DOI:
10.1039/D0RA09074C
(Paper)
RSC Adv., 2021,
11, 3470-3475
Host–guest modes and supramolecular frameworks of complexes of tetramethyl cucurbit[6]uril with 4-chloroaniline and 4,4′-diaminostilbene†
Received
24th October 2020
, Accepted 21st December 2020
First published on 15th January 2021
Abstract
Since the first reportal on decamethylcucurbit[5]uril (Me10Q[5]) in 1992, substituted cucurbit[n]urils have attracted considerable research interest. In this study, the host–guest modes between the tetramethyl cucurbit[6]uril (TMeQ[6]) as a host and 4-chloroaniline and 4,4′-diaminostilbene (G1 and G2) as guests were investigated by single-crystal X-ray diffraction, NMR, ITC, UV-Vis spectrum, and MALDI-TOF mass spectrometry analyses. The experimental results showed that TMeQ[6] formed a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 inclusion compound with G1, and the carbonyl portal of TMeQ[6] formed a 1
1 inclusion compound with G1, and the carbonyl portal of TMeQ[6] formed a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 self-assembly with G2. Further, multi-dimensional supramolecular frameworks were formed driven by weak interaction forces in the system (hydrogen bonding, C–H⋯π interactions, ion–dipole interactions, and dipole–dipole interactions).
1 self-assembly with G2. Further, multi-dimensional supramolecular frameworks were formed driven by weak interaction forces in the system (hydrogen bonding, C–H⋯π interactions, ion–dipole interactions, and dipole–dipole interactions).
1. Introduction
Various porous materials, namely, inorganic porous materials1,2 (such as molecular sieves), organic–inorganic hybrid porous materials3–5 (such as metal–organic frameworks, MOFs), and porous organic framework (POF) materials6,7 have been developed. These porous materials have been widely used in adsorption, heterogeneous catalysis, and ion exchange processes.8–10 Interestingly, in recent years, the use of weak forces as the driving force to construct supramolecular frameworks has been explored,11,12 so the selection of suitable construction units has also become crucial. Cucurbit[n]uril is a fourth-generation supramolecular macrocyclic compound after crown ether, cyclodextrin, and calixarene; there are two main types: ordinary cucurbit[n]urils13–18 and modified cucurbit[n]urils (decamethylcucurbit[5]uril, cyclopentyl-modified cucurbit[n]urils and so on).19–24 Their structure has a hydrophobic cavity, two carbonyl portals, and an outer surface. The charge distribution in cucurbit[n]uril itself is not uniform, which makes cucurbit[n]uril a permanent dipole25 (the cavity of the cucurbit[n]uril has an electrically neutral potential, the outer surface has a positive potential, and the carbonyl portal shows a negative potential). This is conducive to the study of the host–guest chemistry26–30 and coordination chemistry31–35 of the cucurbit[n]uril, and to the construction of good reversible supramolecular organic frameworks36,37 (SOFs).
Based on A–H⋯B (A and B are oxygen, nitrogen or fluorine) hydrogen bond theory.38 There was ion–dipole interaction between [ZnCl4]2− and [CdCl4]2− anions and positive potential outer surface of cucurbit[n]urils, which makes them conducive to the construction of self-assembly complexes based on cucurbit[n]uril, so considered to be known to function as effective structure directing agents.39 In this work, [ZnCl4]2− (formed from ZnCl2 in an aqueous HCl solution) was used as structure directing agents, TMeQ[6]-based SOFs, namely, C92H110Cl17N50O28Zn4 (1) and C68H76Cl8N28O12Zn2 (2) were constructed by the supramolecular collaborative assembly. The driving force of complexes 1 and 2 was studied through single-crystal X-ray diffraction analysis, and the host–guest interaction between TMeQ[6] and guest molecules was studied through NMR, ITC, UV-Vis spectrum and MALDI-TOF mass spectrometry analyses. The experimental results showed that TMeQ[6]-based host–guest SOFs are promising for several applications, e.g., in selective adsorption, heterogeneous catalysis, and ion exchange (Scheme 1).
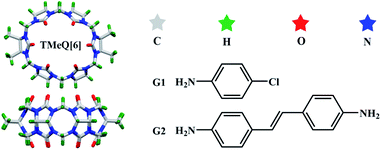 |
| | Scheme 1 TMeQ[6] and guest molecules used in this study. | |
2. Experimental
2.1 Experimental reagents
All the reagents were purchased from Aladdin Industrial Corporation (AR, Shanghai, China). TMeQ[6] was synthesized according to the method reportaled in a previous paper.40
2.2 Synthesis of the complexes
TMeQ[6] (10 mg, 9.51 μmol) and G1 (2.55 mg, 20 μmol) were placed in a crystal bottle and 3 mol L−1 hydrochloric acid (5 mL) was added; the mixture was heated until dissolution. Then, a small amount of the inducing agent ZnCl2 was added, and the solution was heated and stirred for 5–10 min in a water bath at 35 °C. Finally, the solution was allowed to stand at room temperature for 7–10 days to obtain single crystals of complex 1 (35% yield). Complex 2 was synthesized (28% yield) in the same manner as complex 1.
2.3 Crystal structure determination
A crystal with the appropriate size and transparency was selected and fixed on a glass probe with petroleum jelly. A Bruker Smart Apex II single-crystal X-ray diffractometer, with Mo-Kα radiation (λ = 0.071073 nm), was used for the characterization. The SHELX-97 (ref. 41) program packages were used for data refinement by the full-matrix least-squares method, and the SQUEEZE routine of the PLATON program was used to process solvent molecules in the crystal. The X-ray crystallographic data for structures reportaled in this study have been deposited in the Cambridge Crystallographic Data Center under accession numbers CCDC: 2039065 (1) and 2039066 (2).† The crystal parameters, data acquisition conditions, and parameters of the complexes 1 and 2 are listed in Table 1.
Table 1 Crystallographic data for complexes 1 and 2a
| |
1 |
2 |
| Note: [a] Conventional R on Fhkl: ∑‖F0| − |Fc‖/∑|F0|; [b] weighted R on |Fhkl|2: ∑[w(F02 − Fc2)2]/∑[w(F02)2]1/2. |
| Empirical formula |
C92H110Cl17N50O28Zn4 |
C68H76Cl8N28O12Zn2 |
| Formula weight |
3228.42 |
1891.90 |
| Crystal system |
Triclinic |
Triclinic |
| Space group |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) |
| a [Å] |
18.070(3) |
11.664(5) |
| b [Å] |
21.050(3) |
14.379(7) |
| c [Å] |
21.095(5) |
15.558(7) |
| α [°] |
105.503(9) |
101.417(14) |
| β [°] |
104.705(8) |
100.443(15) |
| γ [°] |
104.621(5) |
107.344(14) |
| V [Å3] |
7023(2) |
2359.4(19) |
| Z |
2 |
1 |
| Dcalcd. [g cm−3] |
1.527 |
1.331 |
| T [K] |
273.15 |
273.15 |
| μ [mm−1] |
1.082 |
0.802 |
| Parameters |
1730 |
536 |
| Rint |
0.0444 |
0.1199 |
| R [I > 2σ(I)]a |
0.0531 |
0.1182 |
| wR [I > 2σ(I)]b |
0.1542 |
0.3377 |
| R (all data) |
0.0783 |
0.2050 |
| wR (all data) |
0.1702 |
0.3755 |
| GOF on F2 |
1.051 |
1.110 |
2.4 Determination by 1H NMR
G1, G2, and TMeQ[6] were each dissolved in neutral D2O. The guest solution concentration was fixed at 0.1 mM in all analyses. The TMeQ[6] solution was added dropwise until it was in excess of the guest solution, and the 1H NMR spectra were recorded at appropriate intervals using a JEOL JNM-ECZ400s spectrometer at 25 °C. D2O was used as a field-frequency lock, and the observed chemical shifts are reportaled in parts per million (ppm) relative to D2O as an internal standard (δ = 4.67 ppm).
2.5 Determination by isothermal titration calorimetry
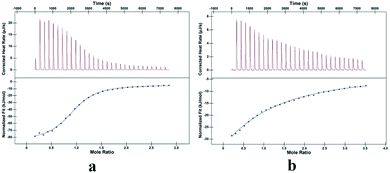
The neutral aqueous solution of TMeQ[6] (1.0 × 10−4 mol L−1, 1.00 mL) was placed in the sample cell, and a 1.00 × 10−3 mol L−1 G1 solution was drawn into a 250 mL syringe. The temperature was set at 25 °C, and the titration was conducted by adding 30 aliquots (8 μL) of the G1 solution at intervals of 300 s. The same method was used for TMeQ[6] and G2. The thermodynamic parameters of each system were determined on a nano ITC isothermal calorimeter. After deleting the first one unwanted data points, the data were analyzed with Launch NanoAnalyze software using an independent model.
2.6 Determination by UV-Vis spectrum
The neutral aqueous solution of TMeQ[6] (1.0 × 10−4 mol L−1) and G1 (1.0 × 10−3 mol L−1) were prepared as stock solutions, and the UV-Vis absorption spectrum of each substance was measured by the molar ratio. Add 300 μL of G1 stock solution to eleven 10 mL volumetric bottles, then add corresponding volume of TMeQ[6] stock solution to these volumetric bottles, and maintain molar ratios of 0, 0.2, 0.4, 0.6, 0.8, 1.0, 1.2, 1.4, 1.6, 1.8 and 2.0 after volume determination with doubly-distilled water. The measurements were performed at room temperature with an UV-2700 external spectrometer. The same method was used for TMeQ[6] and G2.
2.7 Determination by mass spectrometry
The neutral aqueous solution of TMeQ[6] (1.0 × 10−4 mol L−1) was prepared, and 1 mL was pipetted into a centrifuge tube. Then, 100 μL of the G1 aqueous solution with a concentration of 1 × 10−3 mol L−1 was added. The solution was filtered into a chromatographic sample bottle using aqueous 0.22 μm syringe filters. Mass spectra were recorded on an Agilent 6545 Q-TOF mass spectrometer. The same method was used for TMeQ[6] and G2.
3. Results and discussion
3.1 Description of the crystal structures of complexes 1 and 2
3.1.1 Description of the crystal structure of complex 1. Complex 1 exhibits the triclinic P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) space group. As shown in Fig. 1a, TMeQ[6] and protonated 4-chloroaniline formed a 1
space group. As shown in Fig. 1a, TMeQ[6] and protonated 4-chloroaniline formed a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 inclusion compound. In this system, hydrogen bonds are one of the main forces. N49 forms three hydrogen bonds with O2, O4, and O6, and N50 forms three hydrogen bonds with O16, O18, and O20; the N–H⋯O hydrogen bond length is 2.760–2.834 Å. In addition, hydrogen bonds form between O1 and O2 and between O15 and O16 (O2–H2A⋯O1 and O16–H16A⋯O15), with the bond lengths being 2.859 and 2.855 Å, respectively (Fig. 1b). One counterion [ZnCl4]2− connects with three adjacent outer surfaces of TMeQ[6]s via ion–dipole interactions (Fig. 1c). A negatively potential TMeQ[6] portal is connected to another positively potential outer surface of TMeQ[6] by the dipole–dipole interaction (Fig. 1d). In summary, these weak interactions (hydrogen bonding, ion–dipole interaction, and dipole–dipole interaction) caused complex 1 to form an ordered multi-dimensional layered supramolecular framework (Fig. 2a–d).
1 inclusion compound. In this system, hydrogen bonds are one of the main forces. N49 forms three hydrogen bonds with O2, O4, and O6, and N50 forms three hydrogen bonds with O16, O18, and O20; the N–H⋯O hydrogen bond length is 2.760–2.834 Å. In addition, hydrogen bonds form between O1 and O2 and between O15 and O16 (O2–H2A⋯O1 and O16–H16A⋯O15), with the bond lengths being 2.859 and 2.855 Å, respectively (Fig. 1b). One counterion [ZnCl4]2− connects with three adjacent outer surfaces of TMeQ[6]s via ion–dipole interactions (Fig. 1c). A negatively potential TMeQ[6] portal is connected to another positively potential outer surface of TMeQ[6] by the dipole–dipole interaction (Fig. 1d). In summary, these weak interactions (hydrogen bonding, ion–dipole interaction, and dipole–dipole interaction) caused complex 1 to form an ordered multi-dimensional layered supramolecular framework (Fig. 2a–d).
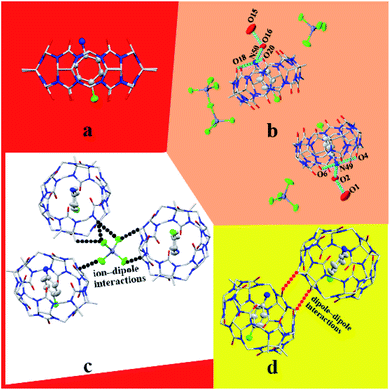 |
| | Fig. 1 Structure of complex 1: (a) 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 inclusion compound of TMeQ[6] and G1, (b) hydrogen bond interaction, (c) ion–dipole interaction, and (d) dipole–dipole interaction. 1 inclusion compound of TMeQ[6] and G1, (b) hydrogen bond interaction, (c) ion–dipole interaction, and (d) dipole–dipole interaction. | |
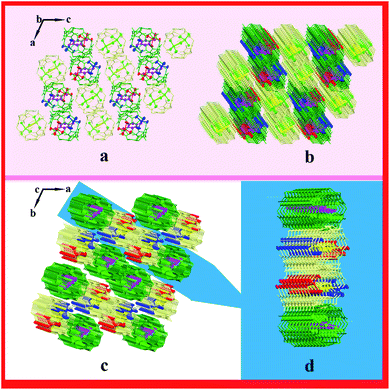 |
| | Fig. 2 Supramolecular framework of complex 1: (a) two-dimensional framework structure viewed along the b-axis, (b and c) three-dimensional supramolecular framework structure viewed along b-axis and c-axis, respectively, and (d) three-dimensional layered framework. | |
3.1.2 Description of the crystal structure of complex 2. Complex 2 exhibits the triclinic P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) space group. The asymmetric unit contains half of TMeQ[6], one counterion [ZnCl4]2−, and two half of protonated 4,4′-diaminostilbenes, as shown in Fig. 3a. Counterions [ZnCl4]2− connects with adjacent TMeQ[6]s by ion–dipole interactions (Fig. 3b). In fact, protonated 4,4′-diaminostilbene also connects with the methylene hydrogen (–CH) and bridging “waist” methylene (–CH2) of TMeQ[6]s by C–H⋯π interactions (Fig. 3c). Further, TMeQ[6] and protonated 4,4-diaminostilbene form a 1
space group. The asymmetric unit contains half of TMeQ[6], one counterion [ZnCl4]2−, and two half of protonated 4,4′-diaminostilbenes, as shown in Fig. 3a. Counterions [ZnCl4]2− connects with adjacent TMeQ[6]s by ion–dipole interactions (Fig. 3b). In fact, protonated 4,4′-diaminostilbene also connects with the methylene hydrogen (–CH) and bridging “waist” methylene (–CH2) of TMeQ[6]s by C–H⋯π interactions (Fig. 3c). Further, TMeQ[6] and protonated 4,4-diaminostilbene form a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 assembly via hydrogen bonds, where N1 of protonated 4,4-diaminostilbene forms two hydrogen bonds with O1 and O2 of TMeQ[6], and N14 of protonated 4,4-diaminostilbene forms a hydrogen bond with Cl2 of counterions [ZnCl4]2−. These hydrogen bonds—i.e., N1–H1A⋯O1, N1–H1B⋯O2, and N14–H14A⋯Cl2—have bond lengths of 2.789, 2.734, and 3.277 Å, respectively (Fig. 3d). Hydrogen bonding, C–H⋯π, and ion–dipole interactions were the driving forces in the construction of the supramolecular assembly. As shown in Fig. 3e, the extension of supramolecular assembly at different angles can result in different shapes. By taking the angle of view along the b-axis as an example and by only considering the hydrogen bond between the portal of TMeQ[6] and 4,4′-diaminostilbene, a one-dimensional supramolecular chain is formed (Fig. 4a). Interestingly, when multiple forces coexist, 4,4′-diaminostilbene forms a “walled” supramolecular framework enclosing TMeQ[6]s, eventually forming the two-dimensional supramolecular framework (Fig. 4b) and the three-dimensional framework (Fig. 4c).
1 assembly via hydrogen bonds, where N1 of protonated 4,4-diaminostilbene forms two hydrogen bonds with O1 and O2 of TMeQ[6], and N14 of protonated 4,4-diaminostilbene forms a hydrogen bond with Cl2 of counterions [ZnCl4]2−. These hydrogen bonds—i.e., N1–H1A⋯O1, N1–H1B⋯O2, and N14–H14A⋯Cl2—have bond lengths of 2.789, 2.734, and 3.277 Å, respectively (Fig. 3d). Hydrogen bonding, C–H⋯π, and ion–dipole interactions were the driving forces in the construction of the supramolecular assembly. As shown in Fig. 3e, the extension of supramolecular assembly at different angles can result in different shapes. By taking the angle of view along the b-axis as an example and by only considering the hydrogen bond between the portal of TMeQ[6] and 4,4′-diaminostilbene, a one-dimensional supramolecular chain is formed (Fig. 4a). Interestingly, when multiple forces coexist, 4,4′-diaminostilbene forms a “walled” supramolecular framework enclosing TMeQ[6]s, eventually forming the two-dimensional supramolecular framework (Fig. 4b) and the three-dimensional framework (Fig. 4c).
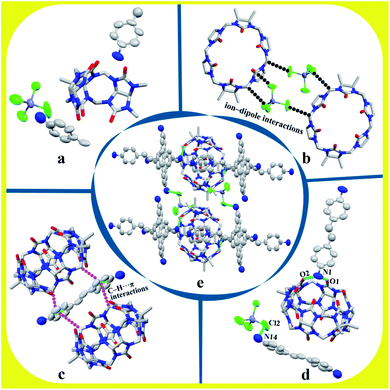 |
| | Fig. 3 Structure of complex 2: (a) asymmetric unit, (b) ion–dipole interaction, (c) C–H⋯π interaction, (d) hydrogen bond interaction, and (e) assembly of unit cell. | |
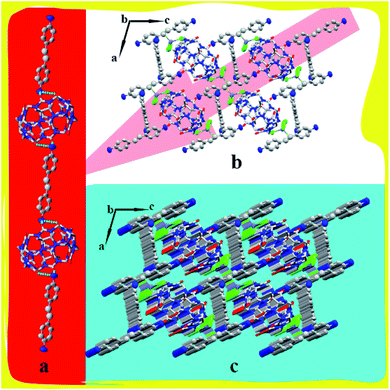 |
| | Fig. 4 Supramolecular framework of complex 1: (a) one-dimensional framework chain, (b) two-dimensional supramolecular framework structure viewed along b-axis, and (c) three-dimensional framework along b-axis. | |
3.2 1H NMR, ITC, UV-Vis spectrum, and MALDI-TOF mass spectrometric analysis of cucurbiturils and guests
3.2.1 1H NMR spectra of TMeQ[6] and guests. The cavity of TMeQ[6] has a shielding effect on proton signals, whereas outside of the portals, in the vicinity of the carbonyl oxygen atoms, the proton signals are subjected to a deshielding effect. Therefore, nuclear magnetic resonance (NMR) is one of the most commonly used techniques to investigate host–guest interactions. Fig. 5 and 6 show the 1H NMR results of interactions of G1 and G2 with TMeQ[6] in D2O, respectively.
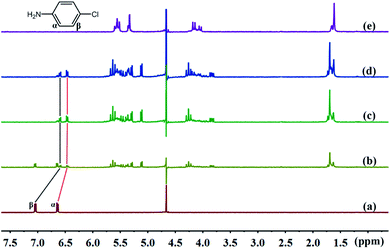 |
| | Fig. 5 Titration 1H NMR spectra of G1 (0.1 mM) with (a) 0.00, (b) 0.51, (c) 1.08, and (d) 1.36 equiv. of TMeQ[6], and of (e) neat TMeQ[6]. | |
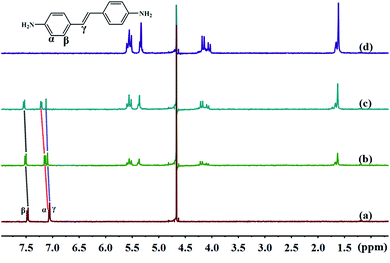 |
| | Fig. 6 Titration 1H NMR spectra of G2 (0.1 mM) with (a) 0.00, (b) 0.71, and (c) 1.39 equiv. of TMeQ[6], and of (d) neat TMeQ[6]. | |
Fig. 5a shows the 1H NMR spectrum of G1; Fig. 5b, c, and d show the 1H NMR spectra of TMeQ[6]–G1 with an equivalent ratio of 0.51, 1.08, and 1.36, respectively; and Fig. 5e shows the 1H NMR spectrum of TMeQ[6]. When TMeQ[6] was added to the system, the chemical shift of Hα and Hβ protons of G1 moved upfield by about δ 0.175 and 0.449 ppm, respectively. This indicates that G1 penetrated into the cavity of TMeQ[6] and was shielded by TMeQ[6]. Moreover, when TMeQ[6] was greater than 1 equiv., the chemical shift of protons Hα and Hβ remained unchanged, indicating that TMeQ[6] and G1 formed a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 inclusion compound, and the existence of the amino group and Cl atom in G1 resulted in the splitting of proton peaks of TMeQ[6].
1 inclusion compound, and the existence of the amino group and Cl atom in G1 resulted in the splitting of proton peaks of TMeQ[6].
Further, Fig. 6a shows the 1H NMR spectrum of G2; Fig. 6b and c show the 1H NMR spectra of TMeQ[6]–G2 with an equivalent ratio of 0.71 and 1.39, respectively; and Fig. 6d shows the 1H NMR spectrum of TMeQ[6]. When TMeQ[6] was added to the system, the chemical shifts of the protons Hα, Hβ, and Hγ of the guest G2 moved downfield, indicating that G2 was at the portal of TMeQ[6] and was subject to the deshielding effect of TMeQ[6]. The proton peaks of TMeQ[6] did not split, indicating that the protons of TMeQ[6] were in a symmetrical environment. By comparing Fig. 6b and c, when the amount of TMeQ[6] increased, the protons Hα, Hβ and Hγ of the guest G2 moved up to the downfield, indicating that TMeQ[6] and G2 was an unstable reversible dynamic interaction.42
3.2.3 UV-Vis spectrum analysis. The binding behaviors of TMeQ[6]@G1 and TMeQ[6]@G2 were studied by employing UV-Vis absorption in deionized water (Fig. 8). The wavelengths of the one strong absorption peak for G1 was observed at 238 nm, and TMeQ[6] exhibited no absorption at these wavelengths. The absorbance of G1 shows a gradually downward trend upon increasing the ratio of n(TMeQ[6])/n(G1). When the ratio of n(TMeQ[6])/n(G1) reaches 1, the absorbance becomes stable as the ratio increases. Therefore, the absorbance (A) vs. the molar ratio of the TMeQ[6] and G1 {n(TMeQ[6])/n(G1)} data can be fitted to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding (Fig. 8a). In addition, the wavelengths of the one strong absorption peak for G2 was observed at 306 nm, and TMeQ[6] exhibited no absorption at these wavelengths. The absorbance of G2 shows a gradually downward trend upon increasing the ratio of n(TMeQ[6])/n(G2). According to the tangent intersection, the absorbance (A) vs. the molar ratio of the TMeQ[6] and G2 {n(TMeQ[6])/n(G2)} data can be fitted to a 1
1 binding (Fig. 8a). In addition, the wavelengths of the one strong absorption peak for G2 was observed at 306 nm, and TMeQ[6] exhibited no absorption at these wavelengths. The absorbance of G2 shows a gradually downward trend upon increasing the ratio of n(TMeQ[6])/n(G2). According to the tangent intersection, the absorbance (A) vs. the molar ratio of the TMeQ[6] and G2 {n(TMeQ[6])/n(G2)} data can be fitted to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding (Fig. 8b).
1 binding (Fig. 8b).
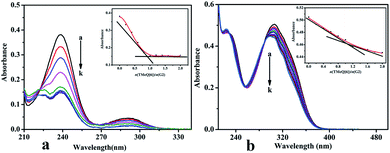 |
| | Fig. 8 UV-Vis spectrum of (a) TMeQ[6]@G1, and (b) TMeQ[6]@G2. | |
3.2.4 Mass spectrometry. In order to further investigate the host–guest interaction modes, as shown in Fig. 9, matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry was used to characterize the complex structures. The molecular ion peaks are visible at m/z 1180.3840 and 1263.4626, and these could correspond to [TMeQ[6] + G1 + H]+ and [TMeQ[6] + G2 + H]+, respectively (calculated m/z values were 1180.3832 and 1263.4811, respectively). Therefore, TMeQ[6] and guests interacted at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, which is consistent with the above analysis results.
1, which is consistent with the above analysis results.
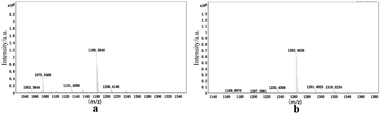 |
| | Fig. 9 Mass spectrometry of (a) TMeQ[6]@G1, and (b) TMeQ[6]@G2. | |
4. Conclusions
In this study, the interaction modes of TMeQ[6] with G1 and G2 were investigated by performing single-crystal X-ray diffraction, NMR, ITC, UV-Vis spectrum, and MALDI-TOF mass spectrometry analyses. The experimental results showed that TMeQ[6] and G1 formed a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 inclusion complex, and the carbonyl portal of TMeQ[6] formed a 1
1 inclusion complex, and the carbonyl portal of TMeQ[6] formed a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 self-assembly with G2. The single crystal structures of complexes 1 and 2 exhibited the ion–dipole interaction, dipole–dipole interaction, C–H⋯π interaction, and hydrogen bonds. These weak interaction forces caused complexes 1 and 2 to form ordered multi-dimensional supramolecular frameworks. They are expected to have a wide range of applications in nanotechnology, molecular sieves, sensors, gas adsorption and separation, ion or molecular transportal, and heterogeneous catalysis.
1 self-assembly with G2. The single crystal structures of complexes 1 and 2 exhibited the ion–dipole interaction, dipole–dipole interaction, C–H⋯π interaction, and hydrogen bonds. These weak interaction forces caused complexes 1 and 2 to form ordered multi-dimensional supramolecular frameworks. They are expected to have a wide range of applications in nanotechnology, molecular sieves, sensors, gas adsorption and separation, ion or molecular transportal, and heterogeneous catalysis.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
This work was financially supportaled by the National Natural Science Foundation of China (Grant No. 21762011) and Guizhou Science and Technology Planning Project (Guizhou Science and Technology Cooperation Platform Talent [2017]5788).
Notes and references
- J. M. Thiebaut and C. Akyel, IEEE Trans. Instrum. Meas., 1988, 37(1), 114–120 CrossRef CAS.
- L. Zhao, R. Y. Ma and X. L. Meng, Adv. Mater. Res., 2012, 450, 1419–1424 Search PubMed.
- X. Zhao, D. Liang and S. Liu, Inorg. Chem., 2008, 47(16), 7133–7138 CrossRef CAS.
- Y. Cu, Y. Yue and G. Qian, Chem. Rev., 2012, 112(2), 1126–1162 CrossRef.
- J. R. Li, R. Kuppler and H. C. Zhou, J. Cheminf., 2009, 38(5), 1477–1504 CAS.
- H. Furukawa and O. M. Yagh, J. Am. Chem. Soc., 2009, 131(25), 8875–8883 CrossRef CAS.
- F. Beuerle and B. Gole, Angew. Chem., Int. Ed., 2018, 57, 4850–4878 CrossRef CAS.
- C. Shuai, Z. Mo and X. Niu, J. Alloys Compd., 2020, 847, 156514 CrossRef CAS.
- Y. Su, Z. Li and H. Zhou, Chem. Eng. J., 2020, 402, 126205 CrossRef CAS.
- C. H. Lin, Z. F. Zao and Z. W. Lei, Spectrochim. Acta, Part A, 2020, 242, 118739 CrossRef CAS.
- W. Yang, A. Greenaway, X. Lin, R. Matsuda, A. J. Blake, C. Wilson, W. Lewis, P. Hubberstey, S. Kitagawa and M. Schröder, J. Am. Chem. Soc., 2010, 132, 14457–14469 CrossRef CAS.
- J. Tian, H. Wang, D. W. Zhang, Y. Liu and Z. T. Li, Natl. Sci. Rev., 2017, 4, 426–436 CrossRef CAS.
- W. A. Freeman, W. L. Mock and N. Y. Shih, J. Am. Chem. Soc., 1981, 103(24), 7367–7368 CrossRef CAS.
- K. Kim, N. Selvapalam and D. H. Oh, J. Inclusion Phenom. Macrocyclic Chem., 2004, 50(1–2), 31–36 CAS.
- A. Day, A. P. Arnold, R. J. Blanch and B. Sunshall, J. Org. Chem., 2001, 66(24), 8094–8100 CrossRef CAS.
- A. I. Day, R. J. Blanch, A. P. Arnold and G. R. Lewis, Angew. Chem., 2002, 41, 275–277 CrossRef CAS.
- S. Liu, P. Y. Zavalij and L. Isaacs, J. Am. Chem. Soc., 2005, 127(48), 16798–16799 CrossRef CAS.
- X. J. Cheng, L. L. Liang, K. Chen, N. N. Ji, X. Xiao, J. X. Zhang, Y. Q. Zhang, S. F. Xue, Q. J. Zhu, X. L. Ni and Z. Tao, Angew. Chem., Int. Ed., 2013, 52(28), 7252–7255 CrossRef CAS.
- A. Flinn, G. C. Hough, J. F. Stoddart and D. J. Williams, Angew. Chem., Int. Ed., 1992, 31(11), 1475–1477 CrossRef.
- J. H. Zhao, H. J. Kim, J. H. Oh, S. Y. Kim, J. W. Lee, S. Sakamoto, K. Yamaguchi and K. Kim, Angew. Chem., 2001, 113(22), 4363–4365 CrossRef.
- F. Wu, L. H. Wu, X. Xiao, Y. Q. Zhang, S. F. Xue, Z. Tao and A. I. Day, J. Org. Chem., 2012, 77, 606–611 CrossRef CAS.
- L. H. Wu, X. L. Ni, F. Wu, Y. Q. Zhang, Q. J. Zhu, S. F. Xue and Z. Tao, J. Mol. Struct., 2009, 920(1–3), 183–188 CrossRef CAS.
- S. Y. Jon, N. Selvapalam, D. H. Oh, J. K. Kang, S. Y. Kim, Y. J. Jeon, J. W. Lee and K. Kim, J. Am. Chem. Soc., 2003, 125(34), 10186–10187 CrossRef CAS.
- H. Isobe, S. Sato and E. Nakamura, Org. Lett., 2002, 4(8), 1287–1289 CrossRef CAS.
- X. L. Ni, X. Xiao, H. Cong, Q. J. Zhu, S. F. Xue and Z. Tao, Acc. Chem. Res., 2014, 47, 1386–1395 CrossRef CAS.
- S. Sinn, A. Prabodh and L. Grimm, Chem. Commun., 2020, 82 10.1039/d0cc03715j.
- A. Sowa and J. Voskuhl, Int. J. Pharm., 2020, 586, 119595 CrossRef CAS.
- Q. Ai, L. Jin L and Z. Gong, Chem. Soc. Rev., 2020, 18, 10581–10585 Search PubMed.
- J. Murray, K. Kim, T. Ogoshi, W. Yao and B. C. Gibb, Chem. Soc. Rev., 2017, 46, 2479–2496 RSC.
- J. Mosquera, Y. Zhao, H. Jang, N. Xie, C. Xu, N. A. Kotov and L. M. Liz-Marzán, Adv. Funct. Mater., 2019, 190208 Search PubMed.
- X. L. Ni, X. Xiao, H. Cong, L. L. Liang, K. Chen, X. J. Cheng, N. N. Ji, Q. J. Zhu, S. F. Xue and Z. Tao, Chem. Soc. Rev., 2013, 42, 9480–9508 RSC.
- J. Lü, J. X. Lin, M. N. Cao and R. Cao, Coord. Chem. Rev., 2013, 257, 1334–1356 CrossRef.
- X. L. Ni, S. F. Xue, Z. Tao, Q. J. Zhu, L. F. Lindoy and G. Wei, Coord. Chem. Rev., 2015, 287, 89–113 CrossRef CAS.
- W. W. Zhao, L. T. Wie, K. Z. Zhou and P. H. Ma, ChemistrySelect, 2019, 4(40), 11674–11677 CrossRef CAS.
- Y. X. Qu, K. Z. Zhou, K. Chen, Y. Q. Zhang, X. Xiao, Q. D. Zhou, Z. Tao, P. H. Ma and G. Wei, Inorg. Chem., 2018, 57, 7412–7419 CrossRef CAS.
- R. X. Cheng, F. Y. Tian and Y. Q. Zhang, J. Mater. Sci., 2020, 55, 16497–16509 CrossRef CAS.
- L. Mei, F. Z. Li, J. H. Lan, C. Z. Wang, C. Xu, H. Deng, Q. Y. Wu, K. Q. Hu, L. Wang, Z. F. Chai, J. Chen, J. K. Gibson and W. Q. Shi, Nat. Commun., 2019, 10, 1532 CrossRef.
- M. H. Abraham and J. A. Platts, J. Org. Chem., 2001, 66(10), 3484–3491 CrossRef CAS.
- X. J. Cheng, L. L. Liang, K. Chen, N. N. Ji, X. Xiao, J. X. Zhang, Y. Q. Zhang, S. F. Xue, Q. J. Zhu, X. L. Ni and Z. Tao, Angew. Chem., Int. Ed., 2013, 52(28), 7252–7255 CrossRef CAS.
- Y. J. Zhao, S. F. Xue, Q. J. Zhu, Z. Tao, J. X. Zhang, Z. B. Wei, L. S. Long, M. L. Hu, H. P. Xiao and A. I. Day, Chin. Sci. Bull., 2004, 49, 1111 CrossRef CAS.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Crystallogr., 2008, 64, 112–122 CrossRef CAS.
- X. D. Zhang, T. Sun and X. L. Ni, Org. Chem. Front., 2021, 8, 32–38 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2039065 (1) and 2039066 (2). For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d0ra09074c |
|
| This journal is © The Royal Society of Chemistry 2021 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *
*
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 inclusion compound with G1, and the carbonyl portal of TMeQ[6] formed a 1
1 inclusion compound with G1, and the carbonyl portal of TMeQ[6] formed a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 self-assembly with G2. Further, multi-dimensional supramolecular frameworks were formed driven by weak interaction forces in the system (hydrogen bonding, C–H⋯π interactions, ion–dipole interactions, and dipole–dipole interactions).
1 self-assembly with G2. Further, multi-dimensional supramolecular frameworks were formed driven by weak interaction forces in the system (hydrogen bonding, C–H⋯π interactions, ion–dipole interactions, and dipole–dipole interactions).![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif)
![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif)
![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) space group. As shown in Fig. 1a, TMeQ[6] and protonated 4-chloroaniline formed a 1
space group. As shown in Fig. 1a, TMeQ[6] and protonated 4-chloroaniline formed a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 inclusion compound. In this system, hydrogen bonds are one of the main forces. N49 forms three hydrogen bonds with O2, O4, and O6, and N50 forms three hydrogen bonds with O16, O18, and O20; the N–H⋯O hydrogen bond length is 2.760–2.834 Å. In addition, hydrogen bonds form between O1 and O2 and between O15 and O16 (O2–H2A⋯O1 and O16–H16A⋯O15), with the bond lengths being 2.859 and 2.855 Å, respectively (Fig. 1b). One counterion [ZnCl4]2− connects with three adjacent outer surfaces of TMeQ[6]s via ion–dipole interactions (Fig. 1c). A negatively potential TMeQ[6] portal is connected to another positively potential outer surface of TMeQ[6] by the dipole–dipole interaction (Fig. 1d). In summary, these weak interactions (hydrogen bonding, ion–dipole interaction, and dipole–dipole interaction) caused complex 1 to form an ordered multi-dimensional layered supramolecular framework (Fig. 2a–d).
1 inclusion compound. In this system, hydrogen bonds are one of the main forces. N49 forms three hydrogen bonds with O2, O4, and O6, and N50 forms three hydrogen bonds with O16, O18, and O20; the N–H⋯O hydrogen bond length is 2.760–2.834 Å. In addition, hydrogen bonds form between O1 and O2 and between O15 and O16 (O2–H2A⋯O1 and O16–H16A⋯O15), with the bond lengths being 2.859 and 2.855 Å, respectively (Fig. 1b). One counterion [ZnCl4]2− connects with three adjacent outer surfaces of TMeQ[6]s via ion–dipole interactions (Fig. 1c). A negatively potential TMeQ[6] portal is connected to another positively potential outer surface of TMeQ[6] by the dipole–dipole interaction (Fig. 1d). In summary, these weak interactions (hydrogen bonding, ion–dipole interaction, and dipole–dipole interaction) caused complex 1 to form an ordered multi-dimensional layered supramolecular framework (Fig. 2a–d).

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 inclusion compound of TMeQ[6] and G1, (b) hydrogen bond interaction, (c) ion–dipole interaction, and (d) dipole–dipole interaction.
1 inclusion compound of TMeQ[6] and G1, (b) hydrogen bond interaction, (c) ion–dipole interaction, and (d) dipole–dipole interaction.![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) space group. The asymmetric unit contains half of TMeQ[6], one counterion [ZnCl4]2−, and two half of protonated 4,4′-diaminostilbenes, as shown in Fig. 3a. Counterions [ZnCl4]2− connects with adjacent TMeQ[6]s by ion–dipole interactions (Fig. 3b). In fact, protonated 4,4′-diaminostilbene also connects with the methylene hydrogen (–CH) and bridging “waist” methylene (–CH2) of TMeQ[6]s by C–H⋯π interactions (Fig. 3c). Further, TMeQ[6] and protonated 4,4-diaminostilbene form a 1
space group. The asymmetric unit contains half of TMeQ[6], one counterion [ZnCl4]2−, and two half of protonated 4,4′-diaminostilbenes, as shown in Fig. 3a. Counterions [ZnCl4]2− connects with adjacent TMeQ[6]s by ion–dipole interactions (Fig. 3b). In fact, protonated 4,4′-diaminostilbene also connects with the methylene hydrogen (–CH) and bridging “waist” methylene (–CH2) of TMeQ[6]s by C–H⋯π interactions (Fig. 3c). Further, TMeQ[6] and protonated 4,4-diaminostilbene form a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 assembly via hydrogen bonds, where N1 of protonated 4,4-diaminostilbene forms two hydrogen bonds with O1 and O2 of TMeQ[6], and N14 of protonated 4,4-diaminostilbene forms a hydrogen bond with Cl2 of counterions [ZnCl4]2−. These hydrogen bonds—i.e., N1–H1A⋯O1, N1–H1B⋯O2, and N14–H14A⋯Cl2—have bond lengths of 2.789, 2.734, and 3.277 Å, respectively (Fig. 3d). Hydrogen bonding, C–H⋯π, and ion–dipole interactions were the driving forces in the construction of the supramolecular assembly. As shown in Fig. 3e, the extension of supramolecular assembly at different angles can result in different shapes. By taking the angle of view along the b-axis as an example and by only considering the hydrogen bond between the portal of TMeQ[6] and 4,4′-diaminostilbene, a one-dimensional supramolecular chain is formed (Fig. 4a). Interestingly, when multiple forces coexist, 4,4′-diaminostilbene forms a “walled” supramolecular framework enclosing TMeQ[6]s, eventually forming the two-dimensional supramolecular framework (Fig. 4b) and the three-dimensional framework (Fig. 4c).
1 assembly via hydrogen bonds, where N1 of protonated 4,4-diaminostilbene forms two hydrogen bonds with O1 and O2 of TMeQ[6], and N14 of protonated 4,4-diaminostilbene forms a hydrogen bond with Cl2 of counterions [ZnCl4]2−. These hydrogen bonds—i.e., N1–H1A⋯O1, N1–H1B⋯O2, and N14–H14A⋯Cl2—have bond lengths of 2.789, 2.734, and 3.277 Å, respectively (Fig. 3d). Hydrogen bonding, C–H⋯π, and ion–dipole interactions were the driving forces in the construction of the supramolecular assembly. As shown in Fig. 3e, the extension of supramolecular assembly at different angles can result in different shapes. By taking the angle of view along the b-axis as an example and by only considering the hydrogen bond between the portal of TMeQ[6] and 4,4′-diaminostilbene, a one-dimensional supramolecular chain is formed (Fig. 4a). Interestingly, when multiple forces coexist, 4,4′-diaminostilbene forms a “walled” supramolecular framework enclosing TMeQ[6]s, eventually forming the two-dimensional supramolecular framework (Fig. 4b) and the three-dimensional framework (Fig. 4c).



![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 inclusion compound, and the existence of the amino group and Cl atom in G1 resulted in the splitting of proton peaks of TMeQ[6].
1 inclusion compound, and the existence of the amino group and Cl atom in G1 resulted in the splitting of proton peaks of TMeQ[6].![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln
ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ka = ΔH − TΔS), it can be seen that the two systems are enthalpy-driven, and the driving force is determined by the hydrogen bond interaction and the hydrophobic effect. By comparing the binding constants of G1·TMeQ[6] and G2·TMeQ[6], the stability of G1 entering the cavity of TMeQ[6] is greater than that of G2 at the portals of TMeQ[6]. G2·TMeQ[6] has a weak binding affinity, resulting in a reversible dynamic interaction of G2·TMeQ[6] (Table 2).
Ka = ΔH − TΔS), it can be seen that the two systems are enthalpy-driven, and the driving force is determined by the hydrogen bond interaction and the hydrophobic effect. By comparing the binding constants of G1·TMeQ[6] and G2·TMeQ[6], the stability of G1 entering the cavity of TMeQ[6] is greater than that of G2 at the portals of TMeQ[6]. G2·TMeQ[6] has a weak binding affinity, resulting in a reversible dynamic interaction of G2·TMeQ[6] (Table 2).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding (Fig. 8a). In addition, the wavelengths of the one strong absorption peak for G2 was observed at 306 nm, and TMeQ[6] exhibited no absorption at these wavelengths. The absorbance of G2 shows a gradually downward trend upon increasing the ratio of n(TMeQ[6])/n(G2). According to the tangent intersection, the absorbance (A) vs. the molar ratio of the TMeQ[6] and G2 {n(TMeQ[6])/n(G2)} data can be fitted to a 1
1 binding (Fig. 8a). In addition, the wavelengths of the one strong absorption peak for G2 was observed at 306 nm, and TMeQ[6] exhibited no absorption at these wavelengths. The absorbance of G2 shows a gradually downward trend upon increasing the ratio of n(TMeQ[6])/n(G2). According to the tangent intersection, the absorbance (A) vs. the molar ratio of the TMeQ[6] and G2 {n(TMeQ[6])/n(G2)} data can be fitted to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding (Fig. 8b).
1 binding (Fig. 8b).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, which is consistent with the above analysis results.
1, which is consistent with the above analysis results.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 inclusion complex, and the carbonyl portal of TMeQ[6] formed a 1
1 inclusion complex, and the carbonyl portal of TMeQ[6] formed a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 self-assembly with G2. The single crystal structures of complexes 1 and 2 exhibited the ion–dipole interaction, dipole–dipole interaction, C–H⋯π interaction, and hydrogen bonds. These weak interaction forces caused complexes 1 and 2 to form ordered multi-dimensional supramolecular frameworks. They are expected to have a wide range of applications in nanotechnology, molecular sieves, sensors, gas adsorption and separation, ion or molecular transportal, and heterogeneous catalysis.
1 self-assembly with G2. The single crystal structures of complexes 1 and 2 exhibited the ion–dipole interaction, dipole–dipole interaction, C–H⋯π interaction, and hydrogen bonds. These weak interaction forces caused complexes 1 and 2 to form ordered multi-dimensional supramolecular frameworks. They are expected to have a wide range of applications in nanotechnology, molecular sieves, sensors, gas adsorption and separation, ion or molecular transportal, and heterogeneous catalysis.