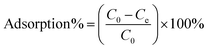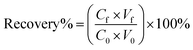Open Access Article
Open Access ArticleA one step method for isolation of genomic DNA using multi-amino modified magnetic nanoparticles†
Jia Xu‡
 a,
Dan Chen‡b,
Yuan Yang‡a,
Hongjian Gonga,
Wenqi Gaoa and
Han Xiao*a
a,
Dan Chen‡b,
Yuan Yang‡a,
Hongjian Gonga,
Wenqi Gaoa and
Han Xiao*a
aInstitute of Maternal and Child Health, Wuhan Children's Hospital (Wuhan Maternal and Child Healthcare Hospital), Tongji Medical College, Huazhong University of Science & Technology, Wuhan, 430016, China. E-mail: tjxiaohan1980@163.com
bWuhan Institute for Food and Cosmetic Control, Wuhan, 430012, China
First published on 15th January 2021
Abstract
A simple and efficient approach for the rapid extraction of genomic DNA from blood using various amino-modified magnetic nanoparticles (AMNPs) has been described. The salmon sperm DNA was isolated from aqueous solution based on electrostatic interaction between the positively charged amino-groups of AMNPs and the negatively charged phosphate groups of the DNA. The results of ultraviolet-visible (UV-Vis) spectrometry showed that increasing number of amino groups on the AMNPs surface resulted in an improvement in DNA adsorption efficiency. Several variables including the extraction pH, adsorption time, ionic strength and quantity of AMNPs were optimized to achieve the best extraction efficiency with the proposed method. Acceptable adsorption efficiency of 92% and recovery of 91% were achieved using multi-amino modified MNPs (mAMNPs) with an extraction time of 10 min and an overall processing time of 30 min. The mAMNPs enabled genomic DNA capture from human whole blood, and the resulting mAMNP/DNA complexes could be directly used as templates for PCR amplification without the need for complex and time-consuming DNA elution and purification steps. Our results imply that this method can be used as an effective strategy for genomic DNA extraction and may be extended to other types of biological samples.
Introduction
Deoxyribonucleic acid (DNA) is a type of nucleic acid that carries genetic information necessary for the synthesis of proteins in biological cells and is essential for the development and normal operation of organisms. DNA is highly attractive as an important biomarker, tool and genetic fingerprint in the life sciences.1–3 DNA analysis is crucial for a wide array of applications related to clinical diagnosis, genomics and food safety because of its inherent specificity and technical capabilities, enabling sensitive and rapid detection.4–6 Unfortunately, DNA is present in relatively low amounts (relative to other components) in complex biological samples, which contain proteins, polysaccharides, phospholipids and metabolites that can significantly interfere with DNA detection. Additionally, damage to the primary structure of DNA during purification often leads to decreased sensitivity, poor repeatability, or complete loss of utility in downstream applications, including polymerase chain reaction (PCR) amplification and sequencing.7 Therefore, it is necessary to obtain high quality of DNA templates from such complex matrices.Considering the complexity of biological samples and the low concentration of DNA in biological matrixes, a suitable DNA purification method is essential. Various methods have been applied for DNA extraction, such as phenol/chloroform extraction, isopropanol precipitation, and formamide cleavage. These conventional methods are usually followed by precipitation, centrifugation and filtration steps, which are time-consuming and labor-intensive. Recently, efforts to develop new DNA retrieval methods, such as liquid–liquid extraction,8 liquid–liquid microextraction,9,10 solid phase extraction,11 and magnetic solid-phase microextraction (MSPE), have increased.7,12–14 Magnetic nanomaterial-based extraction is more widely accepted than other DNA retrieval methods in practical applications, because it requires less organic solvent consumption, has a lower cost, involves simple procedures, and has a rapid processing time.15,16 The magnetic nanoparticles (MNPs) used in MSPE can be rapidly collected using an external magnetic field, thereby avoiding centrifugation steps, which generate shear forces that may lead to nucleic acid degradation. Moreover, the surface functionality of MNPs can be easily modified to improve the selectivity of the sorption procedure. The DNA backbone contains negative charged phosphate groups. Therefore, by modifying the MNPs surface with positively charged groups, DNA can be effectively adsorbed through electrostatic interaction and protected from enzymatic cleavage.17
Materials used to modify the surface of MNPs include amino-groups,6,18–21 dimercaptosuccinic acid,22 poly(acrylic acid),23 chitosan with polyaniline hybridization,24 polydopamine25 and polyethylenimine.26 These modifiers are generally silane-based reagents or polymers with amino, hydroxyl or carboxyl groups, whereas hybrid magnetic particles are coated with inorganic materials (e.g., silica and gold) to achieve high loading efficiency. Among these distinct types of chemically modified MNPs, amino-functionalized magnetic beads are most commonly used for DNA capture, due to their powerful adsorption capability, pH-responsive characteristics and simple adsorption/desorption requirements.11 The adsorption/desorption between hydroxyl- and carboxyl-modified MNPs with DNA occurs in the presence of high polyethylene and salt concentrations, and DNA can bind to or be released from the surface of amino-functionalized MNPs through pH-induced charge switching of the amino groups on MNPs.6 Additionally, the number of amino groups present on the silane compounds affects the adsorption capacity of MNPs, and increasing the number of amino groups can improve the adsorption efficiency.21
Herein, we compared the efficiencies of several amino-modified silica-coated magnetic nanoparticles (AMNPs) in the recovery of DNA from human whole blood samples. Silica can be used as an inorganic support to modify and protect the surface of iron oxide (Fe3O4) for promotion of the biocompatibility, water dispersibility and chemical stability of MNPs under acidic conditions.15 Separation was achieved using magnetite coated with modifiers that enable silanization with increasing number of amino groups present on the silane compounds. DNA adsorption experiments were performed with these particles to evaluate their adsorption capability, and the maximum adsorption capacity was measured using an UV-Vis spectrophotometer. The aim of this study was to engineer multi-amino-modified magnetic nanoparticles (mAMNPs) for the purpose of retrieving DNA from human whole blood samples with sufficiently high purity for direct PCR amplification without the need for additional processing steps, such as target DNA elution or purification. The structures and properties of the tested MNPs were investigated by transmission electron microscopy (TEM), Fourier transform infrared spectroscopy (FT-IR) and vibrating sample magnetometry (VSM), and the amino groups on their surfaces were quantified using the p-nitrobenzaldehyde measurement method.
Materials and methods
Reagents
3-(2-Aminoethylamino)propyltriethoxysilane (EPTES, 96%, w/v), tris(hydroxymethyl)aminomethane (Tris–base), N-[3-(trimethoxysilyl)propyl]ethylenediamine (EDPS, 95%, w/v) and iron oxide (Fe3O4) magnetic nanoparticles (20 nm) were purchased from Aladdin Chemistry Co., Ltd. (Shanghai, China). N,N-Dimethylformamide (DMF), (3-aminopropyl)triethoxysilane (APTES, 99%, w/v), 3-[2-(2-aminoethylamino)ethylamino]propyl-trimethoxysilane (AEEA, 95%, w/v) and salmon sperm DNA sodium salt were purchased from Macklin (Shanghai, China). Tetraethyl orthosilicate (TEOS), sodium chloride (NaCl), methanol, ethanol, hydrochloric acid (HCl), ammonia solution (25–28%, w/v), ethylenediaminetetraacetic acid disodium salt dehydrate (EDTA-Na2·2H2O), glacial acetic acid, and ammonium acetate were purchased from Sinopharm Chemical Reagent, Co., Ltd. (Shanghai, China). Tris–hydrochloride (Tris–HCl) was purchased from Guangzhou Saiguo Biotech Co., Ltd (Guangzhou, China). Deionized water (18.25 MΩ) was used in all experiments. All other chemicals were of analytical grade and were used without further treatment. Primers for the amplification of human whole blood DNA were synthesized by Shanghai Bio-tech Corporation (Shanghai, China).Human whole blood samples from healthy volunteers were provided by the Wuhan Children's Hospital (China). Appropriate amounts of anticoagulant were added to the blood samples which were then stored at 20 °C until future use.
Instruments
The size and morphology of nanoparticles were observed by TEM (JEM-2100, JEOL, Japan). Surface modification of obtained magnetic nanoparticles was investigated using a Nicolet 470FT-IR instrument (FT-IR) (Thermo Fisher Scientific, USA) in the frequency range of 500 cm−1 to 4000 cm−1 with a resolution of 2.0 cm−1, at room temperature. The magnetic properties of obtained magnetic nanoparticles were characterized by using VSM instrument (Physical Properties Measurement System, Quantum Design, USA) at room temperature. Agitation and extraction were accomplished with the UXI orbital shaker (Huxi, Shanghai, China). The concentration of DNA solution was determined by a UV-1600PC UV-Vis spectrophotometer (XIPU, Shanghai, China) with a 1.0 cm quartz cell. DNA amplification was performed in the T100™ thermal cycler (Bio-Rad Laboratories, USA). The PCR products were analyzed using 2% agarose gel electrophoresis with ethidium bromide staining following by visualization on a ChemiDoc XRS+ Imaging System (Bio-Rad Laboratories, USA). Real-time DNA amplification reaction was carried out in a 7500 real-time PCR system (Applied Biosystems, USA).Preparation of silica-coated MNPs
Silica-coated MNPs (SMNPs) were prepared using the sol–gel approach, with some modifications.27 Briefly, 1 g of Fe3O4 nanoparticles were suspended in a mixture of 100 mL deionized water and 300 mL ethanol by ultrasonication for 10 min. The mixture was transferred to a reactor, and the temperature was raised to 80 °C. Then, 10 mL of aqueous ammonia was added to the mixture, followed by the dropwise addition of 5 g TEOS (dissolved in 50 mL ethanol). The reaction proceeded for 10 h with vigorous stirring. After cooling to room temperature, the brown precipitate was collected by magnetic separation and washed three times with an ethanol/water solution (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). Subsequently, the products were dried under a vacuum at 60 °C for 12 h.
1, v/v). Subsequently, the products were dried under a vacuum at 60 °C for 12 h.
Preparation of single amino group modified MNPs (sAMNPs)
The sAMNPs were prepared according to a previously reported method, with some modifications.20 Briefly, 0.5 g of SMNPs was dispersed for 30 min in a mixture of 475 mL ethanol and 25 mL deionized water, with the aid of ultrasonication. Then, the mixture was poured into a two-neck round-bottom flask, and 10 mL APTES was added dropwise, with mechanical agitation. The mixture was constantly stirred for 12 h at room temperature. The obtained sAMNPs were isolated using a permanent magnet and carefully washed three times with a mixture of ethanol and deionized water, then dried under a vacuum at 60 °C for 12 h.Preparation of diamino group modified MNPs (dAMNPs)
Two kinds of dAMNPs (dAMNPs1 and dAMNPs2) were synthesized based on the previously reported methods, with little modification.19,28 The dAMNPs1 was synthesized by first dispersing 0.5 g dried SMNPs in 90 mL DMF for 30 min, with ultrasonication. Then, 0.9 mL of ammonium hydroxide was quickly injected into the mixture, after which 12.5 mL EPTES was added dropwise. The reaction continued for 12 h at room temperature with vigorous mechanical stirring. Finally, the nanoparticles were magnetically recovered, washed sequentially with DMF and ethanol, and then dried under a vacuum at 60 °C for 12 h.The dAMNPs2 was synthesized by first dispersing 0.5 g dried SMNPs in 90 mL DMF for 30 min. Subsequently, 0.9 mL aqueous ammonia and 10 mL EDPS were added with mechanical agitation. The mixture was stirred at room temperature for 12 h. The product was washed sequentially three times with DMF and ethanol, and dried under a vacuum at 60 °C for 12 h.
Preparation of multi-amino group modified MNPs (mAMNPs)
The synthesis of mAMNPs was similar to that of dAMNPs except that the silane regent was replaced by AEEA. Briefly, 0.5 g of SMNPs was dispersed in 90 mL DMF with ultrasonication for 30 min. Subsequently, 0.9 mL ammonium hydroxide and 12.5 mL AEEA were added sequentially with mechanical agitation. After allowing the reaction to proceed for 12 h at room temperature, the product was collected by magnetic separation, rinsed with DMF and ethanol, and dried under a vacuum at 60 °C for 12 h.Measurement of the density of amino groups on the surface of nanoparticles by spectrophotometry
Amino groups on the surface of MNPs were quantified via UV-Vis spectrum analysis based on the reaction between the amino groups with p-nitrobenzaldehyde, with some modifications.29 SMNPs and AMNPs (1 mg) were weighed into 5 mL centrifuge tubes and washed three times with coupling solution (500 mL of methanol containing 4 mL of glacial acetic acid), using a magnet for separation. Then, the washed AMNPs were resuspended into 1 mL of p-nitrobenzaldehyde solution (1% p-nitrobenzaldehyde in coupling solution). After vigorous shaking for 3 h, the MNPs were collected using an applied magnetic field and the supernatant was removed. The MNPs were washed three times with coupling solution and dispersed in 1 mL hydrolysis solution (75 mL of 50% methanol containing 0.1 mL of glacial acetic acid). The mixture was shaken for an additional hour. Finally, the MNPs were collected using a magnetic field and the absorbance of the resulting supernatant was directly measured at 267 nm. SMNPs were used as negative control nanoparticles, as they barely interact with DNA because of the absence of surface amino groups. Calibration curves were generated by diluting a stock solution of p-nitrobenzaldehyde in coupling solution prior to UV detection.DNA adsorption experiments
DNA adsorption experiments were performed as follows. First, a certain amount of salmon sperm DNA was dissolved in a solution containing 10 mM Tris–HCl and 1 mM EDTA at pH 3.0 as a DNA standard solution. The pH of the solution was adjusted using Tris–base or HCl. Second, 2 mL DNA solution with 50 μg mL−1 DNA was added to a 2 mL centrifuge vial. Then, 2.5 mg of mAMNPs was added to the vial. To facilitate DNA adsorption onto the nanoparticles, the mixture was vigorously mixed for 10 min at 500 rpm on an orbital shaker. The temperature was controlled at 25 °C. Finally, a magnet was placed close to the bottom of the vial to collect the magnetic absorbent, the supernatant was transferred to a clean flask, and its absorbance was measured. For quantitative analysis of residual DNA remaining in the supernatant solution following the adsorption process, we determined the intensity of the characteristic absorption peak of DNA at 260 nm. At least three individual experiments were performed to determine the average extraction efficiency.The adsorption capacity of magnetic nanoparticles was calculated as follows:
DNA desorption experiments
We tested the DNA desorption ability of the mAMNPs used in the DNA adsorption experiments. The saturated mAMNPs were washed with 70% ethanol, then mixed with different desorption solution containing 10 mM Tris–HCl and 1 mM EDTA at pH 10.0, with different NaCl concentration. The mixtures were subjected to shaking conditions in an orbital shaker at 500 rpm at 70 °C for 20 min. After elution and subsequent magnetic separation, the DNA concentration of the desorption solutions were analyzed by measuring the absorbance at 260 nm with a UV-Vis spectrophotometer.The extraction recovery was calculated using the following equations:
Results and discussion
Characterization of AMNPs
Each of the prepared AMNPs (sAMNPs, dAMNPs1, dAMNPs2, and mAMNPs) was characterized by FT-IR and VSM analysis. In the FT-IR shown in Fig. 1a, the Si–O stretching vibrations observed at 1095 cm−1 and 799 cm−1 revealed the formation of covalent Fe–O–Si bond during silanization by the modifiers. The amino-groups coated MNPs exhibited absorption bands at 2914 cm−1 and 2837 cm−1, which were attributed to CH2 and CH3 groups of the amino silane coupling agent. Additionally, the intensities of the bands at 3425 cm−1 and 1618 cm−1 probably reflected bending vibrations of the amino groups. Comparison of the IR spectra of all AMNPs revealed that all four types of nanoparticles were modified with amino groups.The magnetization curves of AMNPs (sAMNPs, dAMNPs1, dAMNPs2, and mAMNPs) were analyzed by VSM. No hysteresis following magnetization was observed. Neither coercivity nor remanence was observed, suggesting that all four types of nanoparticles were superparamagnetic. The saturation magnetization was 31.35 emu g−1 for sAMNPs, 31.43 emu g−1 for dAMNPs1, 32.65 emu g−1 for dAMNPs2, and 31.68 emu g−1 for mAMNPs (Fig. 1b). We also measured the magnetization curves of naked MNPs and SMNPs, which were 57.67 emu g−1 and 39.36 emu g−1 respectively. Although the magnetization values revealed a decreasing trend with SiO2 increased coating layers and increased amino groups on the surface of MNPs, they were sufficient rapid separation of the MNPs from the complex matrix using an external magnetic field.
The size and morphology of the magnetic nanoparticles were characterized using TEM. As shown in Fig. S1,† the obtained SMNPs and mAMNPs had core–shell structure with uniform size distribution, and mainly spherical. The naked Fe3O4 nanoparticles were approximately about 20 nm in diameter. After modification, the average diameter of SMNPs was approximately 40 nm, and mAMNPs had a size distribution of 50–100 nm, respectively.
Comparison of different types of AMNPs
IR spectroscopy and VSM measurement cannot quantify the density of amino-groups modifications on the surface of MNPs. Fig. 2a shows UV-Vis-based detection of the quantities of amino groups conjugated to different types of AMNPs using the chemical combination method for formation of covalent bonds between amino groups and p-nitrobenzaldehyde. Based on these results, the density of amino groups on the surface of MNPs increased dramatically when the number of amino group on the silylation modifier was increased from 1 to 3. Our results clearly indicate that mAMNPs possessed significantly the highest amino group density among all four types of AMNPs tested. It should be noted that although ETPES and EDPS both contain two amino groups, there were slightly fewer amino groups on dAMNPs1 than dAMNPs2. It is predicted that the chemical structure of amino precursors may also affect the synthesis of AMNPs. No amino groups were detected on the surface of naked SMNPs (the negative control), which barely reacted with p-nitrobenzaldehyde.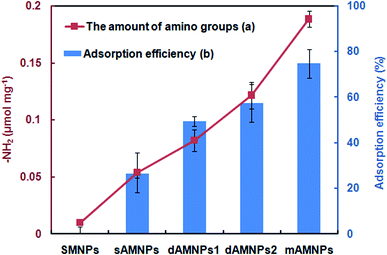 | ||
| Fig. 2 The quantity of amino groups (a) and adsorption efficiency (b) of the sMNPs, mAMNPs, dAMNPs1, dAMNPs2, and sAMNPs. The error bars indicate the standard deviation from the mean (n = 3). | ||
To determine the maximum DNA adsorption on the surface of MNPs, we evaluated the DNA adsorption capability of sAMNPs, dAMNPs1, dAMNPs2, and mAMNPs using salmon sperm DNA in aqueous solution both before and after magnetic separation, using the same extraction conditions. To study DNA extraction, we mixed 2 mg of each MNP type with 2 mL of salmon sperm DNA solution (50 μg mL−1) at pH 3.0. After vigorous mixing of the samples for 10 min and magnetic separation, the DNA adsorption capacity and efficiency were calculated based on the absorbance at 260 nm. The mAMNPs had higher DNA adsorption capacity when than the other three AMNPs types (Fig. 2b), indicating that more DNA was adsorbed when the number of amino groups was greater. Moreover, the adsorption capacity could be improved by changing the structure of amino-precursors. However, the number of amino groups on the surface of the AMNPs had a greater influence on their DNA adsorption capacity than the amino precursor structure. DNA was barely adsorbed on the naked SMNPs, because of electrostatic repulsion between the silica surface and the DNA phosphate groups. These results demonstrate a positive correlation between the amino group density of the MNPs surface and the adsorption capacity of DNA. A maximum value of 74.8% adsorption was observed with mAMNPs.
DNA adsorption experiments
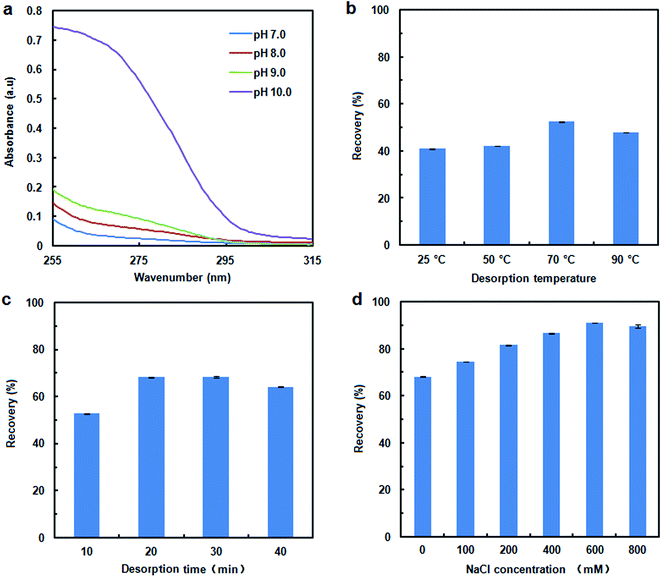
To assess the DNA adsorption capacity of the mAMNPs, we first prepared a model system by dissolving salmon sperm DNA in aqueous solution at a fixed concentration of 50 μg mL−1. It is difficult to directly determine the DNA contents of mAMNP/DNA complexes; however, the amount of DNA adsorbed on the MNP surface can be indirectly determined by detecting the residual DNA remaining in solution after the mAMNP/DNA complexes are separated from solution.The effect of pH on the adsorption of DNA to mAMNPs was studied by varying the pH of the binding solution from 2.0 to 9.0 (Fig. 3a). The highest extraction efficiency was obtained at pH 3.0. Extraction recoveries increased as the pH increased from 2.0 to 3.0, but decreased dramatically with further increases. The results suggest that electrostatic interaction might be one of main factors affecting the binding of DNA to mAMNPs. The effect of the sample pH is consistent with the results of a previous report.11 DNA molecules were selectively adsorbed on the mAMNPs by electrostatic interaction at acidic pH because the DNA moiety is negatively charged and the amino groups on mAMNPs are positively charged. Thus, at a lower pH, strong electrostatic attraction between the DNA moiety and mAMNPs promoted DNA adsorption, resulting in good adsorption efficiency. Increasing pH reduced the positive surface charges of the mAMNPs, which decreased the electrostatic attraction between DNA and the mAMNPs and significantly decreased the adsorption efficiency. At pH 2.0, the extraction recovery was slightly lower than that at pH 3.0 and the baseline UV-absorbance spectra of residual DNA after magnetic separation was slightly elevated, which would affect the accuracy of detection. Therefore, pH 3.0 was used for subsequent experiments.
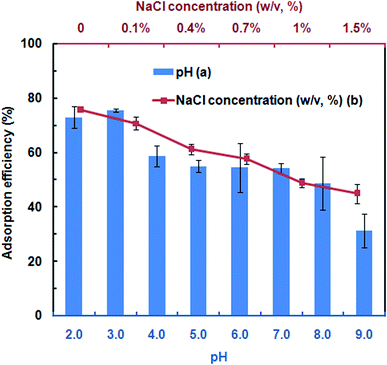 | ||
| Fig. 3 Effects of pH (a) and NaCl concentration (b) on adsorption efficiency of mAMNPs. The error bars indicate the standard deviation from the mean (n = 3). | ||
To investigate the effect of ionic strength on DNA adsorption, different concentrations of NaCl were added to the sample solution. The DNA adsorption capacity decreased with increasing NaCl concentration in the range of 0–1.5% (w/v) (Fig. 3b). Increasing the ionic strength in solution was not beneficial for DNA adsorption due to the decreased electrostatic attraction between the DNA molecule and mAMNPs and the competition between sodium cations and the amino groups for the polyanionic DNA moiety. The results further indicated that DNA adsorption to AMNPs was mainly regulated by electrostatic interaction and that an increased ionic strength weakened the electrostatic force between them.
The quantity of MNPs is one of the most important factors affecting the sorption efficiency and reproducibility. The effect of the MNPs quantity was investigated, using a range of 0.5 mg to 4 mg. The adsorption efficiency increased with increasing amounts of MNPs, from 0.5 mg to 2.5 mg, probably because lower MNP quantities were insufficient for complete DNA adsorption from the samples (Fig. S2†). Beyond 2.5 mg, the adsorption efficiency capacity reached saturation. Thus the optimal MNPs quantity was determined as 2.5 mg.
To obtain higher extraction efficiency, the adsorption time from 0 to 120 min were tested. Rapid adsorption was observed in the first 10 min, during which time >92% adsorption capacity was achieved (Fig. S3†). When the adsorption time was increased from 10 min to 120 min, the adsorption essentially reached equilibrium. Thus, 10 min was selected as the optimal adsorption time for subsequent experiments.
A series of solutions containing different DNA concentrations was used to determine the adsorption capacity of mAMNPs for DNA. The Langmuir adsorption model is represented by the following equation:
DNA desorption experiments
The recovery of adsorbed DNA is important for DNA extraction. DNA desorption is strongly dependent on the pH, ionic strength and temperature.20 Fig. 4a shows the UV-absorbance spectra for DNA desorption in TE buffer with different pH values (adjusted with Tris–base) after 10 min of separation. There were no distinct effect on DNA desorption as the pH of desorption solution was changed over the range of 7 to 9. When the pH of desorption solution was increased to 10.0, some DNA could be eluted from the MNPs, although the desorption efficiency was remained unsatisfactory (41% recovery). Elevating the temperature and prolonging the elution time also appeared to weaken the interaction between DNA and MNPs, and to improve the desorption efficiency. A higher recovery of 68% occurred at 70 °C for a time of 20 min. However, a considerable amount of the DNA remained on the MNPs. To evaluate the effect of the ionic strength on the desorption efficiency, the NaCl concentration in desorption solution was varied from 0 mM to 800 mM. The presence of NaCl improved the DNA desorption efficiency (Fig. 4d); therefore, electrostatic interactions play important roles in the desorption process. DNA recovery was increased to 91% by adjusting the ionic strength with NaCl, with the highest recovery achieved with 600 mM NaCl.Recovery of DNA from human whole blood sample
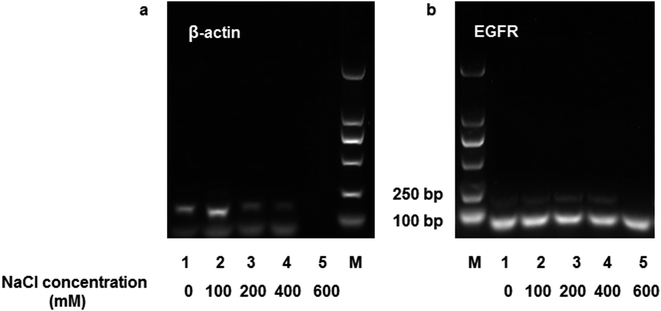
Human whole blood was used to evaluate the practical applicability of DNA extraction using the MNPs. A 200 μL human whole blood sample was mixed with 20 μL of proteinase K (200 μg mL−1) and 20 μL of Triton X-100, after which the mixture was incubated at 56 °C for 10 min. Then, the lysate was diluted to 2 mL in TE buffer (pH 3.0), and 2.5 mg of mAMNPs was resuspended in the solution for DNA isolation. After washing three times with an aqueous ethanol solution, DNA was eluted from the mAMNP–DNA complexes in the presence of different NaCl concentrations (0–600 mM), and the DNA recovered from the samples was directly used as templates for amplification by conventional PCR and real-time PCR (RT-PCR). The A260/A280 ratio of DNA recovered from human whole blood ranged from 1.8 to 1.99, indicating that the extracted DNA templates had high purity and negligible protein contamination.A 153 bp β-actin gene fragment was amplified using the forward primer 5′-GGCATGGGTCAGAAGGATT-3′ and the reverse primer 5′-CACACGCAGCTCATTGTAGA-3′. A 184 bp fragment of the EGFR gene was amplified using the forward primer 5′-CAGGAGGTGGCTGGTTATGT-3′ and the reverse primer 5′-AGCTCCTTCAGTCCGGTTTT-3′. The PCR mixtures (25 μL) consist of 1 μL of recovered DNA, 0.5 μL of both the forward and reverse primers (10 μmol L−1), and 12.5 μL of Taq PCR Master Mix. The therm cycling conditions used were 10 min at 95 °C for denaturation, followed by 35 cycles: 95 °C for 30 s, 58 °C for 30 s, and 72 °C for 1 min. This was followed by a final hold step at 72 °C for 5 min. The band intensities and concentrations of DNA recovered from the supernatants were not positively correlated (Fig. 5). When the NaCl concentration of the desorption solution increased, the intensity of the DNA bands weakened (lane 3–5, Fig. 5). These results suggest that the presence of NaCl negatively affected DNA amplification, even though a high concentration of DNA template was added to the system.
For RT-PCR, magnetically collected DNA was suspended in 500 μL TE buffer (pH 10.0) without additional NaCl, and the supernatant after magnetic separation was directly used as DNA template. Specifically, each reaction mixture (20 μL) consisted of 3 μL supernatant after magnetic separation, 0.4 μL of both the forward and reverse primers (10 μmol L−1), 0.4 μL of ROX Reference Dye II, and 10 μL of TB Green® Premix Ex Taq™ (Takara, Japan). The thermocycling conditions used were 30 s at 95 °C for denaturation, followed by 40 cycles of 95 °C for 5 s and 60 °C for 34 s. DNA recovered from a commercial DNA-extraction kit (Tiangen Biotech, Beijing, China) was used as a positive-control template, and no DNA was added to the negative-control samples. DNA recovered from the salt-free desorption solution could be directly amplified by RT-PCR after magnetic separation, i.e., without additional purification steps (Fig. S5 and S6†). Adding NaCl to the desorption solution inhibited amplification by RT-PCR, even though a sufficiently high concentration of DNA was used. Thus, a negative correlation was found between the ionic strength of the desorption solution and the cycle-threshold value by RT-PCR.
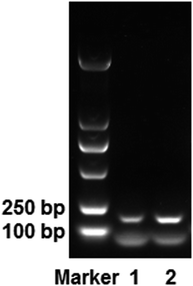
The elution process not only increased the isolation time required, but the NaCl used in the desorption solution also interfered with DNA detection. Therefore, we explored the use of mAMNP/DNA complexes as DNA templates for direct PCR amplification. The EGFR gene was directly amplified using mAMNP/DNA complexes as templates without altering the thermocycling protocol or the reaction composition. Fig. 6 illustrates an electrophoretogram of the PCR products. The 184 bp product from the EGFR gene was clearly amplified from whole blood. The quality of mAMNP/DNA complexes isolated from the whole blood sample was comparable to that isolated using commercial DNA extraction kits. Successful PCR amplifications further demonstrate that the DNA isolated from human whole blood was of high purity, indicating that mAMNPs can be used to isolate genomic DNA from human whole blood and amplify human genes through simplified PCR procedures.
Conclusion
In this study, we synthesized a series of amino modified core–shell structure MNPs. The different types obtained had different numbers of amino groups on the surface formed by several amino-silylation reagents. A positive correlation was found between the amount of DNA adsorbed and the amino group density of the AMNPs, as indicated by the increase in the number of amino groups on amino-precursors. The mAMNPs with the highest surface amino densities showed the best DNA capture efficiency. In the DNA adsorption experiments, the AMNPs showed the highest efficiency at a low pH, which indicated that the adsorption of DNA on mAMNPs was driven by the electrostatic interaction between the negatively charged polyanionic DNA moiety and the positively charged amino groups on the surface of mAMNPs. The adsorbed DNA could be readily recovered using TE buffer at pH 10.0 and manipulation of the ionic strength, resulting in 91% recovery. The PCR and RT-PCR results showed that the mAMNPs appeared to have a greater DNA capture efficiency. Likewise, the mAMNP/DNA complexes could be readily used as DNA templates for DNA amplification, without time-consuming and laborious elution and purification steps. The presence of proteins and polysaccharides in human whole blood was not observed to interfere with DNA extraction. Therefore, the use of mAMNPs for magnetic separation of DNA is a promising method for DNA extraction. These findings provide support for continued efforts to optimize the components of the desorption solution and to explore the possibility of reusing the MNPs.Author contributions
Jia Xu: conceptualization, methodology, formal analysis, validation, writing-original draft. Dan Chen: methodology, formal analysis, validation. Yuan Yang: methodology, formal analysis, validation. Hongjian Gong: writing – review & editing, supervision. Wenqi Gao: writing – review & editing, supervision. Han Xiao: project administration.Ethical statement
The authors state that all experiments were performed in compliance with the relevant laws and institutional guidelines. The human blood used in here was the remaining blood sample after routine clinical examination and the informed consent is exempted with the consent of the ethics committee of Wuhan Children's Hospital (Wuhan Maternal and Child Healthcare Hospital). This study has been approved by the ethics committee of this hospital (No. 2020R024-E01).Conflicts of interest
The authors report no declarations of interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (81802109) and Wuhan Municipal Health Commission of Hubei of China (WX18Y12).References
- F. Garlan, P. Laurent-Puig, D. Sefrioui, N. Siauve, A. Didelot, N. Sarafan-Vasseur, P. Michel, G. Perkins, C. Mulot, H. Blons, J. Taieb, F. D. Fiore, V. Taly and A. Zaanan, Clin. Cancer Res., 2017, 23, 5416–5425 CrossRef CAS.
- Y. van der Pol and F. Mouliere, Cancer Cell, 2019, 36, 350–368 CrossRef CAS.
- A. Campos-Carrillo, J. N. Weitzel, P. Sahoo, R. Rockne, J. V. Mokhnatkin, M. Murtaza, S. W. Gray, L. Goetz, A. Goel, N. Schork and T. P. Slavin, Pharmacol. Ther., 2020, 207, 107458 CrossRef CAS.
- K. Jangpatarapongsa, D. Polpanich, V. Yamkamon, Y. Dittharot, J. Peng-On, R. Thiramanas, S. Hongeng, S. Jootar, L. Charoenmak and P. Tangboriboonrat, Analyst, 2011, 136, 354–358 RSC.
- T. Tangchaikeeree, P. Sawaisorn, S. Somsri, D. Polpanich, C. Putaporntip, P. Tangboriboonrat, R. Udomsangpetch and K. Jangpatarapongsa, Talanta, 2017, 164, 645–650 CrossRef CAS.
- Y. Bai, D. Roncancio, Y. Suo, Y. Shao, D. Zhang and C. Zhou, Colloids Surf., B, 2019, 179, 87–93 CrossRef CAS.
- M. N. Emaus, M. Varona, D. R. Eitzmann, S. Hsieh, V. R. Zeger and J. L. Anderson, TrAC, Trends Anal. Chem., 2020, 130, 115985 CrossRef CAS.
- T. Maruyama, T. Hosogi and M. Goto, Chem. Commun., 2007, 43, 4450–4452 RSC.
- M. N. Emaus, C. Zhu and J. L. Anderson, Anal. Chim. Acta, 2020, 1094, 1–10 CrossRef CAS.
- J. Chen, Y. Wang, X. Wei, R. Ni, J. Meng, F. Xu and Z. Liu, Microchim. Acta, 2020, 187, 7 CrossRef CAS.
- L. Hu, B. Hu, L. Shen, D. Zhang, X. Chen and J. Wang, Talanta, 2015, 132, 857–863 CrossRef CAS.
- Z. Gu, S. Zhao, G. Xu, C. Chen, Y. Wang, H. Gu, Y. Sun and H. Xu, Sens. Actuators, B, 2019, 298, 126953 CrossRef CAS.
- X. Zheng, L. Zhao, D. Wen, X. Wang, H. Yang, W. Feng and J. Kong, Talanta, 2020, 207, 120290 CrossRef CAS.
- C. Li, Y. Huang and Y. Yang, Sens. Actuators, B, 2021, 326, 128845 CrossRef CAS.
- M. Dinc, C. Esen and B. Mizaikoff, TrAC, Trends Anal. Chem., 2019, 114, 202–217 CrossRef CAS.
- Y. Chen, Y. Liu, Y. Shi, J. Ping, J. Wu and H. Chen, TrAC, Trends Anal. Chem., 2020, 127, 115912 CrossRef CAS.
- X. He, K. Wang, W. Tan, B. Liu, X. Lin, C. He, D. Li, S. Huang and J. Li, J. Am. Chem. Soc., 2003, 125, 7168–7169 CrossRef CAS.
- B. Yoza, M. Matsumoto and T. Matsunaga, J. Biotechnol., 2002, 94, 217–224 CrossRef CAS.
- W. Sheng, W. Wei, J. Li, X. Qi, G. Zuo, Q. Chen, X. Pan and W. Dong, Appl. Surf. Sci., 2016, 387, 1116–1124 CrossRef CAS.
- Z. Shan, Y. Jiang, M. Guo, J. Craig Bennett, X. Li, H. Tian, K. Oakes, X. Zhang, Y. Zhou, Q. Huang and H. Chen, Colloids Surf., B, 2015, 125, 247–254 CrossRef CAS.
- Y. Bai, Y. Cui, G. C. Paoli, C. Shi, D. Wang, M. Zhou, L. Zhang and X. Shi, Colloids Surf., B, 2016, 145, 257–266 CrossRef CAS.
- J. H. Min, M. Woo, H. Y. Yoon, J. W. Jang, J. H. Wu, C. Lim and Y. K. Kim, Anal. Biochem., 2014, 447, 114–118 CrossRef CAS.
- S. Khadsai, N. Seeja, N. Deepuppha, M. Rutnakornpituk, T. Vilaivan, M. Nakkuntod and B. Rutnakornpituk, Colloids Surf., B, 2018, 165, 243–251 CrossRef CAS.
- B. G. Maciel, R. J. Silva, A. E. Chávez-Guajardo, J. C. Medina-Llamas, J. J. Alcaraz-Espinoza and C. P. Melo, Carbohydr. Polym., 2018, 197, 100–108 CrossRef CAS.
- Y. Wang, X. Ma, C. Ding and L. Jia, Anal. Chim. Acta, 2015, 862, 33–40 CrossRef CAS.
- L. Ma, N. Sun, J. Zhang, C. Tu, X. Cao, D. Duan, A. Diao and S. Man, Nanoscale, 2017, 9, 17699–17703 RSC.
- Y. Yamini, M. Faraji and M. Adeli, Microchim. Acta, 2015, 182, 1491–1499 CrossRef CAS.
- P. Ashtari, X. He, K. Wang and P. Gong, Talanta, 2005, 67, 548–554 CrossRef CAS.
- M. Waterbeemd, T. Sen, S. Biagini and I. J. Bruce, Micro Nano Lett., 2010, 5, 282–285 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra09409a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |