Boolean logic gate based on DNA strand displacement for biosensing: current and emerging strategies
Shuang
Zhao†
a,
Lianyu
Yu†
a,
Sha
Yang
a,
Xiaoqi
Tang
a,
Kai
Chang
*a and
Ming
Chen
 *abc
*abc
aDepartment of Clinical Laboratory Medicine, Southwest Hospital, Army Medical University, 30 Gaotanyan, Shapingba District, Chongqing 400038, China. E-mail: chming1971@126.com; changkai0203@163.com; Fax: +86-23-68716530; Tel: +86-23-68754429
bCollege of Pharmacy and Laboratory Medicine, Army Medical University, 30 Gaotanyan, Shapingba District, Chongqing 400038, China
cState Key Laboratory of Trauma, Burn and Combined Injury, Army Medical University, 30 Gaotanyan, Shapingba District, Chongqing 400038, China
First published on 24th March 2021
Abstract
DNA computers are considered one of the most prominent next-generation molecular computers that perform Boolean logic using DNA elements. DNA-based Boolean logic gates, especially DNA strand displacement-based logic gates (SDLGs), have shown tremendous potential in biosensing since they can perform the logic analysis of multi-targets simultaneously. Moreover, SDLG biosensors generate a unique output in the form of YES/NO, which is contrary to the quantitative measurement used in common biosensors. In this review, the recent achievements of SDLG biosensing strategies are summarized. Initially, the development and mechanisms of Boolean logic gates, strand-displacement reaction, and SDLGs are introduced. Afterwards, the diversified input and output of SDLG biosensors are elaborated. Then, the state-of-the-art SDLG biosensors are reviewed in the classification of different signal-amplification methods, such as rolling circle amplification, catalytic hairpin assembly, strand-displacement amplification, DNA molecular machines, and DNAzymes. Most importantly, limitations and future trends are discussed. The technology reviewed here is a promising tool for multi-input analysis and lays a foundation for intelligent diagnostics.
1. Introduction
Over the past few decades, computers have rapidly developed but their information storage capacity and parallel computing capabilities have presented major challenges. Hence, novel methods, such as DNA,1 quantum,2 optical,3 and neural network computing,4 continue to emerge. DNA computing was first proposed by Adleman in 1994.5 It is the most promising modality because of its powerful information encoding and storage capacity, biodegradability, and molecular recognition ability.6–8 DNA computers could analyze several targets simultaneously by performing logic gates, and thus show great potential in biosensing. By setting the threshold, the generated signal can be converted into a YES/NO form that is contrary to the quantitative measurement used in common biosensing strategies. These decisive answers eliminate human error and generate relatively more valuable and instructive output.9 Logic elements generate digital output in the form of YES and NO in response to the input. This property meets the critical immediate response requirement of an ideal “fast-screening” diagnostic tool.10Various DNA technologies have been developed to construct Boolean logic gates to effect DNA computing, including DNA strand displacement,11 DNA self-assembly,12 and DNAzymes.13 DNA strand displacement technique is widely used one because the “invading chain-to-displaced chain” model fits well with the “input-to-output” model of logic gates. Moreover, the advantages of its easy-operation and being self-triggered make it promising in practical use.14 However, the input and output of strand displacement-based logic gates (SDLGs) are nucleic acids and the “DNA-to-DNA” model does not support the construction of universal biosensors that can detect multiple types of targets or have different signal transmission methods.15 Besides, the high DNA input concentration required for strand-displacement reactions (SDRs) are always equal to or higher than the nanomolar level.16,17 Also, the sensitivity of the DNA strand displacement-based detection method is limited. In order to remove limitations, scholars have integrated various emerging materials and technologies into SDLG biosensing platforms in an attempt to detect other input signals, such as ATP,18 cancer cells,19 and metal ions.20 In addition, in combination with fluorescent materials,21,22 electrochemical technology,23 and gold nanoparticles (AuNPs),24 SDLGs can generate fluorescent, electrical, and naked-eye signals that accommodate the application requirements of various biosensing platforms. Furthermore, the application of emerging materials and signal-amplification strategies that are suitable for diagnostics enhance SDLG biosensing platform sensitivity; also meeting actual application requirements. The implementation of these strategies augment the practicality, maturity, and diversity of SDLG biosensors.
In view of the vigorous SDLG biosensing platform development in recent years, the present review furnishes a map of the current SDLG biosensor landscape. First, we introduce the development and mechanism of the logic gate, SDR, and SDLG. Second, the diversified input and output of SDLG biosensors are recommended by noting some representative researches. Then, we focus on the highly sensitive SDLG biosensors integrated with signal-amplification methods, such as rolling circle amplification (RCA), catalytic hairpin assembly (CHA), strand-displacement amplification (SDA), DNA molecular machines, and DNAzymes. Finally, the limitations and future prospects of SDLGs are discussed in the review.
2. Boolean logic gate and strand-displacement reaction
Boolean logic gates are physical devices that receive single or multiple inputs, exert logical operations, and yield a single digital output. The principle of logical operations is the foundation of all silicon-based computing,25 and comprises input signals, logic components, and output signals. The input and output signals are converted into binary signals. The true and false signals may be distinguished by setting the thresholds. If the signal value exceeds the threshold, it is regarded as true or “1”; if the signal value is less than the threshold, then it is assumed false or “0”. Different logic gates have various logic operation complexities. For the single-input YES and NO logic gate, the presence or absence of the input directly determines the result of the output (“0 to 0”, “1 to 1” mode). When the input signal information is increased, a series of logic gates may be constructed to execute different logic functions. The common logic gates include AND, OR, INHIBIT, XOR, and those with contrary functions (NAND, NOR, IMPLICATION, and NXOR). Table 1 shows the truth values and symbols for certain dual-input logic gates.For the AND gate, only when all the inputs are present will the “1” output be generated; otherwise, the output will be “0”. For an OR gate, as long as any one input exists, the “1” output will be produced, and only if all the inputs are absent will the “0” output be generated. For the INHABIT gate, one of the inputs is used as a prohibition factor, and only when it does not exist and the other inputs are present will the “1” output be produced. For XOR gates, the existence of any input “10 or 01” will result in a “1” output, whereas when none or both of the inputs exist “00 or 11” or “0” outputs will be produced. This unique input-to-output pattern makes XOR usually hard to be realized at the molecular level. NAND, NOR, IMPLICATION, and XNOR are the opposite of these logic gates. Remarkably, these basic logic gates can evolve into advanced logic devices through various combinations and integrations, such as half/full adder and half/full subtractor.
SDR is the most extensively used dynamic DNA nanotechnology to construct Boolean logic gates and DNA circuits.26 It utilizes the characteristics of molecular hybridization systems to establish stable energy levels. SDR could be induced by the invading chain to release another DNA chain. By way of specific recognition, release, capture, and other manipulations can be accomplished.27 The basic principle of toehold-mediated SDR is shown in Fig. 1A-a chain B′/C′ is a partial double-stranded chain composed of two partially complementary DNA strands. Chain A′ is a single strand whose sequence is fully complementary to that of chain B′/C′. Chain A′ recognizes and binds the toehold and migrates to the most stable branch. The chain B′/C′ base pair is then gradually replaced by the chain A′/C′ base pair until the chain B′ is released. Different from the fully displaced mode, a free toehold domain overhanging ensures the reversibility of the toehold exchange reaction (Fig. 1A-b).28 The reaction process is actually a redistribution of the reactant concentration. The thermodynamic basis of SDR has been discussed in previous research,26,29 which has reported that the binding free energy of the toehold domain determines the displacement rate and equilibrium concentration. The proper design of toeholds can tune the rate of strand displacement over 106 orders of magnitude.30–32 Therefore, the length and base sequence of a toehold are most critical for the control of the SDR reaction rate.33 However, as this rate changes exponentially with the binding strength,31,32 it is difficult to achieve a precise adjustment only by changing the length and sequence. Fortunately, researchers have successively developed a series of strategies for the accurate adjustment of the SDR rate, which greatly enriches the SDR toolbox. As the distance between the branch migration (BM) domain and toehold is crucial for the SDR rate, the continuous displacement rate can be precisely adjusted by changing the distance between the BM domain and the toehold. In 2011, Genot et al. introduced a remote toehold mode (Fig. 1B),34 where the toehold and strand-displacement domains are separated by a spacer. By controlling the length and hybridization state of the spacer, the SDR rate could be changed by over three orders of magnitude. Similarly, in the intramolecular conformational motion strategy (Fig. 1C),35 the author cleverly designed the spacer into a reconfigurable stem-loop structure. SDR is more likely to occur when the stem-loop structure is formed by an accelerating internal diffusion rate and favorable thermodynamics. When the stem-loop structure and complementary sequences form a rigid double helix structure, SDR cannot occur. In this way, by controlling the structural length of the stem-loop and its complementary sequence, the SDR kinetics was allowed to be reversible and real-time regulated. Compared with the method that separated the toehold and BM domain by an additional sequence or structures in a single strand, designing them on two chains may simplify the design. Chen et al. proposed a toehold activation strategy, in which the toehold and BM domain are separated on the two strands and then hybridized together (Fig. 1D).36 Two bulged bases at the three-way junction ensure high efficiency, and thus this method is feasible for practical applications. Similarly, the allosteric DNA toehold was proposed by Yang et al.,37 in which the toehold and BM domains are also separated. The toehold was designed as an regulator, which not only simplifies the DNA sequence design but also dynamically and selectively controls DNA strand displacement (Fig. 1E). In addition, this unique design is compatible with other activation mechanisms, thus making it possible to design more complex DNA circuits. Robust regulation strategies for enzyme-free DNA circuits, such as nonlinear regulation with ultrasensitive switches, have also been proposed.38 These methods supplement the control of the SDR rate and greatly enrich the SDR toolbox. Therefore, a more controllable, flexible, and compatible SDR can be developed.
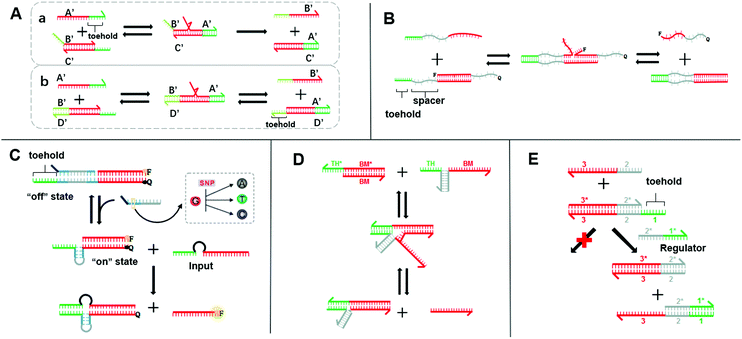 | ||
| Fig. 1 (A) Schematic diagram of two models for the toehold-mediated strand-displacement reaction: (a) toehold displacement reaction, (b) toehold exchange reaction. (B) Mechanism of the remote toehold mode.34 (C) Intramolecular conformational motion strategy for kinetic modulation in strand displacement.35 (D) Scheme of hybridization-based associative toehold activation.36 (E) Schematic illustration of the principles of allosteric toehold-mediated DNA strand displacement.37 In all the panels, the toeholds are shown as red lines and BM domains are shown as green lines. | ||
In short, the advantages of its dynamic nature, high control, and freedom make SDR a powerful and promising tool for Boolean logic gates construction.39 Ogihara et al. applied the DNA strand-displacement theory to construct logic gates.40 In 1996, they successfully constructed “AND” and “OR” gates using a designed DNA strand as the input and output. In 2006, Winfree's group constructed a serious of digital logic circuits using SDR.41 The more complex full/half adder based on DNA SDR was also successfully constructed in 2016.42 More importantly, a series of in-depth research studies were conducted to improve the performance of SDR-based DNA circuits. For example, the signal leakage in the DNA circuits was reduced by using a digital droplet assay.43 Switching s SDR allowed implementing faster digital computing.44 The optically controlled DNA switching circuits broaden the DNA circuit functions.45 In particular, some strategies were proposed to improve the specificity of nucleic acids hybridization in DNA circuits, which further ensured the accuracy when applied in biosensing.46–48 Therefore, the in-depth mechanism research for SDR logic circuits has paved the way for realizing more complex functions in biosensing and other fields.
3. Diversified SDLG biosensing platform input and output
Embedding logic analysis in biosensors will make them possible to perform multi-target intelligent analysis. However, since both invading and displaced strands are nucleic acids in SDLG, this “DNA to DNA” model cannot meet the various input and output requirements of biosensing. Fortunately, the application of aptamers, DNA enzymes, fluorescent-labeling technology, electrochemical technology, and spherical nucleic acids can make the input and output of SDLG more diversified. Thus, SDLG has the potential to become a general biosensing strategy.3.1 Diversified input of SDLG biosensors
It is easy for SDLG to detect nucleic acids. The target nucleic acids could be designed as an invading chain, which could trigger the SDR and drive the logical analysis. This strategy has been shown to be effective and efficient.21,23,49 Notably, the addition of some identification devices makes it not only capable of detecting nucleic acids but also ATP, cancer cells, and metal ions.Aptamers are screened from in vitro oligonucleotide libraries via systematic ligand evolution through exponential enrichment technology.50 They consist of 20–80 bases and have complex spatial structures that bind enzymes, peptides, metal ions, and cells. The features of high specificity and high affinity make them a good choice for SDLG biosensors. Zhu et al. introduced a three-way junction-driven strand-displacement mode to detect nucleic acids and ATP in the presence of aptamers.18 They built a logic circuit composed of AND and NOR logic gates using ATP and DNA strands as the input (Fig. 2A). The molecular platform could be applied to a wider field by altering the aptamer sequence. The detection limits of ATP and DNA were 40 μM and 6.9 nM, respectively. However, these detection limits are unremarkable for practical applications. By combining aptamers and toehold-mediated strand displacement, Tan's group realized the logic analysis of multiple markers on the cell membrane to identify cancer cells and built AND, OR, and NOT gates19 (Fig. 2B shows the “A AND B NOT C” gate). However, taking the “A AND B” gate as an example, if two neighboring cells express A and B markers respectively, this platform will produce a “joint-positive” result. To overcome this false-positive result, a simple but powerful molecular device named a “nano-claw” was designed in their other research51 (Fig. 2C). Two-input and three-input AND gates were achieved with similar mechanisms. The above two platforms could be highly integrated to achieve a more powerful cell recognition function.
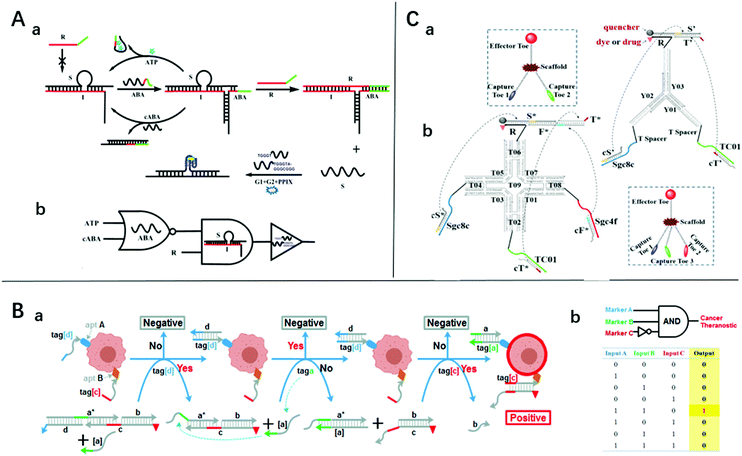 | ||
| Fig. 2 (A) (a) Schematic diagram of the sensing platform based on three-way junction-driven toehold-mediated SDR and (b) logic circuit with strands R, cABA, and ATP as input. Reprinted with permission from ref. 18, Copyright 2014, American Chemical Society. (B) (a) Experimental schemes, (b) symbols, and (c) truth tables and of 3-input “a AND b NOT c” gate for identifying cancer cells. Reprinted with permission from ref. 19, Copyright 2015, American Chemical Society. (C) The symbols and construction schemes of (a) 2-input trivalent “Y”-shaped Nano-Claw and (b) 3-input tetravalent “X”-shaped Nano-Claw. Reprinted with permission from ref. 51, Copyright 2014, American Chemical Society. | ||
Metal ion-dependent DNAzyme can catalyze the cutting of DNA to detect metal ions. The first DNA-based enzyme was isolated by Breaker and Joyce in 1994.52 As early as 2009, Willner used two DNAzymes that relied on UO2+ and Mg2+, respectively, to construct AND and OR gates.53 When UO2+ and Mg2+ exist, the corresponding DNAzyme cuts the chain and the product of cleavage can form a G-quadruplex with hemin to generate a visible signal to achieve the purpose of detection. Using two DNA enzymes to implement AND logic gates in the same system may make the design more complicated. The application of multi-ion-dependent DNAzyme may simplify the design. Although not described as an AND gate, the DNAzyme Ce13d is indeed such an enzyme that requires both Na+ and Ce3+ to be activated.54 The Hg2+ sensor based on the UO22+ DNAzyme is also such an example.55 Through the clever design of the circular substrate, Bi et al. used DNAzymes to construct a complete set of logic gates (OR, AND, INHIBIT, XOR, NOR, NAND, and XNOR) to realize the rapid colorimetric detection of metal ions.20 This further demonstrates the application potential of DNAzymes in the SDLG strategy. Because of the designability of DNAzymes, it could unite with other DNA nanotechnology to achieve more incredible functions. Yue et al. integrated the DNAzyme structure into DNA tweezers,56 and the changes of the K+ concentration in solution could reconstruct the orthogonal balance of the tweezers. This input-guided continuous dynamic network had the potential for biological sensing and logical operation applications. Markedly, the addition of metal ion-dependent DNA enzymes makes the SDLG strategy expandable for detecting metal ions. The unique catalytic properties and adjustability also make these DNAzymes more widely used in SDLG. This point will be discussed further in later chapters.
3.2 Diversified output of SDLG biosensors
In practical application, biosensing often requires different signal output strategies. For example, in the laboratory, the output of fluorescent or electrochemical signals can meet the requirements, but in a point-of-care testing environment, visual color comparison may be more suitable. The following are typical examples of SDLG biosensors producing three main types of output.Fluorescent signals are mostly created using fluorescent dyes or labels. Chen et al. used the displaced chain of SDLG as a primer of RCA, whereby the fluorescence signal could be detected when SYBR Green I bound the amplified product.21 Fu's group reported another method, in which the input of the target molecule opened the DNA lock by driving the SDR,57 then the cofactor was exposed and combined with the enzyme to generate a fluorescent signal. Meanwhile, Peng et al. used SDR to separate the fluorescent agent and quencher to generate a fluorescent signal,58 which has also been widely used. The characteristics of high sensitivity, high spatial resolution, and high detection speed of fluorescent signals make it widely used in SDLG biosensors.
The most common electrochemical sensing platform immobilizes nucleic acid probes labeled with methylene blue on the electrode. When the reaction occurs on the electrode, the probe state will trigger the changes of the electrochemical signals. For example, Wang et al. fixed a DNA nanorobot on the electrode. SDLG activated this nanorobot to cleave a methylene blue-labeled probe that was fixed on a gold electrode, generating electrochemical signals.23 As the immobilization of the nucleic acid chain on the electrode will inevitably produce steric hindrance, the signal-to-noise ratio is low. To avoid this situation, Dai et al. combined novel electrochemical active–inactive conversion molecular beacons, such as ferrocene or hemin molecular beacons, as an electrochemical platform that was not fixed on the electrode surface, resulting in a higher signal-to-noise ratio.59
As the naked-eye colorimetric method does not require bulky inspection equipment, it is most suitable for point-of-care testing. Spherical nucleic acid (SNA) is a functionalized AuNP composed of a gold core and an oriented oligonucleotide shell. It has the same physicochemical properties as AuNPs and the powerful programming and algorithm functions of the nucleic acid shell.60,61 Besides, the disassembly of SNA aggregates can be easily detected by the naked eye through a visible color change of the supernatant because of its the distinct optical properties. These features demonstrated SNA conjugate's superiorities in colorimetric biosensing. Wei et al. combined DNA strand-displacement circuits with SNA and established a smart strategy for visible SNA aggregate disassembly that was programmed by entropy-driven SDR.62 The initiation of visible SNA aggregate disassembly relied upon a trigger strand released from the upstream DNA circuit through successive SDRs. The SNA may be compatible with any upstream circuit and there is no need to change any DNA strand in the downstream system (Fig. 3A). In this manner, costs are lowered and it is not necessary to design new SNA aggregates. By uniting two parallel catalytic DNA circuits upstream with a fixed SNA disassembly system downstream, an OR gate was constructed for multiplexed target detection. Thus, the SNA aggregate-based disassembly strategy is simple and universal.
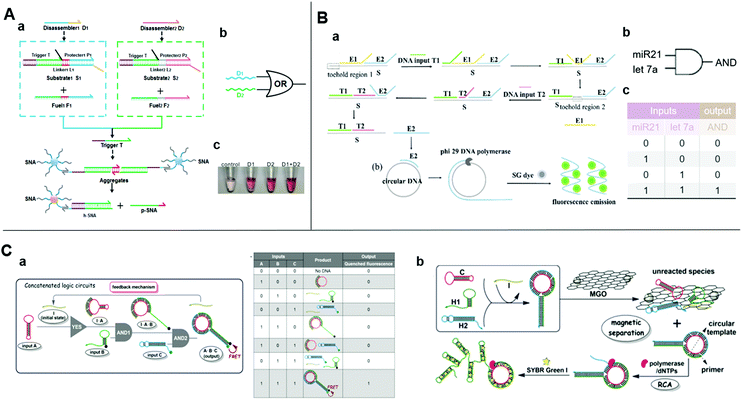 | ||
| Fig. 3 (A) (a) Schematic diagram, (b) symbol, and (c) image of the visible of OR logic gate based on upstream SDR and downstream SNA. Reprinted with permission from ref. 62, Copyright 2019, American Chemical Society. (B) (a) Schematic diagram and (b) symbol, and (c) truth table of two-input AND logic gate detecting DNA T1 and T2 inputs simultaneously with entropy-driven SDR and RCA. Reprinted with permission from ref. 21; permission conveyed through Copyright Clearance Center, Inc. (C) (a) Schematic illustration of three-input concatenated YES–AND–AND logic circuits based on catalytic self-assembly of duplex-looped DNA hairpin. (b) Truth table of the concatenated logic circuits. (c) Schematic illustration of using RCA and MGO reduce background, amplifying signal. Reprinted from ref. 65, Copyright 2016, with permission from Elsevier. | ||
4. SDLG-integrated signal-amplification strategy for biosensing
4.1 SDR initiates rolling circle amplification for logic-controlled biosensing
Rolling circle amplification (RCA) is an isothermal enzymatic reaction in which DNA or RNA polymerases, such as phi29 DNA polymerase,63 are used to produce single-stranded DNA or RNA molecules connected by numerous complementary units in a template. The reaction is simple and productive since the signal of a single binding event could be magnified exponentially over a thousand-fold.64 Thus, RCA is optimal for the required ultrasensitive detection. An effectively amplified DNA AND logic gate platform based on an SDR and RCA strategy was designed for the ultrasensitive detection of low-abundance DNA fragment inputs21 (Fig. 3B). It consisted of entropy-driven, toehold-mediated SDR that released the strand and the RCA reaction to amplify the released strand. This process allowed a highly sensitive simultaneous detection of two nucleic acids. Fluorescence of the SYBR Green I could reduce the detection limit to 500 aM. The detection of endogenous miR-21 and let-7a in total RNA samples extracted from HeLa cells showed comparable good performance and minimal interference due to the biological sample heterogeneity. Therefore, this approach is apt for clinical use. Similarly, Bi et al. also used RCA to amplify the product of a logic circuit for achieving a lower detection limit.65 They applied SDR-based programming for the catalytic self-assembly of the duplex-looped DNA hairpin and developed a three-input YES–AND–AND logic circuit (Fig. 3C-a). When all the inputs exist, the duplex-looped DNA hairpin will be formed to trigger RCA. Recycling the initiator in the cascade reaction and the presence of magnetic graphene oxide further reduced the background and enhanced the sensitivity (Fig. 3C-b). The proposed delicate DNA SDR-based methodology with engineering dynamic functions could have numerous applications in the creation of controllable DNA nanostructures, functional biosensing platforms, and complex DNA circuits.4.2 SDR initiates catalytic hairpin assembly for logic-controlled biosensing
Catalytic hairpin assembly (CHA) is a toehold-mediated SDR originally introduced by Yin et al. in 2008.66 Li et al. later modified it for use in various analytical applications.67 The CHA reaction is usually executed by using two stable hairpin structures with complementary sequences. The complementary domains are caged within hairpin stems. Thus, spontaneous interactions between DNA strands are kinetically blocked. As a toehold is present, two hairpins successively open and the most thermodynamically favorable DNA duplexes are rapidly formed.68 This mechanism results in an enzyme-free, high-efficiency, isothermal amplification method.59 Yue et al. proposed a chemiluminescence resonance energy-transfer (CERT) biosensing platform based on a cross-CHA amplification strategy and established a logic network (Fig. 4A).69 The system consists of a crossed two-layer CHA cascade that generates exponential signal amplification and provides high-sensitivity DNA detection in <1.5 h. The detection range was five orders of magnitude. The experiments of Yue et al. demonstrated its excellent specificity and good performance with biological samples. A multilevel circuit was established and it consisted of one YES and three AND gates with a feedback mechanism. This kind of logic network offered a new perspective in the design of diagnostic hierarchical molecular circuits.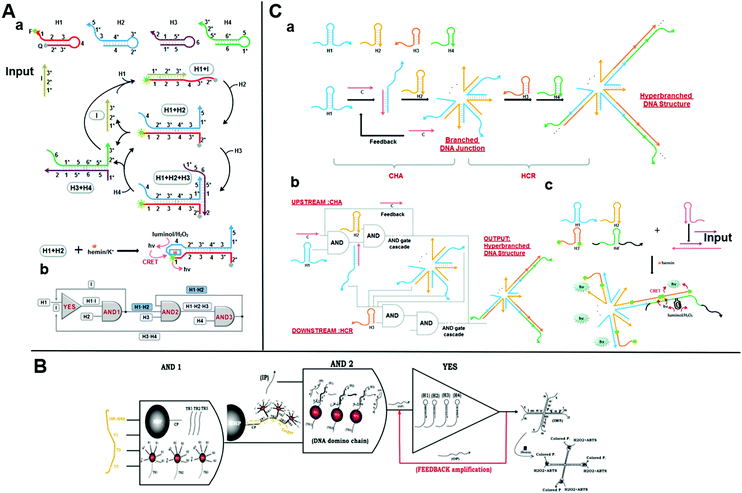 | ||
| Fig. 4 (A) (a) Schematic illustration of the reaction pathways and (b) the logic network based on CHA. Reprinted from ref. 49, Copyright 2017, with permission from Elsevier. (B) Representation of AND–AND–YES DNA domino-based nanoscale logic circuit. Reprinted with permission from ref. 73, Copyright 2017, American Chemical Society. (C) (a) Graphical representation and (b) logic scheme of the integrated DNA logic circuitry comprising CHA with a feedback mechanism and HCR based on toehold-mediated SDR to form a hyperbranched DNA structure as output. Reprinted with permission from ref. 74; permission conveyed through Copyright Clearance Center, Inc. | ||
The catalysts initiate successive CHA reactions between stable hairpins and form branched junctions with different numbers of arms. The wise sticky end design gives full play to the branched DNA junctions. Sensing platforms based on branched DNA junctions amplify the signals and dramatically improve the sensitivity as the ends of each arm can be engineered to form DNAzymes,70 converted to other amplification methods,71 and bound to nanoparticles for further signal amplification.72 Based on this strategy, Ravan et al. designed a DNA domino-based nanoscale logic circuit composed of three logic gates (AND–AND–YES), which could analyze nucleic acid biomarkers simultaneously.73 The triple signal-amplification strategy based on AuNP, a hairpin assembly, and DNAzymes provided about a tenfold signal improvement and a detection limit of 100 aM (Fig. 4B). This nanodevice could analyze and compute several nucleic acid disease biomarkers by expanding the DNA domino chain as a multiplexed platform. In 2016, a cascaded DNA nanocircuit integrating CHA with a hybridization chain reaction (HCR) was proposed for hyperbranched DNA structure self-assembly.74 HCR is also a simple, effective, widely used isothermal amplification strategy based on SDR. The integrated DNA circuit consists of cascaded AND gates, four hairpins (HP1-4), and one catalyst DNA (Fig. 4C). The entire DNA logic circuit includes an upstream CHA and a downstream HCR. A single-stranded C opens the HP1 of the upstream circuit and releases its single-stranded region. The HCR then triggers HP3 and HP4 cross-opening and forms a self-assembled hyperbranched DNA structure as the output. When HCR was combined with CHA for miR-122 detection, the dynamic range was 1 pM–10 nM and the detection limit was 0.72 pM. This high sensitivity was attributed to the amplification effect of the DNA logic circuit and the highly sensitive chemiluminescence resonance energy-transfer system. Therefore, the integrated DNA logic circuits could efficiently amplify the signals for the construction of various molecular diagnosis platforms.
4.3 SDR initiates strand-displacement amplification for logic-controlled biosensing
Strand-displacement amplification (SDA) was inspired by isothermal nucleic acid amplification and first proposed in 2009.75 SDA consists of a hairpin-shaped molecular beacon, a polymerase, and a primer. The target triggers the SDA reaction and the primer binds. The polymerase extends the primer on the molecular beacon template and the hybridized target is then released and enters another cycle reaction to generate a considerably enhanced signal. In 2018, Xue et al. concatenated three logic gates to perform multiple SDA and screen cancer-related point mutations.22 The design included a hairpin probe, which was a self-folding molecular beacon whose overhangs exhibited a near-inverted mirror image. Cascade SDAs occur on this hairpin probe and promote each other so the molecular beacon is successively opened and the signal is amplified (Fig. 5A). The targets could be detected at concentrations as low as 10 pM. Xue et al. used two primers and target genes as indispensable input signals and proposed a series of logic gate circuits consisting of a YES and an AND gate. For the YES gate, the target opens the hairpin structure of the mirror image probe and the two primers combine to produce the “AND” gate. The three concatenated logic gates circuit change from “off” to “on” only when two primers and a complementary target DNA co-exist in the sample. This process yields a strong fluorescent signal. If mismatched DNA is used, then the circuit is “off” regardless of the number of mismatched bases. This feature enables the sensitive screening of single base mutations and deletions. Blind tests proved that the distinction of mutant from wild type genes was feasible with this complex biological matrix. This innovative biosensing strategy demonstrated the cooperation between SDA and SDLG biosensing.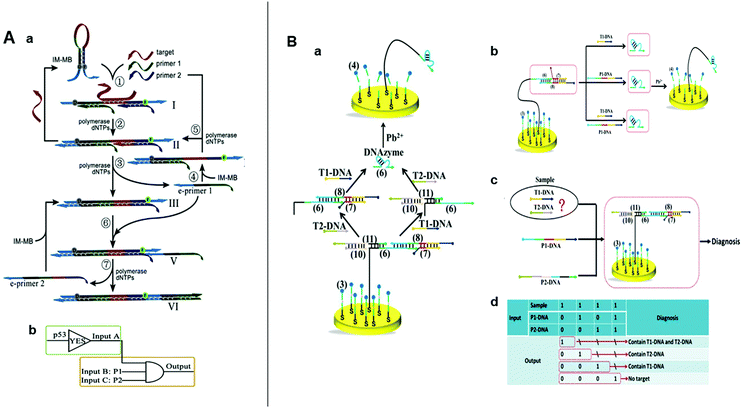 | ||
| Fig. 5 (A) (a) Schematic illustration and (b) symbol of the SDA logic circuits. Reprinted from ref. 22, Copyright 2019, with permission from Elsevier. (B) (a) Schematic illustration of intelligent detection of T1-DNA and T2-DNA using DNA walker-based strategy. (b) Illustration of AND, (c) OR gate, (d) and the truth table for the combinatorial logic gates. Reprinted from ref. 23, Copyright 2018, with permission from Elsevier. | ||
4.4 SDR initiates DNA walker for logic-controlled biosensing
In recent decades, DNA machines have aroused widespread attention because of their structural simplicity, uncomplicated construction, ample sequence design space, and predictable molecular behavior.30,66,76 DNA machines include tweezers,34 motors,77 robots,78 and walkers.79 DNA walkers run autonomously on their designated tracks and have great applicability toward biosensing platforms.80 In 2018, Wang et al. developed an electrochemical nucleic acid detection platform by combining SDR with DNA walker.23 Here, a series of SDRs are triggered by the target to release the DNA walker, which then walks autonomously by cleaving the DNA probe modified on the gold electrode. This reaction generates a strong electrochemical signal and could achieve the detection of 36 fM. The cooperation between SDR and DNA walker obviated the requirements for nucleases and hairpin self-assemblies and boosted the signal-to-noise ratio and sensitivity. Then, the aforementioned SDR-driven nanorobot was used to construct AND OR and combinatorial logic gates (OR and AND) for the intelligent analysis of multi-targets in a sample (Fig. 5B). This innovative sensing strategy combined SDLG and DNA walker perfectly, and laid a good foundation for the development of high-sensitive SDLG electrochemical biosensors.4.5 SDR activates enzymes for logic-controlled biosensing
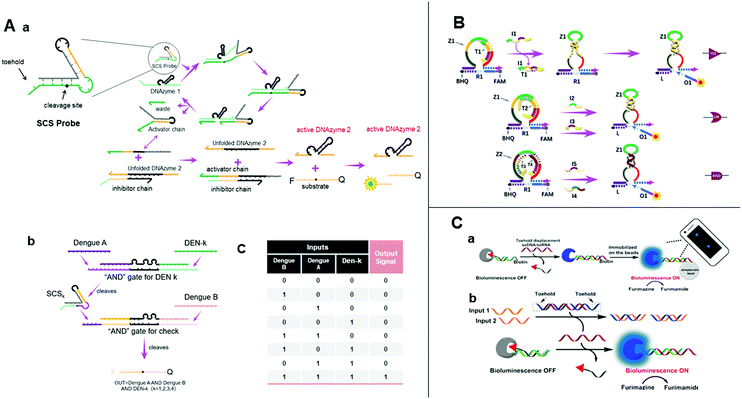 | ||
| Fig. 6 (A) (a) Illustration of the design of a structured chimeric substrate (SCS), mechanism of cleavage of the SCS by an upstream DNAzyme and DNAzyme displacement reaction. (b) Design of multi-layer diagnostic logic circuits for the detection of sequences from the genomes of all four dengue serotypes. (c) Truth table of dengue serotypes detection logic circuits.83 (B) Schematic diagram of YES, OR, and AND gates using the allosteric strategy of E6-type DNAzymes.84 (C) (a) Nucleic acid detection by smartphone imaging. (b) Schematic illustration of two analytes AND gate based on a semisynthetic luciferase. Reprinted with permission from ref. 86, Copyright 2020, American Chemical Society. | ||
5. Conclusion and prospects
In this review, we focused on the application of SDLG strategies in the field of biosensing. After decades of development, SDLG has achieved fruitful results. Table 2 compares some representative SDLG biosensing platforms. SDLG biosensors integrate the stringency of logical operations with the controllability and biocompatibility of strand displacement perfectly. In addition, SDLG biosensors have been shown to cooperate well with some emerging materials and technologies in enabling the highly-sensitive detection of multiple types of targets and the production of multiple types of output signals. For many diseases, it is difficult to obtain an accurate diagnosis using a single or limited number of disease molecular markers, and it is necessary to conduct a comprehensive analysis of different permutations and combinations of multiple targets. Since some biomarkers are related to the pathophysiological conditions in specific diseases, an SDLG strategy could be designed based on this relationship and could complete the intelligent analysis of multi-targets, giving a unique YES or NO output. This is a revolutionary change in biosensing brought about by the development of computer science and DNA nanotechnology, making it is not only a promising tool for clinical use, but also in ecotoxicological analyses and environmental risk assessments, particularly in terms of measuring the levels of xenobiotics and anthropogenic chemicals and evaluating their impact on living organisms in an ecosystem.| Sensitivity enhancing strategy | Targets | Read-out | LOD | Logic gate | Ref. |
|---|---|---|---|---|---|
| LOD: limit of detection, PPIX: Protoporphyrin IX, AuNP: Au nanoparticles, RCA: rolling circle amplification, MGO: magnetic graphene oxide, TMSDR: toehold-mediated strand-displacement reaction, CHA: catalytic hairpin assembly, CRET: chemiluminescence resonance energy-transfer, HCR: hybridization chain reaction, SDA: strand-displacement amplification, NIR Ag2S QDs: near-infrared Ag2S quantum dots. | |||||
| — | DNA&ATP | Fluorescence of PPIX + G-quadruplex | DNA 40 μM ATP 6.9 nM | YES AND NOR | 18 |
| — | Cancer cells | Fluorescence of FAM | — | AND OR NOT | 19 |
| — | Ions | Colorimetric signals of AuNPs | — | OR AND INHIBIT XOR NOR NAND XNOR | 20 |
| RCA | miRNA | Fluorescence of SYBR Green I | 0.05 nM | AND | 21 |
| Feedback + RCA + MGO | DNA | Fluorescence of SYBR Green I | 0.84 nM | YES–AND–AND network | 65 |
| TMSDR feedback | DNA | Colorimetric signals of AuNPs | 2 nM in 20 min, 0.2 nM in 4 h | OR | 62 |
| CHA | DNA&miRNA | CRET signals | 0.67 pM | YES AND | 49 |
| CHA + AuNP + DNAzyme | DNA | Absorbance values | 100 aM | AND YES | 73 |
| CHA + HCR + DNAzyme | miRNA | CRET signals | 0.72 pM | AND | 74 |
| SDA | DNA | Fluorescence of FAM | 10 pM | YES–AND | 22 |
| TMSDR feedback + DNA walker | DNA | Electrical signals | 36 fM | AND OR combinatorial logic gate (OR and AND) | 23 |
| DNAzyme | DNA | Fluorescence of FAM | — | AND | 83 |
| Semisynthetic luciferase | miRNA | Bioluminescence signals | 400 pM | AND | 86 |
| NIR Ag2S QDs | miRNA | Fluorescence of NIR Ag2S quantum QDs | 100 fM | YES AND OR | 87 |
However, SDLG has some limitations and development bottlenecks: (1) various enzymes, labeled nucleic acid strands, and nucleic acid amplification methods are frequently used for high sensitivity, making the detection system complicated and time-consuming. A simpler and low-cost logic detection platform needs to be developed. In this process, certain materials with excellent functions, such as near-infrared Ag2S quantum dots,87 may be able to effect the measurement of low-abundance molecules without time-consuming radiolabeling or complex amplification;88,89 (2) SDLG strategies have achieved various functions at the cell level. In addition to the aforementioned analysis of biomarkers on the cell surface to identify cancer cells,19,51,90 there are also logical operations on the cell surface to regulate cell adhesion behavior.91In situ detection in living cells has the unique advantage of locating the source of targets. Owing to the biocompatibility of DNA nanomaterials, SDLG has the potential to enter the cell for in situ logical operations. Recently, Bai et al. reported a DNA circuit that could identify cancer cells by the logical analysis of telomerase and microRNAs in situ.92 This is an area that “silicon-based computing cannot reach” and an important direction that needs to be studied more deeply in the future; (3) higher-order logic circuits that could perform comprehensive analysis of more than three targets are difficult to design at the DNA molecular level. These circuits need to be better designed and conceived; (4) the output form of YES/NO is nonlinear, the boundary value of the transition in the biological monitoring process thus needs to be evaluated. Namely, for the YES/NO index of multi-target comprehensive logic analyses, a related evaluation and judgment system needs to be established.
In conclusion, SDLG has shown its magic power in “real-smart” biosensing. Though it is still in the stage of principle verification and in-tube detection, we believe that SDLG-based diseases diagnosis will definitely reach a new level in the not-too-distant future.
Conflicts of interest
The authors declare that they have no competing interests.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 81972027, 82030066) and Medical pre-research project of the Army Medical University (2018XYY04).References
- D. E. Rozen, S. McGrew and A. D. Ellington, Curr. Biol., 1996, 6, 254–257 CrossRef CAS PubMed.
- Y. Cao, J. Romero, J. P. Olson and M. Degroote, Chem. Rev., 2019, 119, 10856–10915 CrossRef CAS PubMed.
- C. Qian, X. Lin, X. Lin, J. Xu, J. Xu, Y. Sun, E. Li, B. Zhang and H. Chen, Light: Sci. Appl., 2020, 9, 59 CrossRef CAS PubMed.
- I. A. Basheer and M. Hajmeer, J. Microbiol. Methods, 2000, 43, 3–31 CrossRef CAS.
- L. M. Adleman, Science, 1994, 266, 1021–1024 CrossRef CAS PubMed.
- K. Sakamoto, H. Gouzu, K. Komiya, D. Kiga, S. Yokoyama, T. Yokomori and M. Hagiya, Science, 2000, 288, 1223–1226 CrossRef CAS PubMed.
- W. E. Arter, Y. Yusim, Q. Peter, C. G. Taylor, D. Klenerman, U. F. Keyser and T. P. J. Knowlee, ACS Nano, 2020, 14, 5763–5771 CrossRef CAS PubMed.
- X. Song and J. Reif, ACS Nano, 2019, 13, 6256–6268 CrossRef CAS PubMed.
- Y. H. Lai, S. C. Sun and M. C. Chuang, Biosensors, 2014, 4, 273–300 CrossRef PubMed.
- J. Wang and E. Katz, Anal. Bioanal. Chem., 2010, 398, 1591–1603 CrossRef CAS PubMed.
- L. Qian and E. Winfree, Science, 2011, 332, 1196–1201 CrossRef CAS PubMed.
- A. Carbone and N. C. Seeman, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 12577–12582 CrossRef CAS PubMed.
- L. Wang, J. G. Hall, M. Lu, Q. Liu and L. M. Smith, Nat. Biotechnol., 2001, 19, 1053–1059 CrossRef CAS PubMed.
- T. Fu, Y. Lyu, H. Liu, R. Peng, X. Zhang, M. Ye and W. Tan, Trends Biochem. Sci., 2018, 43, 547–560 CrossRef CAS PubMed.
- E. A. Barnoy, R. Popovtzer and D. Fixler, J. Biophotonics, 2020, 13(9), e202000158 CrossRef CAS PubMed.
- G. Seelig, D. Soloveichik, D. Y. Zhang and E. Winfree, Science, 2006, 314, 1585–1588 CrossRef CAS PubMed.
- J. Zhu, L. Zhang and E. Wang, Chem. Commun., 2012, 48, 11990–11992 RSC.
- J. Zhu, L. Zhang, Z. Zhou, S. Dong and E. Wang, Anal. Chem., 2014, 86, 312–316 CrossRef CAS PubMed.
- M. You, G. Zhu, T. Chen, M. J. Donovan and W. Tan, J. Am. Chem. Soc., 2015, 137, 667–674 CrossRef CAS PubMed.
- S. Bi, Y. Yan, S. Hao and S. Zhang, Angew. Chem., Int. Ed., 2010, 49, 4438–4442 CrossRef CAS PubMed.
- Y. Chen, Y. Song, F. Wu, W. Liu, B. Fu, B. Feng and X. Zhou, Chem. Commun., 2015, 51, 6980–6983 RSC.
- C. Xue, S. Xiao, C. H. Ouyang, C. C. Li, Z. H. Gao, Z. F. Shen and Z. S. Wu, Anal. Chim. Acta, 2019, 1051, 179–186 CrossRef CAS PubMed.
- K. Wang, M. Q. He, F. H. Zhai, J. Wang, R. H. He and Y. L. Yu, Biosens. Bioelectron., 2018, 105, 159–165 CrossRef CAS PubMed.
- T. Song and H. Liang, J. Am. Chem. Soc., 2012, 134, 10803–10806 CrossRef CAS PubMed.
- S. Erbas-Cakmak, S. Kolemen, A. C. Sedgwick, T. Gunnlaugsson, T. D. James, J. Yoon and E. U. Akkaya, Chem. Soc. Rev., 2018, 47, 2228–2248 RSC.
- D. Y. Zhang and G. Seelig, Nat. Chem., 2011, 3, 103–113 CrossRef CAS PubMed.
- W. Tang, W. Zhong, Y. Tan, G. A. Wang, F. Li and Y. Liu, Top Curr. Chem., 2020, 378, 10 CrossRef CAS PubMed.
- D. Y. Zhang, A. J. Turberfield, B. Yurke and E. Winfree, Science, 2007, 318, 1121–1125 CrossRef CAS PubMed.
- F. C. Simmel, B. Yurke and H. R. Singh, Chem. Rev., 2019, 119, 6326–6369 CAS.
- B. Yurke and A. P. Mills, Genet. Program. Evol. Mach., 2003, 4, 111 CrossRef.
- D. Y. Zhang and E. Winfree, J. Am. Chem. Soc., 2009, 131, 17303 CrossRef CAS PubMed.
- B. Yurke, A. J. Turberfield, A. P. Mills, Jr., F. C. Simmel and J. L. Neumann, Nature, 2000, 406, 605–608 CrossRef CAS PubMed.
- B. R. Wolfe, N. J. Porubsky, J. N. Zadeh, R. M. Dirks and N. A. Pierce, J. Am. Chem. Soc., 2017, 139(8), 3134–3144 CrossRef CAS PubMed.
- A. J. Genot, D. Y. Zhang, J. Bath and A. J. Turberfield, J. Am. Chem. Soc., 2011, 133(7), 2177–2182 CrossRef CAS PubMed.
- W. Lai, L. Ren, Q. Tang, X. Qu, J. Li, L. Wang, L. Li, C. Fan and H. Pei, ACS Nano, 2018, 12(7), 7093–7099 CrossRef CAS PubMed.
- X. Chen, J. Am. Chem. Soc., 2012, 11,134(1), 263–271 CrossRef PubMed.
- X. Yang, Y. Tang, S. M. Traynor and F. Li, J. Am. Chem. Soc., 2016, 138(42), 14076–14082 CrossRef CAS PubMed.
- W. Lai, X. Xiong, F. Wang, Q. Li, L. Li, C. Fan and H. Pei, ACS Synth. Biol., 2019, 8(9), 2106–2112 CrossRef CAS PubMed.
- Y. Dai, A. Furst and C. C. Liu, Trends Biotechnol., 2019, 37, 1367–1382 CrossRef CAS PubMed.
- M. Ogihara and A. Ray, Algorithmica, 1999, 25, 239–250 CrossRef.
- G. Seelig, D. Soloveichik, D. Y. Zhang and E. Winfree, Science, 2006, 314, 1585–1588 CrossRef CAS PubMed.
- W. Li, F. Zhang, H. Yan and Y. Liu, Nanoscale, 2016, 8, 3775–3784 RSC.
- W. E. Arter, Y. Yusim, Q. Peter, C. G. Taylor, D. Klenerman, U. F. Keyser and T. P. J. Knowles, ACS Nano, 2020, 14(5), 5763–5771 CrossRef CAS PubMed.
- F. Wang, H. Lv, Q. Li, J. Li, X. Zhang, J. Shi, L. Wang and C. Fan, Nat. Commun., 2020, 11(1), 121 CrossRef CAS.
- X. Xiong, M. Xiao, W. Lai, L. Li, C. Fan and H. Pei, Angew. Chem., Int. Ed., 2020, 60, 3397–3401 CrossRef PubMed.
- D. Y. Zhang and E. Winfree, Nucleic Acids Res., 2010, 38(12), 4182–4197 CrossRef CAS PubMed.
- J. S. Wang and D. Y. Zhang, Nat. Chem., 2015, 7(7), 545–553 CrossRef CAS PubMed.
- J. S. Wang, Y. H. Yan and D. Y. Zhang, Nat. Chem., 2017, 9(12), 1222–1228 CrossRef CAS PubMed.
- S. Yue, T. Zhao, H. Qi, Y. Yan and S. Bi, Biosens. Bioelectron., 2017, 94, 671–676 CrossRef CAS PubMed.
- N. A. Skrypina, L. P. Savochkina and R. Beabealashvilli, Nucleosides, Nucleotides Nucleic Acids, 2004, 23, 891–893 CrossRef CAS PubMed.
- M. You, L. Peng, N. Shao, L. Zhang, L. Qiu, C. Cui and W. Tan, J. Am. Chem. Soc., 2014, 136, 1256–1259 CrossRef CAS PubMed.
- R. R. Breaker and G. F. Joyce, Chem. Biol., 1994, 1(4), 223–229 CrossRef CAS PubMed.
- M. Moshe, J. Elbaz and I. Willner, Nano Lett., 2009, 9(3), 1196–1200 CrossRef CAS PubMed.
- P. J. Huang, J. Lin, J. Cao, M. Vazin and J. Liu, Anal. Chem., 2014, 86(3), 1816–1821 CrossRef CAS PubMed.
- J. Liu and Y. Lu, Angew. Chem., Int. Ed., 2007, 119, 7731–7734 CrossRef.
- L. Yue, S. Wang, A. Cecconello, J. Lehn and I. Willner, ACS Nano, 2017, 11(12), 12027–12036 CrossRef CAS PubMed.
- S. W. Oh, A. Pereira, T. Zhang, T. Li, A. Lane and J. Fu, Angew. Chem., Int. Ed., 2018, 57(40), 13086–13090 CrossRef CAS PubMed.
- R. Peng, X. Zheng, Y. Lyu, L. Xu, X. Zhang, G. Ke, Q. Liu, C. You, S. Huan and W. Tan, J. Am. Chem. Soc., 2018, 140(31), 9793–9796 CrossRef CAS PubMed.
- S. Dai, Y. Zhou, G. Cheng, P. He and Y. Fang, Talanta, 2020, 217, 121079 CrossRef CAS PubMed.
- J. Zheng, X. Ji, M. Du, S. Tian and Z. He, Nanoscale, 2018, 10, 17206–17211 RSC.
- Y. Yang, J. Huang, X. Yang, K. Quan, H. Wang, L. Ying, N. Xie, M. Ou and K. Wang, J. Am. Chem. Soc., 2015, 137, 8340–8343 CrossRef CAS PubMed.
- B. Wei, D. Yao, B. Zheng, X. Zhou, Y. Guo, X. Li, C. Li, S. Xiao and H. Liang, ACS Appl. Mater. Interfaces, 2019, 11, 19724–19733 CrossRef CAS PubMed.
- L. Blanco, A. Bernad, J. M. Lázaro, G. Martín, C. Garmendia and M. Salas, J. Biol. Chem., 1989, 264, 8935–8940 CrossRef CAS.
- H. Dong, C. Wang, Y. Xiong, H. Lu, H. Ju and X. Zhang, Biosens. Bioelectron., 2013, 41, 348–353 CrossRef CAS PubMed.
- S. Bi, S. Yue, Q. Wu and J. Ye, Biosens. Bioelectron., 2016, 83, 281–286 CrossRef CAS PubMed.
- P. Yin, H. M. Choi, C. R. Calvert and N. A. Pierce, Nature, 2008, 451, 318–322 CrossRef CAS PubMed.
- B. Li, A. D. Ellington and X. Chen, Nucleic Acids Res., 2011, 39, e110 CrossRef CAS PubMed.
- Q. Li, F. Zeng, N. Lyu and J. Liang, Analyst, 2018, 143, 2304–2309 RSC.
- H. Wang, H. Wang, I. Willner and F. Wang, Top Curr. Chem., 2020, 378, 20 CrossRef CAS PubMed.
- Y. Long, C. Zhou, C. Wang, H. Cai, C. Yin, Q. Yang and D. Xiao, Sci. Rep., 2016, 6, 23949 CrossRef CAS PubMed.
- Y. Nie, X. Yuan, P. Zhang, Y. Q. Chai and R. Yuan, Anal. Chem., 2019, 91, 3452–3458 CrossRef CAS PubMed.
- J. Wen, J. Chen, L. Zhuang and S. Zhou, Biosens. Bioelectron., 2016, 79, 656–660 CrossRef CAS PubMed.
- H. Ravan, M. Amandadi and S. Esmaeili-Mahani, Anal. Chem., 2017, 89, 6021–6028 CrossRef CAS PubMed.
- S. Bi, S. Yue, Q. Wu and J. Ye, Chem. Commun., 2016, 52, 5455–5458 RSC.
- Q. Guo, X. Yang, K. Wang, W. Tan, W. Li, H. Tang and H. Li, Nucleic Acids Res., 2009, 37, e20 CrossRef PubMed.
- F. Zhang, J. Nangreave, Y. Liu and H. Yan, J. Am. Chem. Soc., 2014, 136, 11198–11211 CrossRef CAS PubMed.
- T. G. Cha, J. Pan, H. Chen, J. Salgado, X. Li, C. Mao and J. H. Choi, Nat. Nanotechnol., 2014, 9, 39–43 CrossRef CAS PubMed.
- K. Lund, A. J. Manzo, N. Dabby, N. Michelotti, A. Johnson-Buck, J. Nangreave, S. Taylor, R. Pei, M. N. Stojanovic, N. G. Walter, E. Winfree and H. Yan, Nature, 2010, 465, 206–210 CrossRef CAS PubMed.
- M. Q. He, K. Wang, W. J. Wang, Y. L. Yu and J. H. Wang, Anal. Chem., 2017, 89, 9292–9298 CrossRef CAS PubMed.
- H. Zhang, M. Lai, A. Zuehlke, H. Peng, X. F. Li and X. C. Le, Angew. Chem., Int. Ed., 2015, 54, 14326–14330 CrossRef CAS PubMed.
- L. Gong, Z. Zhao, Y. F. Lv, S. Y. Huan, T. Fu, X. B. Zhang, G. L. Shen and R. Q. Yu, Chem. Commun., 2015, 51, 979–995 RSC.
- C. W. Brown 3rd, M. R. Lakin, D. Stefanovic and S. W. Graves, ChemBioChem, 2014, 15, 950–954 CrossRef PubMed.
- C. W. Brown 3rd, M. R. Lakin, E. K. Horwitz, M. L. Fanning, H. E. West, D. Stefanovic and S. W. Graves, Angew. Chem., Int. Ed., 2014, 53, 7183–7187 CrossRef PubMed.
- X. Zheng, J. Yang, C. Zhou, C. Zhang, Q. Zhang and X. Wei, Nucleic Acids Res., 2019, 47, 1097–1109 CrossRef CAS PubMed.
- J. Yang, R. Wu, Y. Li, Z. Wang, L. Pan, Q. Zhang, Z. Lu and C. Zhang, Nucleic Acids Res., 2018, 46, 8532–8541 CrossRef CAS PubMed.
- D. Chang, K. T. Kim, E. Lindberg and N. Winssinger, ACS Sens., 2020, 5, 807–813 CrossRef CAS PubMed.
- Y. Zhang, G. Hong, Y. Zhang, G. Chen, F. Li, H. Dai and Q. Wang, ACS Nano, 2012, 6, 3695–3702 CrossRef CAS PubMed.
- X. He, Z. Li, M. Chen and N. Ma, Angew. Chem., Int. Ed., 2014, 53, 14447–14450 CrossRef CAS PubMed.
- P. Miao, Y. Tang, B. Wang and F. Meng, Anal. Chem., 2016, 88, 7567–7573 CrossRef CAS PubMed.
- T. Song, S. Shah, H. Bui, S. Garg, A. Eshra, D. Fu, M. Yang, R. Mokhtar and J. Reif, J. Am. Chem. Soc., 2019, 141(42), 16539–16543 CrossRef CAS PubMed.
- X. Qu, S. Wang, Z. Ge, J. Wang, G. Yao, J. Li, X. Zuo, J. Shi, S. Song, L. Wang, L. Li, H. Pei and C. Fan, J. Am. Chem. Soc., 2017, 139(30), 10176–10179 CrossRef CAS PubMed.
- M. Bai, F. Chen, X. Cao, Y. Zhao, J. Xue, X. Yu, C. Fan and Y. Zhao, Angew. Chem., Int. Ed., 2020, 59(32), 13267–13272 CrossRef CAS PubMed.
Footnote |
| † Shuang Zhao and Lianyu Yu contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |