Open Access Article
Open Access ArticleQuantifying genome DNA during whole-genome amplification via quantitative real-time multiple displacement amplification†
Jing Tu *,
Yi Qiao
*,
Yi Qiao ,
Yuhan Luo,
Naiyun Long
,
Yuhan Luo,
Naiyun Long and
Zuhong Lu
and
Zuhong Lu
State Key Laboratory of Bioelectronics, School of Biological Science and Medical Engineering, Southeast University, Nanjing, 210096, China. E-mail: jtu@seu.edu.cn; Tel: +86-25-83792396
First published on 22nd January 2021
Abstract
DNA quantification is important in the research of life sciences. In an independent quantification process, the extracted part of a DNA sample is usually difficult to be recycled for further use while the widely used real-time PCR is used to count the copies with certain sequences. Based on the popular multiple displacement amplification (MDA), we proposed and performed quantitative real-time MDA to obtain the information of template amount based on fluorescence signals while amplifying whole-genome DNA. The detection limit of real-time MDA was as low as 0.5 pg μl−1 (5 pg DNA input), offering the whole-genome research a promising tool to quantify the entire DNA during amplification without sacrificing sample completeness or introducing redundant steps.
Introduction
Multiple displacement amplification (MDA) has become one of the most popular whole genome amplification (WGA) methods since it was first proposed by Dean et al.1 In MDA, strand displacement DNA polymerase such as phi29 DNA polymerase or Bst DNA polymerase amplifies genome DNA under isothermal conditions through the extension of a random hexamer primer. With a much higher coverage breadth and fidelity than its rivals2,3 and simplicity in protocol, MDA has been widely used in low initial amount DNA amplifying, particularly in the preparation step for whole genome sequencing4,5 and forensic analysis.6 Particularly, MDA has been routinely used in single cell genomic research as the DNA amplifying method, with proven fidelity in conserving single-cell heterogeneity in the sample preparation step of high-throughput sequencing.7,8However, in most of existing protocols, the initial DNA amount in MDA is obtained additionally before amplification or maintained undetermined all along. Although there are commercial kits that quantify DNA directly by measuring the fluorescence and have achieved fairly low detection level (e.g. 25 pg ml−1 in Quant-iT Picogreen, ThermoFisher, US), a large sample amount is required (>50 pg in Quant-iT Picogreen). When dealing with rare and precious samples, the part extracted for quantification is hard to recycle for further use in any downstream sections and that introduces unnecessary waste of sample.
Quantification during amplification could avoid sample waste in the quantifying process where the amount of the initial DNA template is calculated through the efficiency of amplification. As a classical quantification strategy, real-time PCR (also known as quantitative PCR, qPCR)9,10 measures the original concentration of the template through the Ct value (defined as the cycle number before the fluorescence reaching a certain threshold).11 Its accuracy and sensitivity in quantification (several copies of the template are enough for the reaction12) make it an extremely common tool in gene expression research13,14 and disease diagnosis.15,16 In particular, the 2019 novel coronavirus was first detected by real-time PCR,17 which is of great significance. Nonetheless, the primers in qPCR are unique; therefore, only the copy number of specific sequence is counted and the non-target molecules are not taken into account. As a result, PCR-based methods are not suitable for quantifying the total amount of DNA, and cannot be used in the whole-genome research.
Some methods based on WGA are realized to quantify trace amounts of DNA with principle similar to qPCR. Kang et al.18 successfully detected sub-picogram amounts of DNA via DOP-PCR using the SYBR dye, and the digital counting principle was also used in LAMP.19 In the aspect of MDA, monitoring MDA during amplification is proposed and carried out using a fluorescent dye with an initial concentration as low as 5 pg μl−1,20 but the detection level and detailed description of the amplifying process can be further investigated. Considering the versatility of MDA, accurate quantification would contribute to the simplicity and certainty of varieties of DNA analyzing processes.
Here, we proposed a DNA quantification method based on real-time MDA. The process of the MDA reaction was monitored via obtaining fluorescence signal, and the baseline time was measured to predict the initial DNA amount in the solution (Fig. 1). The influence Of the dye and threshold setting was also compared to ensure the quality of amplified DNA. A sequential dilution was performed to evaluate the accuracy and detection limit of this method. This real-time MDA method was proven to become a promising DNA quantification tool in the whole-genome research with the detection limit as low as the pictogram (sub-single cell) level.
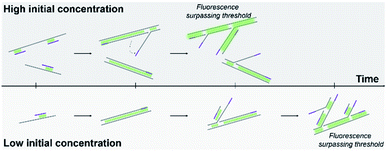 | ||
| Fig. 1 Schematic of real-time MDA. Higher initial DNA concentration requires shorter time to reach the fluorescent threshold. | ||
Experimental
Real-time MDA
Genomic DNA extracted from the GM12878 cell line was used as the template for real-time MDA, and the amount of template was controlled by adding a diluted DNA solution. The MDA system contained 1× phi29 DNA polymerase reaction buffer (New England Biolabs, USA), 50 μM random hexamer primer (Sangon, China), 4 mM dNTP, 1.5 U μl−1 phi29 DNA polymerase (New England Biolabs, USA), 0–4× EVAgreen dye (Biotium, USA) and 1× ROX dye (Takara, Japan). The reaction was carried out under 30 °C and the polymerase was inactivated with 65 °C after amplification using a real time thermal cycler (ABI 7500, Life Technologies, USA). Fluorescence intensity was detected and recorded every 9 min. The threshold was defined in different values, as described in the results section. Plots and fitting curves were generated using Origin Pro 2017.Genomic DNA extracted from 3 different tumor cell lines (U937, A375 and ZR-75) was adjusted to 125 pg μl−1 and then diluted to 1/5 sequentially (5 times) with water. The samples of different DNA concentrations were then amplified via real-time MDA. The threshold time was measured and the dilution time was calculated accordingly.
DNA electrophoresis
Agarose gel electrophoresis was performed to demonstrate the state of DNA samples amplified via quantitative real-time MDA. Amplified samples (3 μl) mixed with the loading buffer (0.6 μl) were loaded into lanes in a 1% agarose gel stained by GelRed (Biotium, USA) and were photographed after 30 min of electrophoresis. A DL15000 DNA ladder (Takara, Japan) was also added as the length reference.Results and discussion
Influence of the EVAgreen dye on MDA efficiency
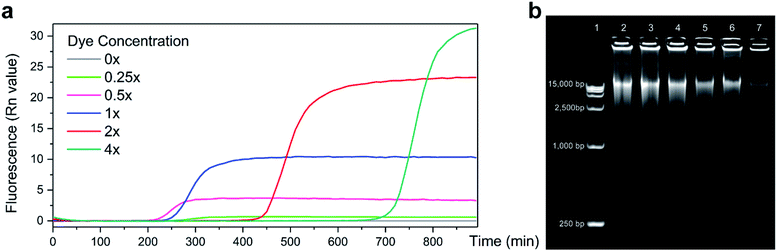
A real-time MDA protocol was designed to monitor DNA concentration increase through fluorescence change during amplification. EVAgreen was chosen as the indicating dye for its higher dynamic range and lower amplifying inhibition [9] compared to SYBR green, which is a traditional dye used in qPCR. To test its influence on MDA and find a proper concentration setup, performances of different concentrations of EVAgreen were compared (Fig. 2a). Keeping other conditions same, the baseline time and saturating plateau of MDA varied with the dye concentration. Agreed with the common knowledge, the dynamic range dropped, whereas the reaction efficiency rose as the amount of dye reduced until the amount of dye was too low to clearly indicate the amplification process (0.25×). The time before the reaction entering its exponential period was also influenced by the dye. It is clear that for the dye concentration higher than 1×, the efficiency of amplification decreased with the increase in dye, indicating that the dye truly inhibited the MDA reaction. MDA with 0.25× EVAgreen was detected to be less efficient than expectation, and we attribute it to the fact that the dye was too little to indicate the dynamic change in the nucleic acid concentration.The electrophoresis results (Fig. 2b) show that the length peaks of samples are similar (above 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 bp), but the distribution of sample length varied. It is clear that the dye inhibited MDA, exhibiting both in the total amount and in the length range of the product. A high dye concentration had significantly more impact on the product than a low dye concentration.
000 bp), but the distribution of sample length varied. It is clear that the dye inhibited MDA, exhibiting both in the total amount and in the length range of the product. A high dye concentration had significantly more impact on the product than a low dye concentration.
MDA could proceed under 0.5× EVAgreen with rather high efficiency yet got a dynamic range in fluorescence lower than the recommended concentration 1×. We set 1× and 0.5× as the concentrations of the EVAgreen dye in the following experiments aiming at a broad dynamic range and low amplifying inhibition, respectively.
Quantification of the DNA amount through real-time MDA
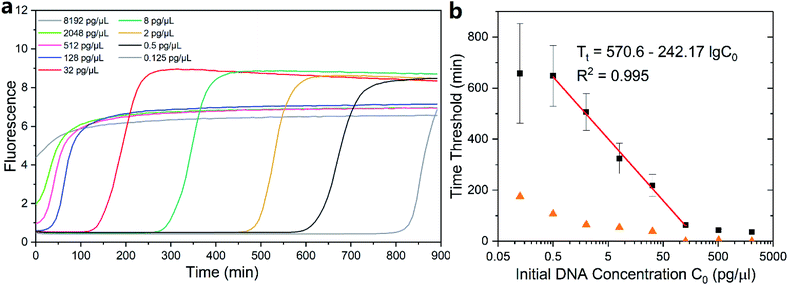
Genomic DNA extracted from the GM12878 cell line was dilute and quantified via real-time MDA (1× EVAgreen). The initial concentration of the template was set to 8192 pg μl−1 and was reduced to one fourth in the neighboring group until reaching the lowest concentration of 0.125 pg μl−1 (Fig. S1†). One fluorescence curve among each concentration group is shown in Fig. 3a. Interestingly, the curves of MDA with a high initial DNA concentration (no less than 128 pg μl−1 in this case) ended up with slightly low intensity repeatedly (Fig. S1†) than other groups. We assume that it was because the low magnifying time in those groups hindered the further growth of DNA branches in the MDA reaction, which contributed to the increase in the fluorescence.First, we defined 30% of total fluorescence increase as the threshold and then measured the reaction time below the threshold. The threshold of 30% ensured that MDA had entered into its exponential period, significantly surpassing the baseline. Since MDA was isothermal, which was maintained at 30 °C and cannot be divided into temperature cycles, amplifying time before the threshold was utilized to describe the amplifying speed instead of the Ct value used in qPCR. The time before threshold was inversely proportional to the log DNA initial concentration (Fig. 3b) with a slope of −242.17 in the range of 0.5–128 pg μl−1. Consequently, the detection limit of real-time MDA was determined to be as low as 0.5 pg μl−1, and the initial amount of DNA in a sample could be relatively quantified according to the performance of known samples and the fitting line (red line in Fig. 3b). Results of other thresholds are also obtained (Fig. S2†). The time threshold increased with a decrease in the initial concentration but still exhibited similar trends. It is worth noting that the conventional method of defining threshold in real-time PCR (i.e. 10 times of the standard deviation of first several signals) was also suitable for measurement but was difficult in rather high initial concentration (Fig. S3†). This was because with a high DNA concentration the fluorescence signal may rise immediately after the initiation, leaving no baseline, as shown in Fig. 3a, for deviation calculation. Moreover, the inhibition of high initial DNA concentration on the final intensity may also impact the detection limit.
While the fitted points exhibited a nice predictable trend, some of the data outside the 0.5–128 pg μl−1 range did not. The concentration higher than the range did not follow the inversely proportional trend despite their low deviations because the initial fluorescence was so high that it occupied a considerable amount of the total dynamic range of the dye (Fig. 3a), which made them more suitable for direct quantification. The standard variation of each group increased when the initial DNA concentration went down (orange triangles in Fig. 3b), suggesting a higher accuracy of quantification in the relatively high initial amount. As a result, the initial concentration of 0.125 pg μl−1 and below was excluded for their low accuracy and high deviation.
Based on the slope of the fitting line, the efficiency E of MDA in its exponential period could be calculated according to the principle of exponential amplification: XT = X0 × (1 + E)T. The efficiency E of MDA was 1.0% per minute. Alternatively, the amount of DNA in the present MDA reaction doubled every ∼70 min. Given an initial DNA concentration of 0.5 ng μl−1, it would take ∼10 h to reach ∼200 ng μl−1, which generally agrees with the common knowledge on the MDA reaction time. Further, under the consideration that dye would inevitably hinder the amplification progress, the actual reaction time would not be longer than the expectation.
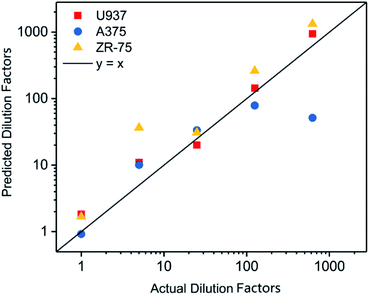
After dilution to 125 pg μl−1, the concentration of DNA extracted from tumor cell lines was adjusted based on the results quantified via Qubit 2.0 (Table S1†). The threshold time in the real-time MDA of DNAs was measured (Table S2†). The dilution factor of samples was calculated via the formula, as shown in Fig. 3. The predicted dilution factors showed a strong linear relationship with the actual dilution factors (Fig. 4 and Table S3†), indicating the reproducibility and generalization of this quantification method. Results of the high dilution factor where the initial DNA concentration was estimated to be ∼0.2 pg drifted rather far from the line, so the application in an extremely low DNA concentration should be concerned with more care.
We also tested the performance of quantitative real-time MDA with a 0.5× EVAgreen dye (Fig. S4, and S5†). The lower detection limit was clearly not as low as that with 1× EVAgreen (∼2 pg μl−1 vs. 0.5 pg μl−1). The ∼4 times difference may be attributed to the lower amount of dye that could maintain enough sensitivity even after hours (>9 h) of stand-by time. However, the fitting line of plots within the reliable range still showed a high R-square (0.989 for threshold = 30% total increasing).
With the optimized parameters, the detection level of the real-time MDA could be down to 5 pg (0.5 pg μl−1 DNA in 10 μl solution); however, the reaction setting is preferable on amplifying human genome. Care must be taken during quantifying samples from different species since fluctuation in the GC content may alter the performance of MDA.21 In addition, direct fluorescent measurement rather than droplet digital MDA (ddMDA) was chosen to quantify DNA because the observed positive fraction was significantly higher than predicted in ddMDA, which may be contributed by the binary characteristic of the digital analysis and the high sensitivity of MDA.22
Conclusions
MDA has been proven to be a promising tool for whole genome amplification and is wildly used in genome research. In this study, we demonstrated the ability of real-time MDA in quantifying the DNA amount during the whole sequence amplification. Briefly, the amplifying time before a certain threshold of real-time MDA exhibits a clear logarithmic trend similar to routinely used qPCR yet entire template molecules was amplified during the quantifying process. The detection limit of real-time MDA could be as low as the picogram (sub-single cell) level. The quantification in this method has low standard variation among a relatively wide range. Based on the results and with proper optimization, real-time MDA should be a powerful quantification tool in whole-genome related researches.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key Research and Development Program of China (No. 2016YFA0501604), National Natural Science Foundation of China (No. 61971125) and Six Talent Peaks Project of Jiangsu Province (2019-SWYY-004).References
- F. B. Dean, J. R. Nelson, T. L. Giesler and R. S. Lasken, Genome Res., 2001, 11, 1095–1099 CrossRef CAS.
- C. F. A. de Bourcy, I. D. Vlaminck, J. N. Kanbar, J. Wang, C. Gawad and S. R. Quake, PLoS One, 2014, 9, e105585 CrossRef.
- E. Borgström, M. Paterlini, J. E. Mold, J. Frisen and J. Lundeberg, PLoS One, 2017, 12, e0171566 CrossRef.
- M. Rhee, Y. K. Light, R. J. Meagher and A. K. Singh, PLoS One, 2016, 11, e0153699 CrossRef.
- R. Stepanauskas, E. A. Fergusson, J. Brown, N. J. Poulton, B. Tupper, J. M. Labonté, E. D. Becraft, J. M. Brown, M. G. Pachiadaki and T. Povilaitis, et al., Nat. Commun., 2017, 8, 1–10 CrossRef.
- E. Giardina, I. Pietrangeli, C. Martone, S. Zampatti, P. Marsala, L. Gabriele, O. Ricci, G. Solla, P. Asili and G. Arcudi, et al., BMC Genomics, 2009, 10, 1–9 CrossRef.
- L. Huang, F. Ma, A. Chapman, S. Lu and X. S. Xie, Annu. Rev. Genomics Hum. Genet., 2015, 16, 79–102 CrossRef CAS.
- Y. Fu, F. Zhang, X. Zhang, J. Yin, M. Du, M. Jiang, L. Liu, J. Li, Y. Huang and J. Wang, Commun. Biol., 2019, 2, 147 CrossRef.
- S. Choi and S. C. Jiang, Appl. Environ. Microbiol., 2005, 71, 7426–7433 CrossRef CAS.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef CAS.
- J. S. Yuan, D. Wang and C. N. Stewart Jr, Biotechnol. J., 2008, 3, 112–123 CrossRef CAS.
- C. A. Heid, J. Stevens, K. J. Livak and P. M. Williams, Genome Res., 1996, 6, 986–994 CrossRef CAS.
- L. Xu, Y. Liu, Y. Sun, B. Wang, Y. Xiong, W. Lin, Q. Wei, H. Wang, W. He and G. Li, Stem Cell Res. Ther., 2017, 8, 1–11 CrossRef CAS.
- J.-T. Yao, S.-H. Zhao, Q.-P. Liu, M.-Q. Lv, D.-X. Zhou, Z.-J. Liao and K.-J. Nan, Pathol., Res. Pract., 2017, 213, 453–456 CrossRef CAS.
- A. Heim, C. Ebnet, G. Harste and P. Pring-\AAkerblom, J. Med. Virol., 2003, 70, 228–239 CrossRef CAS.
- J. Liu, J. A. Platts-Mills, J. Juma, F. Kabir, J. Nkeze, C. Okoi, D. J. Operario, J. Uddin, S. Ahmed and P. L. Alonso, et al., Lancet, 2016, 388, 1291–1301 CrossRef CAS.
- C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao, Y. Hu, L. Zhang, G. Fan, J. Xu and X. Gu, et al., Lancet, 2020, 395, 497–506 CrossRef CAS.
- M.-J. Kang, H. Yu, S.-K. Kim, S.-R. Park and I. Yang, PLoS One, 2011, 6, e28661 CrossRef CAS.
- Q. Zhu, Y. Gao, B. Yu, H. Ren, L. Qiu, S. Han, W. Jin, Q. Jin and Y. Mu, Lab Chip, 2012, 12, 4755–4763 RSC.
- B. Bruijns, F. Costantini, N. Lovecchio, R. Tiggelaar, G. Di Timoteo, A. Nascetti, G. de Cesare, J. Gardeniers and D. Caputo, Sens. Actuators, B, 2019, 293, 16–22 CrossRef CAS.
- R. Marine, C. McCarren, V. Vorrasane, D. Nasko, E. Crowgey, S. W. Polson and K. E. Wommack, Microbiome, 2014, 2, 3 CrossRef.
- A. M. Sidore, F. Lan, S. W. Lim and A. R. Abate, Nucleic Acids Res., 2016, 44, e66 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra09021b |
| This journal is © The Royal Society of Chemistry 2021 |