Open Access Article
Open Access ArticleTuning DNA–nanoparticle conjugate properties allows modulation of nuclease activity†
Jeff C.
Hsiao
 a,
Tomas
Buryska
a,
Tomas
Buryska
 a,
Eunjung
Kim
a,
Eunjung
Kim
 b,
Philip D.
Howes
b,
Philip D.
Howes
 *a and
Andrew J.
deMello
*a and
Andrew J.
deMello
 *a
*a
aInstitute for Chemical and Bioengineering, Department of Chemistry and Applied Biosciences, ETH Zürich, Vladimir Prelog Weg 1, 8093 Zürich, Switzerland. E-mail: philip.howes@chem.ethz.ch; andrew.demello@chem.ethz.ch
bDivision of Bioengineering and Department of Bioengineering and Nano-Bioengineering, Incheon National University, Incheon 22012, Republic of Korea
First published on 25th February 2021
Abstract
Enzyme–nanoparticle interactions can give rise to a range of new phenomena, most notably significant enzymatic rate enhancement. Accordingly, the careful study and optimization of such systems is likely to give rise to advanced biosensing applications. Herein, we report a systematic study of the interactions between nuclease enzymes and oligonucleotide-coated gold nanoparticles (spherical nucleic acids, SNAs), with the aim of revealing phenomena worthy of evolution into functional nanosystems. Specifically, we study two nucleases, an exonuclease (ExoIII) and an endonuclease (Nt.BspQI), via fluorescence-based kinetic experiments, varying parameters including enzyme and substrate concentrations, and nanoparticle size and surface coverage in non-recycling and a recycling formats. We demonstrate the tuning of nuclease activity by SNA characteristics and show that the modular units of SNAs can be leveraged to either accelerate or suppress nuclease kinetics. Additionally, we observe that the enzymes are capable of cleaving restriction sites buried deep in the oligonucleotide surface layer and that enzymatic rate enhancement occurs in the target recycling format but not in the non-recycling format. Furthermore, we demonstrate a new SNA phenomenon, we term ‘target stacking’, whereby nucleic acid hybridization efficiency increases as enzyme cleavage proceeds during the beginning of a reaction. This investigation provides important data to guide the design of novel SNAs in biosensing and in vitro diagnostic applications.
1 Introduction
Beyond its genetic function, DNA has become firmly established as a functional polymeric material with diverse applications.1 Intertwined with the advancement of DNA-based materials is the development of colloidal nanoparticles. The two have so often been used together that they essentially comprise a field in themselves,2 with their combination giving rise to hybrid nanomaterials that blend the recognition, structural and/or catalytic functions of DNA with the myriad functionalities of different colloidal nanoparticles, including strong photoluminescence, superparamagnetism, intense plasmonics and efficient photothermal heating. Highlight applications of such hybrid constructs include theranostics,3 cellular sensing and imaging,4 and crystal engineering.5Nanoparticles coated with a dense layer of highly oriented oligonucleotides—a class of spherical nucleic acid (SNA)—exhibit unique phenomena that are distinct in nature from the isolated oligonucleotides and nanoparticles.6 For instance, SNAs show higher binding constants and sharper melting transitions with their complements than free oligonucleotides,7,8 and they readily enter cells without the need for transfection agents.6,9,10 SNAs are highly tunable, as various sizes of nanoparticles (typically between 10 and 100 nm), lengths of DNA or RNA (typically up to 50 bases), spacer lengths/compositions and oligonucleotide modifications can serve as tunable modular units.6,11 Herein, we use the term SNA to refer exclusively to oligonucleotide-gold nanoparticle (AuNP) conjugates. In general terms, biomolecular binding and activity at the nanoparticle surface–solution interface can be significantly affected by interfacial factors including binding hindrance or enhancement, reversible adsorption, valence, orientation and curvature.12–15 For enzymes acting on nanoparticle-bound substrates,16 this means a transition from a bulk regime where enzyme activity is dominated by molecular colliding, to a new nano-interfacial regime where the above factors can significantly alter (and sometimes enhance) enzyme activity.16–18
A curious aspect of SNAs is their interaction with nucleases, and specifically how these enzymes interact with DNA/RNA when densely displayed on a nanoparticulate scaffold (as opposed to free in solution). Contemporary studies suggest that SNAs are resistant to degradation by nucleases.19–21 Indeed, this is a critical characteristic for the survival of these constructs in cellular milieu.22 This phenomenon has been rationalized by the fact that the dense nucleic acid shell sterically resists nuclease binding and that the high ion concentration around the nanoparticle core inhibits some nucleases.19,23 However, other reports have shown that the activity of certain nucleases can actually be enhanced when acting on SNAs.23,24 Such an enhancement is not unique to SNAs, with a growing number of reports of enzymatic acceleration when they act at the nanoparticle interface.16–18
Regarding SNAs specifically, Prigodich et al. demonstrated that ribonuclease H (RNase H, a non-sequence-specific endonuclease that cleaves the RNA in an RNA/DNA substrate) shows a 2.5-fold enhanced activity on an RNA/SNA substrate (13 nm, 86 DNA per AuNP) when compared to RNA/DNA in free solution, whereas serum nucleases from fetal bovine serum (FBS) are severely inhibited by the SNA.23 Further, they modulated the coverage of DNAs around the AuNP cores by introducing spacers (polyA, polyT or propyl phosphodiester) and observed that the activity of the RNase H increased as a function of DNA coverage. Additionally, Qu et al. investigated the interaction of three different nuclease enzymes with an SNA (15 nm AuNP, 100 DNA per NP).24 They qualitatively observed that deoxyribonuclease I (DNase I, an endonuclease that non-specifically cleaves ssDNA and dsDNA) was significantly slower at digesting SNA compared to free DNA, with exonuclease I (ExoI, which cleaves ssDNA in the 3′ to 5′ direction) cleaving at a similar rate on SNAs and free DNAs. Interestingly, they observed that exonuclease III (ExoIII, which performs a uni-strand 3′ to 5′ digestion in dsDNA on blunt or recessed 3′ ends) exhibited significantly higher activity towards SNA compared to free DNA. In contrast to the above results, Degliangeli et al. observed that duplex specific nuclease (DSN, which non-specifically cleaves DNA, making double strand breaks in dsDNA, and single strand breaks on the DNA of DNA/RNA duplex) had an initial reaction rate 1.7-fold lower for SNAs compared to free substrates.25 The SNAs used therein had a lower surface coverage (13 nm AuNP, 26 DNA per NP) compared to conventional SNAs, which are typically of higher coverage (e.g. 50–250 on 15 nm AuNPs).26,27 Collectively, these results suggest that the nature of the interaction between SNAs and nucleases is highly dependent on the characteristics of the different enzymes and on the SNAs themselves. With the exception of the Prigodich et al. study,23 investigations that have remarked upon the interaction of specific nucleases with SNAs have tended to do so as a sideline to a different goal. Given the interesting observations to date, it is important to dedicate studies to specifically examine the nature of the interactions of SNAs with different nucleases. Better characterization of SNA behavior relative to free DNA will aid in the development of systems that co-employ nanoparticles and enzymes. To our knowledge, no such systematic study has been conducted before.
In this study, we perform an assessment of the interactions between nucleases and SNAs, with a view to uncovering useful phenomena for evolution into future functional nanosystems. Specifically, we studied two nucleases, an exonuclease (ExoIII) and an endonuclease (Nt.BspQI), using fluorescence-based kinetic assays. We investigated their interactions with SNAs and compared their activity directly with a free DNA analog system in non-recycling and target recycling28 detection formats. We then tuned various SNA features (DNA sequence, DNA surface coverage and AuNP size) to examine their influence on enzyme kinetics. We demonstrate that the catalytic activity of both nucleases can be controllably regulated for both inhibition and enhancement depending on the target detection format and SNA design. Analysis of the full progress curves, in addition to the initial rates, provides insight into kinetic parameters and the mechanisms underlying nuclease inhibition and enhancement. Further, we demonstrate for the first time a phenomenon we term ‘target stacking’, where the SNA exhibits increased hybridization efficiency as the nucleases act.
2 Experimental
2.1 Materials
15, 20 and 40 nm AuNPs were purchased from BBI Solutions (Crumlin, UK). All oligonucleotides were obtained from Microsynth AG (Balgach, Switzerland). Both ExoIII and Nt.BspQI were supplied by New England Biolabs (Massachusetts, US). Nuclease-free water was obtained from Thermo Fisher Scientific (Massachusetts, US). The mineral oil and 96 well half-area microplates used in the fluorescence-based kinetic assays were both purchased from Sigma-Aldrich (Buchs, Switzerland).2.2 Oligonucleotide sequences
The probe sets were designed according to the following criteria: (1) six bases on either side of the endonuclease recognition sequences to allow binding, (2) restricted proximity of guanines and dyes to minimize fluorescence quenching, (3) minimized self-complementarity to stop hairpin formation hindering enzyme access, (4) minimized dimerization to stop SNA aggregation, and (5) flanking sequences were not palindromic or similar to the recognition sequence to also prevent SNA aggregation. Phosphorothioate linkages between select bases and 3′ overhangs were used to inhibit the ExoIII from degrading the non-target sequences. Sequences and their functionalization are shown in the corresponding figures, and also summarized in Table S1.† Note that we refer to the dye-labelled strand as the ‘target probe’, and the quencher-labelled/nanoparticle-bound strand as the ‘quencher probe’. See Fig. S1† for an explanation of the quenching mechanisms used in the experiments described herein.2.3 SNA synthesis and characterization
SNAs were synthesized using a freeze–thaw method developed by Liu et al.29,30 For a typical experiment, thiol-modified oligonucleotides were added to 15, 20, or 40 nm citrate-capped AuNPs at a ratio of 6 × 1013 DNA per cm2 of AuNP surface area, and incubated overnight at room temperature. Next, the mixture was placed in a −80 °C freezer for 2 hours, followed by thawing at room temperature. The SNAs were purified through three successive rounds of centrifugation (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 817g, 20 min, 4 °C) to remove free DNA. After each centrifugation cycle, the supernatant was removed, and the pellet resuspended in nuclease-free water. Finally, the SNA solution was filtered using a Millex-GV 0.22 μm syringe filter (Sigma-Aldrich, Buchs, Switzerland). SNA concentrations were determined by measuring the absorption at 506 nm and applying the following extinction coefficients: 15 nm AuNPs (3.97 × 108 M−1 cm−1), 20 nm AuNPs (9.11 × 108 M−1 cm−1), and 40 nm AuNPs (6.12 × 109 M−1 cm−1).31 SNA hydrodynamic diameter was measured by dynamic light scattering (DLS) at 25 °C using a Zeta Nano ZS (Malvern Instruments, Malvern, UK). An average of three measurements, with each measurement constituting 10 runs of 10 seconds, was taken on 0.18 nM SNA samples. All SNAs were stored at 4 °C.
817g, 20 min, 4 °C) to remove free DNA. After each centrifugation cycle, the supernatant was removed, and the pellet resuspended in nuclease-free water. Finally, the SNA solution was filtered using a Millex-GV 0.22 μm syringe filter (Sigma-Aldrich, Buchs, Switzerland). SNA concentrations were determined by measuring the absorption at 506 nm and applying the following extinction coefficients: 15 nm AuNPs (3.97 × 108 M−1 cm−1), 20 nm AuNPs (9.11 × 108 M−1 cm−1), and 40 nm AuNPs (6.12 × 109 M−1 cm−1).31 SNA hydrodynamic diameter was measured by dynamic light scattering (DLS) at 25 °C using a Zeta Nano ZS (Malvern Instruments, Malvern, UK). An average of three measurements, with each measurement constituting 10 runs of 10 seconds, was taken on 0.18 nM SNA samples. All SNAs were stored at 4 °C.
2.4 Surface coverage measurement
The average number of DNA molecules per AuNP, comprising the SNAs, was calculated by comparison with a standard curve. The standard curve was constructed by treating a solution of known concentration of thiolated DNA to the exact same purification procedure as applied to the SNAs. A comparison of the curves generated by the SNAs and thiolated DNA was then performed. The concentration of thiolated DNA for the calibration curve was chosen based on an estimate of the DNA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) AuNP ratio, to ensure that the test and calibration curves traversed a similar concentration range. To prepare the test and calibration samples, the samples (SNAs or thiolated DNA) were incubated in aqueous solutions of 500 mM DTT overnight at 4 °C (displacing the thiolated DNA from the AuNP cores). After centrifugation, supernatants were collected and used to produce a dilution series in a 96-well plate. Diamond Nucleic Acid Dye (Promega AG, Dübendorf, Switzerland) was applied to stain the oligonucleotides in both sets of dilutions according to the manufacturer's protocol. 100 μL of each solution was transferred into a microtiter plate followed by the addition of 50 μL of mineral oil. After incubation for 30 minutes, fluorescence from each well was read in a Spark™ 10M multimode microplate reader (Tecan, Männedorf, Switzerland). This generated two curves: one from the displaced DNA sample and one from the sample of known DNA concentration. The ratio of the slopes between the curves of the unknown and known samples multiplied by the initial estimate of the SNA surface coverage yielded an estimate of the mean surface coverage.
AuNP ratio, to ensure that the test and calibration curves traversed a similar concentration range. To prepare the test and calibration samples, the samples (SNAs or thiolated DNA) were incubated in aqueous solutions of 500 mM DTT overnight at 4 °C (displacing the thiolated DNA from the AuNP cores). After centrifugation, supernatants were collected and used to produce a dilution series in a 96-well plate. Diamond Nucleic Acid Dye (Promega AG, Dübendorf, Switzerland) was applied to stain the oligonucleotides in both sets of dilutions according to the manufacturer's protocol. 100 μL of each solution was transferred into a microtiter plate followed by the addition of 50 μL of mineral oil. After incubation for 30 minutes, fluorescence from each well was read in a Spark™ 10M multimode microplate reader (Tecan, Männedorf, Switzerland). This generated two curves: one from the displaced DNA sample and one from the sample of known DNA concentration. The ratio of the slopes between the curves of the unknown and known samples multiplied by the initial estimate of the SNA surface coverage yielded an estimate of the mean surface coverage.
2.5 Surface coverage tuning
System 1 SNAs with different DNA surface coverages were synthesized by incubating the thiolated oligonucleotides with 15 nm AuNPs at different ratios prior to the freeze–thaw cycle. The following ratios of thiolated DNA per cm2 of AuNP surface area were used: (1) 6 × 1013, (2) 5 × 1013, (3) 2.5 × 1013. System 2 SNAs with different DNA surface coverages were obtained by displacing the bound DNA on the SNAs with thiolated PEG. This backfilling displacement procedure was performed by adding 800 Da thiolated PEG (Sigma-Aldrich, Buchs, Switzerland) to the SNA solutions at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 500
1 and 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PEG
1 PEG![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SNA ratios. The mixture was then incubated for 1 hour before purifying the SNAs through three successive rounds of centrifugation (20
SNA ratios. The mixture was then incubated for 1 hour before purifying the SNAs through three successive rounds of centrifugation (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 817g, 20 minutes, 4 °C), supernatant removal, and resuspension. The DNA surface coverages of both System 1 and System 2 SNAs after modification were quantified using the protocol stated above.
817g, 20 minutes, 4 °C), supernatant removal, and resuspension. The DNA surface coverages of both System 1 and System 2 SNAs after modification were quantified using the protocol stated above.
2.6 Probe set design
Two probe sets were designed (termed System 1 and System 2), each compatible with both of the enzymes to yield four working modes. These four modes are split into non-recycling and recycling target detection formats. System 1 employs a recycling format while System 2 employs a non-recycling format for ExoIII. The vice versa is true for Nt.BspQI. In the non-recycling case, the enzyme cleaves the target probe. This leads to the release of a dye molecule and the evolution of a fluorescence signal. In the recycling case,28 the enzyme cleaves the quencher probe and releases the target probe, whereupon it can hybridize with remaining quencher probes and initiate further cleavage events. Thus, there is a lag phase in the evolution of fluorescence, which is indicative of the time taken for the enzyme to cleave enough quencher probes such that there is a detectable population of target probes free in solution (i.e. unquenched). Additionally, System 3 was designed where the 3′ recognition site of ExoIII was placed on the inside of the SNA, close to the AuNP surface, rather than outside and away from the surface. System 3 employs a non-recycling system, allowing for direct comparison of ExoIII activity with System 2, where the recognition site is on the outside. In this article, we use the term substrate to refer to hybridized double-stranded target/quencher probe sequences. Schematics are presented alongside the relevant results.2.7 Enzyme kinetic assays
SNAs and free probes were incubated with their complimentary target sequence (1 target per 7 SNA-bound/free DNA quencher probes) at the desired concentrations in 1× NEBuffer™ 3.1 (New England Biolabs, BioConcept AG, Allschwil, Switzerland) comprising 100 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2, and 100 μg ml−1 BSA, at pH 7.9, and referred to herein as NEB 3.1. The solution was allowed to hybridize at room temperature for 30 minutes. A microtiter plate was then prepared with a range of hybridized quencher/target concentrations obtained from appropriate dilutions. Each well contained 20 μL of reaction volume, with 15 μL mineral oil added above to prevent evaporation, and the microtiter plate was placed on ice. To prepare ExoIII and Nt.BspQI for experimentation, the enzymes were spun through a 7 K MWCO Zeba™ Spin Desalting Column (Thermo Fisher Scientific) following the manufacturer's protocol to perform a buffer exchange from the enzyme's storage buffer to 1× NEB 3.1, the reaction buffer. SNAs demonstrated significantly improved stability in 1× NEB 3.1 than in the enzyme storage buffers (Fig. S2†). The enzyme concentration in the filtrate was then determined using a NanoDrop Lite™ (Thermo Fisher Scientific) and diluted to the desired concentration using 1× NEB 3.1. Finally, 5 μL of enzyme solution was added to the reaction solution and mixed using the pipette tip, and the microtiter plate was immediately transferred to a microplate reader for fluorescence measurements. ExoIII kinetic assays were performed in a Spark™ 10M multimode microplate reader (Tecan) maintained at 37 °C, and the fluorescence of each well was measured every 20 seconds over a period of 2 hours, at 520 nm and with excitation at 475 nm. Similarly, Nt.BspQI kinetic assays were performed in a CLARIOstar multimode microplate reader (BMG Labtech) maintained at 37 °C and the fluorescence of each well was measured every 40 or 50 seconds for approximately 5 hours at 520 nm with excitation at 475 nm. All samples were measured in triplicate per experiment and in experimental duplicate (N = 2, n = 3).In the non-recycling study, for the varied enzyme concentration experiments, ExoIII-to-substrate molar ratios were varied between 0.25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 5
1 and 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, while the Nt.BspQI-to-substrate molar ratios tested were between 3.75
1, while the Nt.BspQI-to-substrate molar ratios tested were between 3.75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 7.5
1 and 7.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. We used the obtained results to guide the ExoIII (25 nM) and Nt.BspQI (125 nM) concentrations used in the subsequent non-recycling experiments. In the varied substrate concentration experiments, the substrate-to-ExoIII molar ratios were varied between 0.1
1. We used the obtained results to guide the ExoIII (25 nM) and Nt.BspQI (125 nM) concentrations used in the subsequent non-recycling experiments. In the varied substrate concentration experiments, the substrate-to-ExoIII molar ratios were varied between 0.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2.4
1 and 2.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, while the substrate-to-Nt.BspQI molar ratios were varied between 0.04
1, while the substrate-to-Nt.BspQI molar ratios were varied between 0.04![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 0.6
1 and 0.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.
1.
In the recycling study, for the varied enzyme concentration experiment, ExoIII-to-substrate molar ratios were varied between 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 6
1 and 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, while the Nt.BspQI-to-substrate molar ratios studied were between 3
1, while the Nt.BspQI-to-substrate molar ratios studied were between 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 10
1 and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. We used the obtained results to guide the ExoIII (60 nM) and Nt.BspQI (180 nM) concentrations used in the subsequent recycling experiments. For the varied substrate concentration experiments, substrate-to-ExoIII molar ratios were tested between 0.08
1. We used the obtained results to guide the ExoIII (60 nM) and Nt.BspQI (180 nM) concentrations used in the subsequent recycling experiments. For the varied substrate concentration experiments, substrate-to-ExoIII molar ratios were tested between 0.08![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1.33
1 and 1.33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, while the substrate-to-Nt.BspQI molar ratios were between 0.03
1, while the substrate-to-Nt.BspQI molar ratios were between 0.03![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 0.42
1 and 0.42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.
1.
Maximum reaction rate refers to the slope of the linear region or the maximum reaction velocity in the progress curve and correspond to the steady state conditions. This is normalized to enzyme concentration, therefore expressed in units per s. We do not use initial rate/velocity due to the lag phases observed in various experiments.
2.8 Data analysis
In all experiments, control wells were included to determine the expected fluorescence signal from a fully digested substrate complex. Here, the control reaction solution was identical to the reaction solution, except substituting in a non-complementary dye-labelled target probe (i.e. yielding a fluorescence signal is the un-quenched state, but controlling for any secondary effects in the system, for example non-specific binding). This fluorescence value was then equated to the maximum substrate turnover in a given reaction. The point of minimum fluorescence was equated to zero substrate turnover.Obtained traces were averaged from six measurements, two independent batches of three replicates. The raw fluorescence data traces were analyzed in OriginPro 9.5E (OriginLab Corporation, Northampton, USA) using a logistic sigmoidal model that best fitted the data with R2 > 0.98. Models used in fitting obtained reaction slope values were hyperbolic or cooperative as implemented in the program. All equations are provided in the ESI.†
The calculation of lag time was standardized across all progress curves. These were calculated by fitting a sigmoidal curve to the kinetic data. The reported value is the time at which the function reached 1% of its maximal value, with results quoted as mean ± standard deviation.
3 Results
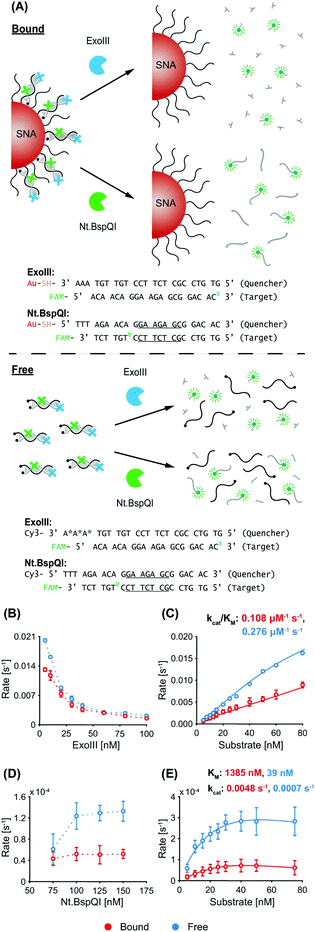
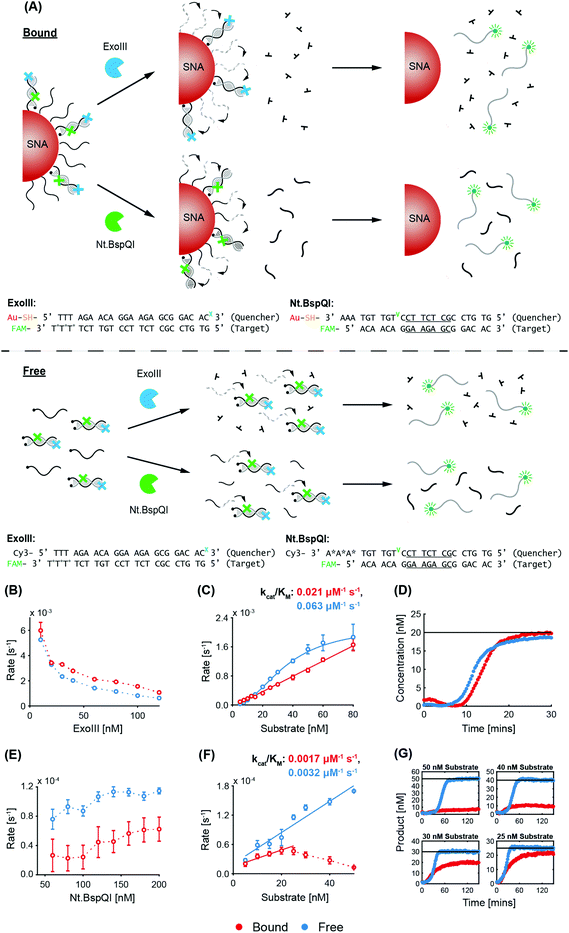
Exonuclease III (ExoIII) was chosen due to its widespread use in biological sensing applications, and in particular, systems co-employing nanomaterials and enzymes.32–35 ExoIII is a non-processive enzyme that acts on double-stranded DNA (dsDNA), where it cleaves mononucleotides from the 3′-hydroxyl terminus of one strand, leaving the other strand intact.36,37 The preferred substrates are blunt or recessed 3′-termini of dsDNA, but the enzyme can also act at nicks or abasic sites. Along with its ease of availability and high catalytic activity, an advantage of ExoIII is its non-sequence specificity, giving it wide applicability (in biosensing,38 for example) compared to site-specific endonucleases. Nt.BspQI was chosen as the representative endonuclease. It is a site-specific DNA nicking endonuclease39 that nicks one strand in a duplex, and has also been used in biosensing applications.40,41 Due to its specificity and non-palindromic restriction sequence, Nt.BspQI is a promising endonuclease for co-employment with SNAs. Unless otherwise stated, the AuNP core of the SNAs was a 15 nm particle. Note that, throughout the text, we refer to the dye-labelled strand as the ‘target probe’, and the quencher-labelled/nanoparticle-bound strand as the ‘quencher probe’ (these are indicated in the corresponding figures).3.1 Non-recycling study
A schematic of the two enzymatic processes (and corresponding DNA sequences) in a non-recycling detection format are depicted in Fig. 1A. In this configuration, ExoIII binds to the blunt-ended duplex substrate and begins non-processive cleavage (i.e. multiple binding/cleavage events) of the target probe which is fluorescently labelled. While initially quenched (via nanosurface energy transfer (NSET) in the bound case and fluorescence resonance energy transfer (FRET) in the free case),42 removal of a sufficient number of nucleotides releases the degraded target probe, with the fluorescent tags now being able to fluoresce. In contrast, Nt.BspQI binds and nicks the target probe of the substrate at its specific restriction site, dividing it into two distinct sequences. The nicked sequences are designed to release from the quencher probe at the reaction temperature, releasing the fluorescently-labelled fragment from NSET- and FRET-based quenching in the bound and free case, respectively. In this non-recycling scenario, the SNA retains its dense coating of oligonucleotides, while nuclease degradation occurs. Therefore, the enzymes experience a similar density of DNA throughout the entire kinetic run. We leveraged this non-recycling design to study how ExoIII and Nt.BspQI activity is affected by (1) enzyme concentration, (2) substrate concentration, (3) recognition site location, (4) SNA surface coverage and (5) AuNP size. In these experiments, System 1 bound quencher probe, free quencher probe, and target probe were used for Nt.BspQI, while System 2 bound quencher probe, free quencher probe (protected), and target probe were used for the ExoIII study (see Fig. 1A and Table S1† for oligonucleotide nomenclature and sequences).ExoIII. With constant quencher and target probe concentrations, an increasing ExoIII concentration resulted in suppressed maximum reaction rates (normalized to enzyme concentration), which is likely due to increasing competition between enzymes for substrates (Fig. 1B). The free case exhibited higher reaction rates than the bound case at lower ExoIII concentrations, but above 30 nM differences were negligible. The progress curves exhibited a lag phase prior to signal increase (Fig. S3A†), which is consistent with previous reports that ExoIII exhibits an intrinsic lag that is compounded by the enzyme's strong temperature dependence and the time taken to heat reaction solutions; previous studies found that it exhibits a 10-fold increase in rate between 22 and 40 °C.43 The most significant effect in our study, however, was the fact that the dye-labelled target would only decouple after a sufficient amount of nucleotides had been removed, leading to a further delay in signal development. Increasing ExoIII concentration led to shorter lag times in both the free and bound cases (Fig. S3A†), with the bound case exhibiting a longer lag phase than the free case (bound 4.7 ± 1.3 minutes, free 3.3 ± 1.0 min). This finding is also evident in the time taken to reach the point-of-inflection (Fig. S3B†). During the lag phase there was a slight initial reduction in fluorescence intensity for the bound case, but not in the free case. We term this phenomenon ‘target stacking’ and detail its mechanism in the Discussion section below.
When the ExoIII concentration was fixed at 25 nM, increasing substrate concentrations yielded increased maximum reaction rates for both the free and bound cases (Fig. 1C). Again, a reduced reaction rate and longer lag phase (bound 5.2 ± 0.1 min, free 3.9 ± 1.4 min) were observed for the bound case versus the free case. The specificity constant (kcat/KM), a measure of the catalytic efficiency in transforming substrate into product, of the free case is approximately 2.5 times that of the bound case.
Nt.BspQI. In contrast to ExoIII, increasing the Nt.BspQI concentration did not affect the maximum reaction rate for the bound case, and actually increased it for the free case (Fig. 1D). Nt.BspQI exhibited higher maximum reaction rates on free DNA for all enzyme concentrations tested. As with the ExoIII reaction, the Nt.BspQI progress curves exhibit a lag phase (bound 4.9 ± 0.7 min, free 4.6 ± 0.1 min). There is no apparent relationship between the lag time and enzyme concentration for both the free and bound cases though (Fig. S3C†). When comparing the bound and free scenarios, higher concentrations of enzyme were needed for the bound case to reach reaction completion—not all of the targets were released from the bound quencher probe sequences even with 75 and 100 nM of enzyme (Fig. S3D†). Dynamic light scattering (DLS) measurements revealed that the hydrodynamic diameter of the SNAs increased from 27.1 to 31.6 nm before and after experimentation with the enzyme (50 nM substrate, 125 nM Nt.BspQI) (Fig. S3E†), suggesting significant adsorption to the AuNP surface during the reaction and potential inhibition of Nt.BspQI.44,45
With a fixed Nt.BspQI concentration of 125 nM, an increasing maximum reaction rate was observed with increasing substrate concentration (Fig. 1E and Fig. S3F†). In the bound case, the rate peaks at 40 nM (at 0.8 s−1) after which the reaction rate decreases slightly. This observation is likely due to a higher substrate concentration correlating to an increased amount of SNA in solution and a corresponding increase in Nt.BspQI adsorption and inactivation. In the free case, the rate plateaus at 2.7 s−1 after 30 nM, having reached its Vmax. Over the substrate concentrations tested, Nt.BspQI performed 70–80% slower in the bound versus free case. Interestingly, analysis of the kinetic parameters reveals that the bound case had a significantly higher kcat (0.0048 s−1), while binding less strongly to the SNA (KM of 1385 nM) compared to the free case (0.0007 s−1 and 39 nM, respectively). These kinetic parameters suggest that Nt.BspQI performs faster when its substrate is densely bound onto an AuNP scaffold rather than free in solution; however, these gains in reaction rate are dramatically reversed by difficulties in substrate binding.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.
1.
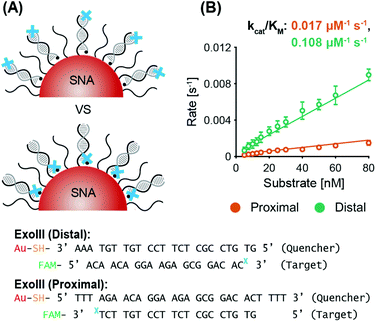
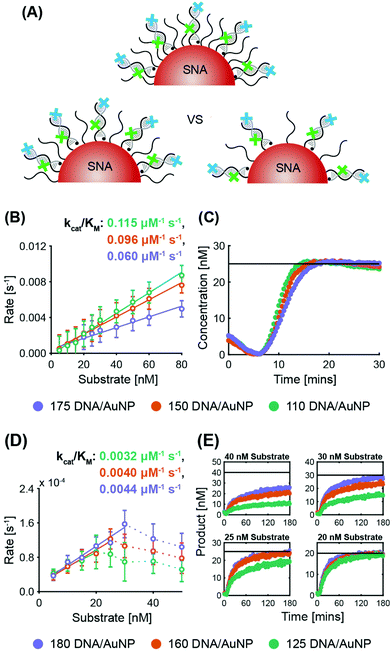
ExoIII. System 2 SNAs were prepared with three different surface coverages (175 DNA per AuNP at 2.5 × 1013 DNA per cm2, 150 DNA per AuNP at 2.1 × 1013 DNA per cm2, and 110 DNA per AuNP at 1.6 × 1013 DNA per cm2). In line with previous observations of the inverse relationship between nuclease activity and sterics, ExoIII maximum reaction rates increased as the surface coverage decreased (Fig. 3B and Fig. S5A†). The SNA with 110 DNA per AuNP had a specificity constant 20% greater than that of the SNAs with 150 DNA per AuNP, and twice that of the SNAs with 175 DNA per AuNP. The three cases exhibit no significant difference in lag time (175 yields 6.2 ± 0.3 min, 150 yields 6.3 ± 0.2 min, and 110 yields 6.4 ± 0.4 min). Once more, target stacking is observed and the characteristic initial dip in fluorescence is clearly demonstrated in the progress curve with 25 nM substrate (Fig. 3C).
Nt.BspQI. Surprisingly, Nt.BspQI demonstrated an opposite trend to that of ExoIII. Using the System 1 quencher probe oligos at three SNA surface coverages (180 DNA per AuNP at 2.5 × 1013 DNA per cm2, 160 DNA per AuNP at 2.3 × 1013 DNA per cm2, and 125 DNA per AuNP at 1.8 × 1013 DNA per cm2), Nt.BspQI performs slightly faster on SNAs with higher surface coverage, despite enhanced steric hindrance (Fig. 3D and Fig. S5B†). SNAs with 180 DNA per AuNP led to a specificity constant approximately 10% greater than that of the SNAs with 160 DNA per AuNP, and approximately 37.5% greater than that of the SNAs with 125 DNA per AuNP. This finding is likely a function of the propensity of Nt.BspQI to adsorb onto the AuNP surface, a phenomenon that would be exacerbated at lower surface densities. Thus, despite reduced steric hindrance, SNAs with lower surface densities substantially reduce the effective concentration of Nt.BspQI acting on available substrates through adsorption. Further highlighting the role of adsorption, we observe that at higher substrate concentrations, Nt.BspQI is unable to liberate all of the fluorescent dyes i.e. the reactions do not appear to go to completion (Fig. 3E). This is reflected in a dramatic drop in reaction rates beyond a corresponding concentration threshold (Fig. 3D), which is ∼25 nM for 125 DNA per AuNP, ∼30 nM for 160 DNA per AuNP, ∼40 nM for 180 DNA per AuNP. We observe that the substrate concentration at which Nt.BspQI cannot complete the reaction is dependent upon the surface coverages and corresponding amount of AuNP surface exposure due to the differences in exposed AuNP surface area. Finally, the three cases exhibit no significant difference in lag time (180 yields 4.3 ± 1.2 min, 160 yields 4.5 ± 1.0 min, and 110 yields 4.4 ± 1.6 min).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and the concentration of quencher probes was kept constant between conditions of different AuNP size. Therefore, in real terms, the molar concentration of SNAs was highest for the 15 nm case and lowest for the 40 nm case since SNAs with smaller cores have less DNA per AuNP.
1, and the concentration of quencher probes was kept constant between conditions of different AuNP size. Therefore, in real terms, the molar concentration of SNAs was highest for the 15 nm case and lowest for the 40 nm case since SNAs with smaller cores have less DNA per AuNP.
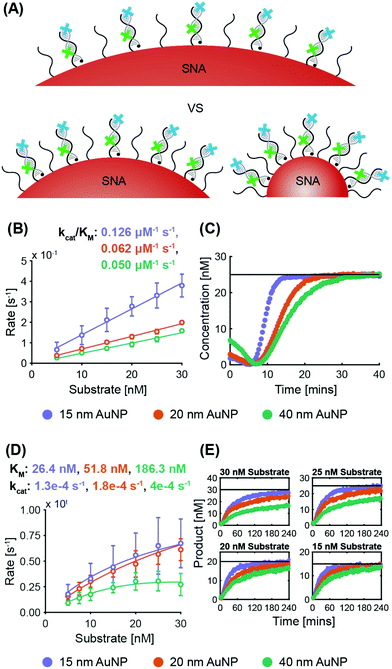
ExoIII. System 2 SNAs were synthesized with three different AuNP cores, obtaining surface coverages and densities of 210 DNA per AuNP at 3.0 × 1013 DNA per cm2 for 15 nm AuNPs, 280 DNA per AuNP at 2.2 × 1013 DNA per cm2 for 20 nm AuNPs, and 700 DNA per AuNP at 1.4 × 1013 DNA per cm2 for 40 nm AuNPs. Fig. 4B shows that the maximum ExoIII reaction rate increased with decreasing particle size. The 15 nm AuNP case yielded a specificity constant twice that of the 20 nm AuNPs, and 2.5 times that of the 40 nm. The smallest SNAs (15 nm) exhibited slightly shorter lag phases (4.5 ± 1.0 min), compared to the 20 nm (5.4 ± 1.2 min) and 40 nm (5.3 ± 1.2 min) SNAs (Fig. S6A†). These results imply that in this case, increased deflection angles between the oligonucleotides on the AuNPs played a more significant role than surface density. Although there are more DNA per surface area on the smaller cores, the increased DNA deflection angle allows easier ExoIII access to its binding sites. The spacing out of DNA at higher deflection angles is more accentuated the further away from the core—an effect that is especially relevant for ExoIII since the 3′ terminal site is at the distal end of the oligonucleotide sequence. Analysis of the progress curves reveals that the initial dip in fluorescence and subsequent recovery is correlated with AuNP size as larger cores exhibited a higher initial fluorescence value followed by a sharper decrease (Fig. 4C).
Nt.BspQI. System 1 SNAs were synthesized with surface coverages and densities of 210 DNA per AuNP at 3.0 × 1013 DNA per cm2 for 15 nm AuNPs, 330 DNA per AuNP at 2.6 × 1013 DNA per cm2 for 20 nm AuNPs, and 860 DNA per AuNP at 1.7 × 1013 DNA per cm2 for 40 nm AuNPs. Fig. 4D shows that the same trend exists as for the ExoIII
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Nt.BspQI maximum reaction rates are higher on SNAs with smaller AuNP cores. The kcat increases with SNA size, which is likely due to there being a larger absolute number of targets per SNA for the larger sizes; if the Nt.BspQI can move from one restriction site to the next on a single SNA, it benefits from an effectively higher local concentration of targets. Nevertheless, small deflection angles between oligonucleotides in larger SNAs sterically hinder the ability of Nt.BspQI to access the restriction sites and bind to the substrate. This results in Nt.BspQI having a KM on SNAs with 40 nm AuNP cores three times that of SNAs with 20 nm AuNP cores, and more than seven times that of SNAs with 15 nm AuNP cores. Such inhibition in substrate binding ability ultimately limits nuclease activity despite higher kcat values. Further, it was observed that at higher substrate concentrations, the larger (20 and 40 nm) AuNP cases failed to go to completion, with the 40 nm case being the worse of the two (Fig. 4E and Fig. S6B†). As the larger SNAs have a lower surface density of DNA and more exposed AuNP surface, enzyme adsorption is likely to increase with increasing particle size. There was no significant difference in lag time between the three cases (ca. 4 minutes for all).
Nt.BspQI maximum reaction rates are higher on SNAs with smaller AuNP cores. The kcat increases with SNA size, which is likely due to there being a larger absolute number of targets per SNA for the larger sizes; if the Nt.BspQI can move from one restriction site to the next on a single SNA, it benefits from an effectively higher local concentration of targets. Nevertheless, small deflection angles between oligonucleotides in larger SNAs sterically hinder the ability of Nt.BspQI to access the restriction sites and bind to the substrate. This results in Nt.BspQI having a KM on SNAs with 40 nm AuNP cores three times that of SNAs with 20 nm AuNP cores, and more than seven times that of SNAs with 15 nm AuNP cores. Such inhibition in substrate binding ability ultimately limits nuclease activity despite higher kcat values. Further, it was observed that at higher substrate concentrations, the larger (20 and 40 nm) AuNP cases failed to go to completion, with the 40 nm case being the worse of the two (Fig. 4E and Fig. S6B†). As the larger SNAs have a lower surface density of DNA and more exposed AuNP surface, enzyme adsorption is likely to increase with increasing particle size. There was no significant difference in lag time between the three cases (ca. 4 minutes for all).
3.2 Recycling system
In the recycling case, where the quencher probe and not the fluorescently-labelled target probe is cleaved, we hypothesized that there would be a difference between the enzyme kinetics in the bound and free case due to the significantly different substrate environments. In the free case, when the target is released, it diffuses through the solution until it finds another free quencher probe to hybridize with, where the nuclease can then bind, cleave, and repeat the process. In the SNA case, the target recycling mechanism from one bound quencher probe to another is not as apparent. Since the target is in an environment densely packed with complementary quencher probes on the AuNP, whether the target rapidly hybridizes to neighboring strands on the same SNA or diffuses into solution to quencher probes on another SNA will alter both quenching efficiency and nuclease kinetics. Additionally, as the reaction proceeds, the SNAs become gradually depleted of their DNA covering. Therefore, we aimed to uncover how target recycling and evolving local conditions on the SNA tunes nuclease activity.To do this, we compared the nuclease kinetics of the bound and free case while studying the effects of varying enzyme and substrate concentrations in this recycling format. A schematic of the scenario with both nucleases is shown in Fig. 5A. It should be noted that the target probes in both the free and bound scenarios will remain quenched until sufficient quencher probes have been degraded. In these experiments, System 1 bound quencher probe, free quencher probe, and target probe (protected) were used for ExoIII while System 2 bound quencher probe, free quencher probe (protected), and target probe were used for Nt.BspQI (see Fig. 5A and the Table S1† for oligonucleotide sequences).
ExoIII. Fig. 5B shows that increasing ExoIII concentrations results in decreased reaction rates per enzyme, similar to the non-recycling format. Analysis of the progress curves demonstrated a longer lag phase than that of the non-recycling system for both the free (by 2.8 ± 1.1 min) and bound (by 5.9 ± 1.6 min) cases (Fig. S7A†). In addition to the lag intrinsic to ExoIII and from the temperature equilibration time, the progress curves from the recycling design exhibited an additional lag time before fluorescence signal detection, as the dye remained quenched while recycling through different quencher probes. In agreement with the non-recycling system, the lag phase time decreased with increasing ExoIII concentration for both the free and bound cases. This is reflected in the time required for the progress curves to reach inflection (Fig. S7B†). Both reaction rate and the time to inflection approached a plateau at higher ExoIII concentrations though. Comparing the bound and free cases, the lag phase is longer for the bound case than the free case (by an average of 4.2 ± 1.1 minutes). However, with a fixed substrate concentration of 20 nM, the bound case exhibited faster reaction rates that the free case at all enzyme concentrations (Fig. 5B). In fact, the progress curves demonstrate that the bound case reaches completion, whereby all target probes are fluorescent upon sufficient quencher probe degradation, before the free case when the ExoIII concentration is sufficiently high (>40 nM) (Fig. 5D and Fig. S7A†). This is also in spite of target stacking only occurring in the bound case. Such enzymatic acceleration implies rapid and efficient target recycling to neighboring quencher probes on the same SNA, requiring ExoIII to diffuse short distances before reaching the next substrate.
In the previous non-recycling studies, we observed that the ExoIII reactions generally go to completion (with the exception of the System 3 proximal recognition site), which we expect is due to a lower propensity of the ExoIII to irreversibly bind to the AuNP, compared to Nt.BspQI. Contrary to those findings, in the recycling case, we observed that at lower ExoIII concentrations the reaction does not go to completion (Fig. S7A†), for both the free and bound cases. This suggests that the ExoIII is limited in the amount of catalytic cycles it can go through, and this effect becomes apparent in the recycling system as many more cleavage events are required, and lowering the enzyme concentration exacerbates this limitation. However, the drop-off in performance was accentuated for the bound case versus the free case, suggesting an extra inhibitory factor involving the SNA. As the enzyme degrades the surface-bound quencher probes of the SNA while the target recycles, the protective effect of the surface-bound DNA is decreased and the AuNP surface is increasingly exposed, resulting in a higher likelihood for enzyme adsorption. DLS characterization of SNA size before and after experimentation (20 nM substrate and 10 nM ExoIII) revealed a size decrease, from 24.2 nm to 21.9 nm (Fig. S7C†). Such a decrease in hydrodynamic size is less than expected, presumably due to adsorption, as the SNA after nuclease degradation of quencher probe sequences should more closely resemble the size of the bare AuNP (13.9 nm). Nevertheless, despite adsorption, the recycling system enables ExoIII to operate more efficiently in the bound system.
To further probe the potential for enzymatic acceleration, 60 nM ExoIII was assessed in a subsequent study. As expected, increasing substrate concentration increases the maximum reaction rate for both cases (Fig. 5C). In comparison, beyond the low substrate concentrations, ExoIII has higher maximum reaction velocities in the free case than in the bound case. Despite this finding, progress curves under such conditions show that the time to completion is not significantly different between the free and bound case because the slowdown of reaction as it approaches completion is much more pronounced in the free case (Fig. S7D†). This indicates that ExoIII can finish the reaction on bound substrates before free substrates without exhibiting a faster maximum reaction velocity. In parallel, although the specificity constant is three times lower for the bound case, all target probes may still be released first under certain reaction conditions, most likely due to rapid target recycling enabled by the densely packed quencher probes scaffolded onto an AuNP.
Nt.BspQI. When the Nt.BspQI concentration was varied at a constant 20 nM substrate concentration, the maximum reaction rate increased with increasing enzyme concentrations for both the free and bound case (Fig. 5E). Due to target recycling, both the free and bound case also showed a longer lag phase relative to non-recycling experiments for Nt.BspQI (Fig. S7E†). Across a comparable range, the bound case exhibited an average lag of 7.2 ± 2.6 minutes, whereas in the non-recycling case it was 4.9 ± 0.7 minutes. Interestingly, the free case exhibited a two-phase reaction, with the progress curves showing an initial region of slow increase in product formation, followed by a shoulder at around 20% reaction completion and then a steep increase upwards towards the eventual plateau (Fig. S7E†). The initial phase of slow increase is most likely due to inefficient recycling of the target across multiple free quencher probes. Only when the majority of free quencher probes have been degraded does the second phase of higher reaction rate occur. Since the bound system only exhibits one phase of high reaction rate, we hypothesize there is rapid and efficient target recycling between quencher probes on a single SNA which in turn allows the bound case to avoid an initial slow phase.
125 nM Nt.BspQI was needed to complete the reaction in a non-recycling design whereas 180 nM Nt.BspQI is needed with this recycling design, likely due to enhanced adsorption. Similar to the recycling detection format with ExoIII, AuNP surface exposure increases as the surface-bound quencher probes are degraded. Looking at the hydrodynamic diameter with DLS, although Nt.BspQI should decrease the SNA size by cleaving the abundant surface-bound quencher probes during the reaction, the SNA shows an insignificant change in diameter before and after experimentation (from 22.3 to 22.7 nm, Fig. S7F†). This indicates a significant amount of Nt.BspQI adsorption counteracting the size decrease from the cleaved oligonucleotides.
With this target recycling design, the bound case is consistently slower than the free case (Fig. 5E), due to the same reasons as those detailed in the non-recycling system; however, the difference between the two cases with the recycling system is less than that of the non-recycling system (cf.Fig. 1D). This is likely due to the benefit afforded by efficient target recycling on the SNA and relatively shorter diffusion distances between each Nt.BspQI binding/nicking event, especially if the endonuclease has already overcome steric hindrance and is within the dense DNA layer. Interestingly, and similar to the non-recycling system, the bound case does not exhibit a relationship between the length of the lag phase and enzyme concentration, while the free case does (Fig. S7E†). In fact, this effect results in the bound case starting its fluorescence increase before the free case. This presents further evidence for the benefit of the locally concentrating effect of the surface-bound scenario.
In order to probe the effects of earlier signal detection in the bound case and to aim for reaction completion with all substrate concentrations tested, we chose a Nt.BspQI concentration of 180 nM. We observed that increasing substrate concentration increased the maximum reaction rate, with the free case consistently faster than the bound case (Fig. 5F). Even with 180 nM of enzyme, inactivation of Nt.BspQI due to adsorption was again a problem for the bound case, especially at higher substrate concentrations, where there is an increased amount of AuNPs present in solution (Fig. 5G). In fact, adsorption was so significant that reaction rates at high substrate concentrations were lower than the rates at low substrate concentrations. Similar to the progress curves of the bound case in other studies, target stacking is present once more (Fig. S7G†).
Overall, Nt.BspQI performs faster in the free case than the bound; however, the difference is less than that seen in the non-recycling system. When normalized to the specificity constant of the respective free cases, Nt.BspQI demonstrates accelerated activity on SNAs for this recycling system relative to the non-recycling system (at concentrations where adsorption does not limit reaction completion). The bound-to-free ratio of the specificity constant in this recycling study is 0.53, while in the non-recycling study it is 0.19, demonstrating the difference to be significantly less here. Again, this suggests efficient target recycling from one bound quencher probe to another and reduced diffusion distances for Nt.BspQI. Another interesting finding is how the length of the lag phase depends on the substrate concentration for the free case while no such relationship exists in the bound case—similar to the ExoIII recycling experiments. In the bound case, the lag times across the changing substrate concentrations are all around 5 minutes, whereas in the free case the lag time varied significantly (between 2 minutes at the lowest substrate concentration and 43 minutes at the highest). Accordingly, at high substrate concentrations, fluorescent signal evolution occurs earlier in the bound than in the free case. When leveraged together with accelerated Nt.BspQI activity, these two observations are promising advantages of the recycling target detection format using SNAs.
4 Discussion
In this work, we demonstrate the tuning of nuclease activity by SNA properties, and show that the modular units of SNAs can be leveraged to accelerate or suppress nuclease kinetics. By systematically investigating different SNA parameters and target detection formats with an exonuclease and endonuclease, our work extends previous observations in literature through a dedicated study on enzyme kinetics. Our findings are summarized in Table 1.| Condition | Trend |
|---|---|
| ExoIII | |
| Bound (relative to free) | k cat/KM↓ |
| Proximal digestion (relative to distal) | k cat/KM↓ |
| Surface coverage ↑ | k cat/KM↓ |
| AuNP diameter ↑ | k cat/KM↓ |
| Bound target recycling (relative to free) | k cat/KM↓ |
| Nt.BspQI | |
| Bound (relative to free) | K M↑, kcat↑ |
| Surface coverage ↑ | k cat/KM↑ |
| AuNP diameter ↑ | K M↑, kcat↑ |
| Bound target recycling (relative to free) | k cat/KM↓ |
In the experiments using ExoIII within a non-recycling detection format, we observe robust trends across the data. ExoIII experiences a longer lag phase and is generally inhibited in its cleavage activity on bound substrates relative to substrates free in solution, as revealed by consistently lower reaction rates and specificity constants. This is likely due to a combination of factors: (1) high SNA binding constants and sharp melting curves, (2) high salt concentrations surrounding the SNA, and (3) steric hindrance from the dense layer of DNA. First, SNAs are reported to have binding constants 100 times higher than free DNA, resulting in a sharper melting curve.46 In our studies, this necessitates ExoIII to remove more nucleotides from the target in the bound case than the free case before the quencher–target probe binding energy is sufficiently reduced to release the fluorophore-labeled target. Second, SNAs exhibit a significant ion cloud due to the negatively charged DNA and AuNP core.19 Since high salt concentrations are known to inhibit ExoIII activity, the high local ion concentrations it encounters in the bound case is unfavorable relative to the free case.45 Furthermore, as ExoIII removes nucleotides sequentially, it will encounter increasing local salt concentrations the closer it moves towards the AuNP core, incrementally decreasing its activity. Finally, steric hindrance from the dense layer of DNA on the AuNP hampers overall ExoIII activity by reducing catalytic rates and/or hindering substrate binding. Steric effects are further compounded by the non-processive nature of ExoIII, which necessitates multiple binding/cleavage events.
Similar to ExoIII, for the non-recycling experiments using Nt.BspQI, we observed slower overall activity and a longer lag phase when the substrate was bound to an AuNP rather than free in solution. To nick the target, Nt.BspQI must first diffuse past a dense layer of DNA prior to reaching its restriction site. If the enzyme manages to do so, it then must overcome steric hindrance from the local environment in order to bind to the target–quencher probe hybrid and catalyze the nicking reaction. Interestingly, kinetic analysis revealed that Nt.BspQI exhibited a higher kcat in the bound case compared to the free case, suggesting the endonuclease is actually acting faster on the SNA due to the high local concentrations of substrate. However, this also suggests that the limiting factor during a nicking event is the difficulty associated with substrate binding, as evidenced by the significantly higher KM in the bound case. Accordingly, while dense packing of oligonucleotides localize high concentrations of substrate onto a scaffold, this also creates a sterically unfavorable environment that severely restricts Nt.BspQI mobility and consequent substrate binding. While decreased overall reaction rates were similarly observed with ExoIII, there are two important distinctions between the nucleases, each with important implications for applications co-employing nucleases with SNAs. Nt.BspQI tended to exhibit a shorter lag time, likely due to its endonuclease mechanism; Nt.BspQI conducts a single nick while ExoIII requires multiple binding and cleaving events to release the target. Another variation is the propensity of Nt.BspQI to adsorb onto the AuNP surface and become inactivated, hence higher concentrations were needed to complete reactions. Significant adsorption is likely to contribute to the slower Nt.BspQI activity as well.
Intriguing effects were found for both ExoIII and Nt.BspQI when a recycling detection format was used, with both ExoIII and Nt.BspQI experiencing accelerated activity. For ExoIII, the bound case had higher reaction rates than the free case and completed some reactions faster. Although Nt.BspQI remained slower in the bound case, the difference between the bound and free case in the recycling format is considerably less than that for the non-recycling format. The evolving SNA surface during a recycling reaction presents a very different environment compared to the relatively consistent SNA surface in the non-recycling scenario. Thus, a discrepancy in trends between the two detection formats is unsurprising. We hypothesize two factors that may cause enzyme acceleration for a recycling detection format in the bound case. First, once a target probe is released upon degradation of the surface quencher probe, it may rapidly hybridize to a neighboring probe on the same AuNP rather than needing to diffuse through solution (as is the case when the quencher probes are free in solution). Second, the nucleases need only diffuse a short distance within the SNA shell, especially if the released target successfully hybridizes to a neighboring quencher probe, to bind to another substrate. Accordingly, we propose that acceleration comes about due to rapid recycling of target probes between neighboring probes on a single SNA and shorter nuclease diffusion distances between substrates.
A notable feature observed in the progress curves of all reactions was an initial drop in fluorescence before a subsequent increase as enzyme digestion proceeded. We attribute this to temperature equilibration as samples were prepared on ice before being put into the plate reader at 37 °C. However, most of the ExoIII experiments exhibited a more pronounced signal drop (see Fig. S3A, S3C, S4A, S5A, S6A, S7A and S7D†). We think that this cannot be explained by temperature equilibration and that another phenomenon is at play, which we refer to as ‘target stacking’. Since SNAs do not exhibit 100% hybridization efficiency at the target![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SNA ratio we employed (30
SNA ratio we employed (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), some targets remain free in solution when the reaction begins, providing an initial level of fluorescence.46 Subsequently, as the ExoIII acts on the oligonucleotides, the net negative charge of the SNA gradually decreases with the release of nucleotides, leading to a drop in electrostatic repulsion between the SNA and free target probes in solution. This decrease in the electrostatic repulsion from the local environment allows for corresponding increases in the hybridization efficiency between the SNA and target. Such a scenario is supported by previous reports that hybridization between free oligonucleotides and SNAs occurs with negative cooperativity due to electrostatic repulsion.7,47 Thus, as the enzyme acts, there is a net flux of targets hybridizing with the SNA—meaning that the fluorescent signal will decrease until all the initially free and unquenched targets bind to the SNA quencher probes.
1), some targets remain free in solution when the reaction begins, providing an initial level of fluorescence.46 Subsequently, as the ExoIII acts on the oligonucleotides, the net negative charge of the SNA gradually decreases with the release of nucleotides, leading to a drop in electrostatic repulsion between the SNA and free target probes in solution. This decrease in the electrostatic repulsion from the local environment allows for corresponding increases in the hybridization efficiency between the SNA and target. Such a scenario is supported by previous reports that hybridization between free oligonucleotides and SNAs occurs with negative cooperativity due to electrostatic repulsion.7,47 Thus, as the enzyme acts, there is a net flux of targets hybridizing with the SNA—meaning that the fluorescent signal will decrease until all the initially free and unquenched targets bind to the SNA quencher probes.
It is important to note that the target stacking behavior with ExoIII was particularly amenable to changes in the AuNP core size (Fig. 4C). In comparison to smaller AuNPs, the larger AuNP cores exhibited a higher initial fluorescence and a sharper decrease due to larger SNAs having more oligonucleotides per AuNP and higher electrostatic repulsion with free targets in solution. Furthermore, the fluorescence drop was not observed in the proximal digestion case in the ‘Varying ExoIII recognition site location’ study, but it was observed in the distal digestion case (see Fig. S4A†). This can be rationalized by the fact that, in the proximal case, removal of nucleotides by ExoIII is occurring in the inner region of the oligonucleotide shell around the SNA, such that the outer shell is maintaining its charge. Thus, the incoming targets are still acted upon by a strong repulsion force. However, in the distal digestion case, the ExoIII is depleting the number of nucleotides in the outer shell, which would significantly reduce the repulsion force to incoming targets.
Collectively, knowledge of these factors can be leveraged to purposefully tune nuclease activity as desired according to SNA features. For example, moving its recognition site proximal to the AuNP still permits ExoIII to penetrate the DNA shell of the SNA and cleave; however, ExoIII will have a significantly slower reaction rate and specificity constant (Fig. 2). We also observed that ExoIII and Nt.BspQI kinetics are dependent on SNA surface coverage. Whereas a higher amount of DNA per AuNP restricts ExoIII activity mainly due to sterics, increased surface densities increase the specificity constant of Nt.BspQI (Fig. 3). It is important to note that Nt.BspQI adsorption onto the AuNP surface most likely plays a significant role. As another means of altering the SNA local environment to modulate nuclease activity, employing SNAs of different AuNP core sizes will impart differential effects due to curvature-dependent properties. We demonstrate that deflection angle has a more dominant role than surface density such that both ExoIII and Nt.BspQI act more quickly on smaller SNAs than larger ones (Fig. 4). Finally, utilizing a target recycling system will speed up the reaction rates for both nucleases (Fig. 5). In fact, ExoIII degradation of SNA oligonucleotides may even perform faster than the free case due to more efficient target recycling and lower enzyme diffusion distances. While the Nt.BspQI does not experience as dramatic an acceleration with a switch in detection format, its reaction kinetics will still be enhanced such that the difference between the free and bound case is not as large as that for the non-recycling case. From these findings, we present a survey of easily tunable SNA features and their corresponding impact on nuclease kinetics. We believe that these observations provide a preliminary design guideline for applications that rely on enzyme activity alongside SNAs and other nucleic acid–nanoparticle conjugates.
In this study we sought to broadly screen the effects of several SNA features and detection formats on nuclease activity and proposed plausible mechanisms of action. In doing so, we have identified specific phenomena that are worthy of further study. Additional work is needed to uncover the exact details of each mechanism, and which kinetic model is most suited to describing the reactions observed herein.16 Furthermore, other parameters and considerations may also be key mediators of nuclease activity. Herein, the buffer conditions applied were a compromise between the enzyme catalytic efficiency and SNA stability. Specific buffer conditions (pH, ionic strength, etc.) catered to an enzyme may result in interesting trends not reported here. Molecular crowding is an additional biologically relevant condition that may result in differential enzyme performance. Another interesting direction will be to explore various SNA surface chemistries and enzyme charge distributions, and their impact on catalytic activity as a function of enzyme adsorption. Minimizing adsorption may also permit the use of lower enzyme concentrations whereby Michaelis–Menten conditions are satisfied. Further, studies similar to those performed herein on SNAs with a smaller core diameter (15 nm was the minimum studied herein) could yield additional interesting and maybe unexpected results, given how the deflection angle is intensified at low particle diameters (deflection angle is inversely proportional to particle radius). Follow-up studies dedicated to exploring how enzyme activity is impacted by modular nanoparticle features will continue to further our understanding of enzyme–nanoparticle interactions. Overall, we speculate studies such as these suggested above could uncover enhanced or otherwise modulated enzyme activity on not just SNAs but other nanomaterial-based systems as well.
5 Conclusions
The application of nanomaterials to a multitude of biologically relevant fields requires a thorough understanding of their interactions with enzymes. Herein, we have presented a systematic study on the nature of such interactions between nucleases and SNAs. Careful design of three distinct sets of oligonucleotides permitted fluorescence-based kinetic assays and direct extraction of kinetic parameters under various SNA designs. We demonstrate that ExoIII (exonuclease) and Nt.BspQI (endonuclease) can be controllably regulated to inhibit or accelerate nucleic acid degradation of SNAs. In a standard non-recycling target detection format, both nucleases are inhibited when their substrates are bound onto AuNPs versus free in solution. Resistance to nuclease degradation can be attributed to the unique physicochemical local environment of the SNA. Furthermore, we hypothesize that enzyme inactivation due to adsorption onto the AuNP surface significantly reduces the effective nuclease concentration and hampers reaction rates, especially in the Nt.BspQI case. However, by tuning the surface coverage and AuNP size of the SNA, the reaction rates of both nucleases can be controlled. In addition, by altering the location of the ExoIII recognition site, we demonstrate the surprising ability of ExoIII to diffuse through the dense DNA layer of the SNA and initiate nucleotide cleavage proximal to the AuNP surface, albeit at a slower rate. Beyond adjusting the tuneable SNA features, we also employed a target recycling detection format and observed accelerated nuclease activity. The enhanced reaction rate can be attributed to the abundant quencher probes densely packed onto the AuNP scaffold, allowing for rapid target recycling and smaller nuclease diffusion distances between substrate binding. Finally, target stacking, a phenomenon where SNA hybridization efficiency is improved as the nucleases act, is observed for the first time. These findings serve to guide the design of future DNA-nanoparticle conjugates, and other nanomaterial bioconjugates. Further studies dedicated to the kinetics of other enzymes on nanomaterial bioconjugates will continue the advancement of these multifaceted nanostructures for biological applications.Author contributions
Jeff Hsiao: Funding acquisition, methodology, investigation, formal analysis, visualization, writing – original draft. Tomas Buryska: Formal analysis, visualization, writing – review & editing. Eunjung Kim: Conceptualization, writing – review & editing. Philip D. Howes: Funding acquisition, project administration, conceptualization, methodology, investigation, writing – original draft, writing – review & editing. Andrew J. deMello: Funding acquisition, supervision, writing – review & editing.Conflicts of interest
The authors have no conflicts of interest.Acknowledgements
J. C. H. thanks the Whitaker Foundation for support through the Whitaker International Fellowship Program. P. D. H. acknowledges support from European Union's Horizon 2020 Research and Innovation Program through the Individual Marie Sklodowska-Curie Fellowship “Ampidots” under grant agreement no. 701994. E. K. acknowledges support from the Technology Innovation Program (or Industrial Strategic Technology Development Program) (20009121) funded by the Ministry of Trade, Industry & Energy (MOTIE, Korea).References
- N. C. Seeman and H. F. Sleiman, DNA nanotechnology, Nat. Rev. Mater., 2017, 3, 17068 CrossRef.
- A. Samanta and I. L. Medintz, Nanoparticles and DNA – a powerful and growing functional combination in bionanotechnology, Nanoscale, 2016, 8, 9037–9095 RSC.
- H. Pei, X. Zuo, D. Zhu, Q. Huang and C. Fan, Functional DNA Nanostructures for Theranostic Applications, Acc. Chem. Res., 2014, 47, 550–559 CrossRef CAS.
- H. Liang, X.-B. Zhang, Y. Lv, L. Gong, R. Wang, X. Zhu, R. Yang and W. Tan, Functional DNA-Containing Nanomaterials: Cellular Applications in Biosensing, Imaging, and Targeted Therapy, Acc. Chem. Res., 2014, 47, 1891–1901 CrossRef CAS.
- C. R. Laramy, M. N. O'Brien and C. A. Mirkin, Crystal engineering with DNA, Nat. Rev. Mater., 2019, 4, 201–224 CrossRef CAS.
- J. I. Cutler, E. Auyeung and C. A. Mirkin, Spherical nucleic acids, J. Am. Chem. Soc., 2012, 134, 1376–1391 CrossRef CAS.
- P. S. Randeria, M. R. Jones, K. L. Kohlstedt, R. J. Banga, M. Olvera de la Cruz, G. C. Schatz and C. A. Mirkin, What Controls the Hybridization Thermodynamics of Spherical Nucleic Acids?, J. Am. Chem. Soc., 2015, 137, 3486–3489 CrossRef CAS.
- L.-K. Fong, Z. Wang, G. C. Schatz, E. Luijten and C. A. Mirkin, The Role of Structural Enthalpy in Spherical Nucleic Acid Hybridization, J. Am. Chem. Soc., 2018, 140, 6226–6230 CrossRef CAS.
- S. P. Narayan, C. H. J. Choi, L. Hao, C. M. Calabrese, E. Auyeung, C. Zhang, O. J. G. M. Goor and C. A. Mirkin, The Sequence-Specific Cellular Uptake of Spherical Nucleic Acid Nanoparticle Conjugates, Small, 2015, 11, 4173–4182 CrossRef CAS.
- P. S. Randeria, W. E. Briley, A. B. Chinen, C. M. Guan, S. H. Petrosko and C. A. Mirkin, Nanoflares as probes for cancer diagnostics, Cancer Treat. Res., 2015, 166, 1–22 CAS.
- S. N. Barnaby, G. A. Perelman, K. L. Kohlstedt, A. B. Chinen, G. C. Schatz and C. A. Mirkin, Design Considerations for RNA Spherical Nucleic Acids (SNAs), Bioconjugate Chem., 2016, 27, 2124–2131 CrossRef CAS.
- B. Liu and J. Liu, Interface-Driven Hybrid Materials Based on DNA-Functionalized Gold Nanoparticles, Matter, 2019, 1, 825–847 CrossRef.
- W. Wang, S. Yu, S. Huang, S. Bi, H. Han, J. R. Zhang, Y. Lu and J. J. Zhu, Chem. Soc. Rev., 2019, 48, 4892–4920 RSC.
- S. D. Mason, G. A. Wang, P. Yang, Y. Li and F. Li, Probing and Controlling Dynamic Interactions at Biomolecule–Nanoparticle Interfaces Using Stochastic DNA Walkers, ACS Nano, 2019, 13, 8106–8113 CrossRef CAS.
- A. M. Carver, M. De, H. Bayraktar, S. Rana, V. M. Rotello and M. J. Knapp, Intermolecular Electron-Transfer Catalyzed on Nanoparticle Surfaces, J. Am. Chem. Soc., 2009, 131, 3798–3799 CrossRef CAS.
- W. R. Algar, T. Jeen, M. Massey, W. J. Peveler and J. Asselin, Small Surface, Big Effects, and Big Challenges: Toward Understanding Enzymatic Activity at the Inorganic Nanoparticle–Substrate Interface, Langmuir, 2019, 35, 7067–7091 CrossRef CAS.
- B. J. Johnson, W. Russ Algar, A. P. Malanoski, M. G. Ancona and I. L. Medintz, Understanding enzymatic acceleration at nanoparticle interfaces: Approaches and challenges, Nano Today, 2014, 9, 102–131 CrossRef CAS.
- J. N. Vranish, M. G. Ancona, S. A. Walper and I. L. Medintz, Pursuing the Promise of Enzymatic Enhancement with Nanoparticle Assemblies, Langmuir, 2018, 34, 2901–2925 CrossRef CAS.
- D. S. Seferos, A. E. Prigodich, D. A. Giljohann, P. C. Patel and C. A. Mirkin, Polyvalent DNA Nanoparticle Conjugates Stabilize Nucleic Acids, Nano Lett., 2009, 9, 308–311 CrossRef CAS.
- N. L. Rosi, D. A. Giljohann, S. Thaxton, A. K. R. Lytton-Jean, M. S. Han and C. A. Mirkin, Oligonucleotide-Modified Gold Nanoparticles for Intracellular Gene Regulation, Science, 2006, 312, 1027–1030 CrossRef CAS.
- D. S. Seferos, D. A. Giljohann, H. D. Hill, A. E. Prigodich and C. A. Mirkin, Nano-flares: Probes for transfection and mRNA detection in living cells, J. Am. Chem. Soc., 2007, 129, 15477–15479 CrossRef CAS.
- A. E. Prigodich, P. S. Randeria, W. E. Briley, N. J. Kim, W. L. Daniel, D. A. Giljohann and C. A. Mirkin, Multiplexed nanoflares: mRNA detection in live cells, Anal. Chem., 2012, 84, 2062–2066 CrossRef CAS.
- A. E. Prigodich, A. H. Alhasan and C. A. Mirkin, Selective Enhancement of Nucleases by Polyvalent DNA-Functionalized Gold Nanoparticles, J. Am. Chem. Soc., 2011, 133, 2120–2123 CrossRef CAS.
- X. Qu, D. Zhu, G. Yao, S. Su, J. Chao, H. Liu, X. Zuo, L. Wang, J. Shi, L. Wang, W. Huang, H. Pei and C. Fan, An Exonuclease III-Powered, On-Particle Stochastic DNA Walker, Angew. Chem., Int. Ed., 2017, 56, 1855–1858 Search PubMed.
- F. Degliangeli, P. Kshirsagar, V. Brunetti, P. P. Pompa and R. Fiammengo, Absolute and direct microRNA quantification using DNA-gold nanoparticle probes, J. Am. Chem. Soc., 2014, 136, 2264–2267 CrossRef CAS.
- H. D. Hill, J. E. Millstone, M. J. Banholzer and C. A. Mirkin, The Role Radius of Curvature Plays in Thiolated Oligonucleotide Loading on Gold Nanoparticles, ACS Nano, 2009, 3, 418–424 CrossRef CAS.
- S. J. Hurst, A. K. R. Lytton-Jean and C. A. Mirkin, Maximizing DNA Loading on a Range of Gold Nanoparticle Sizes, Anal. Chem., 2006, 78, 8313–8318 CrossRef CAS.
- Y. V. Gerasimova and D. M. Kolpashchikov, Enzyme-assisted target recycling (EATR) for nucleic acid detection, Chem. Soc. Rev., 2014, 43, 6405–6438 RSC.
- B. Liu and J. Liu, Freezing Directed Construction of Bio/Nano Interfaces: Reagentless Conjugation, Denser Spherical Nucleic Acids, and Better Nanoflares, J. Am. Chem. Soc., 2017, 139, 9471–9474 CrossRef CAS.
- B. Liu and J. Liu, Freezing-Driven DNA Adsorption on Gold Nanoparticles: Tolerating Extremely Low Salt Concentration but Requiring High DNA Concentration, Langmuir, 2019, 35, 6476–6482 CrossRef CAS.
- X. Liu, M. Atwater, J. Wang and Q. Huo, Extinction coefficient of gold nanoparticles with different sizes and different capping ligands, Colloids Surf., B, 2007, 58, 3–7 CrossRef CAS.
- D. Fan, X. Zhu, Q. Zhai, E. Wang and S. Dong, Polydopamine Nanotubes as an Effective Fluorescent Quencher for Highly Sensitive and Selective Detection of Biomolecules Assisted with Exonuclease III Amplification, Anal. Chem., 2016, 88, 9158–9165 CrossRef CAS.
- B. Li, L. Xu, Y. Chen, W. Zhu, X. Shen, C. Zhu, J. Luo, X. Li, J. Hong and X. Zhou, Sensitive and Label-Free Fluorescent Detection of Transcription Factors Based on DNA-Ag Nanoclusters Molecular Beacons and Exonuclease III-Assisted Signal Amplification, Anal. Chem., 2017, 89, 7316–7323 CrossRef CAS.
- W. Song, N. Zhang, Z. Luan, X. Zhang and P. He, Application of a cation-exchange reaction of CuS nanoparticles and fluorescent copper nanoparticles in a DNA biosensor, RSC Adv., 2018, 8, 15248–15252 RSC.
- Y. Chen, H. T. T. Duong, S. Wen, C. Mi, Y. Zhou, O. Shimoni, S. M. Valenzuela and D. Jin, Exonuclease III-Assisted Upconversion Resonance Energy Transfer in a Wash-Free Suspension DNA Assay, Anal. Chem., 2018, 90, 663–668 CrossRef CAS.
- K. R. Thomas and B. M. Olivera, Processivity of DNA exonucleases, J. Biol. Chem., 1978, 253, 424–429 CrossRef CAS.
- R. Wu, G. Ruben, B. Siegel, E. Jay, P. Spielman and C.-P. D. Tu, Synchronous digestion of SV40 DNA by exonuclease III, Biochemistry, 1976, 15, 734–740 CrossRef CAS.
- Q. Xu, A. Cao, L. Zhang and C. Zhang, Rapid and Label-Free Monitoring of Exonuclease III-Assisted Target Recycling Amplification, Anal. Chem., 2012, 84, 10845–10851 CrossRef CAS.
- P. Zhang, P. H.-M. Too, J. C. Samuelson, S.-H. Chan, T. Vincze, S. Doucette, S. Bäckström, K. D. Potamousis, T. M. Schramm, D. Forrest, D. C. Schwartz and S. Xu, Engineering BspQI nicking enzymes and application of N.BspQI in DNA labeling and production of single-strand DNA, Protein Expression Purif., 2010, 69, 226–234 CrossRef CAS.
- Y. Yu, Z. Chen, W. Jian, D. Sun, B. Zhang, X. Li and M. Yao, Ultrasensitive electrochemical detection of avian influenza A (H7N9) virus DNA based on isothermal exponential amplification coupled with hybridization chain reaction of DNAzyme nanowires, Biosens. Bioelectron., 2015, 64, 566–571 CrossRef CAS.
- C.-H. Lu, F. Wang and I. Willner, Zn 2+ -Ligation DNAzyme-Driven Enzymatic and Nonenzymatic Cascades for the Amplified Detection of DNA, J. Am. Chem. Soc., 2012, 134, 10651–10658 CrossRef CAS.
- C. Chen and N. Hildebrandt, Resonance energy transfer to gold nanoparticles: NSET defeats FRET, TrAC, Trends Anal. Chem., 2020, 123, 115748 CrossRef CAS.
- S. Henikoff, in Methods in Enzymology, Academic Press, 1987, vol. 155, pp. 156–165 Search PubMed.
- E. Petryayeva, T. Jeen and W. R. Algar, Optimization and Changes in the Mode of Proteolytic Turnover of Quantum Dot–Peptide Substrate Conjugates through Moderation of Interfacial Adsorption, ACS Appl. Mater. Interfaces, 2017, 9, 30359–30372 CrossRef CAS.
- M. Wu and W. R. Algar, Acceleration of Proteolytic Activity Associated with Selection of Thiol Ligand Coatings on Quantum Dots, ACS Appl. Mater. Interfaces, 2015, 7, 2535–2545 CrossRef CAS.
- A. K. R. Lytton-Jean and C. A. Mirkin, A thermodynamic investigation into the binding properties of DNA functionalized gold nanoparticle probes and molecular fluorophore probes, J. Am. Chem. Soc., 2005, 127, 12754–12755 CrossRef CAS.
- C. Chen, W. Wang, J. Ge and X. S. Zhao, Kinetics and thermodynamics of DNA hybridization on gold nanoparticles, Nucleic Acids Res., 2009, 37, 3756–3765 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0nr08668a |
| This journal is © The Royal Society of Chemistry 2021 |