Open Access Article
Open Access ArticleConcerted dispersion of Staphylococcus aureus biofilm by bacteriophage and ‘green synthesized’ silver nanoparticles†
Salim Manoharadas *a,
Mohammad Altafad,
Abdulwahed Fahad Alrefaeib,
Rajesh Mamkulatil Devasiac,
Ahmed Yacine M. Badjah Hadjd and
Mohammed Saeed Ali Abuhasile
*a,
Mohammad Altafad,
Abdulwahed Fahad Alrefaeib,
Rajesh Mamkulatil Devasiac,
Ahmed Yacine M. Badjah Hadjd and
Mohammed Saeed Ali Abuhasile
aKing Saud University, Department of Botany and Microbiology, Central Laboratory RM 55A, College of Science Building 5, P.O. Box. 2454, Riyadh 11451, Saudi Arabia. E-mail: smanoharadas@ksu.edu.sa; Fax: +966-14699665; Tel: +966-114689170
bKing Saud University, Department of Zoology, College of Science, P.O. Box. 2454, Riyadh 11451, Saudi Arabia
cSCMS Institute of Bioscience and Biotechnology, South Kalamassery, Kochi, Kerala 682033, India
dKing Saud University, Department of Chemistry, College of Science, P.O. Box. 2454, Riyadh 11451, Saudi Arabia
eKing Saud University, Department of Food Science and Nutrition, College of Agriculture and Food Science, P.O. Box. 2454, Riyadh 11451, Saudi Arabia
First published on 5th January 2021
Abstract
Staphylococcal biofilms predominantly cause persistent nosocomial infections. The widespread antibiotic resistance followed by its ability to form biofilm in biological and inert surfaces often contributes to major complications in patients and veterinary animals. Strategic importance of bacteriophage therapy against critical staphylococcal infections had been predicted ever since the advent of antibiotic resistant staphylococcal strains. The significance of metal nanoparticles in quenching biofilm associated bacteria was previously reported. In this study, we demonstrate a concerted action of ‘green synthesized’ silver nanoparticles and bacteriophages in removing pre-formed Staphylococcus aureus biofilms from an inert glass surface in a time dependent manner. Our results demonstrate, for the first time, the rapid co-operative dispersion of the bacterial biofilm. In addition, the synergistic activity of the nanoparticles and bacteriophages causes the loss of viability of the biofilm entrapped bacterial cells thus preventing establishment of a new infection and subsequent colonization. This work further opens up a platform for the combinational therapeutic approach with a variety of nanoparticles and bacteriophages against mono or poly bacterial biofilm in environmental, industrial or clinical settings.
Introduction
The aggregates of microbes entrapped in a matrix of self-produced extracellular polymeric substances (EPS) are known as biofilms.1 The aggregation is primarily achieved by an irreversible attachment of the microbes to a surface. The attached microbes replicate by cell division and promote the adhesion of other microbes to more diverse sites.2 The presence of EPS provides an optimal survival environment for the microbes by filling the empty spaces between the individual cells of the biofilm. In addition, mechanical stability of the biofilm is also provided by the EPS.3 Apart from polysaccharides and proteins, another major component of the biofilms is the extracellular DNA (eDNA), which confers structural integrity to the biofilm matrix.4 It was recently shown that the eDNA protects the biofilm encased Pseudomonas aeruginosa from aminoglycoside antibiotics and other antimicrobial peptides by directly binding to the positively charged group of antimicrobial peptides and aminoglycosides.4 DNABII proteins were found to stabilize the secondary structure of eDNA.5 Furthermore, the formation of stable filamentous biofilm network of the isolated aquatic bacterium (strain F8) was supported by eDNA.6The biofilm associated chronic infections are severe with Staphylococcus aureus. Habitual biofilm formation of S. aureus in clinical settings predominantly occurs in host tissues or surgical implants such as pacemakers, catheters and prosthetic joints.7–9 Poly intracellular adhesin (PIA) forms the major constituent of S. aureus biofilm matrix. PIA is encoded the genomic locus icaADBC.10 The eDNA in S. aureus biofilms bind to several proteins. For instance, insoluble oligomers of β-toxin are formed upon interaction with eDNA. Interestingly these oligomers act as a bridge to hold the biofilm structure together.11,12 S. aureus biofilms can be dissipated using different strategies. Foremost method using enzymatic processes involve, the use of nuclease, dispersin B or proteases. S. aureus secretes one metalloprotease, two cysteine proteases and seven serine proteases.13 The protease mediated destabilization of biofilms is achieved by the cleavage of critical matrix proteins.14 The enzyme dispersin B was shown to be efficient in dispersing S. aureus and S. epidermidis biofilms, wherein the hydrolysis of the unique glycosidic linkages of PIA lead to the biofilm dispersion.15–17 It was shown that the mutation in staphylococcal accessory regulator (sarA) gene leads to the formation of a weak biofilm by S. aureus mutants, particularly due to a reduction in PIA production.18,19 This result invariably suggests the importance of PIA in biofilm formation by S. aureus.
Nanoparticles have been described to show anti-biofilm activity. In a recent study, the phosphatidylcholine-decorated gold nanoparticle linked with gentamicin was shown to inhibit and disrupt pre-formed biofilms of S. aureus and P. aeruginosa.20 Furthermore, pullulan-mediated silver nanoparticles exerted antibiofilm activity against E. coli and P. aeruginosa.21 Earlier, transition metal dichalcogenides (TMD) has been shown to possess antimicrobial activity.22 Apart from this study, polymer nanoparticle carriers (NPCs) docked with the hydrophobic antibacterial compound, farnesol was shown to disrupt biofilms caused by S. mutans.23 Furthermore, a study also stated that the synthesized AuNPs and AgNPs displayed MIC of 6.25 and 5 μg mL−1 against P. aeruginosa and E. coli biofilms, respectively.24
The double stranded DNA lytic phage ϕ44AHJD was sequenced and characterized earlier.25 Phage ϕ44AHJD is classified in the order Caudovirales, family Podoviridae, genus φ29-like phages. In contrast to phage P68, 44AHJD lacks a 1440 nucleotide stretch spanning from 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 091 to 11
091 to 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 531 base pair of the P68 genome.25
531 base pair of the P68 genome.25
The major objective of this study was to test the efficacy of a combinational approach of silver nanoparticles and bacteriophage ϕ44AHJD against the biofilm formed from a veterinary isolate of S. aureus. Furthermore, we also wanted to study if the combinational therapeutic approach could disperse the biofilm and also eliminate the entrapped bacterial cells. Even though earlier attempts were made to study the enhanced biofilm penetration by using a conjugation of polyvalent phages with magnetic colloidal nanoparticle clusters,26 this is the first instance where an additive action of phage and nanoparticles was studied to disperse S. aureus biofilms.
Materials and methods
Bacterial strains and bacteriophage
The S. aureus strain B68, S. aureus strain Rumba and E. coli strain BL21 DE3 were used to test the formation of biofilms on immersed glass cover slip. S. aureus strain Rumba was isolated from the udder of a cow named ‘Rumba’ with bovine mastitis. E. coli BL21 DE3 is a routinely used lab strain. S. aureus B68 is the host strain used for the propagation of the phage ϕ44AHJD. S. aureus B68 and the bacteriophage ϕ44AHJD was a gift from Dr Udo Blaesi, MFPL, Vienna, Austria.Growth condition of the bacterial strains and bacteriophage
All the bacterial strains used in this study was grown at 37 °C in nutrient broth (NB) medium with shaking conditions (180 rpm) for 16 h. The bacteriophage ϕ44AHJD used in this study was propagated on S. aureus B68 and the phage titer was determined before the experiment by plaque forming unit assay. The sensitivity of propagated phage ϕ44AHJD was further tested on the bacterial strain S. aureus Rumba by spot analysis. The enumeration of bacteriophage by plaque forming unit assay was performed according to the standard protocol.27 Shortly, the propagated crude ϕ44AHJD bacteriophage and the serial dilutions thereof were mixed with the specific host strain, S. aureus Rumba in molten soft agar and were poured onto the surface of nutrient agar that supports bacterial growth. Following incubation at 37 °C for 18 h, the growth of bacteria to an opaque, confluent layer was seen in the area where the ϕ44AHJD bacteriophage was not present. However, in the zone where ϕ44AHJD bacteriophage is present, consistent infection of the host cells by the phage progenies produce a visible clear circular area where no bacterial growth is seen. The plaques are counted, and the phage concentration/titer is commonly expressed as the number of plaque-forming units (PFU) per mL of the assayed preparation. The prepared phage lysate was stored at 4 °C until further use.Biofilm formation assay
The biofilm formation ability of the strains was tested on an immersed glass cover slip. One mL of overnight cultures (approximately 1 × 108 CFU per mL) of S. aureus B68, S. aureus Rumba and E. coli DE3 were inoculated into 25 mL of NB in a petri dish. Sterile glass cover slips were immersed in the culture dish and the inoculated cultures were incubated at 37 °C in shaking conditions. The NB media was changed every 24 h time points. Microbial biomass was analyzed at specific time points. The glass coverslip was withdrawn at particular time point and was washed thrice with distilled water to remove any unattached planktonic bacteria. The glass coverslips encompassing the microbial biomass was stained with 0.1% w/v acridine orange (Sigma-Aldrich, USA) in PBS buffer for 5 min and was analyzed for biofilm formation with confocal microscope (Zeiss, Germany). The Zen software (Zeiss, Germany) was used for image acquisition and further analysis. Acridine orange is excited at 476 nm and gives a broad fluorescence spectrum by intercalating into ds and ssDNA and RNA.28 However, the stain does not differentiate between live or dead cells and will even binds to the eDNA present in the biofilm matrix.Preparation of Allium sativum extract and green synthesized AgNP
In order to prepare the Allium sativum extract, 6 g of fresh garlic was chopped and was soaked in 50 mL of deionized water. The garlic-soaked mixture was incubated at room temperature for 24 h. The resulting solution was decanted to a fresh tube to collect a pale white transparent garlic extract solution. Any undesirable solid garlic pieces were discarded. The concentration of the garlic extract was assessed by evaporating 2 mL of the garlic extract in a vacuum oven at a temperature not exceeding 40 °C. The solid weight of the concentrate was measured and was estimated to be 21.8 ± 0.5 mg mL−1. Even though in our experiments we used chopped garlic for the preparation of garlic extract, crushing the garlic prior to soaking and/or soaking at elevated temperatures was found to increase the extraction efficiency resulting in larger mass concentrations of extract. The nanoparticle synthesis was impacted directly by the variations in Allium sativum extract preparation. Polydisperse population of nanoparticles was observed when larger quantity of Allium sativum was employed during synthesis.AgNP was synthesized by adding varying amount of the of Allium sativum extract (0.5 mL, 1.0 mL, 1.5 mL and 2.0 mL) to 10 mL of 1 mM of AgNO3 (Sigma Aldrich, USA) in DI water. The effective final concentration of the Allium sativum extract in the reaction mixture was calculated to be 11.0, 22.0, 33.0 or 44.0 mg. After addition of AgNO3 to the Allium sativum extract, a color change in the reaction mix from colorless to light orange was observed within 2 h of incubation of the mixture at room temperature at constant shaking at 60 rpm, indicating the presence of AgNP. The reaction mix was further allowed to ‘age’ for 48 h to yield a deep orange/brown color.
The characterization of the yielded AgNP was done by Transmission Electron Microscopy (TEM). The image was acquired using Hitachi 7600 Tem (Japan) with an accelerating voltage of 120 kV. For TEM analysis, the sample was prepared by spotting two drops of the nanoparticle dispersion onto carbon-coated copper TEM grid followed by air drying at ambient conditions. The TEM sample was stored in a desiccator and imaged shortly after collection. The size distributions were determined by image analysis using the ImageJ software package. At least 100 nanoparticles were counted for meaningful and relevant statistics.
Further characterization of the AgNP was done by energy-dispersive X-ray spectroscopy (EDS: JOEL JED-2200 Series, Japan), dynamic light scattering (DLS: Malvern, UK) and UV/vis spectrophotometer (Shimadzu, Japan).
Live-dead staining of biofilm dwelling bacteria
The live dead staining of bacterial cells was done using standard protocol,29 with variations. Briefly, the glass cover slip with pre-formed S. aureus biofilm was washed extensively with double distilled water, followed by washing with PBS, pH 7.0. The staining for viable cells on the cover slips was performed by the addition and 5 μM of Syto9 (ThermoFischer Scientific, USA), diluted in DMSO. The staining was done for 10 min in dark conditions. After incubation, the cover slips were further washed extensively with 1× PBS and final rinsing in double distilled water. In order to stain for nonviable S. aureus cells, propidium iodide (ThermoFischer Scientific, USA) was diluted in 2× SSC buffer (0.3 M NaCl, 0.03 M sodium citrate, pH 7.0) to a final concentration of 500 nm. The diluted propidium iodide was added to the washed and rinsed Syto9 stained cover slip. The staining was done for 10 min, followed by washing with 1× PBS and final rinsing with double distilled water. The imaging was done by Confocal microscope (Zeiss, Germany) at an excitation/emission of 483/503 nm for syto9 and 535/617 nm for propidium iodide. The images were acquired on a Rolera Em-C2 camera with a 63× oil immersion objective (Zeiss, Germany). The acquired image was processed by Zen software (Zeiss, Germany).Results
S. aureus strain Rumba was able to form biofilm matrix on immersed glass cover slips
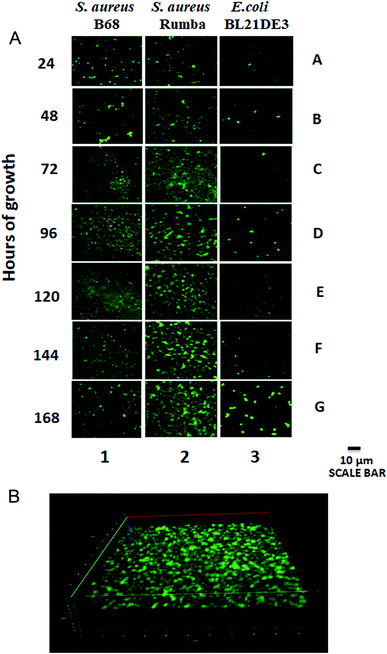
S. aureus is capable of forming biofilms on inert materials. However, there has been hardly any information on the ability of S. aureus to form biofilm on glass surface.30 In order to check the efficacy of S. aureus and E. coli strains to form biofilms on glass surface, 0.17 mm glass cover slips were immersed in the bacterial growth media and the formation of biofilms was monitored over a period of 168 h. Two S. aureus strains, B68 and Rumba were used for the study. A progressive biofilm formation over time was observed with S. aureus Rumba (Fig. 1A; Lane 2). Intact biofilm mass formation was not visually observed with S. aureus B68 even until 168 h of incubation. However, the bacteria were able to attach to the glass surface (Fig. 1A; Lane 1). E. coli BL21 DE3 weakly attached to the glass surface, but did not establish a strong biofilm mesh (Fig. 1A; Lane 3). The establishment of the biofilm matrix by S. aureus Rumba was evident at 96 h of growth, with the biofilm establishment becoming intense until 168 h. At the end of 168 h, a well-established biofilm was noticed. Nutrient media was particularly selected for the formation of biofilm to mimic the in vivo growth conditions of the bacterium. Robust biofilm formation by S. aureus was earlier observed in vitro in the presence of BHI (Brain Heart Infusion) medium.31 It was also reported that the presence of glucose induces biofilm formation in S. aureus in vitro.32 However, the in vivo growth conditions of S. aureus typically lack these components that promote biofilm formation. Hence, we decided to use the nutrient media with basic ingredients for the development of biofilm on glass cover slip. It was noteworthy that only S. aureus Rumba could form a well-established biofilm in these stringent growth conditions. The 3-dimensional image of the biofilm network formed by S. aureus Rumba grown at 37 °C until 168 h of growth is shown in Fig. 1B. Since a well-established biofilm was formed by S. aureus Rumba in comparison with S. aureus B68 and E. coli BL21 DE3 on immersed glass cover slips, further studies on the possible disruption strategies were done with the biofilm formed by S. aureus Rumba at 37 °C for 168 h of growth.Green synthesized AgNP displays antimicrobial activity against S. aureus Rumba
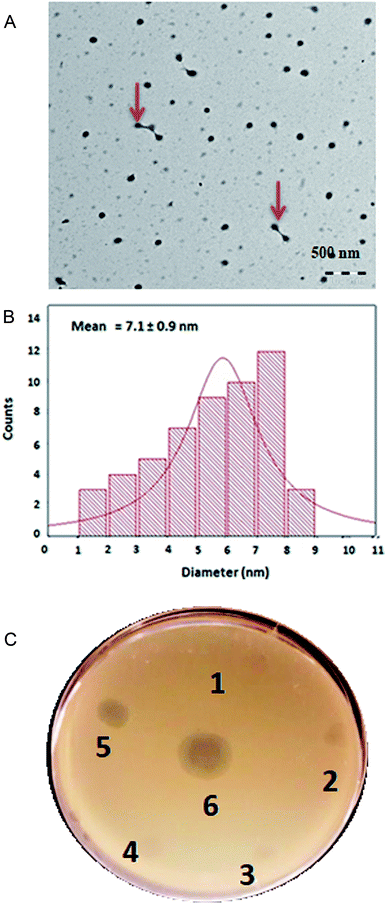
The silver nanoparticles (AgNP) were synthesized by the reduction of silver nitrate (AgNO3) using Allium sativum extract. The green synthesis of nanoparticles typically make use of mixed-valence polyoxometalates, tollens, polysaccharides and biological methods, and do offer distinct advantages over traditional methods involving chemical agents associated with environmental toxicity.33 Apart from rapid synthesis of the nanoparticles using bacterial,34 algal35 and fungal36 extracts, plant based biological methods are also cost effective and can be utilized for the large-scale production of nanoparticles.37 The AgNPs synthesized by Allium sativum extract were characterized by UV/vis spectrophotometry (Fig. S1†), followed by TEM imaging at an accelerating voltage of 120 kV. The TEM image of the synthesized AgNP is shown in Fig. 2A. In addition to the predominantly seen monodisperse individual AgNPs, a few were complexed to each other as shown by red arrow marks (Fig. 2A). Higher magnification TEM images of the synthesized AgNP is shown in ESI (Fig. S2†). The particle size of the AgNPs was calculated based on the number of counts of the particles in comparison to the diameter of the particles. The mean diameter of the synthesized AgNPs was 7.1 ± 0.9 nm (Fig. 2B). AgNP particles of larger size (≈150 nm) was also present, however smaller size particles of less than 10 nm was predominantly present in the mixture. Further characterization of the AgNP was done by EDS. The analysis by EDS was particularly done to estimate the mass percentage of Ag present in the synthesized AgNPs. As shown in Fig. S3,† the concentration of Ag in AgNP was 68.63 ± 1.68 mass percentage. Dynamic light scattering analysis (DLS) of the synthesized AgNP was also done to determine the particle size. The average size of the particles as determined by DLS was 167 nm. Earlier studies have pointed out that the nanoparticle size obtained by DLS is usually greater than that measured by other techniques, like TEM and BET.38 In order to test the antimicrobial activity of the synthesized AgNPs against S. aureus Rumba, varying concentration of the AgNP was spotted on a lawn of S. aureus Rumba with phage ϕ44AHJD serving as the positive control (Fig. 2C; 6). The Allium sativum extract used for synthesis of AgNP was also spot analyzed for any inherent antimicrobial activity. As expected, the Allium sativum extract per se did not show any remarkable antimicrobial effect on the growth of S. aureus Rumba (Fig. 2C; 1). Spot analysis of the synthesized AgNP with a concentration of 1 mM was performed on a lawn of S. aureus Rumba. The AgNP synthesized with 44.0 mg final concentration of Allium sativum extract showed the maximum growth inhibitory activity against S. aureus Rumba in spot analysis (Fig. 2C; 5). However, no inhibitory effect was observed with the AgNP synthesized with lesser concentrations of Allium sativum extract such as 11.0, 22.0 or 33.0 mg (Fig. 2C; 2, 3, 4). The result is particularly encouraging as the Allium sativum extract did not possess any antimicrobial effect and when an optimal concentration was used in the synthesis of AgNP, the inhibitory effect resurfaces. Nevertheless, it cannot be validated from these experiments, as to what specifically contributes to the antimicrobial activity of the synthesized AgNPs. The bacterial growth inhibitory effect of 1 mM AgNP synthesized in the presence of 44.0 mg Allium sativum extract prompted us to test its efficiency against pre-formed biofilm of S. aureus Rumba.Synergistic action of AgNPs and ϕ44AHJD against S. aureus Rumba biofilms
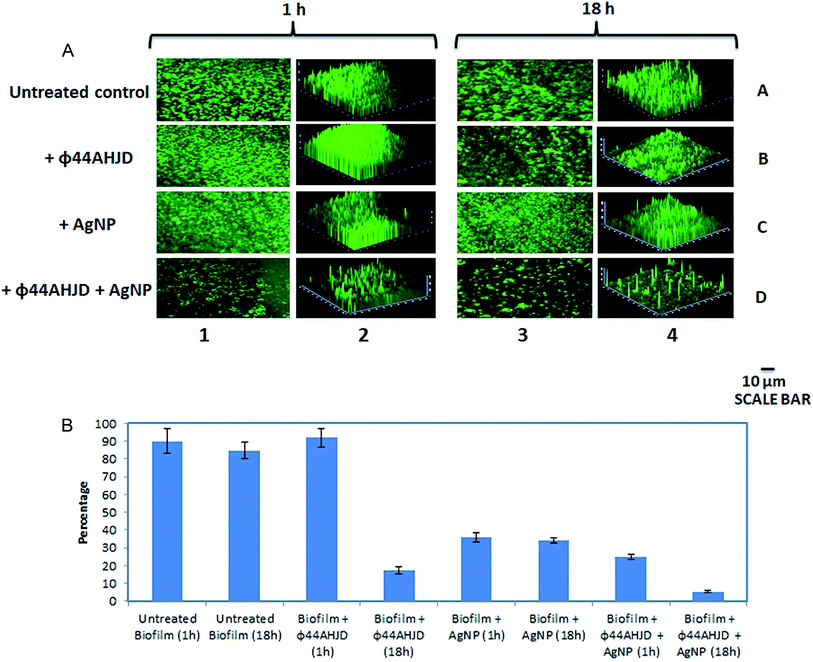
A combination approach of enzyme pronase, cellulase and DnaseI together with benzalkonium chloride (BAC) against a mixed biofilm culture of L. monocytogenes and E. coli was studied earlier.39 However, the dissipated biofilm released a high number of viable cells that is capable of establishing a subsequent infection. We wanted to test the efficiency of the synthesized AgNP and phage ϕ44AHJD against pre-formed S. aureus Rumba biofilms. The objective of this study was two-fold, one the successful dissipation of the biofilm and secondly the annihilation of the biofilm dwelling S. aureus cells. The inhibitory concentration of 1 mM AgNP and 1 × 108 PFU mL−1 phage ϕ44AHJD were added to 25 mL of NA medium and the glass cover slips with 168 h pre-formed S. aureus Rumba biofilm was submerged in the media. The glass slide encompassing the biofilm was further incubated for 1 h or 18 h after addition of AgNP and ϕ44AHJD at 37 °C. The disruption of S. aureus Rumba biofilm following addition of phage ϕ44AHJD and/or AgNP was analyzed by confocal microscopy following staining of the cover slips with 0.01% acridine orange at specific time points. Incubation of the pre-formed biofilm with phage ϕ44AHJD for 1 h did not cause any notable disruption of the biofilm in comparison with the untreated biofilm control (Fig. 3A; Lane 1B vs. Lane 1A). However, the addition of AgNP to the biofilm, followed by incubation at 37 °C for 1 h caused a 65% reduction in the biofilm intensity (Fig. 3A; Lane 1C). Interestingly, the incubation of pre-formed S. aureus Rumba biofilm with AgNP and phage ϕ44AHJD for 1 h caused a reduction of biofilm by 75% (Fig. 3A; Lane 1D). As evident, a clear synergistic action of the AgNP and phage ϕ44AHJD was seen in disrupting the pre-formed S. aureus Rumba Biofilms. We wanted to further analyze for a more profound effect on biofilm dispersion subjected to a longer incubation time with AgNP and phage ϕ44AHJD. Interestingly, a severe disruption of ϕ44AHJD treated biofilm was seen with the biofilm intensity dropping to about 83% as compared to the untreated control with incubation over 18 h (Fig. 3A: Lane 3B vs. Lane 3A). This result is particularly important as the disruption of biofilm was progressive with time, as 1 h incubation with phage ϕ44AHJD showed negligible disruption of the biofilm as compared to 83% disruption with an incubation period of 18 h. In contrast, the AgNP treated disruption of biofilm was comparable on both the time frame of 1 h and 18 h causing a 65% disruption of the pre-formed biofilm (Fig. 3A; Lane 3C). The synergistic action of AgNP and phage ϕ44AHJD against the biofilm at a longer incubation period of 18 h caused a 95% disruption (Fig. 3A; Lane 3D). The experiment clearly shows a concerted action of AgNP and phage ϕ44AHJD on the disruption of biofilms. The percentage decline in biofilm intensity as calculated from the average of the mean fluorescence intensity is shown in Fig. 3B. Only 5% of the pre-formed S. aureus Rumba biofilm was present on the glass cover slips following treatment with phage ϕ44AHJD and AgNPs after 18 h of incubation (Fig. 3B).Live-dead staining of the treated biofilm sample
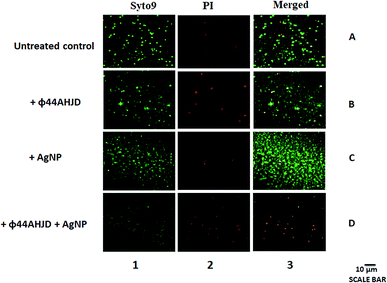
We further wanted to evaluate if the biofilm emancipated S. aureus cells were viable following treatment with AgNP and/or phage ϕ44AHJD until 18 h of incubation. The discrete strategic approach used by us in the dissipation of the S. aureus Rumba biofilm was to damage the biofilm architecture followed by the annihilation of S. aureus Rumba. The live-dead staining of the untreated and treated pre-formed S. aureus Rumba biofilm was performed by staining with Syto9 and propidium iodide. The dead bacteria are stained red by propidium iodide possibly due to permeability of the dye through damaged cell wall. In contrast, the viable cells are stained by Syto9 dye. The untreated control had 91% of viable cells and only 9% dead cells (Fig. 4; Lane 1A, Lane 2A, Lane 3A). The phage ϕ44AHJD treated biofilm had 75% of viable cells and 25% non-viable cells (Fig. 4; Lane 1B, Lane 2B, Lane 3B). The sample treated with AgNP alone, on the other hand had 83% live cells and 17% dead cells (Fig. 4; Lane 1C, Lane 2C, Lane 3C). The most interesting and promising finding was seen in the sample treated with both AgNP and phage ϕ44AHJD, with only 14% of cells in the viable range and 86% cells was seen to be non-viable (Fig. 4; Lane 1D, Lane 2D, Lane 3D). The synergistic action of AgNP and ϕ44AHJD, in addition to the reduction in biofilm intensity also contributes to the killing of the biofilm associated bacteria. Another representative image of the live/dead staining with a higher magnification is shown in ESI (Fig. S5†).Discussion
Biofilm formation of Staphylococcus aureus associated with bovine mastitis has been widely studied.40,41 We have reported the ability of S. aureus Rumba, a diary isolate associated with bovine mastitis to form intense biofilm on glass slides under dynamic growth conditions. The normal cell doubling time of planktonic E. coli and S. aureus under laboratory conditions is 0.2–0.4 h. For instance, E. coli can double every 20 min in the laboratory it only doubles every 15 h in the wild.42 In our analysis, a strong biofilm formation by S. aureus Rumba was formed on glass cover slips by the end of 96 h of primary inoculation. No strong biofilm formation on glass surface was observed with the other two strains (S. aureus B68 and E. coli BL21 DE3) tested.Several strategies including the use of enzyme dispersin B, individually or in combination with antibiotics has been proposed for the dissipation of pre-formed S. aureus biofilms.43–45 The most interesting meticulous chemical approach in the prevention of biofilm formation was making use of nanoparticles, wherein the fabrication of surfaces with nanoparticles prevented bacterial attachment and biofilm establishment,46,47 particularly against S. aureus.48 Besides, silver nanoparticles also display antimicrobial activity against planktonic S. aureus.49 This prompted us to analyze the efficiency of ‘green’ synthesized AgNP against S. aureus biofilm. The synthesized AgNPs were characterized by TEM, DLS, EDS and UV/vis spectrophotometry. The determined size of the particles varied between TEM and DLS analysis. It has been suggested earlier that electron microscopy is the simplest and most widely used technique to directly measure the particle size, size distribution and morphology.38 DLS measurement on the other hand measures the scattering intensity of the particles in Brownian motion. The size of the particles is then calculated by the Stokes–Einstein relation. Particle size calculated by DLS is usually large in comparison with TEM analysis. This is primarily due to the measurement of Brownian motion of the particles and the subsequent size distribution. Furthermore, particles aggregate during DLS measurements in aqueous state, hence, the calculated size of is mostly of clustered particles rather than individual particles.38
The antimicrobial effectiveness of AgNP is thought to be due to the production of ROS upon interaction with the bacterial cells.50 In our study, the effectiveness of 1 mM AgNP synthesized by Allium sativum extract was observed to create a disruption of the pre-formed biofilm architecture. An earlier study has used a varying range of concentration of AgNP (12.5 mM, 25 mM, 50 mM and 100 mM) against S. aureus and observed a growth inhibition zone of 23.56 mm with 12.5 mM of AgNPs.51 We used a further less concentration of AgNP, since in our observation, 1 mM AgNP was able to inhibit growth as determined by spot analysis. Apart from the dissipation of the biofilm, the annihilation of biofilm associated cells was not effective, stating the inefficiency of AgNP in causing death of glass surface attached bacteria. Interestingly, resurgence of resistant S. aureus was not observed on AgNP treated samples, marking the efficiency of AgNP against planktonic cells. This is of particular importance as the cells detached from biofilm are affected by AgNP and incapacitate them in establishing a fresh infection. The inefficiency of AgNP in completely removing the biofilm associated S. aureus even after a longer incubation period triggered us to simultaneously test the effectivity of phage ϕ44AHJD against S. aureus Rumba biofilm. Bacteriophages were earlier tested against bacterial biofilms.52 In addition to the intact phages, phage derived proteins and chimeric variants thereof were shown to be active against planktonic bacterial pathogens and biofilms.53–56 However, the main hurdle that forgoes the use of phages against bacterial biofilms is the high level of adaptability of the bacteria in response to the selective pressure caused by phage treatment, which in turn result in the emergence of phage-resistant variant of the bacteria.57 In our study the viable number of cells following the treatment of the pre-formed S. aureus biofilm with phage ϕ44AHJD amounted to 75% of the total number of cells. In contrast to the AgNP treated biofilm, the reduction in the density of biofilm was observed only after 18 h of incubation of the sample with the phage.
We speculated that the combinational approach with both AgNP and phage ϕ44AHJD could cause a profound reduction of the S. aureus biofilm density at a shorter period of incubation. It has been earlier reported that phage interaction with the biofilm dwelling bacteria is primarily governed by phage mobility through the biofilm.58 Interestingly, it has also been suggested that in certain cultured biofilms, the replication of bacteriophages was more efficient than in planktonic cultures, presumably by making use of self-encoded depolymerases.59,60 Synthetic biology approach has been employed in the engineering of enzymatic bacteriophage with dispersin B, which was able to disrupt biofilms by two orders of magnitude in comparison with non-enzymatic bacteriophage.61 These results indicate that even though bacteriophages can exhibit the activity against bacterial biofilms, a better efficiency of biofilm disruption and bacterial targeting can be achieved with the presence of an external succor. The biofilm matrix formed by S. aureus are held in structure by the presence of eDNA,62 and it has been reported that the biofilm formed by clinical isolates of S. aureus comprise a greater quantity of eDNA, suggesting the profound importance of eDNA in S. aureus biofilms.63 It has been suggested that AgNP can cause intermediate DNA damage by oxidative stress and AgNP more than 10 nm are not actively up taken by bacterial cells.64 In addition to morphological changes, DNA fragmentation was observed in E. coli presumably by the damaging of sulfur and phosphorus moieties by AgNP. In this context, an educated guess suggests that the AgNP could damage the eDNA in S. aureus biofilm, subsequently causing a slackening of the biofilm network. Bacteriophage ϕ44AHJD gain access to the biofilm dwelling S. aureus cells through this disrupted biofilm mesh, thereby causing the lysis of the bacteria. The results corroborated our speculation wherein a concerted action of AgNP and phage ϕ44AHJD was very effective in removing pre-formed S. aureus Rumba biofilm. A drop in biofilm intensity of 75% occurred within 1 h following the treatment with the reduction in the biofilm intensity progressing to 95% after 18 h of treatment. In contrast with ϕ44AHJD or AgNP individually treated biofilms, the synergistic action of both caused a decrease in viable cell count of the biofilm associated S. aureus to less than 15%.
The described methodology of a combinational approach using green synthesized nanoparticles and bacteriophages in sequestering biofilm disruption is a robust technique that can be successfully tested in biofilm associated with wound infections. The marked advantage of this method is that a combination of nanoparticles with different phages with varying host range can be synthesized based on the spectrum of bacteria associated with biofilms. Remarkably, phages will be more stable in clinical settings as compared to their purified endolysins, which may get trapped in the biofilm mesh. In addition, the treatment of intact phages along with nanoparticles is beneficial against biofilm forming Gram-positive and Gram-negative bacterial pathogens, in comparison with enzybiotics or phage endolysins used along with nanoparticles which can be applicable only against Gram-positive bacterial biofilms.
Conclusion
Here, we report the disruption of pre-formed S. aureus biofilm by a combination of silver nanoparticles and bacteriophage. Initially, the ability of three different bacterial species to form biofilms on glass surface was analyzed. The strongest biofilm formation was observed with S. aureus Rumba. In order to test the dissipation of pre-formed S. aureus Rumba biofilm, either silver nanoparticles synthesized using garlic extract or intact bacteriophage ϕ44AHJD or a combination of both was used. The maximum dissipation of the biofilm was observed when both silver nanoparticles and bacteriophages were used together. Apart from the disruption of the biofilm, the treatment was also effective in the annihilation of biofilm encased S. aureus bacterial cells. The mentioned technique of using nanoparticles and bacteriophages will prove to be a unique approach in countering multidrug resistant bacterial biofilms as varying combination of nanoparticles and bacteriophages can be used against different species of bacterial biofilms that could be formed on industrial and clinical settings.Author contributions
Salim Manoharadas: conceptualization, methodology, experimentation, formal analysis, funding acquisition, software, and writing the manuscript. Mohammad Altaf: methodology, data analysis, and experimentation. Abdulwahed Fahad Alrefaei: methodology, data analysis, and microscopy. Rajesh Mamkulatil Devasia: conceptualization and manuscript editing. Ahmed Yacine M. Badjah Hadj: formal analysis and project administration. Mohammed Saeed Ali Abuhasil: experimentation. Mohammad Altaf and Abdulwahed Fahad Alrefaei equally contributed to the work.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors extend their appreciation to the Deanship of Scientific Research at King Saud University for funding this work through research group no (RG-1439-075). The authors would like to thank Prof. Dr Udo Blaesi, Department of Microbiology and Immunobiology, MFPL, Vienna, Austria for the bacterial strains: S. aureus Rumba and bacteriophage ϕ44AHJD. We are grateful to the Confocal Microscopy unit at Central Laboratory, College of Science, King Saud University, Riyadh, Kingdom of Saudi Arabia.References
- H. C. Flemming, et al., Biofilms: an emergent form of bacterial life, Nat. Rev. Microbiol., 2016, 14, 563–575 CrossRef CAS.
- L. Hall-Stoodley, et al., Bacterial biofilms: From the natural environment to infectious diseases, Nat. Rev. Microbiol., 2004, 2, 95–108 CrossRef CAS.
- A. Persat, et al., The mechanical world of bacteria. The mechanical world of bacteria, Cell, 2015, 161, 988–997 CrossRef CAS.
- W. C. Chiang, et al., Extracellular DNA shields against aminoglycosides in Pseudomonas aeruginosa biofilms, Antimicrob. Agents Chemother., 2013, 57, 2352–2361 CrossRef CAS.
- A. Devaraj, et al., DNABII proteins play a central role in UPEC biofilm structure, Mol. Microbiol., 2015, 96, 1119–1135 CrossRef CAS.
- U. Böckelmann, et al., Bacterial extracellular DNA forming a defined network-like structure, FEMS Microbiol. Lett., 2006, 262, 31–38 CrossRef.
- M. R. Parsek and P. K. Singh, Bacterial biofilms: an emerging link to disease pathogenesis, Annu. Rev. Microbiol., 2003, 57, 677–701 CrossRef CAS.
- L. Barrett and B. Atkins, The clinical presentation of prosthetic joint infection, J. Antimicrob. Chemother., 2014, 69, i25–i27 CrossRef CAS.
- S. Chatterjee, et al., Biofilms on indwelling urologic devices: microbes and antimicrobial management prospect, Ann. Med. Health Sci. Res., 2014, 4, 100–104 CrossRef CAS.
- J. P. O'Gara, ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus, FEMS Microbiol. Lett., 2007, 270, 179–188 CrossRef.
- M. J. Huseby, et al., Beta toxin catalyzes formation of nucleoprotein matrix in staphylococcal biofilms, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 14407–14412 CrossRef CAS.
- J. L. Lister and A. R. Horswill, Staphylococcus aureus biofilms: recent developments in biofilm dispersal, Front. Cell. Infect. Microbiol., 2014, 4, 178 Search PubMed.
- L. Shaw, et al., The role and regulation of the extracellular proteases of Staphylococcus aureus, Microbiology, 2014, 150, 217–228 CrossRef.
- A. K. Zielinska, et al., sarA-mediated repression of protease production plays a key role in the pathogenesis of Staphylococcus aureus USA300 isolates, Mol. Microbiol., 2012, 86, 1183–1196 CrossRef CAS.
- J. B. Kaplan, et al., Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms, J. Bacteriol., 2004, 186, 8213–8220 CrossRef CAS.
- Y. Itoh, et al., Depolymerization of beta-1,6-N-acetyl-D-glucosamine disrupts the integrity of diverse bacterial biofilms, J. Bacteriol., 2005, 187, 382–387 CrossRef CAS.
- S. Sugimoto, et al., Broad impact of extracellular DNA on biofilm formation by clinically isolated Methicillin-resistant and -sensitive strains of Staphylococcus aureus, Sci. Rep., 2018, 8, 2254 CrossRef.
- K. E. Beenken, et al., Mutation of sarA in Staphylococcus aureus limits biofilm formation, Infect. Immun., 2003, 71, 4206–4211 CrossRef CAS.
- J. Valle, et al., SarA and not sigmaB is essential for biofilm development by Staphylococcus aureus, Mol. Microbiol., 2003, 48, 1075–1087 CrossRef CAS.
- H. Mu, et al., Potent Antibacterial Nanoparticles against Biofilm and Intracellular Bacteria, Sci. Rep., 2016, 6, 18877 CrossRef CAS.
- P. Kanmani and S. T. Lim, Synthesis and characterization of pullulan-mediated silver nanoparticles and its antimicrobial activities, Carbohydr. Polym., 2013, 97, 421–428 CrossRef CAS.
- V. Agarwal, et al., Recent advances in the field of transition metal dichalcogenides for biomedical applications, Nanoscale, 2018, 10, 16365–16397 RSC.
- K. R. Sims Jr, et al., Enhanced design and formulation of nanoparticles for anti-biofilm drug delivery, Nanoscale, 2019, 11, 219–236 RSC.
- P. Singh, et al., Green synthesis of gold and silver nanoparticles from Cannabis sativa (industrial hemp) and their capacity for biofilm inhibition, Int. J. Nanomed., 2018, 13, 3571–3591 CrossRef CAS.
- D. Vybiral, et al., Complete nucleotide sequence and molecular characterization of two lytic Staphylococcus aureus phages: 44AHJD and P68, FEMS Microbiol. Lett., 2003, 219, 275–283 CrossRef CAS.
- L. L. Li, et al., Enhanced biofilm penetration for microbial control by polyvalent phages conjugated with magnetic colloidal nanoparticle clusters (CNCs), Environ. Sci.: Nano, 2017, 4, 1817–1826 RSC.
- B. Anderson, et al., Enumeration of bacteriophage particles: Comparative analysis of the traditional plaque assay and real-time QPCR- and nanosight-based assays, Bacteriophage, 2011, 1, 86–93 CrossRef.
- T. Bernas, et al., Confocal fluorescence imaging of photosensitized DNA denaturation in cell nuclei, Photochem. Photobiol., 2005, 81, 960–969 CrossRef CAS.
- M. Berney, et al., Assessment and Interpretation of Bacterial Viability by Using the LIVE/DEAD BacLight Kit in Combination with Flow Cytometry, Appl. Environ. Microbiol., 2007, 73, 3283–3290 CrossRef CAS.
- N. K. Archer, et al., Staphylococcus aureus biofilms: Properties, regulation and roles in human disease, Virulence, 2011, 2, 445–459 CrossRef.
- P. Chen, et al., An improved medium for growing Staphylococcus aureus biofilm, J. Microbiol. Methods, 2012, 90, 115–118 CrossRef CAS.
- H. McCarthy, et al., Methicillin resistance and the biofilm phenotype in Staphylococcus aureus, Front. Cell. Infect. Microbiol., 2015, 5, 1 Search PubMed.
- S. Iravani, et al., Synthesis of silver nanoparticles: chemical, physical and biological methods, Results Pharma Sci., 2014, 9, 385–406 CAS.
- A. R. Shahverdi, et al., Rapid synthesis of silver nanoparticles using culture supernatants of Enterobacteria: A novel biological approach, Process Biochem., 2007, 42, 919–923 CrossRef CAS.
- K. Govindaraju, et al., Silver, gold and bimetallic nanoparticles production using single-cell protein (Spirulina platensis) Geitler, J. Mater. Sci., 2008, 43, 5115–5122 CrossRef CAS.
- M. Saravanan, et al., Synthesis of silver nanoparticles from Phenerochaete chrysosporium (MTCC-787) and their antibacterial activity against human pathogenic bacteria, Microb. Pathog., 2018, 117, 68–72 CrossRef CAS.
- S. Iravani, Green synthesis of metal nanoparticles using plants, Green Chem., 2011, 13, 2638–2650 RSC.
- A. Dhawan, et al., Toxicity assessment of nanomaterials: methods and challenges, Anal. Bioanal. Chem., 2010, 398, 589–605 CrossRef CAS.
- P. Rodríguez-López, et al., Removal of Listeria monocytogenes dual-species biofilms using combined enzyme-benzalkonium chloride treatments, Biofouling, 2017, 33, 45–58 CrossRef.
- E. Thiran, et al., Biofilm formation of Staphylococcus aureus dairy isolates representing different genotypes, J. Dairy Sci., 2018, 101, 1000–1012 CrossRef CAS.
- S. Notcovich, et al., Biofilm-Forming Potential of Staphylococcus aureus Isolated from Clinical Mastitis Cases in New Zealand, Vet. Sci., 2018, 5, pii: E8 CrossRef.
- B. Gibson, et al., The distribution of bacterial doubling times in the wild, Proc. Biol. Sci., 2018, 285, 20180789 Search PubMed.
- S. Hogan, et al., Potential use of targeted enzymatic agents in the treatment of Staphylococcus aureus biofilm-related infections, J. Hosp. Infect., 2017, 96, 177–182 CrossRef CAS.
- C. B. Waryah, et al., In Vitro antimicrobial efficacy of tobramycin against Staphylococcus aureus biofilms in combination with or without DNase I and/or dispersin B: A preliminary investigation, Microb. Drug Resist., 2017, 23, 384–390 CrossRef CAS.
- K. Belfield, et al., Evaluation of combinations of putative anti-biofilm agents and antibiotics to eradicate biofilms of Staphylococcus aureus and Pseudomonas aeruginosa, J. Antimicrob. Chemother., 2017, 72, 2531–2538 CrossRef CAS.
- J. Wang, et al., Silver-nanoparticles-modified biomaterial surface resistant to staphylococcus: new insight into the antimicrobial action of silver, Sci. Rep., 2016, 6, 32699 CrossRef CAS.
- R. Thomas, et al., Microbially and phytofabricated AgNPs with different mode of bactericidal action were identified to have comparable potential for surface fabrication of central venous catheters to combat Staphylococcus aureus biofilm, J. Photochem. Photobiol., B, 2017, 171, 96–103 CrossRef CAS.
- S. Gomez-Carretero, et al., Electro enhanced Antimicrobial Coating Based on Conjugated Polymers with Covalently Coupled Silver Nanoparticles Prevents Staphylococcus aureus Biofilm Formation, Adv. Healthcare Mater., 2017, 6, 1700435 CrossRef.
- M. Qasim, et al., Antimicrobial activity of silver nanoparticles encapsulated in poly-N-isopropylacrylamide-based polymeric nanoparticles, Int. J. Nanomed., 2018, 13, 235–249 CrossRef CAS.
- S. Qayyum, et al., Obliteration of bacterial growth and biofilm through ROS generation by facilely synthesized green silver nanoparticles, PLoS One, 2017, 12, e0181363 CrossRef.
- E. Pazos-Ortiz, et al., Dose-Dependent Antimicrobial Activity of Silver Nanoparticles on Polycaprolactone Fibers against Gram-Positive and Gram-Negative Bacteria, J. Nanomater., 2017, 4752314 Search PubMed.
- D. Gutiérrez, et al., Bacteriophages as Weapons Against Bacterial Biofilms in the Food Industry, Front. Microbiol., 2016, 7, 825 CrossRef.
- S. Manoharadas, et al., Antimicrobial activity of a chimeric enzybiotic towards Staphylococcus aureus, J. Biotechnol., 2009, 139, 118–123 CrossRef CAS.
- D. Gutiérrez, et al., Effective removal of staphylococcal biofilms by the endolysin LysH5, PLoS One, 2014, 9, e107307 CrossRef.
- H. Haddad Kashani, et al., Recombinant Endolysins as Potential Therapeutics against Antibiotic-Resistant Staphylococcus aureus: Current Status of Research and Novel Delivery Strategies, Clin. Microbiol. Rev., 2017, 31, e00071 CrossRef.
- D. Gutiérrez, et al., Are Phage Lytic Proteins the Secret Weapon to Kill Staphylococcus aureus?, mBio, 2018, 9, e01923 CrossRef.
- D. P. Pires, et al., A Genotypic Analysis of Five P. aeruginosa Strains after Biofilm Infection by Phages Targeting Different Cell Surface Receptors, Front. Microbiol., 2017, 8, 1229 CrossRef.
- M. Simmons, et al., Phage mobility is a core determinant of phage-bacteria coexistence in biofilms, ISME J., 2018, 12, 531–543 CrossRef.
- D. R. Harper, et al., Bacteriophages and Biofilms, Antibiotics, 2014, 3, 270–284 CrossRef.
- D. P. Pires, et al., Bacteriophage-encoded depolymerases: their diversity and biotechnological applications, Appl. Microbiol. Biotechnol., 2016, 100, 2141–2151 CrossRef CAS.
- T. K. Lu and J. J. Collins, Dispersing biofilms with engineered enzymatic bacteriophage, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 11197–11202 CrossRef CAS.
- A. S. DeFrancesco, et al., Genome-wide screen for genes involved in eDNA release during biofilm formation by Staphylococcus aureus, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E5969–E5978 CrossRef CAS.
- B. Zatorska, et al., Does Extracellular DNA Production Vary in Staphylococcal Biofilms Isolated from Infected Implants versus Controls?, Clin. Orthop. Relat. Res., 2017, 475, 2105–2113 CrossRef.
- K. S. Butler, et al., Silver nanoparticles: correlating nanoparticle size and cellular uptake with genotoxicity, Mutagenesis, 2015, 30, 577–591 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra09725j |
| This journal is © The Royal Society of Chemistry 2021 |