 2012, 51, 2041–2044 CrossRef CAS PubMed.
- A. R. Gosmanov and E. C. Fontenot, Diabetes Care, 2012, 35, e60 CrossRef.
- S. Desai, A. Brinker, J. Swann and S. Iyasu, Arch. Intern. Med., 2010, 170, 1169–1171 CrossRef PubMed.
- R. Nirogi, V. Kandikere, K. Mudigonda, P. Komarneni, R. Aleti and R. Boggavarapu, Biomed. Chromatogr., 2008, 22, 214–222 CrossRef CAS PubMed.
- R. N. Rao, P. K. Maurya and S. Khalid, Talanta, 2011, 85, 950–957 CrossRef CAS PubMed.
- E. Uçaktürk, J. Pharm. Biomed. Anal., 2013, 74, 71–76 CrossRef PubMed.
- S. Bonde, R. Bhadane, A. Gaikwad, D. Katale, S. Gavali and A. Narendiran, Int. J. Pharm. Pharm. Sci., 2013, 5, 463–470 CAS.
- W. Zeng, D. G. Musson, A. L. Fisher, L. Chen, M. S. Schwartz, E. J. Woolf and A. Q. Wang, J. Pharm. Biomed. Anal., 2008, 46, 534–542 CrossRef CAS PubMed.
- R. I. El-Bagary, E. F. Elkady and B. M. Ayoub, Talanta, 2011, 85, 673–680 CrossRef CAS PubMed.
- R. I. El-Bagary, E. F. Elkady and B. M. Ayoub, Int. J. Biomed. Sci., 2011, 7, 62 CAS.
- C. B. Sekaran and A. P. Rani, Ecletica Quim., 2010, 35, 45–53 CrossRef.
- R. I. El-Bagary, E. F. Elkady and B. M. Ayoub, Int. J. Biomed. Sci., 2011, 7, 55 CAS.
- M. Salim, N. El-Enany, F. Belal, M. Walash and G. Patonay, Anal. Chem. Insights, 2012, 7, 31–46 CAS.
- A. Barseem, H. Ahmed, Y. El-Shabrawy and F. Belal, Microchem. J., 2019, 146, 20–24 CrossRef CAS.
- S. Caglar, A. Onal and S. Toker, Curr. Pharm. Anal., 2012, 8, 278–285 CrossRef CAS.
- W. Zeng, Y. Xu, M. Constanzer and E. J. Woolf, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2010, 878, 1817–1823 CrossRef CAS PubMed.
- M. Al Bratty, H. A. Alhazmi, S. A. Javed, K. G. Lalitha, M. Asmari, J. Wölker and S. El Deeb, Chromatographia, 2017, 80, 891–899 CrossRef CAS.
- L. Burugula, R. Mullangi, N. R. Pilli, A. Makula, D. S. Lodagala and R. Kandhagatla, Biomed. Chromatogr., 2013, 27, 80–87 CrossRef CAS PubMed.
- R. M. Ahmed, G. M. Hadad, A. E. El-Gendy and A. Ibrahim, Anal. Chem. Lett., 2018, 8, 813–828 CrossRef CAS.
- R. I. El-Bagary, E. F. Elkady and B. M. Ayoub, Eur. J. Chem., 2013, 4, 360–365 CrossRef CAS.
- O. S. Ahmad, T. S. Bedwell, C. Esen, A. Garcia-Cruz and S. A. Piletsky, Trends Biotechnol., 2019, 37, 294–309 CrossRef CAS.
- S. A. Piletsky and A. P. Turner, Electroanalysis, 2002, 14, 317–323 CrossRef CAS.
- L. Ye and K. Haupt, Anal. Bioanal. Chem., 2004, 378, 1887–1897 CrossRef CAS.
- M. Hadi, H. Poorgholi and H. Mostaanzadeh, S. Afr. J. Chem., 2016, 69, 132–139 CAS.
- J. Narang, N. Malhotra, C. Singhal, G. Singh and C. S. Pundir, Int. J. Nanomed., 2018, 13, 117 CrossRef CAS PubMed.
- M. B. Gholivand and L. Mohammadi-Behzad, Anal. Biochem., 2013, 438, 53–60 CrossRef CAS PubMed.
- P. S. Sharma, A. Garcia-Cruz, M. Cieplak, K. R. Noworyta and W. Kutner, Curr. Opin. Electrochem., 2019, 16, 50–56 CrossRef CAS.
- S. A. Piletsky, N. W. Turner and P. Laitenberger, Med. Eng. Phys., 2006, 28, 971–977 CrossRef PubMed.
- R. S. Gaster, D. A. Hall, C. H. Nielsen, S. J. Osterfeld, H. Yu, K. E. Mach, R. J. Wilson, B. Murmann, J. C. Liao and S. S. Gambhir, Nat. Med., 2009, 15, 1327 CrossRef CAS PubMed.
- G. Selvolini and G. Marrazza, Sensors, 2017, 17, 718 CrossRef PubMed.
- H. Munawar, A. H. Safaryan, A. De Girolamo, A. Garcia-Cruz, P. Marote, K. Karim, V. Lippolis, M. Pascale and S. A. Piletsky, Food Chem., 2019, 298, 125044 CrossRef CAS PubMed.
- E. Mazzotta, R. Picca, C. Malitesta, S. Piletsky and E. Piletska, Biosens. Bioelectron., 2008, 23, 1152–1156 CrossRef CAS PubMed.
- M. Wei, X. Geng, Y. Liu, H. Long and J. Du, J. Electroanal. Chem., 2019, 842, 184–192 CrossRef CAS.
- H. Munawar, A. Garcia-Cruz, M. Majewska, K. Karim, W. Kutner and S. A. Piletsky, Sens. Actuators, B, 2020, 321, 128552 CrossRef CAS.
- K. Smolinska-Kempisty, O. S. Ahmad, A. Guerreiro, K. Karim, E. Piletska and S. Piletsky, Biosens. Bioelectron., 2017, 96, 49–54 CrossRef CAS PubMed.
- D. Garcia-Mutio, A. Guerreiro, A. Gomez-Caballero, R. Gutierrez-Climente, S. Piletsky, M. Goicolea and R. Barrio, Procedia Eng., 2015, 120, 1132–1136 CrossRef CAS.
- J. Wackerlig and P. A. Lieberzeit, Sens. Actuators, B, 2015, 207, 144–157 CrossRef CAS.
- V. M. Ekomo, C. Branger, A.-M. Gavrila, A. Sarbu, D. A. Koutsouras, C. Stolz, G. G. Malliaras and H. Brisset, MRS Commun., 2020, 10, 324–331 CrossRef CAS.
- V. M. Ekomo, C. Branger, R. Bikanga, A.-M. Florea, G. Istamboulie, C. Calas-Blanchard, T. Noguer, A. Sarbu and H. Brisset, Biosens. Bioelectron., 2018, 112, 156–161 CrossRef CAS PubMed.
- D. Udomsap, H. Brisset, G. Culioli, P. Dollet, K. Laatikainen, H. Siren and C. Branger, Mater. Today Commun., 2018, 17, 458–465 CrossRef CAS.
- A. Garcia-Cruz, O. Ahmad, K. Alanazi, E. Piletska and S. Piletsky, Microsyst. Nanoeng., 2020, 6, 1–9 CrossRef.
- P. S. Sharma, A. Garcia-Cruz, M. Cieplak, K. R. Noworyta and W. Kutner, Curr. Opin. Electrochem., 2019, 16, 50–56 CrossRef CAS.
- G.-C. Alvaro, T. Cowen, A. Voorhaar, E. Piletska and S. Piletsky, Analyst, 2020, 145(12), 4224–4232 RSC.
- K. Alanazi, A. G. Cruz, S. Di Masi, A. Voorhaar, O. S. Ahmad, T. Cowen, E. Piletska, N. Langford, T. J. Coats and M. R. Sims, Sens. Actuators, B, 2020, 129128 Search PubMed.
- K. Alanazi, A. G. Cruz, S. Di Masi, A. Voorhaa, O. S. Ahmad, T. Cowen, E. Piletska, N. Langford, T. J. Coats and M. R. Sims, Sens. Actuators, B, 2020, 129128 Search PubMed.
- D. Garcia-Mutio, A. Gomez-Caballero, A. Guerreiro, S. Piletsky, M. Goicolea and R. Barrio, Sens. Actuators, B, 2016, 236, 839–848 CrossRef CAS.
- E. Mazzotta, A. Turco, I. Chianella, A. Guerreiro, S. A. Piletsky and C. Malitesta, Sens. Actuators, B, 2016, 229, 174–180 CrossRef CAS.
- A. R. Leach, Molecular Modelling: Principles and Applications, Pearson Education, 2001 Search PubMed.
- A. G. Cruz, I. Haq, T. Cowen, S. Di Masi, S. Trivedi, K. Alanazi, E. Piletska, A. Mujahid and S. A. Piletsky, Biosens. Bioelectron., 2020, 169, 112536 CrossRef PubMed.
- S. A. Piletsky, K. Karim, E. V. Piletska, C. J. Day, K. W. Freebairn, C. Legge and A. P. F. Turner, Analyst, 2001, 126, 1826–1830 RSC.
- T. Cowen, K. Karim and S. Piletsky, Anal. Chim. Acta, 2016, 936, 62–74 CrossRef CAS PubMed.
- H. Kim, K. Kim and S. J. Lee, NPG Asia Mater., 2017, 9, e445 CrossRef CAS.
- A. Gandhi, A. Paul, S. O. Sen and K. K. Sen, Asian J. Pharm. Sci., 2015, 10, 99–107 CrossRef.
- M. Zhu, M. Z. Lerum and W. Chen, Langmuir, 2012, 28, 416–423 CrossRef CAS PubMed.
- F. Paul and T. R. Weikl, PLoS Comput. Biol., 2016, 12, e1005067 CrossRef PubMed.
- A. D. Vogt and E. Di Cera, Biochemistry, 2012, 51, 5894–5902 CrossRef CAS PubMed.
- M. Salim, N. El-Enany, F. Belal, M. Walash and G. Patonay, Anal. Chem. Insights, 2012, 7, S9940 CrossRef PubMed.
- L. Burugula, R. Mullangi, N. R. Pilli, A. Makula, D. S. Lodagala and R. Kandhagatla, Biomed. Chromatogr., 2013, 27, 80–87 CrossRef CAS PubMed.
- M. R. Safa'a, M. R. Rezk, G. Y. Mahmoud, A.-A. El Bayoumi and A. Aleem, Anal. Bioanal. Electrochem., 2013, 5, 416–425 Search PubMed.
- A. M. El-Kosasy, L. Abd El Aziz and Y. A. Trabik, J. Appl. Pharm. Sci., 2012, 2(8), 51 Search PubMed.
- A. H. Kamel and H. R. Galal, Int. J. Electrochem. Sci., 2014, 9, 4361–4373 Search PubMed.
- A. Florea, T. Cowen, S. Piletsky and K. De Wael, Talanta, 2018, 186, 362–367 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1na00194a |
| This journal is © The Royal Society of Chemistry 2021 |