Open Access Article
Open Access ArticleImmune response and possible therapeutics in COVID-19
Anindya Dutta†
a,
Ananya Roy†a,
Laboni Roy†a,
Samit Chattopadhyay*b and
Subhrangsu Chatterjee *a
*a
aDepartment of Biophysics, Bose Institute, Centenary Campus, P-1/12 C.I.T. Scheme VIIM, Kolkata-700054, India. E-mail: subhro_c@jcbose.ac.in; subhrangsu@gmail.com; Tel: +91-033-25693340
bBITS Pilani Goa Campus, NH-17B, Zuarinagar, Goa 403726, India. E-mail: samitc@goa.bitspilani.ac.in; Tel: +91-0832-2580856
First published on 4th January 2021
Abstract
COVID-19 has emerged as a pandemic affecting about 213 countries in all the continents of the globe, resulting in more than 37.8 million individuals getting infected and over 1.08 million deaths worldwide, jeopardizing global human health and the economy. This presents an urgent need to develop therapies that target the SARS-CoV2 virus specifically. This review aims at presenting the available information on the coronavirus disease 2019 along with various drugs that are having widespread use until a vaccine candidate is available to aid in the development of therapeutic strategies against COVID-19.
1. Introduction
In December 2019, several people were diagnosed with a severe pneumonia of unknown origin in Wuhan, Hubei province of China. Analysis of the bronchoalveolar lavage fluid (BALF) from these patients led to the isolation of a novel coronavirus, SARS-CoV2 (Severe Acute Respiratory Syndome coronavirus 2; previously named 2019-nCoV). SARS-CoV2 is a positive-sense single-stranded RNA virus that belongs to the betacoronavirus genus, believed to have zoonotic origins since it has high genetic similarity to the coronavirus RaTG13, isolated from bats. On 30th January 2020, the World Health Organization (WHO) declared the outbreak as a Public Health Emergency of International Concern and officially named the viral disease as coronavirus disease 2019 (COVID-19) on February 12, 2020 and on 11th March 2020, COVID-19 was assessed as a pandemic. As of 12th October 2020, COVID-19 has affected over 214 countries and territories around the world infecting >37.7 million people and proving fatal to >1.08 million individuals, thus endangering global health and the economy.1–4Corona viruses (order Nidovirales, family coronaviridae, subfamily coronavirinae) are a large family of spherical, enveloped, non-segmented RNA viruses with size ranging from 60 nm to 140 nm in diameter with external solar spike like projections on its surface which are the peplomers of viral S protein, gives it a crown like appearance under electron microscope (corona in Latin is coronam, which means crown).2,3,5–7 This virus possesses approximately 27–32 kb of single stranded positive sense RNA packed inside a helical capsid and further surrounded by an envelope.7,8 Apart from infecting birds and mammals, these viruses have crossed the animal–human species barrier causing disease like upper respiratory tract infections (URTIs) resembling the common cold, to lower respiratory tract infections (LRTIs) such as bronchitis, pneumonia and severe acute respiratory syndrome (SARS).9
Corona viruses (CoV) are classified/divided into four genera, Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus. Alpha and beta CoV carries the potential to infect mammals, while gamma and delta CoV infect birds although delta CoV infect both mammalian and avian species.1,7,8 Among them three coronaviruses, SARS-CoV, MERS-CoV and SARS-CoV2 belonging to beta-coronavirus group (Orthocoronavirinae subfamily) possess the potential to cause severe respiratory tract infection.1,2
SARS-CoV originated in Guangdong, China in November 2002, spreading to Hong Kong in February 2003 and subsequently to various Asian countries. It infected 8422 people globally, mostly in China and Hong Kong causing 916 deaths (mortality rate being 11%) until the epidemic came to an end in June 2003.2 MERS (Middle-East Respiratory Syndrome) outbreak took place in April 2012 in Jordan. As of February 29 2020, there have been 2494 confirmed cases, with majority of cases reported in Saudi Arabia, causing 859 deaths worldwide (34% fatality rate).10
The ongoing SARS CoV-2 outbreak has rapidly evolved and spreading globally. On January 7 2020, this virus was identified to have >95% homology with the bat coronavirus HKU9-1 and this virus is identified to have 79.5% genome sequence identity to SAR-CoV >70% similarity with the SARS-CoV.1 To date, the closest relative of SARS-CoV-2 is RaTG13, identified from a Rhinolophus affinis sampled in Yunnan province in 2013. This virus shared 96.1% nucleotide identity and 92.9% identity in the S gene, suggesting the role of bats as coronavirus reservoirs.11 The events leading to the transmission of the virus from animals to humans is still largely unknown, though it has been confirmed that it originated in bats. It appears that SARS-CoV2 has a higher rate of human to human transmission as compared to SARS and MERS (Basic Reproduction Number R0 has been estimated to be between 1.4 and 3.9) but has a lower case fatality rate of about 2.3%, affecting mainly the elderly and persons with severe health complications.12 Currently there are no clinically approved drugs or vaccines available in the market that specifically target SARS-CoV2. Currently, the therapeutic strategies to deal with COVID-19 are symptomatic treatment with respiratory support and prevention aimed at reducing community transmission. The exponential rise in infection have led to serious public health control measures, international travel restrictions and severe economic crisis worldwide.
2. Ultrastructure of SARS-CoV2
SARS-CoV2 is a positive sense single stranded RNA virus having a diameter of 60–140 nm.13 On 10th January 2020, the first whole genome sequence of SARS-CoV2 was released aiding its diagnosis by quick identification of the virus in suspected patients using reverse transcription polymerase chain reaction (RT-PCR) methods.3 So far 178 genome sequences of SARS-CoV2 from different labs have been submitted to GISAID database.4,8SARS-CoV2 (genome of ∼30 kb) consists of a 5′-untranslated region (UTR), a replicase complex (orf1ab) encoding polyproteins (pp1a and pp1ab), 16 non-structural proteins (NSPs) including RNA-dependent RNA polymerase (RdRP), a spike protein (S) gene, envelope protein (E) gene, a membrane protein (M) gene that helps in virus assembly, a nucleocapsid protein (N) gene, 3′-UTR and several5–7,9–11 unidentified non-structural open reading frames.1,4 Studies have highlighted that SARS-CoV2 genes share ∼80% nucleotide identity and 89.10% nucleotide similarity with SARS-CoV genes. In a study, the mutation in NSP2 and NSP3 proteins have been suggested as a potential mechanism differentiating COVID-19 from SARS and may account for the high transmissibility of the virus.14
Transcriptionally frame shifted ORF1a and ORF1b produce pp1a and pp1ab polypeptides which upon proteolytic cleavage generate various proteins. The proteolytic processing is mediated by papain-like protease (PLpro) and 3-chymotrypsin-like protease (3CLpro).4,12 The 3CLpro cleaves the polyprotein at 11 distinct sites that generates 16 non-structural proteins (NSPs) that are involved in viral replication.4 Other ORFs encode for structural proteins that includes spike protein, membrane, envelope, and nucleocapsid proteins. 3CLpro play a critical role in the replication of virus particles and unlike structural/accessory protein-encoding genes, it is located at the 3′ end which exhibits excessive variability. Multiple sequence alignment results revealed that 3CLpro was conserved, with 100% identity among all SARS-CoV2 genomes. Analysis of physicochemical parameters revealed that the SARS-CoV2 3CLpro polypeptide is 306 amino acids long with a molecular weight of 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 796.64 Da categorising the protein as a stable, hydrophilic molecule capable of establishing hydrogen bonds. Therefore, it is essential to discover novel compounds that may inhibit SARS-CoV2 3CLpro and serve as potential anti-COVID-19 drug compounds.15
796.64 Da categorising the protein as a stable, hydrophilic molecule capable of establishing hydrogen bonds. Therefore, it is essential to discover novel compounds that may inhibit SARS-CoV2 3CLpro and serve as potential anti-COVID-19 drug compounds.15
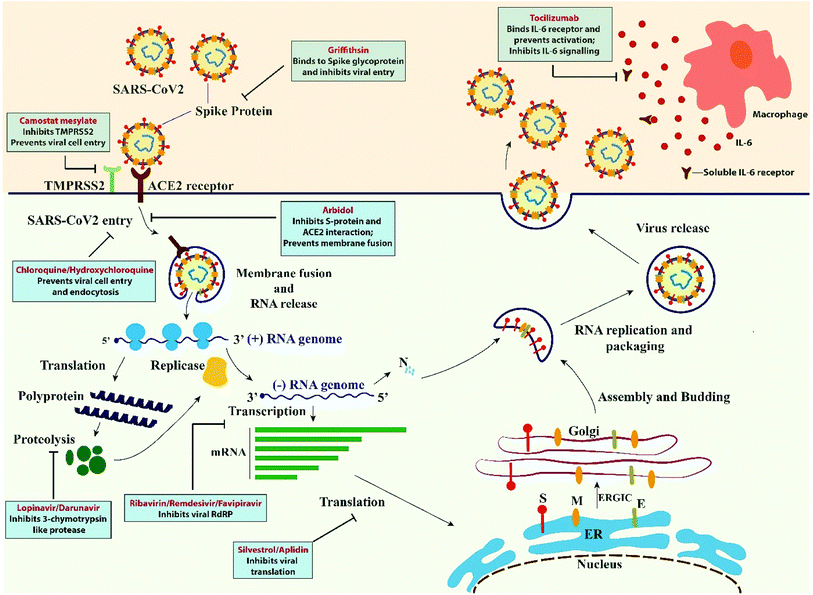
3. Mechanism of human pathogenicity
Spike (S) glycoprotein of SARS-CoV/SARS-CoV2 and host cell zinc peptidase angiotensin-converting enzyme 2 (ACE2) receptor recognition and affinity binding is a critical step for virus entry into the lower respiratory tract of humans.1 Spike protein is a clove shaped trimer with S1 and S2 subunits.7 S1 sub-unit with receptor binding domain (RBD) is involved in cell type selection/tissue tropism and ACE2 receptor recognition and attachment, while virus–host cell membrane fusion is mediated by the S2 subunit allowing the entry of viral RNA inside host cell cytoplasm. The invasion process being primed by TMPRSS21 (transmembrane serine protease) produced by the host cell.16 S proteins of SARS-CoV2 share about 76% and 97% of amino acid identities with SARS-CoV and RaTG13 respectively. Whereas the amino acid sequence of RBD of SARS-CoV2 is only about 74% and 90.1% homologous to that of SARS-CoV and RaTG13 respectively.17 The NTDs (N terminal domain of S protein) of SARS-CoV2 share only 53.5% of homology in amino acid sequence with SARS-CoV S protein. Binding of SARS corona virus to cellular receptor ACE2 results in uptake of virions into endosomes, where the spike protein is activated by the pH dependent cysteine protease cathepsin L. SARS-CoV2 S protein is capable of triggering protease-independent and receptor-dependent syncytium formation that possess the capability to enhance virus spreading through cell–cell fusion.17 Upon entry inside host cell, the uncoated viral genome encodes pp1a and pp1ab polyproteins which in turn participates in formation of replication transcription complex (RTC). RTC synthesizes a nested subset of sub-genomic RNA (sgRNAs) from which the accessory proteins and structural proteins are encoded utilizing the host ER and Golgi complex. Finally, the newly synthesized viral RNA, nucleocapsid proteins and envelope glycoproteins are assembled and packaged in multiple vesicles which fuse with the plasma membrane to release the viruses in the host system, ready to infect more cells.4. Clinical symptoms, diagnosis and mode of transmission
SARS-CoV2 primarily affects the respiratory system in humans. The virus passes through the nasal and larynx mucosal membranes and enters the lungs through the respiratory tract. It may utilize the circulating blood to attack other organs that express ACE2, such as the heart, renal, gastrointestinal tract.18 Clinical features vary from asymptomatic state to acute respiratory distress and multi-organ dysfunction. ACE2 is highly expressed at the protein level on lung alveolar epithelial cells. As mentioned earlier, viral S protein and host cell ACE2 receptor interaction are critical to viral entry.1 Its correlation with higher expression of ACE2 in the lungs may help us understand the route of infection and disease manifestation.The primary mode of infection is human-to-human transmission through close contact occurring via droplets from infected person's cough or sneeze.19 COVID-19 has asymptomatic incubation period between 2 and 14 days during which the virus is transmissible.19 For this reason, the rapid spread of SARS-CoV2 has occurred with the basic reproduction number R0 of 1.4–3.9.19 The typical clinical features are cough, sore throat, fever, headache, fatigue and breathlessness with characteristic pulmonary ground glass opacity changes on chest CT scan.3,6,20 In severe cases the disease rapidly progresses to pneumonia, respiratory failure, multi organ dysfunction (like cardiac and kidney injury) and death. Nasopharyngeal and oropharyngeal swab, bronchoalveolar lavage fluid (BALF) and sputum samples from suspected/symptomatic patients are used for clinical diagnosis of SARS-CoV2.8 Serological test employed are ELISA and western blot for detection of specific COVID-19 proteins.2,21 Viral antigen detection is done by using direct immunofluorescent assay (IFA), viral nucleic acid detection is done by real-time reverse transcriptase (RT)-PCR (based on RdRP gene, N gene, E gene and ORF gene) and further detection confirmed by next-generation sequencing.1,21,22 Although RT-PCR is regarded as the reference standard, recent studies have addressed the importance of chest computed tomography (CT) examination in COVID-19 patients with false negative RT-PCR results and reported the CT sensitivity to be 98%.20 Additionally, National Health Commission of China has declared CT examination to be of great significance not only in diagnosing COVID-19 but also in monitoring disease progression and evaluating therapeutic efficacy. Series of lab tests confirmed that most patients had low white blood cell counts and lymphocytopenia.3 In acute/severe cases-neutrophil count, blood urea, creatinine levels, D-dimer, CPK, LDH, cytokines were significantly higher along with steady decrease in lymphocyte count.2
Adverse outcomes and death are more common in senior citizens (age > 60), people with underlying co-morbidities such as hypertension, cardiovascular disease diabetes and immune-compromised patients (fatality rate: 50–75%), while the overall case fatality rate ranges between 2 to 3%.
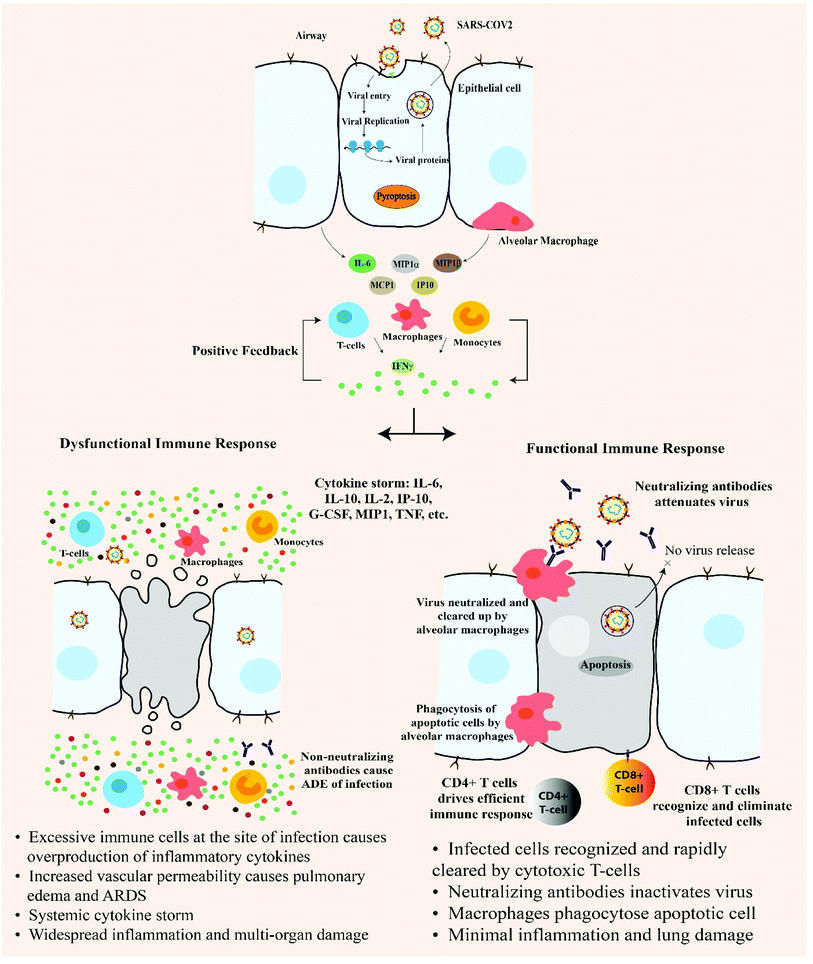
5. Innate immune response
When the foreign genome (viral RNA) enters the host cell, it is detected as Pathogen Associated Molecular Patterns (PAMPs) by the host cell pattern recognition receptors (PRRs). Endosomal RNA receptor TLRs (TLR 3, 7, 8, 9) and cytosolic RNA receptors- RIG-1, MDA5, cGAS are involved in sensing and recognizing viral RNA in the host cell.19 These molecules coordinate a complex signalling that results in recruitment of several adaptors like MAVS, STING which in turn triggers MyD88 and a series of downstream cascade molecules which finally leads to the production of IFNα and IFNβ, activation of NFκB, IRF3. Nuclear translocation of these molecules leads to expression of type I IFN and other pro-inflammatory cytokines and this initial response comprise the first line defence against viral infection. Type 1 IFN activates JAK-STAT pathway which results in transcriptional initiation of IFN stimulated genes (ISGs).19 Successful type 1 IFN response should be able to suppress viral replication and dissemination at an early stage. But at the step of type 1 IFN induction, SARS-CoV disrupts the downstream signalling by degrading RNA sensor adaptor molecules like MAVS and TRAF3/6 and inhibits IRF3 nuclear translocation resulting in dampening of type 1 IFN response and viral control failure.19 In the later stages, active viral replication results in the hyperproduction of type 1 IFN and influx of neutrophils and macrophages, which are the major sources of pro-inflammatory cytokines. COVID-19 patient's deteriorating condition is associated with increase in C-Reactive Protein (CRP) and inflammatory factors in the plasma, like the interleukins IL-6, IL-10, IL-2, IL-7, 4, 12, 13, 17, TNFα, GCSF, MCSF, IP-10, MCP-1, MIP-1α, HGF, producing a cytokine storm in the body of critical COVID-19 patients1–3,19 (Fig. 1).6. Adaptive immune response
In coronavirus disease, most of the immune response (70%) was found against the structural proteins (spike, envelope, membrane and nucleocapsid). In studied SARS-CoV patients, long-lasting specific IgG and neutralizing antibodies were reported even after 2 years after infection. T cell response was extensively investigated in SARS-CoV and it was reported that CD8+ T cell response was heightened compared to CD4+ T cell response. Higher magnitude of T cell response was found to be correlated with significantly higher neutralizing antibody. In severely infected SARS CoV patients, polyfunctional CD4+ T cells (IFNγ, TNFα and IL2) and CD8+ T cells (IFNγ, TNFα and degranulated state) were detected at higher frequency accompanied by high serum availability of Th2 cytokines (IL4, IL5, IL10).19In MERS-CoV, early rise of CD+ T cells were detected in acutely infected stage, while in the recovery stage, Th1 type helper T cells were found to be dominant. When macrophages and dendritic cells were infected by MERS-CoV, T cell activation markedly diminished due to disrupted/suppressed antigen presentation by MHC class I and II molecules.
Coronaviruses are adapted to evade immune detection and dampen the immune response in infected individuals. The viral proteins (M proteins, NSPs (NS4a, NS4b, NS15)) being the key molecules helping in host cell modulation. Early immune evasion by SAR-CoV2 is the reason for the long incubation period of COVID-19. Th1 type response is a key to successful control of SARS-CoV and MERS-CoV and recent evidences strongly indicate the same to be true for SARS-CoV2 as well.19
7. Treatment and therapeutic potentials
Current treatments are mainly focused on symptomatic and respiratory support. There is yet no effective antiviral drug against SARS-CoV2.Two therapeutic measures are proposed by L. Lin et al. based on anticoagulant therapy to enhance the immune function and block the inflammatory storm in COVID-19 patients. When T lymphocyte and B lymphocyte count significantly starts to fall accompanied by abnormal increase in inflammatory cytokines and D-dimer in COVID-19 patients, they are recommended with IVIg treatment, which may bring down the inflammatory factors at an early stage and enhance the immunity. Low Molecular Weight Heparin (LMWH) anticoagulation therapy is also recommended in early stage of the disease when the infection and inflammation lead to excessive activation of coagulation factor such as the D-dimer.18 Detailed clinical evidence of COVID-19 patients are required for understanding the efficacy of the treatment.
Cross reactive neutralizing activity has been detected in serum/plasma of SARS-CoV recovered patient. Isolated monoclonal antibodies targeted against SARS-CoV RBD (like m396, S230, 80R and CR3014) failed to bind and neutralize SARS-COV2. The loss of reactivity points towards the structural difference and intrinsic sequence mismatch between RBDs of SARS-CoV and SARS-CoV2. RBD is the critical region for receptor binding, thus targeting the conserved epitopes in the RBD may have high potential in development of antibodies and promising targeted drug against SARS-CoV2.23
SARS-CoV2 RBM has undergone structural changes in the hACE2-binding ridge compared to SARS-CoV, due to major involvement of 4 amino acid residues 482–485: Gly-Val-Glu-Gly. Leu in the hydrophobic pocket of SARS-CoV RBM is replaced by Phe-486 in SARS-CoV2 RBM. These structural changes evolved to stabilize the hotspots and heightened the binding efficiency. Identification of functionally important epitopes in SARS-CoV2 RBM may lead to structure-based design of highly efficacious vaccines against viral RBD.24
M protein is a key SARS coronavirus enzyme that plays the pivotal role in mediating viral replication and transcription making it an attractive target for drug design against SARS-CoV2. Through virtual drug screening, structure assisted drug design and high throughput sequencing, a Michael acceptor inhibitor (N3) has been designed which has the ability to inhibit CoV M protein enzyme. Animal model study has revealed that N2 possesses potent antiviral activity. Homology modelling and molecular docking of SARS-CoV2 M protein and N3 has proved N3 to be an irreversible inhibitor of M protein viral enzyme. The development of antiviral agent targeting M protein may provide an effective first line defence against COVID-19.25
Binding of viral S protein to ACE2 leads to ACE2 downregulation which in turn leads to overproduction of angiotensin II. High level of angiotensin is hypothesized to be responsible for increased pulmonary vascular permeability that adversely affects lung condition. An increased level of ACE2 might protect against SARS-CoV-2 induced lung injury.26 There is no clinical evidence that confirms the adverse effects of ACE inhibitors in COVID-19 patients. Thus, patients already on ACE2 inhibitor drug medication should continue to take the medicines as sudden stop of these medicine intake may lead to stroke and other complications.27
Binding of SARS corona virus to cellular receptor ACE2 results in uptake of virions into endosomes, where the spike protein is activated by the pH dependent cysteine protease cathepsin L.28 Activation of the spike protein by cathepsin L can be blocked by lysomotropic agents, like bafilomycin A1 and ammonium chloride, which indirectly inhibit cathepsin L activity by interfering with endosomal acidification, or by compounds which directly block the proteolytic activity of cathepsin L, like MDL28170.
SARS-CoV-2 RNA dependent RNA polymerase is highly dynamic and one of the most attractive targets in the virus life cycle. Fragment-Based Drug Design (FBDD) strategy and docking experiments revealed a new compound MAW 22. Docking and MD simulation studies indicated MAW-22 to have higher ability to inhibit SARS-CoV-2 polymerase than Remdesvir.29 Implementation of more of such structure-based drug design strategies can lead to development of new drug against COVID-19 that can enter clinical trial.
In silico screening revealed six potential druggable pockets on the surface of the central β-sheet core of the S-protein RBD of SARS-CoV2 and virtual screening campaign on FDA-approved drug library identified several steroidal compounds, bile acids and their derivatives as potential hit against two pockets (pocket 1 and pocket 5). Incubation of viral spike RBD with betulinic acid, glycyrrhetinic acid, oleanolic acid and potassium canrenoate resulted in concentration depended reductions of the binding of S Spike RBD to the ACE2 receptors.30 This approach might have some efficacy in preventing virus entry only in the case of low viral load.
7.1 Immunomodulatory agents
Chloroquine and hydroxychloroquine (4-aminoquinoline) are proven drugs for malarial treatment.22,31 It acts as anti-inflammatory and immunomodulating agent for the treatment of malaria, rheumatoid arthritis and lupus erythematosus by suppressing the production/release of TNF-α and IL-6.19 It also works as a novel class of autophagy inhibitor, which may interfere with viral replication, proteolytic processing of M protein, virion assembly and budding. Studies revealed that it also has potential broad-spectrum antiviral activities by increasing endosomal pH required for virus/cell fusion thus inhibiting pH dependent viral replication as well as interfering with the glycosylation of cellular receptors of SARS-CoV preventing it from binding to host cell ACE2 receptor.32 Since SARS-CoV2 utilizes the same receptor for host cell entry, it is believed that chloroquine may interfere with ACE2 receptor glycosylation thus preventing viral–host cell attachment. In an in vitro study, chloroquine has been found to function at both entry and at post entry stages of COVID-19 infection in Vero E6 cells.8 The anti-viral and anti-inflammatory activities of chloroquine may account for its potent efficacy in treating patients with COVID-19 pneumonia. The structure and mechanism of action of chloroquine and hydroxychloroquine (HCQ) are the same except an additional hydroxy moiety in one terminal in HCQ. Due to the hydroxy group, HCQ has lesser retinal toxicity compared to chloroquine.28 The long-term clinical safety profile and higher potency of HCQ makes it a better option in treating COVID-19 over chloroquine. In a recent trial with patients on COVID-19 treatment, 100% of patients treated with hydroxychloroquine in combination with the macrolide antibiotic azithromycin were virologically cured.33,34 These drugs were authorized for emergency use by the FDA during the COVID-19 pandemic. However, the FDA withdrew the authorization when data analysis showed that the drugs are unlikely to be effective causing serious heart problems.The in vitro studies showed a supportive role of chloroquine and hydroxychloroquine but all of the clinical trials did not show the same results. Chen et al. who performed a pilot study about the role of hydroxychloroquine in viral clearance. This study included two groups, one receiving hydroxychloroquine and supportive treatment while the other group received supportive treatment alone. No significant difference of percentage of viral clearance was observed between the two groups, (86.7% vs. 93.3% P > 0.05).35 A randomized trial which was also performed by Chen et al. showed that HCQ treatment shortened clinical recovery.36 Another study which was performed by Megagnoli et al. with 368 patients divided into three groups. The death rates in HCQ group, non HCQ group and HCQ plus azithromycin were 27.8%, 11.4% and 22.1% which indicate that death rate was much higher in the patients who received only hydroxychloroquinine.37 So more randomized and controlled trial with varying drug doses and adequate sample size with placebo in control are needed to be performed to understand the role of chloroquine and hydroxychloroquinine in SARS CoV2 infection.38
Tocilizumab is a monoclonal antibody which target IL-6 receptor and plays a great role against rheumatoid arthritis. There are several reports of good results with tocilizumab for COVID-19 treatment. In a study reported on 21 COVID-19 patients who were treated with a dose of 400 mg of tocilizumab, 91% of the patients showed improved respiratory function. One report contain 21 patients of COVID-19 treated with tocilizumab and it was seen that 91% of the patients provided with a dose of 400 mg improved respiratory function.39
The main target of neutralizing antibodies is the S glycoprotein of coronaviruses. Neutralizing human mAbs from the memory B cells of SARS-CoV orMERS-CoV patients have previously been isolated. mAb S309 potently neutralized SARS-CoV2 (2019n-CoV/USA_WA1/2020) with an IC50 of 79 ng mL−1.40 S309 recognizes a proteoglycan epitope on the SARS-CoV2, distinct from the RBM. The epitope is accessible in both the open and closed conformation of the S glycoprotein, which explains the stoichiometric binding of Fab to the trimer of the S glycoprotein. S309 has the potential to be a promising drug candidate. Fc variants of S309 with increased half-life and effector functions have entered an accelerated development path towards clinical trials.
Baricitinib is approved by FDA for the treatment of rheumatoid arthritis which is a chronic inflammatory disorder. Baricitinib act as a Janus kinase 2 inhibitor. Binding to the receptor of the host cell, SARS-CoV2 undergo endocytosis which is mediated by numb-associated kinase (NAK) family—including AAK1 and GAK. Baricitinib targets these and also reduces inflammatory response.41 Ruxolitinib is another JAK-STAT inhibitor and a potent target towards HIV suppression and reduced inflammation. According to some report this drug is under phase III trial for the treatment of COVID 19.42
Nitazoxanide was approved for the treatment of human protozoan infection and possess therapeutic activity against soil transmitted helminths.43 When the SARS-COV2 enter the host cell it directly inhibits innate interferon pathway thus increasing the viral load and in that way escape the immune system. Nitazoxanide inhibits the SARS-CoV2 through the effect on interferon pathway, reduces the viral load and prevent the viral escape.43,44
Sirolimus, an inhibitor of mammalian target of rapamycin (mTOR) is reported to be an efficient blocker of viral protein expression and viral release. This drug is reported to have shown improvement in MERS-CoV infected patients and in H1N1 pneumonia infected patients. Sirolimus and dactinomycin (RNA synthesis inhibitor) in combination have been suggested as repurposable drug for HCoV treatment.45
7.2 Protease inhibitors
Lopinavir/ritonavir are anti-retroviral protease inhibitors that have been used for years for treatment of HIV-1 patients.46 In historical control study, lopinavir/ritonavir with ribavirin given to SARS-CoV patients resulted in significant clinical improvement with decrease in viral load and increase in lymphocyte count.47 It has reported that β-coronavirus viral loads of a COVID-19 patient in Korea significantly decreased after lopinavir/ritonavir (Kaletra®, AbbVie, North Chicago, IL, USA) treatment.48 In a randomized controlled open label trial on lab confirmed severe COVID-19 adult patients, treatment with lopinavir-ritonavir was not associated with clinical improvement compared to standard care patients. Future trials in COVID symptomatic patients may help to confirm or stop the use of this drug combination.49Darunavir was approved by FDA for the treatment of HIV as it is a protease inhibitor. It possesses a high binding affinity (Kd = 4.5 × 10–12 M) to the protease active sites and the presence of hydrogen bond increases its ability to fit exactly.50 Darunavir is often administrated with ritonavir which increases its potential towards viral suppression. Due to its high binding affinity, it become more resistant and build a genetic barrier against multidrug resistant HIV. Darunavir is often administrated with cobicistat which inhibit the 3C like protease and often inhibit the viral RNA synthesis an active agent against HIV and AIDS.51 There are some in vitro studies of darunavir against SARS-CoV2 but no human trial is performed. Some researchers of China declared that darunavir inhibited SARS-CoV-2 infection.52
Camostat mesylate approved in Japan for the treatment of pancreatic inflammation as it is a protease inhibitor.52 Corona viruses enter the cell through its spike protein and binds to the ACE 2 receptor and a host type 2 transmembrane serine protease (TMPRSS2) which facilitate their entry. Camostat mesylate causes the inhibition of TMPRSS2 and prevent the SARS-CoV-2 entry into host cell. Complete inhibition with no cytotoxic effect was obtained when Camostat mesylate and E-64d were administered.53
Fosamprenavir is a phosphate ester pro-drug which is converted to amprenavir, the active ingredient, after administration. It is used in the treatment of HIV as a HIV-1 protease inhibitor. The combination of fosamprenavir and ritonavir possess a great antiviral effect.54
7.3 RNA polymerase inhibitors
Ribavirin gained the FDA approval in 1970 for the treatment of respiratory syncytial virus and in combination with interferon and alpha-2b for hepatitis C. Ribavirin is a purine nucleoside analogue and it can prevent the replication of large number of RNA and DNA viruses. It not only interferes with polymerases, but also alters RNA capping of guanosine which normally causes RNA degradation. It directly inhibits inosine monophosphate dehydrogenase which is the precursor of guanosine.55 The elimination of ribavirin include two phases first phase of half-life about 2 hours and second phase takes longer half-life around 16 to 164 hours. Ribavirin also include two metabolic pathways-first one is the reversible phosphorylation pathway and second one is the degradative pathway include de-ribosylation and amide hydrolysis. Ribavirin administration results in adverse effects like conjunctivitis, headache, nausea, rashes and it may also cause haemolytic anaemia and bone marrow suppression. Ribavirin is applied with several combinations – ribavirin + lopinavir/ritonavir + interferon-β1b; ribavirin + lopinavir/ritonavir + IFN-α1b; ribavirin + IFN-α1b.56Remdesiviris a 1′-cyano-substituted adenosine nucleotide analogue molecule known to inhibit RdRP of Ebola virus.57 It also has a broad-spectrum antiviral activity against several RNA viruses. Based on in vitro cell line and mouse model studies, remdesivir could interfere with the nsp12 polymerase even in the setting of intact ExoN proofreading activity. Wang et al. 2020 presented data showing that remdesivir functions at host cell post entry stage and is effective against the COVID-19 in Vero E6 cells.33 Remdesivir has been reported to treat several cases of COVID-19 in the United States successfully.58 The first randomised, placebo controlled trial which was performed in China with 237 patients (158 in the remdesivir group and 19 in placebo control group). After the remdesivir administration the result revealed there was no significant reduction of time to achieve the clinical improvement in comparison with the placebo controlled group and mortality rate and viral clearance time was same as the placebo group. In this case remdesivir showed poor clinical benefits.59 In another trial where remdesivir was used along with immunosuppressants, faster clinical improvement was observed than the placebo group. More clinical trials needed to be performed to evaluate the efficacy of this drug.60
EIDD-2801 is an antiviral isopropylester prodrug of N4-hydroxycytidine having reports of preventive and therapeutic improvements in murine models of SARS-CoV and MERS-CoV when administered orally. It gets incorporated in the viral genome during RNA synthesis and drives mutagenesis leading to viral error catastrophe. Recent studies have reported the inhibitory action of EIDD-2801 on the replication of SARS-CoV2 in human respiratory epithelial cells. It is being prepared for clinical trials with the potential to become an effective antiviral against SARS-CoV2 in future.61
Favipiravir which is a pro-drug and potentially inhibit RNA dependent RNA polymerase.62 In that way it inhibits the viral replication. This drug is effective against influenza and Ebola virus and some other virus also. Favipiravir is easily incorporated into the viral RNA and thereby inhibiting the viral RNA duplication and beside dysregulation of duplication it also causes mutagenesis of viral RNA. Favipiravir is approved in India, China and Japan as a medication for COVID 19 treatment and reported favipiravir administration causes faster recovery for mild to moderate infected patients.63
Galidesivir is also an adenosine analogue and can inhibit the RNA polymerase activity through the unwanted RNA chain termination.64 It shows antiviral activity against many viruses and 100% protection against Marburg virus disease. Experimental study is going on to treat COVID 19 with galidesivir.65 Sofosbuvir is a uridine nucleotide analogue and a potent inhibitor of NS5B polymerase of hepatitis C virus. The docking experiment performed with Sofosbuvir showed that it can bind with both SARS-CoV and SARS-CoV2RdRPs and also possess a good binding energy (−6.5 up to −9.0 kcal mol−1). This contradict with the function of viral protein and leads towards viral eradication.66,67
7.4 Viral entry inhibitors
Arbidol is approved in Russia and China for the treatment of influenza virus.68 There is a short trimerization domain found in both haemagglutinin protein of Influenza virus and SARS-CoV2 glycoprotein where arbidol interacts with both of these drugs. This interaction leads to the formation of naked immature and less infectious virus.69 Arbidol possess its broad antiviral activity towards enveloped and non-enveloped viruses such as influenza, respiratory syncytial (RSV), Coxsackie B5, parainfluenza, adenovirus, Ebola (EBOV), and hepatitis B and hepatitis C.70 The mechanism of action that the arbidol follow that it target S protein/ACE2 interaction and in that way it inhibit the membrane fusion. 200 mg of arbidol is given orally for every 8 hours for influenza treatment and in COVID 19.71Griffithsin (GRFT) is a red alga derived lectin and possess a most potent activity for the inhibition of viral entry.72 GRFT was found to possess anti-HIV activity and it was led by the researchers at National Cancer Institute (NCI).73 GRFT may also be a potential agent towards this global health problem that is SARS-CoV2.74 Most of the viruses are enveloped with diverse array of glycoproteins for their successful entry and posses their different receptor association. GRFT binds specifically to the terminal mannose residue of N linked glycan that is found on the surface of corona virus and from collaborative studies it was found to have a broad spectrum of anti-viral activity against corona viruses including SARS-CoV.75
7.5 Translation inhibitors
Silvesterol is a natural compound isolated from plants belongs to the genus Aglaia.76 It possesses a great antiviral activity as this compound is a specific inhibitor of DEAD-box RNA helicase eIF4A which allow the translation through unwinding RNA secondary structure at the 5 prime untranslated region (UTR) of mRNA. Silvesterol specificity towards eIF4A is proved by CRISPR based studies. Corona virus is also a plus stranded RNA virus and use the 5′-cap dependent mRNA translation initiation strategies. It was found that silvesterol act as a potent inhibitor of mRNA translation in CoV-infected human embryonic lung fibroblast (MRC-5) cells. Silvesterol binding to eIF4A increases its affinity for mRNA and in that way causing the stalling of helicase to its substrate.77Aplidin was previously used to treat multiple myeloma and was approved in Australia in 2018.78 Plitidepsin is another translation inhibitor and it mainly target EF1A (eukaryotic translation elongation factor 1 alpha 1) thus multiplication and spread of virus gets affected. Human hepatoma cell line was used to analyse the activity of this drug which was infected with HCoV-229E-GFP virus, similar to SARS-CoV2. Some promising result was found but more clinical trials need to be performed (Fig. 2 and Table 1).78,79
8. The COVID-19 vaccine landscape
The complete genetic sequence of SARS-CoV2 was published on 11th January 2020, triggering a massive worldwide effort in vaccine development against COVID-19. The current situation has pulled us into unchartered territory of vaccine development during an ongoing pandemic. The impact of which on global health and economy has compelled the scientific community to explore novel paradigm with next generation vaccine development platforms. The distinct feature of the COVID-19 vaccine development programme is the diversity of vaccine technology platforms being investigated globally ranging from nucleic acids (DNA/RNA), recombinant proteins/peptides, virus-like particles, viral vectors, live attenuated virus and inactivated virus strategies. Many of these vaccine platforms offer unprecedented flexibility in terms of antigen manipulation and speed of development and manufacturing. The World Health Organization (WHO) has launched the solidarity trial to evaluate promising drug treatments against COVID-19 in countries around the world. According to the WHO, as of 12th October 2020, there are 42 candidate vaccines in clinical phase and 151 candidate vaccines in pre-clinical evaluation all across the globe against COVID-19. The global outlook for COVID-19 vaccine development during the ongoing pandemic has forced us to reiterate the traditional vaccine development pathway, which takes roughly about a decade, to develop and manufacture vaccines in a short timescale during such emergency situations. One important aspect of an effective vaccine is the use of adjuvants that enhance the immune response, making low doses more viable and aids in large scale immunization programmes. Several vaccine developers have expressed interest in developing novel adjuvants to be used for COVID-19 vaccines.Information on the SARS-CoV2 antigen being targeted for vaccine development has limited availability, most of the vaccines seems to direct neutralizing antibody production against the viral Spike (S) protein to hinder it's interaction with the ACE2 receptor of the host, blocking viral uptake. Although this indicates a promising target, further optimization of the antigen is required to trigger efficient immune response. From in vitro and animal model studies, two potential safety issues prevail for immunized animals upon viral challenge: one being cellular immunopathology and antibody-dependent enhancement (ADE). It has been noted in many immunized animals that upon viral infection there is enhanced lung and liver histopathology with increased tissue infiltration of monocytes, lymphocytes and eosinophils. This suggests a TH2 and TH17 immune response linking with IL-6, a cytokine upregulated in COVID-19 patients experiencing cytokine storm. It has also been found that alum, an adjuvant, which promotes TH2 immune response and reduces immunopathology. Such studies indicates the need to develop efficient vaccine delivery platforms and adjuvants that does not trigger a TH17-type immune response.80–82
The development of vaccines typically requires about 5–10 years with critical evaluation of immunogenic responses and safety issues during the pre-clinical and clinical phases. COVID-19 triggered the rapid development of vaccines across the globe based on different vaccine platforms. One such vaccine developed by Moderna Therapeutics and National Institutes of Health, mRNA-1273 based on the stabilized prefusion spike protein, showed promising neutralizing antibody responses in clinical trials.83 Another vaccine candidate developed by scientists at the Oxford University, AstraZeneca and Serum Institute of India, ChAdOx1-S based on adenoviral vector backbone also encoding the wild type spike protein, was tested in prime-boost regimen in rhesus macaques model, also showed promising immune response and recently entered phase III trials in several countries. Sinovac recently published results for their inactivated viral vaccine for Phase I/II trials. They used two doses, adjuvanted with aluminium hydroxide, which showed excellent safety profile.84,85
9. Conclusion
The current pandemic has severely affected human lives and jeopardized global economy. In this desperate situation, scientists across the world is looking for a vaccine to prevent the further loss of lives. Although there is raising awareness and approved guidelines by government authorities despite which new strategies should be developed to break the chain of community spread.86 There are a few drugs with effective therapeutic potential, but the efficacy of these drugs remains controversial which needs more experiments on large, randomized, controlled trials to be performed. Besides drug treatment, vaccination, immunomodulatory therapy, plasma therapy are of great importance in finding the novel treatment. Scientist from different fields like basic sciences, engineering and pharmacy need to work unitedly to enrich the probability of success.34 There has been a huge surge in information regarding COVID-19 since its origin in China. Although SARS which emerged in 2002 in Guangdong province of China was combatable through isolation and travel restrictions, but the current pandemic has become a challenge with no effective, safe or reliable drugs. Many of these drugs causes adverse effects like mutagenesis of viral DNA, low viral clearance, inflammation, headache, nausea and rashes. Among the FDA approved drugs chloroquine, hydroxychloroquine, remdesivir and favipiravir showed great result.86 Apart from finding the reliable drug, scientists are also concerned about patients suffering from other complications like central and peripheral neuropathies, delirium, pulmonary fibrosis. In addition to this, there is an ongoing race-against-time to develop successful vaccine candidates that is supported by the solidarity trial initiative of WHO. Compiling the available information on COVID-19 and the putative repurposable drugs that are in widespread use, presents us with a promising avenue to understand the pathophysiology and disease progression and aid in the development of novel therapeutics and vaccines against COVID-19.| Name | Structure | Year of approval for particular disease | Mode of action | Application in SARS-CoV2 | Citation |
|---|---|---|---|---|---|
| 1. Camostat mesylate | 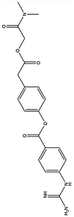 |
Chemostat mesylate approved in Japan for the treatment of pancreatic inflammation on Aug 14 of 2019 | Chemostat mesylate which is a serine protease inhibitor used for the treatment of pancreatitis as it inhibits trypsin which trigger the reaction for the disease | As it is a serine protease inhibitor chemostat mesylate inhibit TMPRSS2 which is required for the entry of virus thus it may regarded as a therapeutical agent against SARS-CoV2 | 53 and 87 |
| 2. Griffithsin | — | — | Griffithsin possess potent inhibitory activity towards viral entry | Griffithsin the plant derived lectin possess anti-viral activity towards SARS-CoV2 by binding towards terminal mannose residue of N derived lectin which is found on the surface of the corona virus | 72, 73 and 75 |
| 3. Arbidol | 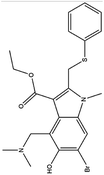 |
Arbidol is approved in Russian and China for the treatment of influenza virus infection | Arbidol inhibits the entry of the virus through interacting with the surface glycoprotein | Arbidol interacts with the trimerization domain of spike glycoprotein of SARS-CoV2 and leads to the formation of naked virus | 33, 70 and 71 |
| 4. Chloroquinine | 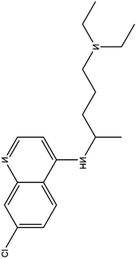 |
Chloroquine is used as a medication to prevent malaria and gained the FDA approval in the year of 1949 | In case of malaria this drug gets accumulated inside acidic food vacuoles of intraerythrocytic trophozoites and in that way prevent the hemoglobin degradation | Chloroquine increases endosomal pH and interfere with the glycosylation of cellular receptor. It also inhibits the quinine reductase II which is involved in sialic acid biosynthesis which makes it a broad antiviral agent. Chloroquine also thought to inhibit MAP kinase which interfere SARS- CoV2 molecular crosstalk and virion assembly, budding and also interfere with proteolytic processing of M protein | 31 and 32 |
| 5. Hydroxychloroquinine | 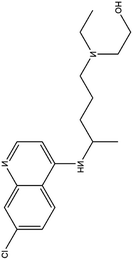 |
This drug was approved for the treatment of type II diabetes from 2014 | Hydroxychloroquinine possess a extra hydroxyl moiety at one terminal which makes it less permeable to blood retinal barrier and allow the faster clearance and thus lesser risk of retinal toxicity | HCQ selectively inhibit entry transport and post entry stages of SARS-CoV2, also reduces pro inflammatory markers and more potent in inhibiting SARS-CoV2 than chloquinine | 28 |
| 6. Ribavirin | 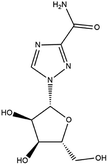 |
Ribavirin is approved by FDA in 1970 for the treatment of hepatitis C | Ribavirin is a purine nucleoside analogue, and it can prevent the replication of large no of RNA and DNA viruses | Ribavirin in combinations with lopinavir/interferon is recommended for the treatment of SARS-CoV2 | 55 and 56 |
| 7. Remdesivir | 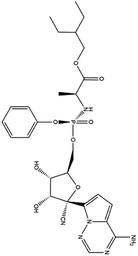 |
Remdesivir is yet not licensed or approved | Remdesivir is a nucleotide pro-drug and is effective against MERS-CoV. It showed a great efficacy in inhibiting the RNA polymerase | Remdesivir goes through a metabolic mechanism and activate nucleoside triphosphate metabolite which can inhibit RNA polymerase. Remdesivir plays a great role in inhibition of replication of SARS-CoV2 with an EC50 23.15 μM and it was the strongest antiviral activity among the tested drugs | 88 and 89 |
| 8. Favipiravir | 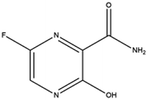 |
Favipiravir is approved by JAPAN in the year of 2014 | Favipiravir is also an RNA polymerase inhibitor | From some studies it was reported favipiravir is successful in shortening the recovery time for Covid-19 patients. Beside dysregulating RNA replication, it also causes mutagenesis in viral RNA | 62 and 90 |
| 9. Sofosbuvir | 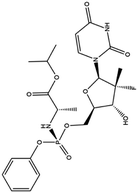 |
Sofosbuvir was approved by FDA on April 7 of 2017 for the treatment of hepatitis C | Sofosbuvir is a potent inhibitor of NS5B polymerase of hepatitis C virus | Sequence analysis and molecular docking experiments were performed against RdRP of COVID-19 with Sofosbuvir and the result suggest this drug bind with a good binding energy which leads towards viral eradication | 66 and 67 |
| 10. Galidesivir | 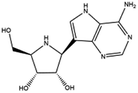 |
— | Galidesivir possess potent activity against hepatitis C, Ebola virus, Marburg virus through inhibition of RNA polymerase | Galidesivir is an adenosine nucleoside analogue which can prevent the RNA polymerase activity through RNA chain termination. Experimental study is going on to treat COVID-19 with galidesivir | 64 and 65 |
| 11. Fosamprenavir | 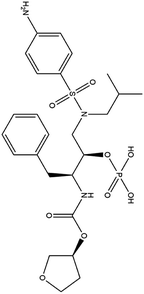 |
Fosamprenavir was approved by FDA and EMA for the treatment of HIV infection | Fosamprenavir is a protease inhibitor approved for the treatment of HIV infection | Fosamprenavir as a protease inhibitor possess a protective role against 3-chymotrypsin-like (3CLpro) protease of SARS-CoV2 | 91 |
| 12. Silvesterol | 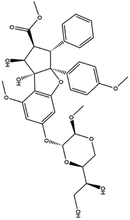 |
— | Silvesterol is a specific inhibitor of RNA helicase eIF4A thus interfere with the viral translation | Silvesterol which is a plant compound act as a potent inhibitor of cap dependent viral mRNA translation and it was found in CoV infected human embryonic lung fibroblast (MRC-5) cell | 76 and 77 |
| 13. Aplidin (plitidepsin) | 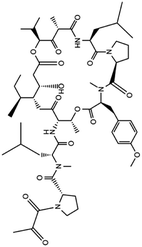 |
Aplidin was approved in 2018 in Australia for the treatment of multiple myeloma | Aplidin which is a anticancer compound potentially inhibited multiple myeloma and induced apoptosis | From the in vitro study it was found that plitidepsin affects EF1A (eukaryotic translation elongation factor 1 alpha 1) through which virus spread and multiply. A promising result was also found through analyzing the activity of this drug in human hepatoma cell line infected with the HCoV-229E-GFP virus, which is similar to SARS-CoV2 | 78 and 79 |
| 14. Lopinavir or ritonavir | 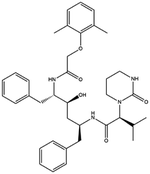 |
The FDA granted early access approval on Sept. 15, 2000 | Ritonavir was originally developed as an inhibitor of HIV protease | Lopinavir or ritonavir showed significant antiviral activity with reducing mortality and throat viral RNA detectability | 46 and 92 |
| 15. Darunavir | 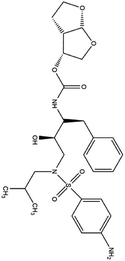 |
Darunavir was approved by FDA for HIV treatment on JUNE 23 of the year 2006 | Darunavir is a protease inhibitor and provides a new therapeutic option for HIV-1 infection | On Feb 4 of 2020 China had announced that darunavir played a great role in inhibition of SARS-CoV2 infection and from the cell study it is revealed that at 300 μM conc. darunavir selectively inhibited SARS-CoV2 infection | 41 and 50 |
| 16. Baricitinib | 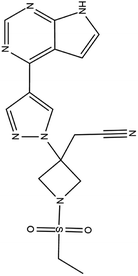 |
Baricitinib was approved by FDA for the treatment of rheumatoid arthritis on JUNE 1 of 2018 | Baricitinib is actually a JAK inhibitor used for the treatment of rheumatoid arthritis as it is a chronic inflammatory disorder | Baricitinib which target numb-associated kinase (NAK) family—including AAK1 and GAK which mediate endocytosis thus effect the entry of SARS-CoV2 and it is also a JAK inhibitor thus reduce inflammatory response | 41 and 93 |
| 17. Ruxolitinib | 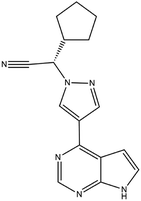 |
In January 11 of 2018 food and drug administration approved ruxolitinib for the treatment of HIV 1 infection | Ruxolitinib which is a JAK inhibitor plays a great role in reducing inflammation in HIV 1 infection | Ruxolitinib which is another JAK-STAT inhibitor plays a great role in reducing inflammation of COVID-19 infection. This drug is also under phase III trial for the treatment of COVID-19 | 94 and 95 |
| 18. Nitazoxanide | 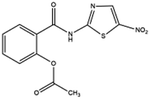 |
Nitazoxanoid approved by food and drug administration (FDA) for the treatment of diarrhea caused by cryptosporidium species and giardia intestinalis in pediatric patients | Nitazoxanoid possess therapeutic activity against soil transmitted helminths thus approved for treating protozoan infection | Nitazoxanide help in viral escape by reducing the inhibitory effect of interferon caused by SARS-CoV2 | 16 and 43 |
| 19. IFN-alpha | — | — | It also acts as an antiviral agent, used to treat hepatitis through inhibiting HBV replication by decreasing RNA transcription | As because SARS CoV and MARS CoV contain Orf-6 and Orf3b protein they are able to inhibit interferon expression but in COVID-19 Orf-6 and Orf3b are truncated thus COVID-19 is more sensitive to IFN 1 and it is reported that interferon alpha possess a great effect in reproduction | 96 and 97 |
| 20. Tocilizumab | — | The European commission has approved tocilizumab as a therapeutic agent for the treatment of moderate to high rheumatoid arthritis | This is an immunosuppressive drug used for the treatment of rheumatoid arthritis | IL-6 which is one of the important cytokines involved COVID-19 induced cytokine storm and thus tocilizumab which is monoclonal antibody against IL-6 plays a great role in the treatment of COVID-19 | 98 and 99 |
| 21. L-163491 | 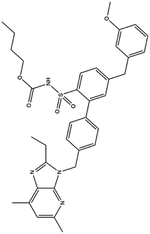 |
— | — | ACE 2 receptor involvement causes overactivation of renin-angiotensin system, which causes the elicitation of inflammatory response, through the catalyzation of degradation of angiotensin II to angiotensin (1 to 7). L-163491 as a partial antagonist of AT1 receptor and partial agonist of AT2 receptor may reduce the severity of corona virus infection | 16 |
| 22. Ivermectin | 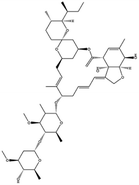 |
Ivermectin was approved by the United States federal food and drug administration (FDA) in 1996 for strongyloidiasis and onchocerciasis | — | IMP α/β heterodimer is responsible for integrase protein nuclear transport which plays a great role in viral replication but ivermectin inhibitory activity interfere the host cell division and effective against SARS-CoV2 | 100 |
Funding
This work was not supported by any funding.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors acknowledge Bose Institute Library for providing literary support. The authors thank Professor Samit Chattopadhyay for providing valuable suggestions and insights to prepare the manuscript. The authors acknowledge DST (Department of Science and Technology, Government of India) and DBT (Department of Biotechnology, Government of India) for providing funding.References
- Y.-R. Guo, Q.-D. Cao, Z.-S. Hong, Y.-Y. Tan, S.-D. Chen, H.-J. Jin, K.-S. Tan, D.-Y. Wang and Y. Yan, The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak – an update on the status, Mil. Med. Res., 2020, 7, 11 CAS.
- T. Singhal, A Review of Coronavirus Disease-2019 (COVID-19), Indian J. Pediatr., 2020, 87, 281–286 CrossRef.
- Z. Y. Zu, M. D. Jiang, P. P. Xu, W. Chen, Q. Q. Ni, G. M. Lu and L. J. Zhang, Coronavirus Disease 2019 (COVID-19): A Perspective from China, Radiology, 2020, 296, E15–E25 CrossRef.
- M. Tahir Ul Qamar, S. M. Alqahtani, M. A. Alamri and L.-L. Chen, Structural basis of SARS-CoV-2 3CL(pro) and anti-COVID-19 drug discovery from medicinal plants, J. Pharm. Anal., 2020, 10, 313–319 CrossRef.
- S. Prasad, V. Potdar, S. Cherian, P. Abraham, A. Basu and I.-N. N. Team, Transmission electron microscopy imaging of SARS-CoV-2, Indian J. Med. Res., 2020, 151, 241–243 Search PubMed.
- T. Bhatnagar, M. Murhekar, M. Soneja, N. Gupta, S. Giri, N. Wig and R. Gangakhedkar, Lopinavir/ritonavir combination therapy amongst symptomatic coronavirus disease 2019 patients in India: Protocol for restricted public health emergency use, Indian J. Med. Res., 2020, 151, 184–189 CAS.
- F. Li, Structure, Function, and Evolution of Coronavirus Spike Proteins, Annu. Rev. Virol., 2016, 3, 237–261 CrossRef CAS.
- J.-M. Kim, Y.-S. Chung, H. J. Jo, N.-J. Lee, M. S. Kim, S. H. Woo, S. Park, J. W. Kim, H. M. Kim and M.-G. Han, Identification of Coronavirus Isolated from a Patient in Korea with COVID-19, Osong Public Health Res. Perspect., 2020, 11, 3–7 CrossRef.
- D. Schoeman and B. C. Fielding, Coronavirus envelope protein: current knowledge, Virol. J., 2019, 16, 69 CrossRef.
- D. Raoult, A. Zumla, F. Locatelli, G. Ippolito and G. Kroemer, Coronavirus infections: Epidemiological, clinical and immunological features and hypotheses, Cell Stress, 2020, 4, 66–75 CrossRef CAS.
- H. Zhou, X. Chen, T. Hu, J. Li, H. Song, Y. Liu, P. Wang, D. Liu, J. Yang and E. C. Holmes, et al., A Novel Bat Coronavirus Closely Related to SARS-CoV-2 Contains Natural Insertions at the S1/S2 Cleavage Site of the Spike Protein, Curr. Biol., 2020, 30, 2196–2203.e2193 CrossRef CAS.
- Q. Li, X. Guan, P. Wu, X. Wang, L. Zhou, Y. Tong, R. Ren, K. S. M. Leung, E. H. Y. Lau and J. Y. Wong, et al., Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia, N. Engl. J. Med., 2020, 382, 1199–1207 CrossRef CAS.
- J. Zheng, SARS-CoV-2: an Emerging Coronavirus that Causes a Global Threat, Int. J. Biol. Sci., 2020, 16, 1678–1685 CrossRef CAS.
- S. Angeletti, D. Benvenuto, M. Bianchi, M. Giovanetti, S. Pascarella and M. Ciccozzi, COVID-2019: The role of the nsp2 and nsp3 in its pathogenesis, J. Med. Virol., 2020, 92, 584–588 CrossRef CAS.
- M. Tahir ul Qamar, S. M. Alqahtani, M. A. Alamri and L.-L. Chen, Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants, J. Pharm. Anal., 2020, 10, 313–319 CrossRef.
- C. Liu, Q. Zhou, Y. Li, L. V. Garner, S. P. Watkins, L. J. Carter, J. Smoot, A. C. Gregg, A. D. Daniels and S. Jervey, et al., Research and Development on Therapeutic Agents and Vaccines for COVID-19 and Related Human Coronavirus Diseases, ACS Cent. Sci., 2020, 6, 315–331 CrossRef CAS.
- X. Ou, Y. Liu, X. Lei, P. Li, D. Mi, L. Ren, L. Guo, R. Guo, T. Chen and J. Hu, et al., Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV, Nat. Commun., 2020, 11, 1620 CrossRef CAS.
- L. Lin, L. Lu, W. Cao and T. Li, Hypothesis for potential pathogenesis of SARS-CoV-2 infection-a review of immune changes in patients with viral pneumonia, Emerging Microbes Infect., 2020, 9, 727–732 CrossRef CAS.
- E. Prompetchara, C. Ketloy and T. Palaga, Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic, Asian Pac. J. Allergy Immunol., 2020, 38(1), 1–9 CAS.
- Z. Ye, Y. Zhang, Y. Wang, Z. Huang and B. Song, Chest CT manifestations of new coronavirus disease 2019 (COVID-19): a pictorial review, Eur. Radiol., 2020, 30, 4381–4389 CrossRef CAS.
- S. Kannan, P. Shaik Syed Ali, A. Sheeza and K. Hemalatha, COVID-19 (Novel Coronavirus 2019) - recent trends, Eur. Rev. Med. Pharmacol. Sci., 2020, 24, 2006–2011 CAS.
- J. Gao, Z. Tian and X. Yang, Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies, BioSci. Trends, 2020, 14, 72–73 CrossRef CAS.
- J. Lan, J. Ge, J. Yu, S. Shan, H. Zhou, S. Fan, Q. Zhang, X. Shi, Q. Wang and L. Zhang, et al., Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor, Nature, 2020, 581, 215–220 CrossRef CAS.
- J. Shang, G. Ye, K. Shi, Y. Wan, C. Luo, H. Aihara, Q. Geng, A. Auerbach and F. Li, Structural basis of receptor recognition by SARS-CoV-2, Nature, 2020, 581, 221–224 CrossRef CAS.
- Z. Jin, X. Du, Y. Xu, Y. Deng, M. Liu, Y. Zhao, B. Zhang, X. Li, L. Zhang and C. Peng, et al., Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors, Nature, 2020, 582, 289–293 CrossRef CAS.
- C. S. Kow, S. T. R. Zaidi and S. S. Hasan, Cardiovascular Disease and Use of Renin-Angiotensin System Inhibitors in COVID-19, Am. J. Cardiovasc. Drugs, 2020, 20, 217–221 CrossRef CAS.
- G. A. FitzGerald, Misguided drug advice for COVID-19, Science, 2020, 367, 1434 Search PubMed.
- A. K. Singh, A. Singh, A. Shaikh, R. Singh and A. Misra, Chloroquine and hydroxychloroquine in the treatment of COVID-19 with or without diabetes: a systematic search and a narrative review with a special reference to India and other developing countries, Diabetes & Metabolic Syndrome: Clinical Research & Reviews, 2020, 14, 241–246 Search PubMed.
- M. A. El Hassab, A. A. Shoun, S. T. Al-Rashood, T. Al-Warhi and W. M. Eldehna, Identification of a New Potential SARS-COV-2 RNA-Dependent RNA Polymerase Inhibitor via Combining Fragment-Based Drug Design, Docking, Molecular Dynamics, and MM-PBSA Calculations, Front. Chem., 2020, 8, 915 Search PubMed.
- A. Carino, F. Moraca, B. Fiorillo, S. Marchianò, V. Sepe, M. Biagioli, C. Finamore, S. Bozza, D. Francisci, E. Distrutti, B. Catalanotti, A. Zampella and S. Fiorucci, Hijacking SARS-CoV-2/ACE2 Receptor Interaction by Natural and Semi-synthetic Steroidal Agents Acting on Functional Pockets on the Receptor Binding Domain, Front. Chem., 2020, 8, 572885 CrossRef.
- A. F. G. Slater, Chloroquine: Mechanism of drug action and resistance in plasmodium falciparum, Pharmacol. Ther., 1993, 57, 203–235 CrossRef CAS.
- M. J. Vincent, E. Bergeron, S. Benjannet, B. R. Erickson, P. E. Rollin, T. G. Ksiazek, N. G. Seidah and S. T. Nichol, Chloroquine is a potent inhibitor of SARS coronavirus infection and spread, Virol. J., 2005, 2, 69 CrossRef.
- S. G. V. Rosa and W. C. Santos, Clinical trials on drug repositioning for COVID-19 treatment, Rev. Panam. Salud Publica, 2020, 44, e40 Search PubMed.
- D. Zhou, S.-M. Dai and Q. Tong, COVID-19: a recommendation to examine the effect of hydroxychloroquine in preventing infection and progression, J. Antimicrob. Chemother., 2020, 75, 1667–1670 CrossRef CAS.
- C. Jun, A pilot study of hydroxychloroquine in treatment of patients with moderate COVID-19, J. Zhejiang Univ., 2020, 49, 215–219 Search PubMed.
- Z. Chen, J. Hu, Z. Zhang, S. Jiang, S. Han, D. Yan, R. Zhuang, B. Hu and Z. Zhang, Efficacy of hydroxychloroquine in patients with COVID-19: results of a randomized clinical trial, 2020, medRxiv, 2020.2003.2022.20040758.
- J. Magagnoli, S. Narendran, F. Pereira, T. Cummings, J. W. Hardin, S. S. Sutton and J. Ambati, Outcomes of hydroxychloroquine usage in United States veterans hospitalized with Covid-19, 2020, medRxiv, 2020.2004.2016.20065920 Search PubMed.
- M. S. Khuroo, Chloroquine and hydroxychloroquine in coronavirus disease 2019 (COVID-19). Facts, fiction and the hype: a critical appraisal, Int. J. Antimicrob. Agents, 2020, 56, 106101 CrossRef CAS.
- X. Xu, M. Han, T. Li, W. Sun, D. Wang, B. Fu, Y. Zhou, X. Zheng, Y. Yang and X. Li, et al., Effective treatment of severe COVID-19 patients with tocilizumab, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 10970–10975 CrossRef CAS.
- D. Pinto, Y.-J. Park, M. Beltramello, A. C. Walls, M. A. Tortorici, S. Bianchi, S. Jaconi, K. Culap, F. Zatta and A. De Marco, et al., Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody, Nature, 2020, 583, 290–295 CrossRef CAS.
- M. E. Dowty, T. H. Lin, M. I. Jesson, M. Hegen, D. A. Martin, V. Katkade, S. Menon and J.-B. Telliez, Janus kinase inhibitors for the treatment of rheumatoid arthritis demonstrate similar profiles of in vitro cytokine receptor inhibition, Pharmacol. Res. Perspect., 2019, 7(6), e00537 Search PubMed.
- C. Gavegnano, M. Detorio, C. Montero, A. Bosque, V. Planelles and R. F. Schinazi, Ruxolitinib and Tofacitinib Are Potent and Selective Inhibitors of HIV-1 Replication and Virus Reactivation In Vitro, Antimicrobial Agents and Chemotherapy, 2014, 58, 1977 CrossRef.
- V. S. Somvanshi, B. L. Ellis, Y. Hu and R. V. Aroian, Nitazoxanide: Nematicidal mode of action and drug combination studies, Mol. Biochem. Parasitol., 2014, 193, 1–8 CrossRef CAS.
- S. Padmanabhan, Potential dual therapeutic approach against SARS-CoV-2/COVID-19 with Nitazoxanide and Hydroxychloroquine, 2020 Search PubMed.
- Y. Zhou, Y. Hou, J. Shen, Y. Huang, W. Martin and F. Cheng, Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2, Cell Discovery, 2020, 6, 14 CrossRef CAS.
- B. Su, Y. Wang, R. Zhou, T. Jiang, H. Zhang, Z. Li, A. Liu, Y. Shao, W. Hua and T. Zhang, et al., Efficacy and Tolerability of Lopinavir/Ritonavir- and Efavirenz-Based Initial Antiretroviral Therapy in HIV-1-Infected Patients in a Tertiary Care Hospital in Beijing, China, Front. Pharmacol., 2019, 10, 1472 CrossRef CAS.
- T. Bhatnagar, M. V. Murhekar, M. Soneja, N. Gupta, S. Giri, N. Wig and R. Gangakhedkar, Lopinavir/ritonavir combination therapy amongst symptomatic coronavirus disease 2019 patients in India: Protocol for restricted public health emergency use, Indian J. Med. Res., 2020, 151, 184–189 CAS.
- Y.-R. Guo, Q.-D. Cao, Z.-S. Hong, Y.-Y. Tan, S.-D. Chen, H.-J. Jin, K.-S. Tan, D.-Y. Wang and Y. Yan, The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status, Mil. Med. Res., 2020, 7, 11 CAS.
- B. Cao, Y. Wang, D. Wen, W. Liu, J. Wang, G. Fan, L. Ruan, B. Song, Y. Cai and M. Wei, et al., A Trial of Lopinavir–Ritonavir in Adults Hospitalized with Severe Covid-19, N. Engl. J. Med., 2020, 382, 1787–1799 CrossRef.
- E. Lefebvre and C. A. Schiffer, Resilience to resistance of HIV-1 protease inhibitors: profile of darunavir, AIDS Rev., 2008, 10, 131–142 Search PubMed.
- C. Harrison, Coronavirus puts drug repurposing on the fast track, Nat. Biotechnol., 2020, 38, 379–381 CrossRef.
- L. Dong, S. Hu and J. Gao, Discovering drugs to treat coronavirus disease 2019 (COVID-19), Drug Discoveries Ther., 2020, 14, 58–60 CrossRef CAS.
- J. Gibo, T. Ito, K. Kawabe, T. Hisano, M. Inoue, N. Fujimori, T. Oono, Y. Arita and H. Nawata, Camostat mesilate attenuates pancreatic fibrosis via inhibition of monocytes and pancreatic stellate cells activity, Lab. Invest., 2005, 85, 75–89 CrossRef CAS.
- L. Flamholc and M. Gisslén, Once-daily fosamprenavir with ritonavir in the treatment of HIV infection in therapy-naïve patients, Ther. Clin. Risk Manage., 2008, 4, 1281–1284 CrossRef CAS.
- J. S. Khalili, H. Zhu, N. S. A. Mak, Y. Yan and Y. Zhu, Novel coronavirus treatment with ribavirin: Groundwork for an evaluation concerning COVID-19, J. Med. Virol., 2020, 92, 740–746 CrossRef CAS.
- G. Koren, S. King, S. Knowles and E. Phillips, Ribavirin in the treatment of SARS: A new trick for an old drug?, CMAJ, 2003, 168, 1289–1292 Search PubMed.
- L. Zhang and R. Zhou, Structural Basis of the Potential Binding Mechanism of Remdesivir to SARS-CoV-2 RNA-Dependent RNA Polymerase, J. Phys. Chem. B, 2020, 124, 6955–6962 CrossRef CAS.
- M. L. Holshue, C. DeBolt, S. Lindquist, K. H. Lofy, J. Wiesman, H. Bruce, C. Spitters, K. Ericson, S. Wilkerson and A. Tural, et al., First Case of 2019 Novel Coronavirus in the United States, N. Engl. J. Med., 2020, 382, 929–936 CrossRef CAS.
- Y. Wang, D. Zhang, G. Du, R. Du, J. Zhao, Y. Jin, S. Fu, L. Gao, Z. Cheng and Q. Lu, et al., Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial, Lancet, 2020, 395, 1569–1578 CrossRef CAS.
- M. Soy, G. Keser, P. Atagündüz, F. Tabak, I. Atagündüz and S. Kayhan, Cytokine storm in COVID-19: pathogenesis and overview of anti-inflammatory agents used in treatment, Clin. Rheumatol., 2020, 39, 2085–2094 CrossRef.
- T. P. Sheahan, A. C. Sims, S. Zhou, R. L. Graham, A. J. Pruijssers, M. L. Agostini, S. R. Leist, A. Schäfer, K. H. Dinnon and L. J. Stevens, et al., An orally bioavailable broad-spectrum antiviral inhibits SARS-CoV-2 in human airway epithelial cell cultures and multiple coronaviruses in mice, Sci. Transl. Med., 2020, 12, eabb5883 CrossRef CAS.
- Y. Furuta, B. B. Gowen, K. Takahashi, K. Shiraki, D. F. Smee and D. L. Barnard, Favipiravir (T-705), a novel viral RNA polymerase inhibitor, Antiviral Res., 2013, 100, 446–454 CrossRef CAS.
- Q. Cai, M. Yang, D. Liu, J. Chen, D. Shu, J. Xia, X. Liao, Y. Gu, Q. Cai, Y. Yang, C. Shen, X. Li, L. Peng, D. Huang, J. Zhang, S. Zhang, F. Wang, J. Liu, L. Chen, S. Chen, Z. Wang, Z. Zhang, R. Cao, W. Zhong, Y. Liu and L. Liu, Experimental Treatment with Favipiravir for COVID-19: An Open-Label Control Study, Engineering, 2020, 6(10), 1192–1198 CrossRef.
- M. Jawaid Akhtar, COVID19 inhibitors: A prospective therapeutics, Bioorg. Chem., 2020, 101, 104027 CrossRef CAS.
- T. Warren, S. MacLennan, A. Mathis, E. Giuliano, R. Taylor and W. Sheridan, Efficacy of Galidesivir against Ebola Virus Disease in Rhesus Monkeys, Open Forum Infect. Dis., 2017, 4, S302 CrossRef.
- A. A. Elfiky, Anti-HCV, nucleotide inhibitors, repurposing against COVID-19, Life Sci., 2020, 248, 117477 CrossRef CAS.
- H. Bhatia, H. Singh, N. Grewal and N. Natt, Sofosbuvir: A novel treatment option for chronic hepatitis C infection, J. Pharmacol. Pharmacother., 2014, 5, 278–284 CrossRef.
- J. Blaising, S. J. Polyak and E.-I. Pécheur, Arbidol as a broad-spectrum antiviral: An update, Antiviral Res., 2014, 107, 84–94 CrossRef CAS.
- R. U. Kadam and I. A. Wilson, Structural basis of influenza virus fusion inhibition by the antiviral drug Arbidol, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 206 CrossRef CAS.
- L. Shi, H. Xiong, J. He, H. Deng, Q. Li, Q. Zhong, W. Hou, L. Cheng, H. Xiao and Z. Yang, Antiviral activity of arbidol against influenza A virus, respiratory syncytial virus, rhinovirus, coxsackie virus and adenovirus in vitro and in vivo, Arch. Virol., 2007, 152, 1447–1455 CrossRef CAS.
- N. Vankadari, Arbidol: A potential antiviral drug for the treatment of SARS-CoV-2 by blocking trimerization of the spike glycoprotein, Int. J. Antimicrob. Agents, 2020, 56, 105998 CrossRef CAS.
- S. Lusvarghi and C. A. Bewley, Griffithsin: An Antiviral Lectin with Outstanding Therapeutic Potential, Viruses, 2016, 8, 296 CrossRef.
- C. Lee, Griffithsin, a Highly Potent Broad-Spectrum Antiviral Lectin from Red Algae: From Discovery to Clinical Application, Mar. Drugs, 2019, 17, 567 CrossRef CAS.
- L. Pereira and A. T. Critchley, The COVID 19 novel coronavirus pandemic 2020: seaweeds to the rescue? Why does substantial, supporting research about the antiviral properties of seaweed polysaccharides seem to go unrecognized by the pharmaceutical community in these desperate times?, J. Appl. Phycol., 2020, 1–3 CAS.
- C. Barton, J. C. Kouokam, A. B. Lasnik, O. Foreman, A. Cambon, G. Brock, D. C. Montefiori, F. Vojdani, A. A. McCormick and B. R. O'Keefe, et al., Activity of and effect of subcutaneous treatment with the broad-spectrum antiviral lectin griffithsin in two laboratory rodent models, Antimicrob. Agents Chemother., 2014, 58, 120–127 CrossRef.
- L. Henss, T. Scholz, A. Grünweller and B. S. Schnierle, Silvestrol Inhibits Chikungunya Virus Replication, Viruses, 2018, 10(11), 592 CrossRef CAS.
- C. Müller, F. W. Schulte, K. Lange-Grünweller, W. Obermann, R. Madhugiri, S. Pleschka, J. Ziebuhr, R. K. Hartmann and A. Grünweller, Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona- and picornaviruses, Antiviral Res., 2018, 150, 123–129 CrossRef.
- J. Delgado-Calle, N. Kurihara, E. G. Atkinson, J. Nelson, K. Miyagawa, C. M. Galmarini, G. D. Roodman and T. Bellido, Aplidin (plitidepsin) is a novel anti-myeloma agent with potent anti-resorptive activity mediated by direct effects on osteoclasts, Oncotarget, 2019, 10(28), 2709–2721 CrossRef.
- S. Drożdżal, J. Rosik, K. Lechowicz, F. Machaj, K. Kotfis, S. Ghavami and M. J. Łos, FDA approved drugs with pharmacotherapeutic potential for SARS-CoV-2 (COVID-19) therapy, Drug Resist. Updates, 2020, 53, 100719 CrossRef.
- T. Thanh Le, Z. Andreadakis, A. Kumar, R. Gómez Román, S. Tollefsen, M. Saville and S. Mayhew, The COVID-19 vaccine development landscape, Nat. Rev. Drug Discovery, 2020, 19, 305–306 CrossRef CAS.
- N. Lurie, M. Saville, R. Hatchett and J. Halton, Developing Covid-19 Vaccines at Pandemic Speed, N. Engl. J. Med., 2020, 382, 1969–1973 CrossRef CAS.
- A. Mullard, COVID-19 vaccine development pipeline gears up, Lancet, 2020, 395, 1751–1752 CrossRef CAS.
- L. A. Jackson, E. J. Anderson, N. G. Rouphael, P. C. Roberts, M. Makhene, R. N. Coler, M. P. McCullough, J. D. Chappell, M. R. Denison and L. J. Stevens, et al., An mRNA Vaccine against SARS-CoV-2 — Preliminary Report, N. Engl. J. Med., 2020 Search PubMed.
- F. Krammer, SARS-CoV-2 vaccines in development, Nature, 2020, 586(7830), 516–527 CrossRef CAS.
- P. M. Folegatti, K. J. Ewer, P. K. Aley, B. Angus, S. Becker, S. Belij-Rammerstorfer, D. Bellamy, S. Bibi, M. Bittaye, E. A. Clutterbuck, C. Dold, S. N. Faust, A. Finn, A. L. Flaxman, B. Hallis, P. Heath, D. Jenkin, R. Lazarus, R. Makinson, A. M. Minassian, K. M. Pollock, M. Ramasamy, H. Robinson, M. Snape, R. Tarrant, M. Voysey, C. Green, A. D. Douglas, A. V. S. Hill, T. Lambe, S. C. Gilbert and A. J. Pollard, Safety and immunogenicity of ChAdOx1 nCoV-19 vaccine administered in a prime-boost regimen in young and old adults (COV002): a single-blind, randomised, controlled, phase 2/3 trial, Lancet, 2020, 396(10249), 467–478 CrossRef CAS.
- V. K. Shah, P. Firmal, A. Alam, D. Ganguly and S. Chattopadhyay, Overview of Immune Response During SARS-CoV-2 Infection: Lessons From the Past, Front. Immunol., 2020, 11, 1949 CrossRef CAS.
- M. Hoffmann, H. Kleine-Weber, S. Schroeder, N. Krüger, T. Herrler, S. Erichsen, T. S. Schiergens, G. Herrler, N.-H. Wu and A. Nitsche, et al., SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor, Cell, 2020, 181, 271–280.e278 CrossRef CAS.
- E. de Wit, F. Feldmann, J. Cronin, R. Jordan, A. Okumura, T. Thomas, D. Scott, T. Cihlar and H. Feldmann, Prophylactic and therapeutic remdesivir (GS-5734) treatment in the rhesus macaque model of MERS-CoV infection, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 6771 CrossRef CAS.
- A. Frediansyah, F. Nainu, K. Dhama, M. Mudatsir and H. Harapan, Remdesivir and its antiviral activity against COVID-19: a systematic review, Clinical Epidemiology and Global Health, 2020, 9, 123–127 CrossRef.
- J. M. Sanders, M. L. Monogue, T. Z. Jodlowski and J. B. Cutrell, Pharmacologic Treatments for Coronavirus Disease 2019 (COVID-19): a Review, JAMA, 2020, 323, 1824–1836 CrossRef CAS.
- S. Becker and L. Thornton, Fosamprenavir: advancing HIV protease inhibitor treatment options, Expert Opin. Pharmacother., 2004, 5, 1995–2005 CrossRef CAS.
- C. M. Chu, V. C. C. Cheng, I. F. N. Hung, M. M. L. Wong, K. H. Chan, K. S. Chan, R. Y. T. Kao, L. L. M. Poon, C. L. P. Wong and Y. Guan, et al., Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings, Thorax, 2004, 59, 252 CrossRef CAS.
- J. Stebbing, A. Phelan, I. Griffin, C. Tucker, O. Oechsle, D. Smith and P. Richardson, COVID-19: combining antiviral and anti-inflammatory treatments, Lancet Infect. Dis., 2020, 20, 400–402 CrossRef CAS.
- C. Harrison and A. M. Vannucchi, Ruxolitinib: a potent and selective Janus kinase 1 and 2 inhibitor in patients with myelofibrosis. An update for clinicians, Ther. Adv. Hematol., 2012, 3, 341–354 CrossRef CAS.
- C. Gavegnano, M. Detorio, C. Montero, A. Bosque, V. Planelles and R. F. Schinazi, Ruxolitinib and tofacitinib are potent and selective inhibitors of HIV-1 replication and virus reactivation in vitro, Antimicrob. Agents Chemother., 2014, 58, 1977–1986 CrossRef.
- A. S. J. Woo, R. Kwok and T. Ahmed, Alpha-interferon treatment in hepatitis B, Ann. Transl. Med., 2017, 5(7), 159 CrossRef.
- L. J. Stockman, R. Bellamy and P. Garner, SARS: Systematic Review of Treatment Effects, PLoS Med., 2006, 3, e343 CrossRef.
- P. Luo, Y. Liu, L. Qiu, X. Liu, D. Liu and J. Li, Tocilizumab treatment in COVID-19: A single center experience, J. Med. Virol., 2020, 92, 814–818 CrossRef CAS.
- T. Gout, A. J. K. Östör and M. K. Nisar, Lower gastrointestinal perforation in rheumatoid arthritis patients treated with conventional DMARDs or tocilizumab: a systematic literature review, Clin. Rheumatol., 2011, 30, 1471 CrossRef.
- L. Caly, J. D. Druce, M. G. Catton, D. A. Jans and K. M. Wagstaff, The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro, Antiviral Res., 2020, 178, 104787 CrossRef CAS.
Footnote |
| † Equally contributed. |
| This journal is © The Royal Society of Chemistry 2021 |