Open Access Article
Open Access ArticleGreen synthesis of silver nanoparticles using plant extracts and their antimicrobial activities: a review of recent literature
Chhangte Vanlalvenia,
Samuel Lallianrawnab,
Ayushi Biswasc,
Manickam Selvaraj d,
Bishwajit Changmai*c and
Samuel Lalthazuala Rokhum
d,
Bishwajit Changmai*c and
Samuel Lalthazuala Rokhum *ce
*ce
aDepartment of Botany, Mizoram University, Tanhril, Aizawl, Mizoram 796001, India
bDepartment of Chemistry, Govt. Zirtiri Residential Science College, Aizawl, 796001, Mizoram, India
cDepartment of Chemistry, National Institute of Technology Silchar, Silchar, 788010, India. E-mail: rokhum@che.nits.ac.in; lr512@cam.ac.uk; bishwajit_rs@che.nits.ac.in
dDepartment of Chemistry, Faculty of Science, King Khalid University, Abha 61413, Saudi Arabia
eDepartment of Chemistry, University of Cambridge, Lensfield Road, Cambridge CB2 1EW, UK
First published on 13th January 2021
Abstract
Synthesis of metal nanoparticles using plant extracts is one of the most simple, convenient, economical, and environmentally friendly methods that mitigate the involvement of toxic chemicals. Hence, in recent years, several eco-friendly processes for the rapid synthesis of silver nanoparticles have been reported using aqueous extracts of plant parts such as the leaf, bark, roots, etc. This review summarizes and elaborates the new findings in this research domain of the green synthesis of silver nanoparticles (AgNPs) using different plant extracts and their potential applications as antimicrobial agents covering the literature since 2015. While highlighting the recently used different plants for the synthesis of highly efficient antimicrobial green AgNPs, we aim to provide a systematic in-depth discussion on the possible influence of the phytochemicals and their concentrations in the plants extracts, extraction solvent, and extraction temperature, as well as reaction temperature, pH, reaction time, and concentration of precursor on the size, shape and stability of the produced AgNPs. Exhaustive details of the plausible mechanism of the interaction of AgNPs with the cell wall of microbes, leading to cell death, and high antimicrobial activities have also been elaborated. The shape and size-dependent antimicrobial activities of the biogenic AgNPs and the enhanced antimicrobial activities by synergetic interaction of AgNPs with known commercial antibiotic drugs have also been comprehensively detailed.
1. Introduction
Nanotechnology is gaining enormous attention as a new area of research dealing with the development of nanomaterials and nanoparticles (NPs) for their utilization in diverse fields such as catalysis, electrochemistry, biomedicines, pharmaceuticals, sensors, food technology, cosmetics, etc.1–3 Nanoparticles (NPs) are nanometer-sized (<100 nm) atomic or molecular scale solid particles having some excellent physical properties compared to the bulk molecules depending on their size and morphology.4,5 Among all types of NPs, metal and metal oxide nanoparticles have been thoroughly examined using science and technology due to their excellent properties such as high surface to volume ratio, high dispersion in solution, etc.6,7 Owing to these, metal and metal oxide nanoparticles display enhanced antimicrobial properties.8,9Currently, modified or fabricated of NPs is widely utilized in industrially manufactured items e.g., cosmetics, electronics, and textiles. Furthermore, the rapid increased in the number of microbes resistant to existing antibiotic drugs that has led to the requirement of novel medicines in the form of bare NPs or in conjunction with existing antibiotics to exert a favourable synergistic effect resulted in the wide spread use of NPs in several medical fields.10,11 Nowadays, NPs have been utilized for molecular imaging to achieve profoundly resolved pictures for diagnosis. In addition, contrast agents are impregnated onto NPs for the tumour and atherosclerosis diagnosis.12–14 Furthermore, nanotherapeutic has been promoted everywhere throughout the world after the first FDA affirmed nanotherapeutic in 1990, to build up different nano-based drugs.15
At the beginning of 20th century, various physical and chemical methodologies such as chemical reduction, milling etc., have been utilized for the synthesis of NPs synthesis as well as to enhance its efficiency.16 However, these conventional techniques involve costly and toxic chemicals and cannot be considered an environmentally benign process.17 Taking into account, nowadays researchers showed great interest on the synthesis of metal and metal oxides NPs employing bio-genic route, that utilized aqueous plant extract and microbes, as they are environment-friendly, stable, clinically adaptable, bio-compatible and cost-effective.16,18 Therefore, bio-inspired technology for NPs synthesis became a significant branch in the field of nanoscience and nanotechnology.19,20 Till now, numerous metal and metal oxide NPs have been synthesized using plant extract and microbes etc.21,22 Owing to their wide availability, renewability and environment-friendly nature, in addition to their vast applications in the synthesis of NPs, plant biomass are also largely targeted by our group and others as a catalyst for chemical synthesis23,24 and biodiesel productions.25,26
Among metal NPs, silver NPs is gaining enormous interest in the research community due to their wide scope of application in microbiology, chemistry, food technology, cell biology, pharmacology and parasitology.27,28 The morphology of the silver NPs is the deciding factor of their physical and chemical properties.28 Basically, several techniques such as sol–gel method, hydrothermal method, chemical vapour deposition, thermal decomposition, microwave-assisted combustion method etc., have been utilized for the synthesis of silver NPs.29–31 Recently, bio-genic synthesis of silver NPs (AgNPs) using biomaterials such as plant extract and microbes as reducing agent and their antimicrobial activity is widely investigated.32,33 AgNPs are produced by oxidation of Ag+ to Ag0 by different biomolecules such as flavonoids, ketones, aldehydes, tannins, carboxylic acids, phenolic and the protein of the plant extracts.
UV-visible spectroscopy is a simple and widely used analytical technique to monitor the formation of AgNPs. Upon interaction with an electromagnetic field, the conducting electrons present in the outermost orbital of metal NPs collectively oscillate in resonance with certain wavelengths to exhibit a phenomenon called surface plasmon resonance (SPR). The excitation of SPR is responsible for the formation of color and absorbance in a colloidal solution of AgNPs. The SPR peaks at around 435 nm are usually taken to confirm the reduction of silver nitrate into AgNPs.34 In general, spherical NPs exhibit only a single SPR band in the absorbance spectra, whereas two or more SPR bands were observed for anisotropic particles depending on the shape.35 The absence of peak in the region 335 and 560 nm in UV-Vis spectra are sometime used as an indication of the absence of aggregation in NPs.32,36
Statistical data analysis in Fig. 1 depicted the increasing trend of published research papers in the field of biogenic synthesis of AgNPs. These data were collected in September 2020 from “SciFinder Database” using the keyword “Green synthesis of silver nanoparticles”. From a meagre 259 publications in the year 2001, it has exponentially increased to 3374 publications in 2019. Thus, in this review, an attempt has been made to inspire the researchers to explore the natural resources to synthesize silver nanoparticles by diverse plants and their organs to interconnect nanotechnology with biotechnology into one, termed as nanobiotechnology. This review will also unlock ideas to utilize different paths for the production of silver nanoparticles, which can help human beings. We have comprehensively discussed the bio-genic synthesis and silver nanoparticles using various plants and their application in antimicrobial activity. We also discussed the effect of the synthesized silver nanoparticles' size and shape in antimicrobial activity towards various pathogenic bacteria. In an attempt to synthesize metals NPs one has to bear in mind that the success of NPs depends not only on the size and shape but also on stability of NPs as they have the tendency to form large aggregates that lead to precipitation, thereby reducing their efficacy.
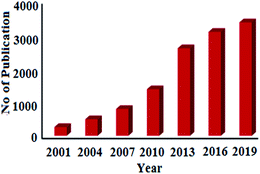 | ||
| Fig. 1 Publications per year for green synthesis of AgNPs during the period 2001 to 2019 (data collected from SciFinder Database). | ||
2. Protocols for the biosynthesis of AgNPs
Biogenic synthesis of AgNPs is an easy single-step protocol without generating harsh and toxic chemicals; hence, they are save, economical and eco-friendly. In recent years, both plant and microbes are extensively investigated for the biosynthesis of AgNPs of varying size, shape, stability, and antimicrobial efficacy.2.1 From plant extract
Various parts of plant such as leaves, roots, flowers, fruits, rhizomes etc., have been successfully utilized for the synthesis of AgNPs.37–39 Different parts of plant are collected from various sources, washed properly with ordinary water followed by distilled water to exclude debris and any other unwanted materials. After that, the portions are dried and ground to make powder or utilized as fresh to make the extract. To prepare the extract, the chopped pieces or the ground powder of the parts of the plant are put in deionized water or alcohol and usually heated below 60 °C for few hours as high-temperature heating long time may leads to the decomposition of phytochemicals in the biomass extract. Plant extract of different pH is added to the solutions having a different concentration of Ag salt as metal precursor followed by heating at different temperature led to the synthesis of AgNPs.40–42 This synthesis process avoids the use of chemical stabilizer as biomaterials present in the extract act as a reducing agent as well as a stabilizing agent for the synthesis of AgNPs.43,44 The progress of the formation of AgNPs can be monitored by visual color changes or using UV-Vis. Spectroscopy, where a sharp peak due to surface plasmon resonance (SPR) of AgNPs at around 430–450 nm is clearly observed.34 After successful synthesis of the AgNPs, the mixture is centrifuged at high rpm to separate the NPs followed by proper washing using solvents and dried in an oven at low temperature.45,46 The different plant parts extracts that have been successfully utilized in the green synthesis of AgNPs are given in Fig. 2.2.2 From microbes
Nowadays, the use of microbial cell for the synthesis of metal NPs has come out as a great approach. Microbial cells turn to be excellent biofactories for the synthesis of AgNPs.47,48 At first, the cultures are allowed to develop as culture suspension in disinfected distilled water having the culture medium. Then, different concentration of precursor of AgNPs is added into the cultured microbial followed by continuous mechanical stirring under dark conditions. The progress of the reaction is monitored by UV-Vis spectrophotometer. Finally, the resultant AgNPs is separated from the mixture via centrifugation at around 3000 rpm for 10–15 min.493. Plant-mediated biogenic synthesis of AgNPs and their antimicrobial activity
Owing to the environmental issue, biogenic synthesis of metal and metal oxide NPs is gaining immense attention from the past decades. Reported literature revealed that various plant parts such as leaf, roots, seed, fruits and stem etc., have been utilized for the biosynthesis of NPs. The synthesis of NPs is fully dependent on the biomaterials/phytochemicals present in the extract. This section aims to discuss the various plant parts extract mediated synthesis of AgNPs and their application as antimicrobial.3.1 From leaf
To date, a numerous number of leaves extract have been utilized for the biosynthesis of AgNPs as shown in Table 1. Skimmia laureola was reported for the synthesis of spherical AgNPs with size 38 ± 0.27 nm and tested against E. coli, K. pneumoniae, P. aeruginosa, P. vulgaris, S. aureus.50 Miri et al.51 have utilized Prosopis farcta extract for the biosynthesis of AgNPs with an average size of 10.8 nm at room temperature (RT). The antimicrobial activity of synthesized AgNPs was tested using disc diffusion method against the Gram-positive (Staphylococcus aureus (PTCC 1431), Bacillus subtilis (PTCC 1420)) and Gram-negative bacteria (Escherichia coli (PTCC 1399), Pseudomonas aeruginosa (PTCC 1074)) and compared with the control. The results showed that the inhibition diameter is increased for every tested pathogen (Fig. 3), indicates that synthesized AgNPs induces cellular damage to the bacteria's, hence can be used as nanoantibiotics. Aloe vera,52 Eclipta alba,53 Momordica charantia,54 Leptadenia reticulata55 are also used for the production of spherical biogenic AgNPs. In another study, AgNPs were synthesized by using tea leaf extract. Bactericidal activity of the synthesized NPs was tested against S. aureus and E. coli showed that inhibition action is more effective in case of S. aureus (89% inhibition rate) compared to E. coli (75% inhibition rate). In addition, treatment of the NPs against the bacteria leads to impairment of bacterial cell–cell adhesion.56 Mukia maderaspatana leave extract was utilized for the biosynthesis of AgNPs with the size range of 58–458 nm. The synthesized nanoparticle was conjugated to the antibiotic ceftriaxone to investigate the antimicrobial activity towards the human pathogens such as B. subtilis, K. pneumonia, S. typhi, S. aureus and compared with the pathogen inhibition efficiency of the free nanoparticle and the antibiotic. The result obtained revealed that the AgNPs conjugated with ceftriaxone showed highest inhibition activity compared to the others.57| No. | Plants | Size and shape | Test microorganisms | Ref. |
|---|---|---|---|---|
| 1 | Skimmia laureola | Spherical; 38 ± 0.27 nm | E. coli, K. pneumoniae, P. aeruginosa, P. vulgaris, S. aureus | 50 |
| 2 | Prosopis farcta | Spherical; 8–11 nm | S. aureus, B. subtilis, E. coli, P. aeruginosa | 51 |
| 3 | Aloe vera | Spherical; 70 nm | Aspergillus sp., Rhizopus sp. | 52 |
| 4 | Eclipta alba | 310 to 400 nm | E. coli, S. aureus, P. aeruginosa | 53 |
| 5 | Momordica charantia | Spherical; 11–16 nm | B. spp., S. spp., P. spp., E. coli, A. niger subsp., A. flavus subsp., P. spp. | 54 |
| 6 | Leptadenia reticulata | Spherical; 50–70 nm | S. pneumonia, K. pneumonia | 55 |
| 7 | Tea leaf | Spherical; 20 nm | S. aureus, E. coli | 56 |
| 8 | Raphanus sativus | Spherical; 6–38 nm | A. fumigatus, C. specifier, F. solani | 40 |
| 9 | Mukia maderaspatana | Spherical; 58–458 nm | B. subtilis, K. pneumonia, S. typhi, S. aureus | 57 |
| 11 | Clitoria ternatea | Spherical; 20 nm | B. subtilis, S. aureus, S. pyogenes, E. coli, P. aeruginosa, K. aerogenes | 58 |
| 12 | Solanum nigrum | Spherical; 28 nm | B. subtilis, S. aureus, S. pyogenes, E. coli, P. aeruginosa, K. aerogenes | 58 |
| 13 | Croton sparsiflorus morong | Spherical; 22–52 nm | S. aureus, E. coli, B. subtilis | 46 |
| 14 | Grewia flaviscences | Spherical; 60 nm | Bacillus, P. aeruginosa | 59 |
| 15 | Terminalia arjuna | Spherical; 8–16 nm | S. aureus, E. coli | 21 |
| 16 | Prunus yedoensis | Spherical, oval; 18–20 nm | P. acnes, S. epidermidis (skin bacteria) | 60 |
| 17 | Justicia adhatoda L. | Spherical; 5–50 nm | P. aeruginosa | 61 |
| 18 | Withania somnifera | 70–110 nm; spherical | S. aureus, P. aeruginosa, C. albicans, P. vulgaris, E. coli, A. tumefaciens | 62 |
| 19 | Pistacia atlantica | Spherical; 10–50 nm | S. aureus | 63 |
| 20 | Tectona grandis Linn | Spherical; 26–28 nm | E. coli and S. aureus | 64 |
| 21 | Ficus virens | Spherical; 4.98–29 nm | B. subtilis, S. epidermidis, E. faecalis, K. pneumoniae, V. cholerae, V. vulnificus | 65 |
| 22 | Azadirachta indica | Spherical; 250–700 nm | E. coli | 66 |
| 23 | Artocarpus altilis | Spherical; 20–50 nm | E. coli, P. aeruginosa, S. aureus, A. versicolor | 67 |
| 24 | Crotalaria retusa | Spherical; 80 nm | E. coli and S. aureus | 68 |
| 25 | Cardiospermum halicacabum | Spherical; 74 nm | P. vulgaris, P. aeruginosa, S. aureus, B. subtilis, S. paratyphi, A. solani, F. oxysporum | 69 |
| 26 | Psidium guajava | Spherical; 10–90 nm | P. aeruginosa | 70 |
| 27 | Cassia fistula | Spherical; 39.5 nm | B. subtilis, S. aureus, E. coli, P. aeruginosa, C. albicans, C. kruseii, C. viswanathii, T. mentagrophytes | 71 |
| 28 | Terminalia chebula | Spherical; 10–30 nm | E. coli, B. subtilis | 72 |
| 29 | Pedalium murex | Spherical; 20–50 nm | E. coli, K. pneumonia, M. flavus, P. aeruginosa, B. subtilis, B. pumilus and S. aureus | 73 |
| 30 | Azadirachta indica | Spherical; 34 nm | E. coli, S. aureus | 74 |
| 31 | Croton bonplandianum | Spherical; 15–40 nm | P. aeruginosa, E. coli, S. aureus | 75 |
| 32 | Tamarix gallica | Spherical; 5–40 nm | E. coli | 76 |
| 33 | Urtica dioica | Spherical; 20–30 nm | B. cereus, B. subtilis, S. aureus and S. epidermidis, E. coli, K. pneumoniae, S. marcescens, S. typhimurium | 77 |
| 34 | Ziziphus oenoplia | Spherical; 10 nm | P. aeruginosa, K. pneumoniae, E. coli, S. typhi | 78 |
| 35 | Lawsonia inermis | Spherical; 25 nm | E. coli, Pseudomonas spp., Bacillus spp., Staphylococcus spp., A. niger, A. flavus, Penicillium spp. | 79 |
| 36 | Lantana camara | Spherical; 20–200 nm | S. aureus, E. coli, P. aeruginosa, K. pneumonia | 81 |
| 37 | Jatropha curcas | Spherical; 20–50 nm | E. coli, P. aeruginosa, B. cereus, S. enterica, L. monocytogenes, S. aureus | 82 |
| 38 | Salvinia molesta | Spherical; 10 nm | S. aureus, E. coli | 83 |
| 39 | Sesbania grandiflora | Spherical; 20 nm | E. coli, Pseudomonas spp., Bacillus spp., Staphylococcus spp., A. niger subsp., A. flavus subsp., Penicillium spp. | 84 |
| 40 | Indoneesiella echioides | Spherical; 29 nm | R. rhodochrous, A. hydrophila, S. aureus, Pseudomonas aeruginosa, C. albicans | 85 |
| 41 | Phlomis | Spherical; 25 nm | S. aureus, B. cereus, S. typhimurium, E. coli | 86 |
| 42 | Hydrocotyle rotundifolia | Spherical; 7.39 nm | E. coli | 87 |
| 43 | Maclura pomifera | Spherical; 6–16 nm | S. aureus, Bacillus cereus, E. coli, P. aeruginosa, A. niger, C. albicans | 88 |
| 44 | Paederia foetida Linn. | Spherical; 5–25 nm | B. cereus, S. aureus, E. coli, A. niger | 89 |
| 45 | Atalantia monophylla | Spherical; 35 nm | B. subtilis, B. cereus, S. aureus, E. coli, P. aeruginosa, K. pneumoniae, C. albicans, A. niger | 90 |
| 46 | Talinum triangulare | Spherical; 13.86 nm | E. coli, S. typhi, B. subtilis, S. aureus, C. albicans | 91 |
| 47 | Ricinus communis | Spherical; 8.96 nm | S. aureus, P. aeruginosa | 92 |
| 48 | Erythrina suberosa | Spherical; 15–34 nm | S. aureus, P. aeruginosa, C. kruseii, T. mentagrophytes | 93 |
| 49 | Lippia citriodora | Spherical; 10–45 nm | S. aureus, B. subtilis, S. typhi, E. coli, C. albicans | 94 |
| 50 | Brassica oleracea L. | Spherical; 30–100 nm | S. aureus, E. coli, C. albicans | 95 |
| 51 | Catharanthus roseus | Spherical; 10–88 nm | E. coli, C. koseri, K. pneumonia, P. aeruginosa, and S. aureus | 96 |
| 52 | Lavandula x intermedia | Spherical; 11–47 nm | E. coli, P. aeruginosa, P. mirabilis, B. cereus, K. oxytoca, S. typhi, S. aureus, C. albicans, A. niger, F. oxysporum | 98 |
| 53 | Canna edulis | Spherical; less than 40 nm | B. cereus, S. aureus, E. coli, S. typhimurium, E faecalis, C. tropicalis, C. kruseii, C. lusitaniae, C. guilliemondii, P. chrysogenum | 99 |
| 54 | Artemisia vulgaris | Spherical; 27–53 nm | E. coli, S. aurous, P. aeruginosa, K. pneumoniae, H. influenza | 100 |
| 55 | Psidium guajava | Spherical; 25 nm | B. aryabhattai, B. megaterium, B. subtilis, A. creatinolyticus, E. coli, Alcaligenes faecalis, S. cerevisiae, A. niger, R. oryzae | 102 |
| 56 | Taraxacum officinale | Spherical; 5–30 nm | X. axonopodis, P. syringae | 104 |
| 57 | Petiveria alliacea L. | Spherical; 16.7–33.74 nm | E. coli, K. pneumoniae, S. aureus | 105 |
| 58 | Nervalia zeylanica | Spherical; 34.2 nm | S. aureus, L. brevis, P. putida, Pseudomonas sp., P. chrysogenum, P. citrinum | 106 |
| 59 | Ficus ingens | Spherical; 81.37 nm | E. coli, S. typhi, B. cereus | 107 |
| 60 | Thymbra spicata | Spherical; 70.2 nm | B. cereus, S. aureus, E. coli, S. typhimurium | 108 |
| 61 | Indigofera tinctoria | Spherical; 9–26 nm | B. pumilis, S. aureus, Pseudomonas sp., E. coli, A. fumigatus, A. niger | 110 |
| 62 | Tecoma stans | Spherical; 2–40 nm | B. subtilis, S. aureus, K. pneumoniae | 114 |
| 63 | Salvia leriifolia | Spherical; 27 nm | P. aeruginosa, E. coli, S. coagulase, C. frurdii, E. aerogenes, A. baumannii, S. marcescens, K. pneumonia, S. pneumoniae | 115 |
| 64 | Leucaena leucocephala L. | Spherical; 25–50 nm | P. aeruginosa, S. pyogenes, S. aureus, E. coli, S. typhi, B. subtilis | 116 |
| 65 | Selaginella bryopteris | Spherical; 5–10 nm | S. aureus, E. coli, A. niger | 117 |
| 66 | Galega officinalis | Spherical; 27.12 nm | E. coli, P. syringae, S. aureus | 118 |
| 67 | Camellia sinensis | Spherical; 30 nm | S. aureus, K. pneumoniae | 119 |
| 68 | Justicia spicigera | Spherical; 86–100 nm | B. cereus, K. pneumoniae, and E. aerogenes, M. phaseolina, A. alternate, Colletotrichum sp., F. solani | 120 |
| 69 | Kleinia grandiflora | Spherical; 20–50 nm | P. aeruginosa, C. albicans | 121 |
| 70 | Eucalyptus citriodora | Spherical; 17.51 nm | C. albicans, A. baumannii, E. coli, K. pneumoniae, P. aeruginosa | 122 |
| 71 | Juniperus procera | Spherical and cubic; 30–90 nm | M. luteus, B. subtilis, P. mirabilis, K. pneumoniae, C. albicans | 123 |
| 72 | Capparis zeylanica | Spherical; 23 nm | S. epidermis, E. faecalis, S. paratyphi, S. dysenteriae, C. albicans, A. niger | 124 |
| 73 | Caesalpinia pulcherrima | Spherical; 9 nm | B. cereus, B. subtilis, S. aureus, C. rubrum, E. coli, P. aeruginosa, S. typhimurium, K. pneumoniae, C. albicans, C. glabrata, C. neoformans | 126 |
| 74 | Ligustrum lucidum | Spherical; 13 nm | S. turcica | 127 |
| 75 | Aesculus hippocastanum | Spherical; 50 ± 5 nm | S. aureus, S. epidermidis, L. monocytogenes, C. renale, M. luteus, B. subtilis, B. cereus, E. faecalis, P. aeruginosa, P. fluorescens, E. coli, E. aerogenes, K. pneumonia, P. mirabilis, C. albicans, C. tropicalis, C. krusei | 128 |
| 76 | Melaleuca alternifolia | Spherical; 11.56 nm | S. aureus, methicillin-resistant Staphylococcus aureus, S. epidermidis, S. pyogenes, K. pneumoniae, P. aeruginosa, T. mentagrophytes, C. albicans | 129 |
| 77 | Carya illinoinensis | Spherical; 12–30 nm | E. coli, P. aeruginosa, S. aureus, L. monocytogenes | 130 |
| 78 | Murraya koenigii | Spherical;; 35–80 nm | E. coli, P. aeruginosa, E. faecalis, C. albicans | 131 |
| 79 | Clerodendrum inerme | Spherical; 5.54 nm | B. subtilis, S. aureus, Klebsiella, E. coli, A. niger, T. harzianum, A. flavus | 132 |
| 80 | Aspilia pluriseta | Spherical; 1–20 nm | B. subtilis, S. aureus, E. coli, P. aeruginosa, C. albicans | 133 |
| 81 | Melia azedarach | Spherical; 18 to 30 nm | V. dahliae | 134 |
| 82 | Scoparia dulcis | Spherical; 3–18 nm | P. aeruginosa, E. coli, B. subtilis, S. aureus, A. niger, C. albicans | 135 |
| 83 | Lantana trifolia | Spherical; 5 and 70 nm | E. coli, P. aeruginosa, C. albicans, S. aureus, B. subtilis | 136 |
| 84 | Mikania micrantha | Spherical; 10–20 nm | B. subtilis, E. coli, P. aeruginosa, S. pneumonia | 137 |
| 85 | Solanum nigrum | Spherical; 3.46 nm | E. coli | 138 |
| 86 | Curcuma longa L. | Spherical; 15–40 nm | S. aureus, P. aeruginosa, S. pyogenes, E. coli, C. albicans | 139 |
| 87 | Syzygium cumini | Spherical; 11–19 nm | S. aureus, A. flavus, A. parasiticus | 140 |
| 88 | Cleistanthus collinus | Spherical however not mentioned in manuscript; 30 to 50 nm | S. sonnei, P. aeruginosa, S. aureus, B. subtilis, S. dysenteriae, V. cholerae, P. mirabilis | 142 |
| 89 | Cestrum nocturnum | Spherical; 20 nm | Citrobacter, E. faecalis, S. typhi, E. coli, P. vulgaris and V. cholerae | 143 |
| 90 | Rice | Spherical; 16.5 nm | R. solani | 144 |
| 91 | Mentha aquatica | Spherical; 8 nm | P. aeruginosa, E. coli, B. cereus, and S. aureus | 146 |
| 92 | Rosmarinus officinalis | Sphere; 29 nm | S. aureus, B. subtilis, E. coli, P. aeruginosa | 147 |
| 93 | Ceropegia thwaitesii | Sphere; 100 nm | S. typhi, B. subtilis, S. aureus, S. epidermis, V. cholerae, S. epidermidis, K. pneumonia, M. luteus, P. mirabilis, P. aeruginosa, S. flexneri | 148 |
| 94 | Ziziphus jujuba | Irregular; 20–30 nm | E. coli | 149 |
| 95 | Ocimum tenuiflorum, Solanum trilobatum, Syzygium cumini, Centella asiatica and Citrus sinensis | Irregular; 28 nm, 26.5 nm, 65 nm, 22.3 nm and 28.4 nm | S. aureus, P. aeruginosa, E. coli, K. pneumoniae | 154 |
| 96 | Amaranthus gangeticus Linn | Globular-shaped; 11–15 nm | S. flexneri, B. subtilis, Sclerotinia sp. | 155 |
| 97 | Andrographis paniculata | Cubic; 40 and 60 nm | P. aeruginosa, E. coli, V. cholerae, S. flexneri, B. subtilis, S. aureus, M. luteus | 156 |
| 98 | Andrographis echioides | Cubic, pentagonal, hexagonal; 68.06–91.28 nm; | E. coli, S. aureus, S. typhimurium, M. luteus, P. aeruginosa | 158 |
| 99 | Azadirachta indica (neem) | Polydispersed; less than 40 nm | P. nitroreducens, A. unguis | 159 |
| 100 | Phyllanthus amarus | Flower-liked; 30 nm to 42 nm | E. coli, P. spp., B. spp., S. spp., A. niger, A. flavus, P. spp. | 160 |
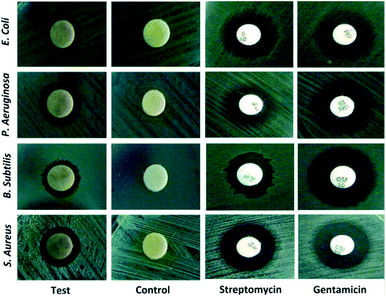 | ||
| Fig. 3 Bactericidal activity of Prosopis farcta extract mediated Ag-NPs against human pathogens. This figure has been reproduced from ref. 51 with permission from Elsevier, copyright 2015. | ||
Clitoria ternatea and Solanum nigrum58 were also reported to synthesize very small-size AgNPs and evaluated against B. subtilis, S. aureus, S. pyogenes, E. coli, P. aeruginosa, K. aerogenes. Interestingly, among the two leaf Clitoria ternatea extract gave smaller nanoparticles, which indicated the important role of extract constituents on the size of the produced nanoparticles. In addition, AgNPs of Clitoria ternatea showed higher activity than the AgNPs of Solanum nigrum against nosocomial pathogens due to its small size. It has been well-documented in literature that smaller size NPs showed higher antimicrobial activities due to larger surface area.56 Grewia flaviscences,59 Prunus yedoensis,60 Justicia adhatoda L,61 Withania somnifera62 produced AgNPs in the range 8–100 nm which mainly are spherical. Numerous microbes such as skin bacteria are responsible for skin infection and body odor, as well as odor in feet, shoes, and/or socks mediated through the breakdown of amino acids present in sweat. Hence proper medication is required for human's wellbeing. Velmurugan et al.60 applied the synthesized AgNPs from Prunus yedoensis to treat P. acnes, S. epidermidis, a well-known skin bacteria, and found that the synthesized NPs are more effective against skin bacteria than commercial AgNPs. The biogenic AgNPs showed 18 mm ZOI (zone of inhibition) in 30 μg scale against P. acnes, whereas commercial AgNPs displayed a lower ZOI of only 12 in the same concentration.
Pistacia atlantica,63 Tectona grandis Linn,64 Ficus virens65 also reported for the synthesis of AgNPs and are evaluated against several microbes. Verma et al.66 reported Azadirachta indica (neem) leaf inspired synthesis of AgNPs and evaluated the effects of pH of the solution on the formation of nanoparticles as change in pH affects the shape and size of the particles by altering the charge of biomolecules, which might affect their capping as well as stabilizing abilities. They have observed that as the pH increases from 9 to 13, the absorption maximum shifts from 383 to 415 nm in the UV-spectrum and detects an increase in absorption intensity with increasing pH. This showed that pH 13 is the most favourable pH for the synthesis of AgNPs leaf extract. The shift in the peak wavelength indicates that the size of the particles increases with increasing pH of the solution. As the particles' diameter gets larger, the energy required for excitation of surface plasmon electrons decreases, as a result the absorption maximum shifted towards the longer wavelength region. Moreover, it was observed that at acidic pH i.e. pH < 7, the formation of nanoparticles is suppressed. At high pH, the bioavailability of functional groups in Azadirachta indica leaf extract promoted the synthesis of nanoparticles. However, at very high pH i.e. pH ∼13, the particles became unstable and agglomerated, when kept for overnight.
AgNPs were also recently synthesized using several leaf extract of plants such as Artocarpus altilis,67 Crotalaria retusa,68 Cardiospermum halicacabum,69 Psidium guajava,70 Cassia fistula71 and Terminalia chebula.72 In 2016, Anandalakshmi et al.73 reported Pedalium murex leaf extract mediated AgNPs. The produced NPs were tested against several microbes and displayed highest ZOI of 10.5 mm (in 15 μL mL−1 scale) against E. coli and P. aeruginosa and least activity against Klebsiella pneumoniae (8.5 mm). The shape and size of the resultant AgNPs were elucidated with the help of TEM. The TEM micrographs showed that the sizes of the particles were around 50 nm and were predominantly spherical in shape. The PXRD pattern showed fcc crystal structure. Azadirachta indica promoted synthesis of AgNPs was reported by Ahmed et al.74 The produced NPs displayed equal efficacy (9 mm ZOI) against E. coli, S. aureus whereas the plant extract show no antimicrobial activity.
Croton bonplandianum mediated AgNPs were also found to be highy active against microbes.75 The minimum inhibitory concentrations of synthesized AgNPs were found to be 50, 45, 75 g mL−1 in case of E. coli, P. aeruginosa, and S. aureus respectively. It was concluded that Gram-negative strains of bacteria with thin cell wall such as E. coli and P. aeruginosa are more susceptible to cell wall damage compared to Gram-positive strain bacteria with a thick cell wall (S. aureus). In another work, Tamarix gallica leaf extract was used for synthesis of AgNPs. To test its activity against E. coli, three sterile filter paper discs (5 mm diameter) were impregnated with 6 μL of AgNPs produced with 5 mL of Tamarix gallica extract and 10 mL of 5 mM AgNO3 solution, López-Miranda et al. studied the green synthesis of AgNPs using and evaluated the effect of extract and AgNO3 concentration on the synthesis.76 They have observed an increase in the intensity of surface plasmon resonance (SPR) with the increase in extract concentration, which is attributed to an increasing number of AgNPs formed. Also, as the AgNO3 concentration increases, many silver ions are increasingly reduced to AgNPs. However, they have seen that the SPR band intensities are nearly independent for 5, 7, and 9 mM AgNO3, which reflected that the reaction is close to an equilibrium system because the reducing compounds and stabilizers from the extract are completely consumed, hence it is impossible to reduce a larger amount of silver ions. Henceforth, from the UV-vis analysis they concluded that the best results were obtained for the sample 0.15 g mL−1 extract with 5 mM AgNO3. The produced showed 9 mm ZOI against E. coli. Similarly, leaf extract of Urtica dioica,77 Ziziphus oenoplia78 and Lawsonia inermis79 are reported for the production of AgNPs with high antimicrobial activities. In 2016, a remarkable work on the synthesis of AgNPs using Urtica dioica leaf extract that showed excellent synergistic effect with known antimicrobial drugs was reported by Jyoti et al.77 Interestingly, the synthesized AgNPs apart from showing high antimicrobial activities against several microbes, showed excellent synergistic effect in combination with antibiotics and displayed higher antibacterial effect as compared with AgNPs alone. A high 17.8 fold increase in ZOI was observed for amoxicillin with AgNPs against S. marcescens proving the synergistic role of AgNPs.77 This work provides helpful insight into the development of new antibacterial agents to fight against several new stain of microbes resistant to existing antibiotic drugs. Fig. 4 displayed the synergistic effect of AgNPs and common antimicrobial drugs. The synergistic interaction between AgNPs and antibiotic drugs has been clearly identified using UV-Vis and Raman spectrometer by McShan et al.80 The authors claimed that this synergistic interaction speed up the ejection of Ag+ from AgNPs which inturn boost its antimicrobial activities.
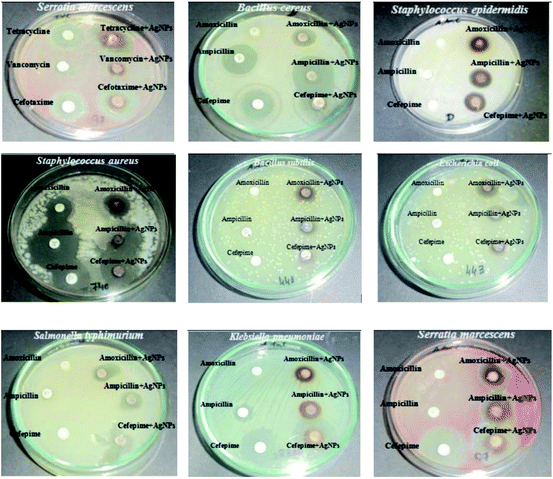 | ||
| Fig. 4 Synergistic effect of Urtica dioica mediated AgNPs with several antibiotics. This figure has been reproduced from ref. 77 with permission from Elsevier, copyright 2016. | ||
Recently, Manjamadha et al.81 have reported ultrasonic-assisted biosynthesis of spherical AgNPs using Lantana camara L. leaf extract. Biosynthesis of AgNPs using ultrasonication improves the reaction conditions such as reducing reaction time and enhancing the reaction rate. Bactericidal activity of the synthesized AgNPs revealed that it shows excellent antibacterial activity against Gram-positive and Gram-negative bacteria. Leaves of Jatropha curcas collected from Micro model complex, Indian Institute of Technology Delhi campus was used for the production of AgNPs.82 The transmission electron microscopy (TEM) analysis showed variation in particle shape and size (20–50 nm), whereas the diameter of NPs was found to be in range of 50–100 nm by scanning electron microscopy (SEM). Complete destruction of the microbial cell was visible using TEM examination. The synthesized NPs were tested for their antimicrobial activities and based on ZOI data, the pattern of sensitivity was observed in the order as E. coli > P. aeruginosa > B. cereus > S. enterica = L. monocytogenes > S. aureus.
Salvinia molesta,83 Sesbania grandiflora,84 Indoneesiella echioides85 and Phlomis86 leaf extract were also useful for the bioreduction of AgNO3 to AgNPs. An ultra-small AgNPs with an average diameter of 7.39 nm were prepared using Hydrocotyle rotundifolia.87 The synthesized AgNPs were tested for its antimicrobial property against E. coli (DH5α). The MIC value was recorded as 5 μg mL−1 and demonstrated significant growth inhibition on agar plate. Formation of spherical AgNPs using Maclura pomifera was achieved in 2017 by Azizian-Shermeh et al.88 The produced NPs (0.1 mg mL−1 concentration) displayed a very high ZOI of 23.4 ± 0.1 mm against E. coli, which is higher than Ampicillin, a well-known antibiotic drug. In the same year, Bhuyan and coworkers at National Institute of Technology Silchar reported Paederia foetida Linn. inspired AgNPs synthesis.89 The order of activities of the AgNPs against tested microbes is B. cereus > E. coli, S. aureus > A. niger. The author claimed that the AgNPs owing to their small size range (5–25 nm) could have easily penetrated the cell membrane, disturbing the metabolism, cause irretrievable damage finally leading to the microbial cell death. Au NPs has also been synthesized but has not shown any antimicrobial activity which testament the higher activity of AgNPs than that of Au NPs. Biosynthesized AgNPs from leaf extract of Atalantia monophylla,90 Talinum triangulare,91 Ricinus communis,92 Erythrina suberosa,93 Lippia citriodora,94 and Brassica oleracea L.95 are also successfully used as an outstanding antimicrobial drug.
In 2017 Al-Shmgani et al. prepared AgNPs using Catharanthus roseus.96 They have used identification by color change, UV-vis spectrum, XRD, FTIR, and AFM techniques to confirm the biosynthesis of AgNPs. The leaf extract color changes from yellowish to reddish-brown after adding 2 mM AgNO3 and exposing to heat at 70 °C for 3 min indicating the formation of the NPs. AFM displays the crystalline NPs with grains sized 10–88 nm in diameter with mean size of about 49 nm. The authors claimed that synthesized AgNPs enter the cell of microbes that resulted in a disruption of adenosine triphosphate (ATP) production and DNA replication, generation of ROS and damage the cell structures as earlier observed by Sahayaraj and Rajesh.97
Spherical shape AgNPs with diameter in the range 11–47 nm (by TEM analysis) were produced using Lavandula x intermedia.98 The AgNPs were found to be most effective against E. coli among all the tested microorganisms shown in Table 1, entry 51. Interestingly, the author also observed that biogenic AgNPs showed ZOI 23 ± 0.0 mm against E. coli whereas streptomycin displayed only 20 ± 0.0 mm under the same concentration. This reflected the high antibacterial efficacy of AgNPs than that of common antimicrobial drug like streptomycin, which could promote its wide use in the future. In another work, a highly crystalline AgNPs were reported to be synthesized from Canna edulis.99 The NPs showed highest antimicrobial activity against S. typhimurium which is closely related to the finding by Sumitha et al.38
In 2017 Artemisia vulgaris mediated AgNPs were reported by Rasheed et al.100 Antimicrobial test revealed that the AgNPs exhibited significant inhibition activities against tested pathogens with the highest value being recorded against S. aureus (18 ± 0.27 mm inhibition zone). Similar to this, earlier in 2016, Thatoi et al.101 reported high activity of AgNPs against S. aureus using AgNPs synthesized from Sonneratia apetala plant extract.
Psidium guajava was applied for the production of spherical AgNPs with average dimension of 25 nm.102 The authors observed that for 100 μg mL−1 Psidium guajava mediated AgNPs, the ZOI were 18.13 ± 0.02 mm and 16.92 ± 0.18 mm against A. faecalis and E. coli, respectively, whereas ZOI of 13.24–14.41 mm were recorded at the same concentration against tested Gram-positive bacteria shown in Table 1, entry 54. This finding clearly testament the higher activity of the synthesized AgNPs towards Gram-negative bacteria than the Gram-positive ones. Similar to this finding, earlier in 2013, Geethalakshmi et al. also reported the higher susceptibility of Gram-negative bacteria to silver nanoparticles compared with Gram-positive bacteria.103 Ironically, Psidium guajava mediated AgNPs is however consistently less sensitive towards tested fungi such as S. cerevisiae, A. niger and R. oryzae as compared to both Gram-positive and Gram-negative bacteria.
Taraxacum officinale leaf extract mediated AgNPs were proved to exhibit an excellent synergistic antibacterial activity with standard antibiotics (such as oxy-tetracycline, tetracycline, ampicillin, and streptomycin) and showed strong positive response against both X. axonopodis, P. syringae, a plant pathogens.104 The combined effect of tetracycline with AgNPs significantly inhibited the growth of selected phytopathogens by increasing ZOI about 40% compared to only antibiotics. The authors are of the opinion that NPs-antibiotic combination and their synergistic action would result in higher penetration in the bacterial cell membrane thereby leads to destruction of various cell organelles and death of bacteria, although the mechanism is not yet fully understood till now.
Lateef et al.105 reported that Petiveria alliacea L. mediated AgNPs showed 100% inhibition against E. coli, K. pneumoniae, S. aureus, A. fumigatus and A. flavus. But only 66.67% inhibition in A. niger. In another work, microwave-assisted synthesis of AgNPs using leaf extract of Nervalia zeylanica was reported.106 The authors observed no formation of NPs (monitored using UV-spectroscopy) even after 5 h under RT stirring of the extract and AgNO3. However, the nanoparticle formation takes place suddenly after 60 s of microwave irradiation. Ficus ingens mediated AgNPs recorded MIC value of 10 μg mL−1 on E. coli and 20 μg mL−1 on both S. typhi and B. cereus107 which is in close agreement with the earlier report of 10 μg mL−1 for E. coli. The AgNPs showed highest inhibition against E. coli and least with S. cereus. Commercial antibiotic Ciprofloxacin showed better activities than the synthesized NPs.
In general, the reduction in the size of the metallic nanoparticles is expected to increase the antibacterial activity due to significantly large surface area of the smaller nanoparticles. However, the results obtained by Erci et al.108 using Thymbra spicata leaf extract is worth discussing. In their study higher antibacterial activity of, say, AgNPs2 (average diameter 70.2 nm) in comparison to AgNPs1 (average diameter 25.1 nm) was recorded. They reasoned that this could be due to the shape of AgNPs2, which have triangles, hexagons, spheres and irregular shapes, whereas AgNPs1 exhibit mostly spherical formation. This interesting finding confirmed the shape-dependent bacterial activity of AgNPs, and support earlier reported protocol.109 The MIC of 50 μg mL−1 was recorded for S. cereus whereas, it was 100 μg mL−1 for E. coli. This finding is in sharp contrast to the work of Kavaz et al.107 mentioned earlier where Gram-negative bacteria has lower MIC than Gram-positive bacteria. However, Erci et al.108 defended their finding of the more pronounced effect of AgNPs against Gram-positive bacteria than Gram-negative bacteria based on the structural difference in cell wall composition of Gram-positive and Gram-negative bacteria. Gram-negative cell wall was covered with an outer lipid membrane (lipopolysaccharide), which is more negatively charged than Gram-positive. As is evident from the zeta value, the biogenic silver nanoparticles were also negatively charged and the electrostatic repulsion between the nanoparticles and Gram-negative bacteria hinders particle attachment and penetration into the cell37 However, this postulate is not yet fully understood. Again, as against the finding of Erci et al.108 the Gram-positive bacteria are less affected by AgNPs (produced from Indigofera tinctoria) than Gram-negative bacteria as reported by Vijayan et al.110 The authors credited the presence of large number of peptidoglycan layers on the walls of Gram-positive bacteria than Gram-negative bacteria that have to some extent prevent the nanoparticles entry to cytoplasmic membrane than Gram-negative bacteria. Hence, the true role of chemicals in the cell wall of bacteria needed to be properly investigated to understand the underlying mechanism of the cell death due to NPs.
Another interesting work on the shape-dependent activity of biogenic AgNPs was reported using Trichoderma viride extract where the authors reported a higher antimicrobial activity of penta- and hexagonal NPs than spherical NPs when the size are of similar range.111 The different shape AgNPs such as pentagonal, hexagonal and spherical were synthesized by manipulating physical parameters, temperature, pH, and reaction time. At neutral pH (7), spherical NPs were observed under all reaction conditions. Delightfully, at pH 5.0 and 9.0, rectangular and penta-/hexagonal NPs were obtained at 40 °C after 72 h of incubation. In general, longer is the reaction, bigger is the size of NPs whereas higher temperature always affords a smaller NP. It was also found that triangular shape AgNPs showed better antimicrobial activity compared to that of spherical and rod shaped as it has high percentage of facet (1 1 1) that possess a high atomic density which increases binding efficiency of Ag to sulfur containing components, whereas spherical and rod shaped particles have a high percentage of (1 0 0) facets.112,113
Recently, Tecoma stans,114 Salvia leriifolia,115 Leucaena leucocephala L.116 and Selaginella bryopteris117 were also reported to produced AgNPs which are mainly spherical in nature. Galega officinalis leaf extract mediated AgNPs with size-dependent activities were reported by Manosalva et al.118 AgNPs with 23 nm and 220 nm recorded MIC of 5 μL mL−1 and 30 μL mL−1 respectively against E. coli showing the higher activity of the smaller NPs. Interestingly MIC of S. aureus (a Gram-positive bacteria) is higher (50 μL mL−1) than E. coli (a Gram-negative bacteria) using 23 nm size AgNPs which implies the higher activity of AgNPs against Gram-negative bacteria.
In the year 2019, antimicrobial fabric tests on the dyed cloths were conducted using AgNPs derived from Camellia sinensis (tea leaf) extract where bleached cotton cloths were dyed using the NPs colloidal solutions. The attachment of AgNPs on the cloths was confirmed by SEM. SEM images of AgNPs with green tea extract also showed the generation of AgNPs. The AgNPs showed excellent antimicrobial activities against S. aureus, K. pneumoniae in the cotton fabric which potentially endorse the suitability of using AgNPs as an effective antimicrobial in cloths.119 Bernardo-Mazariegos et al. used DLS to measure the average hydrodynamic size and zeta potential of the AgNPs synthesized from Justicia spicigera.120 The sample with a mixture of AgNPs of different sizes gave two broad peaks and was weighted toward the larger particles (z-average size of 4.04 μm and 192 nm). The authors are of the opinion that DLS measurement may not be accurate for polydisperse samples due to its nature to respond toward larger particles. Additionally, the zeta potential was of the NPs was found to be 0.2 mV that indicated the less stability and hence, a tendency to agglomerate to form large particles.
In recent times, highly antimicrobial AgNPs were synthesized using Kleinia grandiflora,121 Eucalyptus citriodora,122 Juniperus procera123 and Capparis zeylanica.124 Two different shapes structure in the form of sphere and cubic are observed in SEM analysis of the AgNPs generated from Juniperus procera leaf extract. The produced NPs recorded the highest ZOI against P. mirabilis measured at 29 ± 1.3 mm. The author suggested that the high antimicrobial activity of the NPs is due to the inherent activity of the NPs coupled with the plant particulates attached to the NPs, as the plant which contain high flavonoids and polyphenols are a well-known antimicrobial by themselves.125 Small size AgNPs (9 nm) synthesized using Caesalpinia pulcherrima leaf extract were found to exhibited an MIC as low as 0.078 mg mL−1 and 0.156 mg mL−1 for K. pneumoniae and E. coli respectively. Accordingly, the AgNPs possessed maximum antimicrobial activity against K. pneumoniae and E. coli whereas only moderate effects were shown against C. xerosis, S. mutans, S. aureus, S. viridians, S. pyrogenes, S. viridians and C. diphtheriae that have higher MICs.126
Synergistic antimicrobial activity of Ligustrum lucidum mediated AgNPs and Epoxiconazole under different conjugation ratio was studied against S. turcica, a common maize pathogen.127 The antifungal activity of AgNPs was evaluated alone, and the synergistic inhibition effect was also measured at various conjugation ratios of AgNPs and epoxiconazole, where a prominent synergistic antifungal effect was observed at 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and 9
2 and 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (AgNPs/epoxiconazole) and the inhibition toxicity ratio reached as high as 1.22 and 1.24, respectively.
1 (AgNPs/epoxiconazole) and the inhibition toxicity ratio reached as high as 1.22 and 1.24, respectively.
Aesculus hippocastanum (horse chestnut) mediated special AgNPs with size 50 ± 5 nm was reported to have highest antimicrobial activity (ZOI 20.0 ± 0.00 mm) against a Gram-negative bacteria P. aeruginosa among all the tested microorganisms listed in Table 1, entry 74.128 Interestingly, although the AgNPs have profound effects on all the tested bacteria, it have no effect against fungal strains such as C. albicans ATCC 10231, C. tropicalis ATCC 13803 and C. krusei ATCC 1424. The MIC and MBC of AgNPs for the tested microorganisms were in the range from 0.19–12.5 μg mL−1 and 1.56–25 μg mL−1.
Ramadan et al.129 studied the antiviral activity of green synthesized AgNPs and found that AgNPs greatly enhanced the antiviral activity of M. alternifolia leaf extract, which on its own has no effect on the tested viruses such as herpes simplex virus type 1 (HSV-1), and herpes simplex virus type 2 (HSV-2). In addition, the NPs showed excellent activities against several persistent skin bacteria including S. epidermis and methicillin-resistant Staphylococcus aureus (MRSA). Interestingly, tea tree oil of M. alternifolia itself showed even higher activity than the AgNPs against some tested microbes which is hardly the case in literature. In another work, Carya illinoinensis mediated AgNPs were found to be more efficient against Gram-negative (E. coli) than Gram-positive bacteria (S. aureus)130 in a similar trend reported earlier.118
Although literature revealed that bacterial cell are generally more sensitive to AgNPs, biogenic AgNPs derived from Murraya koenigii leaf extract interestingly shown highly equal activity against Gram-negative bacteria P. aeruginosa (ZOI of 18 mm) and a fungus C. albicans (18 mm ZOI).131 In 2020, several leaf extracts of plant such as Clerodendrum inerme,132 Aspilia pluriseta,133 Melia azedarach,134 Scoparia dulcis,135 and Lantana trifolia.136 All these AgNPs are shown to exhibit an excellent antimicrobial activity against numerous common pathogenic microbes. Mikania micrantha leaf extract mediated AgNPs were also reported to show a high ZOI of 26.17 mm and 26.05 mm against B. subtilis and E. coli respectively.137
More recently, AgNPs of average size 3.46 nm were produced using Solanum nigrum plant leaf extract.138 This is one of the smallest biogenic AgNPs reported so far and NPs as small as 1.74 nm were observed. SPR bands band at 442 nm in UV-visible spectroscopy confirmed the formation of AgNPs. Interestingly, the authors observed a much prominent antimicrobial activity exerted by AgNPs compared to AuNPs and PdNPs potentially due to the more effective capping of AgNPs nanoparticles than either Au or PdNPs which results in well-dispersed small AgNPs without much agglomeration as detected by HRTEM. The authors are of the opinion that polyphenols present in Solanum nigrum extract forms a negative environment around the particles and hence create a repulsive force which overcomes the van der Waals force of attraction and prevent AgNPs agglomeration. The AgNPs showed 22 mm ZOI, while 20 mm and 19 mm ZOI are observed in Au and Pd NPs respectively against E. coli at 10 μL mL−1 concentration. However, although the authors credit the effective capping of AgNPs as a reason for its higher antimicrobial activity, it may also be due to the smaller size of the AgNPs (3.46 nm) as compared to Au (9.39 nm) and Pd NPs (21.55 nm).
Maghimaa et al.139 reported biosynthesis of AgNPs using Curcuma longa leaf extract and investigate their antimicrobial activity in AgNP coated cotton fabric. The loading of AgNPs on the cotton fabric was confirmed by SEM analysis, which was further assisted by the EDX analysis. The authors have reported that the cotton fabric loaded with AgNPs showed great resistance to the growth of pathogenic microorganisms and hence they claimed that the cotton fabric loaded with AgNPs synthesized from Curcuma longa can be used for the diverse application in the medical patient as well as in medical workers to resist microbial infection.
In 2020, green synthesis of spherical AgNPs, CuNPs and FeNPs with size 11–19, 28–35 and 40–52 nm, respectively using Syzygium cumini leaf extract was reported.140 The order of antibacterial property against methicillin- and vancomycin-resistance S. aureus, A. flavus and A. parasiticus microbes was found to be Ag- > Cu- > Fe NPs, which linearly relates with the size of the NPs, thereby reinforcing the size-dependent activity of NPs.141 In addition, the bioproduction of aflatoxins (a family of toxins produced by certain fungi that are found on agricultural crops such as maize (corn), peanuts, cottonseed, and tree nuts) in A. flavus and A. parasiticus was also significantly inhibited by AgNPs when compared with the Fe and Cu NPs. Interestingly, the pH of the plants extract reduced after the formation of NPs in all the cases. Cleistanthus collinus142 and Cestrum nocturnum143 are also known to have produced AgNPs.
In another work, rice leaf extract was utilized for the biosynthesis of AgNPs with size 16.5 nm.144 Antifungal activity of the synthesized NPs was tested against mycelium and sclerotia of R. solani, a fungus that causes sheath blight disease in rice and found that it inhibits the growth of fungus and the growth inhibition is dependent on the concentration of the AgNPs. The MIC values of AgNPs were in the range of 5–10 and 15–20 μg mL−1 towards fungal mycelium and sclerotia, respectively. Results revealed that growth inhibition at 10 μg mL−1 AgNPs is 81.7–96.7% for mycelium and 20 μg mL−1 treatment completely inhibited disease cause by R. solani. In a previous investigation, 43.3–73.6% growth inhibition of R. solani was observed at a higher concentration of 2 mg mL−1 with larger AgNPs with 40–60 nm.145
Recently, an ultra sound-assisted AgNPs of size 8 mm were synthesized using Mentha aquatica leaf extract as reducing and capping agent.146 To the best of our knowledge, this is the smallest biogenic AgNPs reported so far. The production of NPs could occur at RT, but ultrasound greatly reduced the reaction time to 10 min whereas RT took 1 h. The authors highlighted that the phenolic compounds in the Mentha aquatica leaf extract get oxidized to Quinone in an alkaline condition which provides free electrons for reduction of the Ag+ ion to Ag0 to form the desired AgNPs. Largely due to its ultra-small size, the AgNPs displayed a very low MIC of 2.2 μg mL−1 for P. aeruginosa, which showed its high efficacy against the tested microbe.
Rosemary (Rosmarinus officinalis Linn.)147 and Ceropegia thwaitesii148 leaf extract mediated AgNPs which showed consistent higher activities against Gram-negative bacteria were also reported. Interestingly, S. flexneri, S. typhi, B. subtilis, M. luteus, and P. mirabilis are more susceptible to AgNPs than E. coli148 which is not very common in literature.
In the year 2015, Gavade et al. prepared AgNPs using the leaf extract of Ziziphus jujuba under RT.149 The AgNPs have different shapes with 20–30 nm size as revealed by TEM images. The authors investigated the effect of pH on the size and stability of the NPs, and observed form UV-Visible spectroscopic graphs that absorbance value linearly increases with increasing pH increases from 4 to 9, which indicates the rate of formation of AgNPs increases from acidic to basic medium. In addition, at acidic pH, bands were wider and display red shift which is an indication of increase in particle size. However, in basic condition, bands were narrow and display blue shift due to decrease in particle size. The rapid formation of AgNPs in neutral and basic pH this may be due to the ionization of the phenolic groups present in the leaf extract.150 The slow rate of formation and aggregation of AgNPs in acidic pH could be related to electrostatic repulsion of anions present in the solution.151,152 Ironically, at basic pH there is a possibility of AgOH precipitation which need to be avoided.150 Hence, the authors concluded that the optimum condition for the preparation of AgNPs with desired size and stability was neutral medium. The NPs have a zeta potential of −26.4 mV which is an indication of its excellent stability in colloidal state as a zeta potential higher than 30 mV or lesser than −30 mV is indicative of a stable system.153 The AgNPs showed high efficacy against E. coli and found to be stable for more than 6 months probably due an excellent capping of NPs (indicated by FR-IR) and low zeta potential.
Irregular shape AgNPs of average size 28 nm, 26.5 nm, 65 nm, 22.3 nm and 28.4 nm were prepared from O. tenuiflorum, S. cumini, C. sinensis, S. trilobatum and C. asiatica, respectively.154 Among several tested microbes the highest antimicrobial activity of AgNPs synthesized by S. trilobatum and O. tenuiflorum extracts was found against Gram-positive bacteria S. aureus (30 mm ZOI) and Gram-negative bacteria E. coli (30 mm) respectively. Interestingly, C. sinensis, S. trilobatum and C. asiatica derived AgNPs consistently showed higher susceptibility towards a Gram-positive bacteria S. aureus and Gram-negative bacteria E. coli and K. pneumoniae. These findings clearly shown that some AgNPs are more sensitive towards a Gram-positive bacteria whereas some towards a Gram-negative bacteria, hence the question of selective sensitivity of biogenic AgNPs toward Gram-positive or negative bacteria still remains unsolved. Is the selectivity depending on the biomaterial capping agents attached to NPs or the size of NPs? Hence, one may need to consider the biomolecules present in the plant extract or the size of AgNPs to truly understand the selectivity.
A globular shape AgNPs were prepared using Amaranthus gangeticus Linn leaf extract in 2015 which exhibited an inhibitory activity towards Gram-positive, Gram-negative bacteria as well as fungus.155 In another work Andrographis paniculata leaf extract produced a rarely reported cubic shape AgNPs.156 Study on different shape of AgNPs is of great interest due to the shape-dependent activities of AgNPs towards microbes as noted earlier.109 The AgNPs showed a high ZOI of 21.3 ± 0.4 mm for Gram-negative bacteria P. aeruginosa with very low MIC of 3.125 μL mL−1 which testament its high antimicrobial activity. Yao et al.157 noted that the thickness of the peptidoglycan layer of other Gram-negative bacteria such as E. coli is somewhat more than P. aeruginosa, hence the author, in good agreement with Yao's work, observed a lower ZOI (16.6 ± 0.3 mm) in case of E. coli.
Elangovan et al.158 reported the biosynthesis of AgNPs having cubic, pentagonal and hexagonal shape with size range of 68.06–91.28 nm using Andrographis echioides leaf extract and investigate its bactericidal activity against several microbes. The result revealed a high ZOI in the case of E. coli (28 mm) and S. aureus (23 mm) in 100 μg mL−1 concentration of AgNPs. Azadirachta indica (neem) leaf extract was also reported for the green synthesis of polydisperse AgNPs at RT and evaluated as a potent antimicrobial agent against P. nitroreducens, a biofilm-forming bacterium and fungus A. unguis.159
While most biogenic AgNPs are spherical, a flower-like structure was reported by Ajitha et al. in 2017.160 The AgNPs showed very high activity towards bacterial culture Pseudomonas spp. (ZOI of 11 mm) even at very low AgNPs concentration (8 μL mL−1). It is worth note that the AgNPs also consistently displayed a better activity in fungal strain, Penicillium spp. than bacteria such as E. coli and Staphylococcus spp. which is hardly a case in any literature as bacteria are usually considered more sensitive to AgNPs than fungi.
3.2 From seeds
Plant seed extract also well established for the biosynthesis of nanoparticles. Till date, various seeds extract has been utilized for the biosynthesis AgNPs (Table 2, entries 1–27). Sinapis arvensis seeds mediated AgNPs was reported for more than 83% inhibition of mycelium growth of fungus N. parvum. Inductively coupled plasma spectrometry (ICP) analysis revealed complete reduction of Ag+ to Ag0 in more than 95% conversion within 50 days of reaction.161 In another work, grape seed extract was utilized for the biosynthesis of spherical and polygonal AgNPs with size ranging from 25–35 nm. Bactericidal activity of the synthesized NPs was tested against eight different ocean pathogenic bacteria; however, it showed great inhibition activity only against four bacteria such as V. alginolyticus, V. anguillarum, V. parahaemolyticus and A. punctate.162 Sumitha et al.38 reported bio-reduction of AgNO3 to AgNPs using Durio zibethinus seed extract. It is reported that saccharides present in the extract induces the bio-reduction and the amino acids present in the extract stabilized the synthesized AgNPs. Bactericidal activity was tested against different pathogenic bacteria and found that the NPs showed greater activity against S. typhimurium, S. haemolyticus and S. aureus over B. subtilis, E. coli and S. typhi. However, the synthesized NPs showed lesser inhibition compared to the drug Gentamicin against all the mentioned pathogenic bacteria even at a lower dose of Gentamicin.| No. | Plants | Plant parts | Shape and size | Test microorganisms | Ref. |
|---|---|---|---|---|---|
| 1 | Sinapis arvensis | Seeds | Spherical; 1–35 nm | N. parvum | 161 |
| 2 | Grape | Seeds | Spherical and polygonal; 25–35 nm | V. alginolyticus, V. anguillarum, V. parahaemolyticus, A. punctata, E. coli, S. dysenteriae, P. Aeruginosa, S. aureus | 162 |
| 3 | Coffea arabica | Seeds | Spherical and ellipsoidal; 20–30 nm | E. coli and S. aureus | 41 |
| 4 | Durio zibethinus | Seeds | Spherical and rod shaped, 20–75 nm | S. typhi, S. typhimurium, E. coli, S. aureus, S. haemolyticus, B. subtilis | 38 |
| 5 | Pimpinella anisum | Seeds | Spherical; 3.2–16 nm | S. pyogenes, A. baumannii, K. pneumoniae, S. typhi, P. aeruginosa | 163 |
| 6 | Synsepalum dulcificum | Seeds | Spherical; 4–26 nm | P. aeruginosa and K. granulomatis, A. flavus, A. fumigatus, A. niger | 164 |
| 7 | Vigna radiata | Seeds | Spherical; 18 nm | Escherichia coli and Staphylococcus aureus | 165 |
| 8 | Dracocephalum moldavica | Seeds | Spherical; 5–50 nm | E. coli, P. aeruginosa, S. aureus, S. marcescens, S. epidermidis, B. subtilis | 166 |
| 9 | Trifolium resupinatum | Seeds | Spherical; 17 nm | R. solani, N. parvum | 167 |
| 10 | Descurainia sophia | Seeds | Spherical; 1–35 nm | A. rhizogenes, A. tumefaciens, R. solani | 168 |
| 11 | Nigella arvensis | Seeds | Spherical; 2–15 nm | S. pyogenes, B. subtilis, S. aureus, E. coli, P. mirabilis, S. typhimurium | 169 |
| 12 | Linseed | Seeds | Spherical; 10–35 nm | S. mutans, S. epidermidis, P. aeruginosa, E. coli, S. aureus, B. subtilis, A. odontolyticus, A. niger | 170 |
| 13 | Embelia ribes | Seeds | Spherical; 5–35 nm | E. coli, S. aureus | 171 |
| 14 | Melissa officinalis | Seeds | Spherical; 34.64 nm | E. coli, B. subtilis, B. vallismortis | 172 |
| 15 | Leucaena leucocephala | Seeds | Spherical; 6–25 nm | P. gigantea, E. taxodii, E. coli, S. aureus | 173 |
| 16 | Alpinia katsumadai | Seeds | Spherical; 12.6 nm | S. aureus, P. aeruginosa, E. coli | 174 |
| 17 | Myristica fragrans | Seeds | Spherical; 25 nm | Multidrug-resistant (MDR) Salmonella enterica serovar typhi (S. typhi) | 175 |
| 18 | Durio zibethinus | Seeds | Spherical; 20–75 nm | S. aureus, S. typhimurium, E. coli, B. subtilis, S. typhi, S. haemolyticus, S. aureus | 38 |
| 19 | Phoenix sylvestris L. | Seeds | Spherical; 40–50 nm | P. acnes, S. epidermidis | 176 |
| 20 | Phoenix dactylifera | Seeds | Spherical; 14–30 nm | Methicillin-resistant S. aureus | 177 |
| 21 | Tectona grandis | Seeds | Spherical; 10–30 nm | B. cereus, S. aureus, E. coli | 178 |
| 22 | Persea americana | Seeds | Spherical; 50 nm | E. coli | 179 |
| 23 | Salvia hispanica | Seeds | Spherical; 1–27 nm | E. coli, S. aureus | 180 |
| 24 | Trigonella foenum-graecum | Seeds | Spherical; 33.93 nm | E. coli, K. pneumoniae, S. aureus, S. typhi, P. aeruginosa, A. flavus, C. albicans, T. rubrum, P. notatum, T. viridiae | 181 |
| 25 | Sesame (Sesamum indicum, L.) | Seeds | Spherical; 6.6–14.80 nm | P. aeruginosa, K. pneumoniae, B. subtilis, S. aureus | 214 |
| 26 | Hibiscus cannabinus | Seeds | Spherical; 7–11 nm | S. aureus, B. cereus, E. coli | 183 |
| 27 | Carum copticum | Seeds | Spherical; 21.48 nm | P. aeruginosa, S. marcescens, C. violaceum | 184 |
| 28 | Coffea arabica | Seeds | Spherical and ellipsoidal; 20–30 nm | E. coli, S. aureus | 41 |
| 29 | Pimpinella anisum | Seeds | Irregular; 16–48 nm | E. coli, S. aureus, A. flavus, C. albicans | 185 |
| 30 | Marigold (Tagetes erecta) | Flower | Spherical; 46.11 nm | S. aureus, B. cereus, S. coli, P. aeruginosa, C. glabrata, C. albicans, C. neoformans | 188 |
| 31 | Nyctanthes arbortristis | Flower | Spherical and oval; 5–20 nm | E. coli | 189 |
| 32 | Caesalpinia pulcherrima | Flower | Spherical; 12 nm | S. aureus, C. glabrata, B. cereus, E. coli, S. typhimurium, C. albicans, C. neoformans B. subtilis, C. rubrum, Pseudomonas aeruginosa, K. pneumoniae | 190 |
| 33 | Alcea rosea | Flower | Spherical; 7.2 nm | E. coli, S. aureus | 191 |
| 34 | Argemone mexicana | Flower | Spherical; 29.34 nm | S. aureus, P. aeruginosa, E. coli, K. aerogenes | 192 |
| 35 | Turnera ulmifolia | Flower | Spherical; 32.42 nm | S. aureus, P. aeruginosa, E. coli, K. aerogenes | 192 |
| 36 | Tecoma stans | Flower | Spherical; 50–60 nm | E. coli, S. aureus | 193 |
| 37 | Moringa oleifera | Flower | Spherical; 8 nm | K. pneumonia, S. aureus | 194 |
| 38 | Syzygium aromaticum | Flower | Polydisperse; 23 nm | Staphylococcus spp. E. coli, Pseudomonas spp., Bacillus spp., A. flavus, A. niger, Penicillium spp. | 195 |
| 39 | Potentilla fulgens | Root | Spherical; 10–15 nm | E. coli, B. subtilis | 196 |
| 40 | Alpinia calcarata (ginger) | Root | Spherical; 5–15 nm | P. mirabilis, E. coli, B. cereus, S. aureus | 197 |
| 41 | Erythrina indica Lam | Root | Spherical; 20–118 nm | S. aureus, M. luteus, Escherichia coli, B. subtilis, S. typhi, S. paratyphi | 198 |
| 42 | Diospyros paniculata | Root | Spherical; 14–28 nm | P. notatum, A. flavus, A. niger, C. albicans, S. cerevisiae | 199 |
| 43 | Diospyros sylvatica | Root | Spherical; 8 nm | B. pumilis, P. aeruginosa, B. subtilis, S. aureus, K. pneumoniae, E. coli, S. pyogenes, P. vulgaris, A. niger, P. notatum, A. flavus, S. cerevisiae, C. albicans | 200 |
| 44 | Annona muricata | Root | Spherical; 15.08–33.11 nm | K. pneumonia, S. aureus | 201 |
| 45 | Cibotium barometz | Root | Spherical; 23 nm | Escherichia, S. aureus, S. enterica, P. aeruginosa | 202 |
| 46 | Diospyros assimilis | Root | Spherical; 14–28 nm | B. pumilis, B. subtilis, S. aureus, S. pyogenes, K. pneumoniae, E. coli, P. aeruginosa, P. vulgaris, A. niger, A. flavus, C. albicans, P. notatum, S. cerevisiae | 203 |
| 47 | Pelargonium endlicherianum Fenzl. | Root | Spherical; 25–80 nm | P. aeruginosa, E. coli, S. epidermidis | 204 |
| 48 | Rheum palmatum | Root | Spherical; 121 ± 2 nm | S. aureus, P. aeruginosa | 205 |
| 49 | Lepidium draba | Root | Spherical; 20–80 nm | S. aureus, B. cereus, S. typhimurium, E. coli | 207 |
| 50 | Angelica pubescens Maxim | Root | Quasi-spherical; 12.48 nm | E. coli, S. aureus, P. aeruginosa, and S. enterica | 208 |
| 51 | Phoenix dactylifera | Root | Spherical; 15–40 nm | E. coli, C. albicans | 209 |
| 53 | Arctium lappa | Root | Spherical; 21.3 nm | E. coli, A. tumefaciens, L. acidophilus, S. aureus | 210 |
| 53 | Asparagus racemosus | Root | Spherical; 10–17 nm | E. coli, S. aureus, B. subtilis, K. pneumonia, P. fluorescence, A. hydrophila, E. tarda, F. branchiophilum, Y. ruckeri | 211 |
| 54 | Lysiloma acapulcensis | Root | Spherical; 1.2 to 62 nm | E. coli, P. aeruginosa, S. aureus, C. albicans | 212 |
| 55 | Raphanus sativus | Root | Irregular; 3.2–6.0 nm | S. aureus, E. coli, C. albicans, C. glabrata, C. tropicalis | 213 |
Pimpinella anisum,163 Synsepalum dulcificum,164 Vigna radiate,165 Dracocephalum moldavica166 leaf extracts were also successfully applied for the green synthesis of AgNPs. Vigna radiata mediated AgNPs, was found to be more susceptible towards Gram-negative bacteria E. coli (ZOI 20 mm) than Gram-positive S. aureus (ZOI 16 mm) due to the higher thickness of the peptidoglycan layer (approx. 80 nm thick) of the cell wall of Gram positive bacteria which is 10 times thicker than the peptidoglycan Gram-negative bacteria, hence is less susceptible to be destroyed by AgNPs.165
Several reported literatures revealed that the efficiency of AgNPs as antimicrobial agent is extensively dependent on the shape of the nanoparticles. The comparison of spherical, disc like and triangular shaped AgNPs as antimicrobial agent revealed the activity trend follows as spherical AgNPs > disc-like AgNPs > triangular AgNPs.65,136The highest inhibition effect of 94.1% and 84% were observed at 40 ppm concentration of AgNPs against R. solani and N. parvum respectively, using AgNPs derived from Trifolium resupinatum seeds extract.167 In a closely related study, Khatami et al. reported more than 86% inhibition of mycelium growth of R. solani at a concentration 25 μg mL−1 (or 25 ppm) of the biogenic AgNPs.168 Several plant seeds such as Nigella arvensis,169 Linseed,170 Embelia ribes,171 Melissa officinalis172 are applied for the generation of spherical shape AgNPs. While biogenic AgNPs are reported to be more efficient antimicrobial than any other metal NPs in most of the case, it is worth mentioned that the Embelia ribes derived AgNPs is less susceptible to E. coli at showing ZOI of 20 mm against 28 mm ZOI for AuNPs at 250 μL mL−1 concentration.171 Although having small size of NPs (6–25 nm), Leucaena leucocephala mediated AgNPs displayed very low toxicity against both E. coli and S. aureus with ZOI of 18 mm and 22 mm (approx.) respectively at 1000 ppm AgNPs concentration.173 Alpinia katsumadai seeds extract mediated AgNPs showed excellent activities against E. coli and S. aureus than that of P. aeruginosa,174 whereas those derived from Myristica fragrans are found to be highly sensitive to multidrug-resistant (MDR) Salmonella enterica serovar typhi (S. typhi) where a highest ZOI of 16.4 ± 0.45 was observed at 100 μg μL−1 concentration of AgNPs.175
Common skin bacteria such as P. acnes and S. epidermidis are found to be highly inhibited by AgNPs synthesized using Phoenix sylvestris L. The authors also proved that AgNPs is more susceptible to the tested shin bacteria than the seeds extract as well as AgNO3 solution as can be seen from the ZOI.176 The high toxicity of Phoenix dactylifera derived AgNPs against Methicillin-resistant S. aureus is clearly seen in SEM images (Fig. 5a–d) and HRTEM images (Fig. 5e and f). Cells treated with AgNPs undergo deformities and irregular cell surface (red arrow). Attachment and penetration of NPs and deformities of the outer most layers of cell wall and cytoplasmic membrane are also clearly visible in HRTEM.177
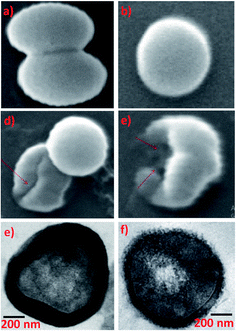 | ||
| Fig. 5 (a and b) Untreated control cells, (c and d) cells treated with 25 and 50 μg mL−1 of AgNPs respectively, SEM images; (e) untreated control cell and (f) treated with 50 μg mL−1 AgNPs TEM images. This figure has been adapted from ref. 177 with permission from Hindawi, copyright 2018. | ||
The seeds extracts of plants such as Tectona grandis,178 Persea americana,179 Salvia hispanica L180 and Trigonella foenum-graecum181 produced AgNPs with high antimicrobial activities. Interestingly the size of AgNPs depends on the concentration of Persea americana extract where a small NPs was recorded at low concentration of aqueous extract, whereas high concentration results in the formation of larger NPs.179 Ironically, the AgNPs from Salvia hispanica L showed lower susceptibility towards antibiotic Ampicillin against E. coli and S. aureus although its high ZOI against E. coli (18.5 mm) and S. aureus (14.9 mm) at 7.7 μL mL−1 concentration.180
The increase in lactate dehydrogenase (LDH) and alkaline phosphatase (ALP) enzyme concentration were used as a means to visualize the change in physiology and inhibition caused to microbes such as S. aureus (263 U L−1) and S. aureus (263 U L−1) by Trigonella foenum-graecum mediated AgNPs.181 This increase in enzyme reflected that bacteria are under stress conditions due to unfavorable environment on treatment with AgNPs.182 Synergistic behavior of ampicillin with Hibiscus cannabinus seeds produced AgNPs against S. aureus, B. cereus, E. coli was investigated by Adnan et al. in 2020.183 Biogenic AgNPs that possessed high inhibitory effect on biofilms formation in P. aeruginosa, C. violaceum and S. marcescens was reported.184 The biofilms of P. aeruginosa was inhibited by 10.6, 18.8, 36.1, 62.0, and 77.6% in presence of 1, 2, 5, 10, and 15 μg mL−1 of Carum copticum mediated AgNPs respectively.
In their effort to synthesize AgNPs from Pimpinella anisum seeds extract, Zayed et al. systematically studied to influence of different parameters such as extraction solvent used, extraction temperature, solvent/plant ratio and extraction time which are crucial for the successful synthesis of AgNPs.185 Hexane, methylene chloride, 70% methanol and water were evaluated as an extraction solvent and 70% methanol was chosen as a best solvent for the fast synthesis of NPs indicated by color change of the reaction solution, whereas this color change is very slow or not visible in other solvent extracts. The high reactivity of 70% aqueous methanol extract towards the reduction of Ag+ to Ag0 NPs is due to an excellent solubility of polyphenols in the plant seeds which is efficiently washed down during the extraction process. The SPR peak intensities of both AgNPs and AuNPs increased as the extraction temperature is raised from 25 to 60 °C. This may be due to increasing the solvent's diffusion rate into plant tissues by destroying the cell structures with raising the temperature.31 They also observed increasing SPR by increasing the extraction temperature 25–60 °C but extraction at 60 to 85 °C resulted in decreasing SPR probably due to decomposition of bioreductant at high temperature. The solvent/plant ratio of 10 mL g−1 was optimized for the AgNPs synthesis. Increasing the ratio from 3–10 mL g−1 inceased the SPR due to increasing solubility of biomolecule, however above 10, SPR went down due to high dilution of the extract. The band intensity reached its maximum value with extracts prepared at 60 min, further increase in contact time caused a decrease in the band intensity. It was observed that as extraction time increases the mass transfer coefficient between the solute and solvent increases that potentially increase the amount of the extracted biomolecule from plants which enhance the formation of the NPs.186 However, prolonged extraction time resulted in the thermal decomposition and oxidation of reactive biomolecules due to prolong heating.187
Similarly, the dried and roasted coffee (Coffea arabica) seed was employed as a reducing and stabilizing agent for the biosynthesis of AgNPs. TEM micrographs of synthesized AgNPs (Fig. 6) were revealed that the nanoparticles are spherical and ellipsoidal in structure with size ranging from (a) 10–40 nm for 0.1 M, (b) 10–50 nm for 0.05 M and (c) 20–150 nm for 0.02 M. The biomolecules that act as a capping agent around the NPs are visible in TEM images. The SAED patterns indicated that the nanoparticles are crystalline in nature with a certain d-spacing corresponds to fcc structure. The authors investigated its bactericidal activity against E. coli and S. aureus. The results revealed that AgNPs solution of 0.05 M and 0.1 M showed a high ZOI in both cases. However, ZOI is higher against E. coli.41
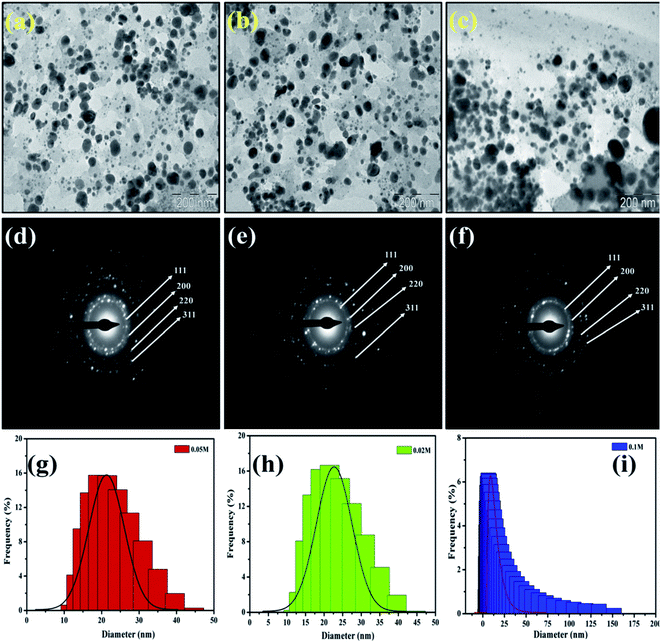 | ||
| Fig. 6 TEM micrographs and SAED pattern of AgNPs with concentration 0.1 M (a and d), 0.05 M (b and e) and 0.02 (c and f) respectively. This figure has been reproduced from ref. 41 with permission from Elsevier, copyright 2016. | ||
3.3 From flowers
Recently, flower extracts have been immensely utilized for the biosynthesis of NPs. There are various literatures available for the bio-reduction of AgNO3 to AgNPs (Table 2, entries 28–37). Padalia et al.188 reported the utilization of flower extract of Tagetes erecta for the bio-reduction of AgNO3 to synthesized AgNPs and investigate their bactericidal activity against both Gram-positive and Gram-negative bacteria such as S. aureus, B. cereus, S. coli, P. aeruginosa, C. glabrata, C. albicans, C. neoformans. The result obtained revealed that the bactericidal activity is greater for E. coli and P. aeruginosa compared to other pathogenic bacteria. Apart from that, the authors reported that the antifungal activity of the AgNPs along with antibiotic against the fungal strain and Gram-negative bacteria showed great activity compared to antibiotic alone. Flower extract of plants such as Nyctanthes arbortristis,189 Caesalpinia pulcherrima,190 Alcea rosea191 and Argemone mexicana192 were also reported from the synthesis of biogenic AgNPs. Tecoma stans flower extract was employed for the biosynthesis of spherical AgNPs with size ranging from 50–60 nm. Antimicrobial activity was tested against S. aureus and E. coli and found that ZOI is higher for S. aureus (24 mm) over E. coli (16 mm).193 Moringa oleifera generated ultra-small AgNPs that showed a high ZOI of 29 mm against S. aureus.194 This is one of the highest ZOI observed at this concentration so far using biogenic AgNPs, probably due to its small size. Another exciting finding in this work is the higher antimicrobial activity of the AgNPs in Gram-positive bacteria (S. aureus) than the Gram-negative one (K. pneumonia), which is a rare case. In another work, Ajitha et al. reported the biosynthesis of polydisperse AgNPs using Syzygium aromaticum flower extract as bio-reducing as well as a capping agent. Antimicrobial activity of the synthesized NPs was explored against several microbes and found that the NPs induces cell disruption of the bacterial strain and it is maximum in case of Pseudomonas spp.1953.4 From roots
Green synthesis of AgNPs and their application as antimicrobial using plants root extract have gained immense attention nowadays (Table 4, entries 38–54). The root extract of Potentilla fulgens was reported as a potent antimicrobial agents against E. coli, B. subtilis showing a ZOI of 9.5 ± 0.2 and 9.7 ± 0.6 respectively.196 Recently, Alpinia calcarata root extract was utilized as a bio-reducing as well as a stabilizing agent for the green synthesis of spherical AgNPs. Antimicrobial activity was tested against P. mirabilis, E. coli, B. cereus, S. aureus and the results showed that Alpinia calcarata root extract assisted synthesized AgNPs have great potential to induce cell disruption of the bacterial strain. Apart from that, the synthesized AgNPs is stable for up to six months.197 AgNPs with microbial activities were also reported to be produced from the root extract of Erythrina indica L.198 Diospyros paniculate199 and Diospyros sylvatica.200Ezealisiji et al. have reported the green synthesis of AgNPs using root bark extract of Annona muricata Linn and investigate their application as an antimicrobial agent against pathogenic bacteria such as B. subtilis, S. aureus, and K. pneumonia, E. coli, and Pseudomonas aeruginosa. The zone of inhibition (ZOI) in diameters were 10.00, 15.00 mm and 12.50, 17.50, 20.00 mm for the five pathogens respectively at AgNPs concentration of 5 μg mL−1. The ZOI is increased to 12.50, 14.50 mm and 14.00, 18.50, and 26.00 mm respectively at AgNPs dose of 10 μg mL−1. Taking into account, the authors have reported that the bactericidal activity of AgNPs is concentration-dependent.201
Cibotium barometz,202 Diospyros assimilis,203 Pelargonium endlicherianum Fenzl.204 roots derived AgNPs were also highly sensitive towards tested microorganisms. Diospyros assimilis derived AgNPs showed high ZOI (18 mm approx. at 100 μL mL−1 AgNPs concentration) against E. coli and S. aureus; however, they showed lower activity than antibiotic chloramphenicol.203
Interestingly the AgNPs derived from Pelargonium endlicherianum Fenzl. seed extract (using 11% ethanol extract contain gallic acid and apocynin as major phytochemicals) are monodisperse, whereas those prepared from 70% methanol extract containing gallic acid, apocynin, and quercetin as major components afforded polydisperse NPs as shown in Fig. 7. These indicated the effect of extract solvent on the composition of the extract and nature of the synthesized AgNPs, which have further bearing on the antimicrobial activities of the NPs.204
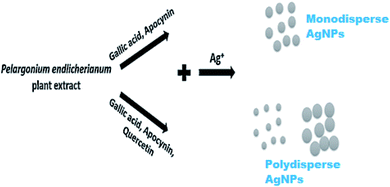 | ||
| Fig. 7 AgNPs formation when Ag + ions were separately mixed with (gallic acid + apocynin) and (gallic acid + apocynin + quercetin). This figure has been adapted from ref. 204 with permission from Elsevier, copyright 2017. | ||
Protein leakage and SEM studies were used as means to study the bactericidal activities of the AgNPs using Rheum palmatum seeds extract.205 SEM images showed abnormality in the cell wall of the tested bacteria, whereas protein was found to leak in high amount due to disruption of membrane in the bacteria when treated with AgNPs which showed the damage caused by AgNPs, which is supported by previous literature.206 The AgNPs showed higher susceptibility towards P. aeruginosa (14.35 ± 0.24 mm ZOI) than S. aureus (10.12 ± 1.81 mm). The antimicrobial activities of AgNPs from seeds extract of Lepidium draba,207 Angelica pubescens Maxim,208 and Phoenix dactylifera209 were also proven. Interestingly, Angelica pubescens mediated AgNPs showed excellent activities whereas AuNPs and root extract do not possess antimicrobial activity against the tested Gram-negative and Gram-positive bacterial strain.208 Green synthesized AgNPs and AuNPs using Arctium lappa as potent antimicrobial agents are of great interest considering the shape and size of the NPs produced. While AgNPs are mainly spherical with average size 21.3 nm, AuNPS are with different shapes such as spherical, hexagonal and triangular geometry with average size of 24.7 nm were seen in TEM. The authors believed these differences in shape and size of AgNPs and AuNPs are due to the difference in reduction potential as well as the capping agents specific to each NPs.210 In another work, the ZOI of Asparagus racemosus mediated AgNPs were 17.0 ± 0.89 and 16.0 ± 0 for S. aureus, B. subtilis respectively. However, the AgNPs showed low ZOI (12.33 ± 0.51 mm) E. coli.211
Lysiloma acapulcensis extract was utilized for the green synthesis of AgNPs with size ranging from 3.2–6.0 nm.212 It is reported that Lysiloma acapulcensis plant is widely used as a traditional medicine in Mexico for the treatment of microbial contamination. Thus, the authors reported that Lysiloma acapulcensis root extract mediated AgNPs have higher antimicrobial activity. Antimicrobial activity was tested against the different microorganisms such as E. coli, P. aeruginosa, S. aureus, C. albicans and found that the inhibition potency is in the order E. coli ≥ S. aureus ≥ P. aeruginosa > C. albicans.
Irregular, triangular nanoplates with nanorods, and spherical with average size 6–20 nm, 50–450 nm, 5–30 nm respectively recorded for seed extract, starch, and CTAB-capped AgNPs from Raphanus sativus, which reflected the crucial influence of capping agents on the size and shape of final NPs. In this study, the average NPs size were measured by dynamic light scattering (DLS) technique. The magnitude of the change in the hydrodynamic radius of CTAB-capped AgNPs lower than both extract and starch-capped ones in DLS measurement; hence, the authors proved CTAB is the best shape-directing agent.213
3.5 From fruit
The plant fruit extract is widely investigated in the field of green synthesis of nanoparticles.214 There are numerous literature available on the green synthesis of AgNPs employing fruit extracts (Table 3, entries 1–36). Emblica officinalis,215 guava,216 carambola,217 Helicteres isora218 and Solanum trilobatum219 fruit extract were utilized successfully for the bio-genic synthesis of spherical AgNPs to investigate its various microbial pathogens such as S. aureus, B. subtilis, E. coli, K. pneumonia, S. mutans, B. cereus and S. typhi. It is reported that, with increase in the concentration of the fruit extract, size of the AgNPs decreases and subsequently antibacterial activity increases.218 Emblica officinalis showed excellent antibacterial activity against Gram-negative bacteria compared to Gram-positive bacteria.215 Due to the very small size of Emblica officinals, guava and Helicteres isora mediated AgNPs, they showed great ZOI against the mentioned pathogens.215,216,218 Apart from that, Lemon,220 Syzygium alternifolium Walp221 and Nothapodytes nimmoniana222 fruit extract were also utilized for the green synthesis of AgNPs. Shape directive CTAB was utilized with lemon extract to control the shape of the AgNPs, which was utilized directly to the various bacterial strains and showed excellent activity against the bacterial strains.220 In contrary, both Syzygium alternifolium Walp221 and Nothapodytes nimmoniana222 fruit extract act as a shape directive and bio-reducing agent for the biosynthesis of AgNPs. In another work, Lemon extract was utilized as a bio-reducing agent for the green synthesis of AgNPs and investigate its antimicrobial activity against four pathogenic bacteria such as P. aeruginosa, E. coli and S. aureus and compared with controls amikacin and lemon extract. The results showed that the synthesized NPs have excellent potency to induce cell disruption of the bacterial strain and ZOI is almost similar to the medicinal antibiotic drug, amikacin.223 Similarly, spherical AgNPs were also prepared using various fruit extract such as apple extract,224 Adansonia digitata L.225 and Momordica charantia226 extract and examined their bactericidal activity against Gram-positive and Gram-negative bacteria. It is reported that, combined extract-AgNPs exert more bacterial cell damage compared to AgNPs and extract alone due to the synergistic effect produced by the phytochemicals capped on the surface of the AgNPs.226 Jayaprakash et al.227 reported microwave-assisted synthesis of spherical AgNPs using Tamarind fruit extract. The produced AgNPs by this method is very stable without the formation of oxide and displayed excellent bactericidal activity. Moreover, this process for AgNPs synthesis is economic, time efficient and straight forward. Phoenix dactylifera,228 strawberry,229 Ginseng-berry,230 Kigelia africana231 and Chaenomeles sinensis232 fruit extract were also successfully utilized for the green synthesis of spherical AgNPs with high antimicrobial activity. Ginseng-berry fruit extract derived AgNPs showed greater ZOI against S. aureus (12.3 mm) compared to E. coli (11 mm).230 A high bactericidal activity against the Gram-positive bacteria was observed by using AgNPs derived from Phoenix dactylifera extract.228| No. | Plants | Plant part | Shape and size | Test microorganisms | Ref. |
|---|---|---|---|---|---|
| 1 | Emblica officinalis | Fruit | Spherical; 15 nm | S. aureus, B. subtilis, E. coli, K. pneumonia | 215 |
| 2 | Psidium guajava | Fruit | Spherical; 2–10 nm | S. mutans, B. cereus, E. coli, S. aureus, and S. typhi | 216 |
| 3 | Carambola | Fruit | Spherical; 10–40 nm | E. coli, P. aeruginos | 217 |
| 4 | Helicteres isora | Fruit | Spherical; 8–20 nm | P. aeruginosa | 218 |
| 5 | Solanum trilobatum | Fruit | Spherical; 12.50–41.90 nm | S. mutans, E. faecalis, E coli, K. pneumoniae | 219 |
| 6 | Lemon | Fruit | Spherical and polyhedral; 15–30 nm | S. aureus, E. coli, Candida albicans, Candida glabrata and Candida tropicalis | 220 |
| 7 | Syzygium alternifolium | Fruit | Spherical; 5–68 nm | B. subtilis, S. aureus, E. coli, K. pneumoniae, P. vulgaris, P. aeruginosa, S. typhimurium, A. solani, A. flavus, A. niger, P. chrysogenum, T. harzianum | 221 |
| 8 | Nothapodytes nimmoniana | Fruit | Spherical; 44–64 nm | B. subtilis, E. coli, S. aureus, S. paratyphi, P. vulgaris, A. hydrophillus, K. pneumoniae | 222 |
| 9 | Citrus lemon | Fruit | Spherical; 2–10 nm | P. aeruginosa, E. coli, S. aureus | 223 |
| 10 | Apple | Fruit | Spherical; 30.25 ± 5.26 nm | E. coli, S. aureus | 224 |
| 11 | Adansonia digitata L. | Fruit | Spherical; 3–57 nm | P. vulgaris, K. pneumoniae, P. aeruginosa, S. typhimurium, E. coli, B. subtilis, S. aureus. T. harzianum, A. niger, A. flavus, P. chrysogenum, A. solani | 225 |
| 12 | Momordica charantia | Fruit | Spherical; 78.5–220 nm | E. coli, S. typhi, S. aureus, P. aeruginosa | 226 |
| 13 | Tamarindus indica (Tamarind) | Fruit | Spherical; 6–8 nm | B. cereus, S. aureus, M. luteus, B. subtilis, Enterococcus sp., P. aeruginosa, S. typhi, E. coli, K. pneumonia | 227 |
| 14 | Phoenix dactylifera | Fruit | Spherical; 25–60 nm | E. coli, K. pneumonia, S. epidermidis, (d) B. cereus, S. aureus | 228 |
| 15 | Strawberry | Fruit | Spherical; 7–65 nm | P. aeruginosa, B. licheniformis | 229 |
| 16 | P. ginseng Meyer | Fruit | Spherical; 10–20 nm | E. coli, S. aureus | 230 |
| 17 | Kigelia africana | Fruit | Spherical; 10 nm | K. pneumoniae, P. aeruginosa, C. albicans | 231 |
| 18 | Chaenomeles sinensi | Fruit | Spherical; 20 nm | S. aureus, E. coli | 232 |
| 19 | Soymida febrifuga | Fruit | Spherical; 14.27 nm | B. subtilis, E. coli, S. aureus, P. putrida | 233 |
| 20 | Ribes nigrum | Fruit | Spherical; 5–10 nm | S. aureus, P. aeruginosa, E. coli, C. albicans, Trichophyton rubrum, A. niger | 234 |
| 21 | Garcinia indica | Fruit | Spherical, hexagonal; 5–30 nm | E. coli, B. subtilis, S. aureus, P. aeruginosa, Salmonella enterica typhi, P. vulgaris, S. marcescens | 235 |
| 22 | Carissa carandas | Fruit | Spherical; 10–60 nm | A. hydrophila, Acinetobacter sp., S. aureus | 236 |
| 23 | Diospyros lotus | Fruit | Spherical; 19 nm | E. coli, S. aureus | 237 |
| 24 | Terminalia bellirica | Fruit | Spherical; 10 nm | P. aeruginosa, K. pneumoniae | 238 |
| 25 | Cordia obliqua Willd | Fruit | Spherical; 7.13 nm | B. circulans, E. coli, P. aeruginosa, S. aureus | 239 |
| 26 | Phyllanthus emblica | Fruit | Spherical; 19.8–92.8 nm | A. oryzae | 240 |
| 27 | Forsythia suspensa | Fruit | Spherical; 47.3 ± 2.6 nm | V. parahaemolyticus, S. aureus | 241 |
| 28 | Rosa canina | Fruit | Spherical; 13–21 nm | B. cereus, E. hirae, S. aureus, E. coli, L. pneumophila, Candida albicans, P. aeruginosa | 242 |
| 29 | Manilkara zapota (Sapota) | Fruit | Spherical; 8–16 nm | E. coli, P. aeruginosa, K. pneumoniae, Bacillus subtilis subsp. Spizizenii, S. aureus | 243 |
| 30 | Abelmoschus esculentus | Fruit | Spherical; 3–11 nm | B. subtilis | 244 |
| 31 | Phyllanthus emblica | Fruit | Spherical; 19 nm to 45 nm | K. pneumoniae, S. aureus | 245 |
| 32 | Aegle marmelos | Fruit | Spherical; 10–200 nm | B. cereus, S. aureus, E. coli, P. aeruginosa, S. typhi, S. dysenteriae Y. pestis | 246 |
| 33 | Nauclea latifolia | Fruit | Irregular, 12 nm | E. coli, C. albicans, Rhizopus sp., A. niger, C. fruendii, S. aureus, Staphylococcus sp. Klebsiella sp. | 247 |
| 34 | Myristica fragrans | Fruit | Irregular; 31.31 nm | E. coli, P. aeruginosa, S. aureus, B. subtilis | 248 |
| 35 | Capsicum frutescens | Fruit | Monodispersed; 20–25 nm | E. coli, B. subtilis | 249 |
| 36 | Areca catechu | Fruit | Polydispersed; 12 nm | E. coli, P. aeruginosa, K. aerogenes, S. aureus | 250 |
| 37 | Azadirachta indica L. | Gum | Spherical; 12.09–29.65 nm | S. enteritidis, B. cereus | 251 |
| 38 | Salmalia malabarica | Gum | Spherical; 7 nm | S. aureus and E. coli | 252 |
| 39 | Styrax benzoin | Gum | Spherical; 12–38 nm | P. aeruginosa, S. aureus, E. coli, C. tropicalis | 253 |
| 40 | Anacardium occidentale L. | Gum | Spherical; 51.9 nm | S. aureus, E. coli | 254 |
| 41 | Araucaria heterophylla | Gum | Spherical; less than 50 nm | E. coli, Streptococcus sp | 255 |
| 42 | Azadirachta indica | Gum | Spherical; less than 50 nm | E. coli, Streptococcus sp | 255 |
| 43 | Prosopis chilensis | Gum | Spherical; less than 50 nm | E. coli, Streptococcus sp | 255 |
| 44 | Buchanania lanzan | Gum | Spherical; 14.74–19.86 nm | E. coli, A. avium, S. intermedius, P. macerans, S. rubidaea, E. mallatovora, E. faecalis, S. haemolyticus, P. mirabilis, S. epidermidis S. chromogenes, E. agglomerans, Staphylococcus capitis ssp. capitis, Staphylococcus capitis ssp. urealyticus | 256 |
| 45 | Mimosa pudica | Gum | Irregular; no report | E. coli, S. commune | 257 |
| 46 | Moringa oleifera | Stem | Spherical; 3–70 nm | E. coli, K. cloacae, S. epidermidis | 258 |
| 47 | Waste grass | Stem | Spherical-oblate; 4–34 nm | P. aeruginosa, A. baumannii, F. solani R. solani | 259 |
| 48 | Swertia paniculate | Stem | Spherical; 31–44 nm | P. aeruginosa, K. pneumoniae, S. aureus | 260 |
| 49 | Caesalpinia pulcherrima | Stem | Spherical; 8 nm | B. cereus, S. aureus, C. rubrum, B. subtilis, E. coli, K. pneumonia, P. aeruginosa, S. typhimurium, C. albicans, C. glabrata, C. neoformans | 261 |
| 50 | Garcinia mangostana | Stem | Spherical; 30 nm | K. planticola, E. coli, B. subtilis | 262 |
| 51 | Dorema ammoniacum D. | Stem | Spherical; 28.4 nm | E. coli, S. typhimurium, S. aureus, B. cereus | 263 |
| 52 | Fumariae herba | Stem | Spherical; 25 nm | S. aureus, B. cereus, B. luteus, B. subtilis, L. monocytogenes, E. coli, P. aeruginosa, K. pneumoniae, P. vulgaris, C. albicans | 264 |
| 53 | Anthemis atropatana | Stem | Spherical; 38.89 nm | S. aureus, S. pyogenes, P. aeruginosa, E. coli | 265 |
| 54 | Afzelia quanzensis | Bark | Spherical; 10–80 nm | E. coli, S. aureus | 266 |
| 55 | Syzygium alternifolium | Bark | Spherical; 4–48 nm | B. subtilis, S. aureus, E. coli, K. pneumoniae, P. vulgaris, P. aeruginosa, S. typhimurium, A. solani, A. flavus, A. niger, P. chrysogenum, T. harzianum | 267 |
| 56 | Cochlospermum religiosum | Bark | Spherical; 20–35 nm | Bacillus, E. coli, Proteus, Pseudomonas, Staphylococcus, A. flavus, Fusarium, C. lunata, Rhizopus, A. niger | 268 |
| 57 | Ficus benghalensis | Bark | Spherical; 85.95 nm | E. coli, P. aeruginosa, B. subtilis | 269 |
| 58 | Azadirachta indica | Bark | Spherical; 90.13 nm | E. coli, P. aeruginosa, B. subtilis | 269 |
| 59 | Plumbago zeylanica | Bark | Spherical; 28.47 nm | B. subtilis, P. aeruginosa, S. aureus, C. tropicalis, E. coli, A. flavus | 270 |
| 60 | Helicteres isora | Bark | Spherical; 16–95 nm | E. coli, V. cholera, S. typhi, P. aeruginosa, B. subtilis and M. luteus | 271 |
| 61 | Terminalia arjuna | Bark | Spherical; 65 nm | E. coli | 272 |
| 62 | Butea monosperma | Bark | Spherical; 18–50 nm | B. subtilis, E. coli | 273 |
| 63 | Prosopis juliflora | Bark | Spherical; 10–50 nm | E. coli, P. aeruginosa | 274 |
| 64 | Garcinia mangostana | Bark | Spherical; 65 nm | E. coli, B. subtilis, S. aureus, B. cereus, K. pneumoniae | 275 |
| 65 | Solanum trilobatum | Bark | Spherical; 25 nm | A. niger, E. coli, Bacillus sp. | 276 |
| 66 | Butea monosperma (Lam.) Taub. | Bark | Spherical; 81 nm | E. coli, S. aureus, A. niger | 277 |
| 67 | Syzygium cumini | Bark | Spherical; 15 nm | E. coli, B. subtilis | 278 |
| 68 | Diospyros montana | Bark | Spherical; 5–40 nm | K. aerogenes, E. coli, B. subtilis, S. aureus | 279 |
| 69 | Handroanthus impetiginosus | Bark | Spherical; 13.4 nm | S. aureus, E. coli | 280 |
Biogenic synthesis of AgNPs are achieved by using various fruit extract such as Soymida febrifuga,233 Ribes nigrum,234 Garcinia indica,235 Carissa caranda berries236 and Diospyros lotus237 and investigate their bactericidal activity against various bacterial pathogens. It is reported that biogenic synthesis of AgNPs depends on the various factors such as AgNO3 concentration, extract to AgNO3 ratio, pH, incubation temperature and time.235 The optimal conditions for green synthesis of AgNPs using Garcinia indica are 1.5 mM AgNO3, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 AgNO3/Kokum fruit extract, pH 10, incubation temperature of 37 °C and 24 h time.235 Carandas berry mediated AgNPs displayed great ZOI against various Gram-negative bacteria. However, to inhibit the growth of bacteria S. aureus, a comparatively high concentration of AgNPs is required.236 There are several reports where different fruit extract such as Terminalia bellirica,238 clammy cherry,239 Phyllanthus emblica,240 Forsythia suspense,241 Rosa canina242 and Manilkara zapota (Sapota)243 have been productively utilized for the green synthesis of AgNPs and evaluate their bactericidal activity against various bacterial pathogens. The results revealed that, AgNPs generated from each fruit extract displayed great antimicrobial activity against both Gram-negative and Gram-positive bacteria. It is reported that microwave-assisted synthesis of AgNPs using cherry extract is a time efficient and cost-effective process. The synthesized NPs are very small with size 7. 13 nm, thus displayed easy cell disruption of various human pathogens.239
1 AgNO3/Kokum fruit extract, pH 10, incubation temperature of 37 °C and 24 h time.235 Carandas berry mediated AgNPs displayed great ZOI against various Gram-negative bacteria. However, to inhibit the growth of bacteria S. aureus, a comparatively high concentration of AgNPs is required.236 There are several reports where different fruit extract such as Terminalia bellirica,238 clammy cherry,239 Phyllanthus emblica,240 Forsythia suspense,241 Rosa canina242 and Manilkara zapota (Sapota)243 have been productively utilized for the green synthesis of AgNPs and evaluate their bactericidal activity against various bacterial pathogens. The results revealed that, AgNPs generated from each fruit extract displayed great antimicrobial activity against both Gram-negative and Gram-positive bacteria. It is reported that microwave-assisted synthesis of AgNPs using cherry extract is a time efficient and cost-effective process. The synthesized NPs are very small with size 7. 13 nm, thus displayed easy cell disruption of various human pathogens.239
Besides, antimicrobial activity of various fruit extract such as Abelmoschus esculentus,244 Phyllanthus emblica,245 Aegle marmelos,246 Nauclea latifolia,247 Myristica fragrans,248 Capsicum frutescens249 and Areca catechu250 mediated AgNPs was also tested and found that the synthesized AgNPs displayed great cell disruption of bacterial strains. The effect of solvent extract of Aegle marmelos on antimicrobial activity was tested by making fruit extract in various solvents such as petroleum, ether, methanol, acetone and chloroform and found that methanol extract of Aegle marmelos displayed highest cell disruption against B. cereus and lowest for E. coli.246
3.6 From gum
Gum extracts of various plants have been widely utilized as a bio-reducing as well as a stabilizing agent for the green synthesis of AgNPs (Table 3, entries 37–45). Velusamy et al.251 have reported the autoclave-assisted green synthesis of AgNPs using gum extract of Azadirachta indica L. AFM analysis of the synthesized AgNPs displayed that the nanoparticles are spherical in shape without any aggregation. Furthermore, the line profile analysis revealed that the average particle size is 23.44 nm. Bactericidal activity test revealed that the synthesized AgNPs can effectively disrupt the cell membranes of S. enteritidis and B. cereus, hence can be exploited in biomedical applications. Similarly, the bactericidal activity of AgNPs generated from various gum extract such as Salmalia malabarica,252 Styrax benzoin253 and Anacardium occidentale L.254 displayed prominent cell damage against both Gram-positive and Gram-negative bacteria. In a comparison study, three different gum extract such as Azadirachta indica, Araucaria heterophylla and Prosopis chilensis have been utilized for the synthesis of AgNPs and compare their antimicrobial activity against both Gram-negative and Gram-positive bacteria. The results revealed that, Azadirachta indica and Prosopis chilensis mediated AgNPs are effective for cell disruption of both Gram-positive and Gram-negative bacterial strains. On contrary, Araucaria heterophylla mediated AgNPs is effective for Gram-negative bacterial strains (E. coli) only.255 In another study, Buchanania lanzan gum extract was utilized for the green synthesis of AgNPs with size ranging from 14.74–19.86 nm. Antimicrobial activity was tested against 14 Gram-negative and 3 Gram-positive bacteria and found that the synthesized AgNPs was more prominent against Gram-negative bacteria over Gram-positive bacteria. Besides, MIC for two Gram-negative bacteria such as E. coli and A. avium was found to be 0.52 μg mL−1 and 0.53 μg mL−1 respectively.256 Beside, Mimosa pudica gum extract was utilized for the green synthesis of AgNPs and investigate its antimicrobial activity against Escherichia coli and Schizophyllum commune, observed that the ZOI for E. coli is higher than the S. commune.2573.7 From stem
Plants stem extract was widely utilized as a reducing agent for the green synthesis of AgNPs (Table 3, entries 46–53). Aqueous extract of Moringa oleifera,258 waste grass259 and Swertia paniculate260 have been utilized for the bio-synthesis of AgNPs to investigate its antimicrobial activity against various bacterial strains. It is reported that the waste grass mediated AgNPs are smaller in size (15 nm) compared to Moringa oleifera and Swertia paniculate mediated AgNPs, hence AgNPs obtained from waste grass extract shows greater antimicrobial activity as it can easily disrupt the bacterial cell wall.259 Similarly, green synthesis of AgNPs by using C. pulcherrima stem extract was reported by Moteriya et al.261 and examine their antimicrobial activity against various pathogenic microorganisms. The results showed that the MIC value for the bacteria is ranging from 0.312 to 2.5 mg mL−1 and for fungi is 2.5 mg mL−1 using AgNPs only. Interestingly, AgNPs together with two antibiotics such as chloramphenicol and amphotericin B recorded a lower MIC value against both bacteria and fungi compared to the bare AgNPs. The combination of AgNPs and chloramphenicol displayed synergistic effect against B. cereus, B. subtilis, S. aureus, C. rubrum and S. typhimurium, while displayed partial synergistic effect against E. coli, P. aeruginosa, K. pneumonia. In addition, green synthesis of AgNPs by using different stem extract of Garcinia mangostana,262 Dorema ammoniacum D.263 and Fumariae herba264 is also reported. The synthesized AgNPs were applied against various bacterial strains of Gram-positive and Gram-negative bacteria and found that the NPs is very active for the cell disruption of Gram-negative bacteria as the Gram-negative bacteria possesses weak cell wall due to the less content of peptidoglycan in the cell wall. In another work, Anthemis atropatana extract was utilized for the biosynthesis of AgNPs and investigated the antimicrobial activity of the produced NPs against various pathogenic bacteria such as S. aureus (ATCC 6538), S. pyogenes (ATCC 19615), P. aeruginosa (ATCC 15442) and E. coli. The result obtained revealed that the highest and lowest MIC value is for P. aeruginosa and S. aureus, respectively.2653.8 From bark
In recent years, bark extract has been widely exploited as a reducing agent as well as a stabilizing agent for the green synthesis of AgNPs (Table 3, entries 54–69). Green synthesis of AgNPs for antimicrobial activity was obtained by using various plant bark extract Afzelia quanzensis,266 Syzygium alternifolium267 and Cochlospermum religiosum.268 Nayak et al.269 have reported the green synthesis of AgNPs with the size of 90.13 nm using bark extract of Ficus benghalensis and Azadirachta indica. Bactericidal activity of the synthesized AgNPs was tested against Gram-positive and Gram-negative bacteria such as E. coli, P. aeruginosa, V. cholera and B. subtilis, observed great inhibition potential of the synthesized AgNPs against the bacterial pathogens. The authors reported that the phytochemicals present around the synthesized AgNPs provide unique surface characteristic and thus, can damage various cell membranes. Apart from that, different plant bark extract such as Plumbago zeylanica,270 Helicteres isora,271 Terminalia arjuna,272 Butea monosperma,273 Prosopis juliflora,274 Garcinia mangostana275 and Solanum trilobatum276 were also utilized for the green synthesis of AgNPs and investigate its antimicrobial activity against various bacterial strains. It is reported that the Butea monosperma bark extract is inefficient for any ZOI, however, the extract mediated AgNPs displayed great ZOI at very low concentration. The ZOI induced by AgNO3 solution is more compared to AgNPs, but required high concentration of AgNO3, which is harmful for the consumer. On contrary, a very small concentration of AgNPs showed good ZOI against the bacterial strains and hence can be used as therapeutic agent.273Recently, Butea monosperma,277 Syzygium cumini278 and Diospyros montana279 bark extract were utilized for the green synthesis of AgNPs and investigate its antimicrobial activity. The resultant AgNPs are displayed prominent cell damage to the various bacterial strains. Syzygium cumini mediated AgNPs displayed greater ZOI against the Gram-negative bacteria compared to Gram-positive bacteria as the cell wall of Gram-negative bacteria is more susceptible for the synthesized AgNPs. The study also revealed that the bactericidal activity of Syzygium cumini mediated AgNPs is more compared to the Syzygium cumini extract and AgNO3 solution, which can attributed to the small size of the AgNPs.278 To increase the rate of the biosynthesis process for AgNPs, the microwave technique was used by Tormena et al.280 where they have used Handroanthus impetiginosus bark extract as a reducing as well as capping agent. Bactericidal activity of the synthesized NPs was tested against two pathogenic bacteria such as S. aureus and E. coli and found good inhibition potential to both bacterial strains with MIC value 3.1 × 102 μg mL−1 and 6.7 × 104 μg mL−1 respectively. However, the pure extract displayed a low MIC value of 2.7 × 103 μg mL−1 and 1.2 × 103 μg mL−1 for S. aureus and E. coli, respectively. Interestingly, the bactericidal activity of AgNPs is higher for S. aureus compared to E. coli (Fig. 8). This is contrary to the generally accepted assumption that AgNPs are more susceptible to Gram-negative bacteria due to their thin cell-wall. However, the authors defended their claim by considering the synergetic effects of biomolecules capping agents and AgNPs.
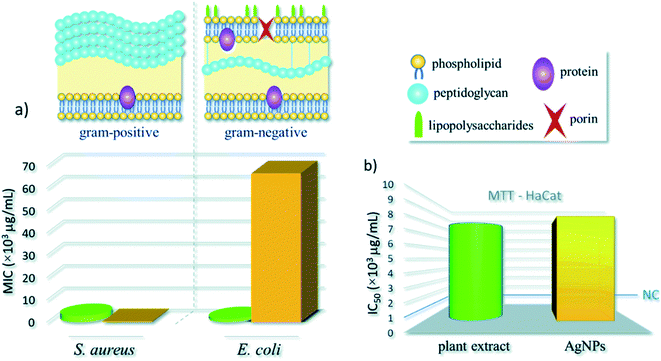 | ||
| Fig. 8 Infographic with (a) minimal inhibitory concentration of AgNPs against Gram-positive (S. aureus) and Gram-negative (E. coli) bacteria models and (b) half maximal inhibitory concentration (IC50) using MTT assay – the blue line indicates the cytotoxicity criteria for preliminary tests of new compounds, as established by the United States' National Cancer Institute (NCI). Green cylinders and yellow bars represent the plant extract and AgNPs results, respectively. This figure has been reproduced from ref. 280 with permission from RSC, copyright 2020. | ||
3.9 From rhizome
Rhizome extract is also utilized in the green synthesis of size-selective AgNPs (Table 4, entries 1–8). Bio-synthesis of spherical AgNPs was obtained by using various rhizome extract such as Bergenia ciliate281 and Dryopteris crassirhizoma.282 The synthesized AgNPs was utilized against different bacterial strains to investigate its antimicrobial activity and found that the NPs displayed excellent cell disruption of the bacterial strains. A wide variety of LED light source such as green, red and blue light have been utilized for the synthesis of AgNPs and examined its bactericidal activity. The authors reported that the ZOI against B. cereus is maximum when the green light mediated AgNPs is used followed by red and blue light mediated AgNPs.282 Rhizome extract of Coptis chinensis was exploited for the green synthesis of AgNPs with size 15 nm. The surface of the synthesized AgNPs was further modified with chitosan. Bactericidal activity of both free AgNPs and chitosan modified AgNPs was tested against E. coli and B. subtilis. The results revealed that the chitosan modified AgNPs showed greater inhibition efficiency compared to free AgNPs against the bacterial strains.37 Recently, rhizome extract of Coptidis,283 Curcuma longa (turmeric),284 Canna indica L.285 and Ferula foetida (asafoetida gum)286 were utilized for the green synthesis of spherical AgNPs with size in the nanometer range. The resultant AgNPs was tested against various bacterial strains to investigate its bactericidal activity and found that the NPs is effective for the bacterial cell damage. It is observed that the Canna indica L. mediated AgNPs displayed greater bactericidal activity against E. coli compared to the other bacteria such as S. aureus and K. pneumoniae. However, on comparison of the bactericidal activity of Canna indica L. mediated AgNPs with conventionally antibiotic drug Gentamicin revealed that the NP is less active compared to Gentamicin.285 In another work, Ginger rhizome extract and sodium citrate were utilized for biosynthesis of B-AgNPs and C-AgNPs respectively followed by investigated their antibacterial activity against six aquatic pathogens such as V. anguillarum, V. alginolyticus, A. punctate, V. parahaemolyticus, V. splendidus, V. harveyi. The results revealed that the chemically synthesized AgNPs showed slightly greater ZOI against the six pathogens compared to Oxford cup indicates that both have very weak bactericidal activity. In contrast, ginger rhizome extract mediated AgNPs (B-AgNPs) showed greater ZOI against the 6 aquatic pathogens, which confirmed that B-AgNPs displayed greater bactericidal activity compared to the chemically synthesized AgNPs.287| No. | Plants | Plant part | Shape and size | Test microorganisms | Ref. |
|---|---|---|---|---|---|
| 1 | Bergenia ciliata | Rhizome | Spherical; 35 nm | M. luteus, S. aureus, E. aerogenes, B. bronchiseptica, A. niger, A. fumigatus, A. flavus, F. solani | 281 |
| 2 | Dryopteris crassirhizoma | Rhizome | Spherical; 5–60 nm | B. cereus and P. aeruginosa | 282 |
| 3 | Coptis chinensis | Rhizome | Spherical; 15 nm | E. coli, B. subtilis | 321 |
| 4 | Coptidis rhizome | Rhizome | Spherical; 30 nm | E. coli, S. aureus | 283 |
| 5 | Curcuma longa (turmeric) | Rhizome | Spherical; 18 ± 0.5 nm | E. coli, L. monocytogenes | 284 |
| 6 | Canna indica L | Rhizome | Spherical; 20–70 nm | S. aureus, K. pneumoniae, E. coli | 285 |
| 7 | Ferula foetida (asafoetida gum) | Rhizome | Spherical; 5.6–8.6 nm | E. coli, K. pneumoniae, C. albicans | 286 |
| 8 | Zingiber officinale (ginger) | Rhizome | Polygonal; 20–80 nm | V. anguillarum, V. alginolyticus, A. punctate, V. parahaemolyticus, V. splendidus, V. harveyi | 287 |
| 9 | Cavendish banana | Peel | Spherical; 23–30 nm | S. aureus, B. subtilis, K. pneumonia, E. coli | 45 |
| 10 | Banana (Musa paradisiaca) | Peel | Spherical; 23.7 nm | B. subtilis, S. aureus, P. aeruginosa, P. aeruginosa, E. coli, C. albicans | 288 |
| 11 | Carica papaya | Peel | Spherical; 16–20 nm | E. coli, S. aureus | 289 |
| 12 | Citrus sinensis | Peel | Spherical; 48.1 ± 20.5 nm | X. axonopodis pv. Citri (Xac) | 290 |
| 13 | Citrus maxima | Peel | Spherical; 4–11 nm | E. coli, S. aureus, F. oxysporum, V. dahliae | 291 |
| 14 | Punica granatum | Peel | Spherical; 6–45 nm | E. coli, S. aureus | 292 |
| 15 | Citrus × clementina | Peel | Spherical; 15–20 nm | E. coli, B. cereus, S. aureus | 293 |
| 16 | Solanum melongena | Peel | Spherical; 92.4 nm | P. fluorescens, B. amyloliquefaciens | 294 |
| 17 | Citrus limetta | Peel | Spherical; 18 nm | C. albicans, C. glabrata, C. parapsilosis, C tropicalis, M. luteus, S. mutans, S. epidermidis, S. aureus, E. coli | 295 |
| 18 | Allium cepa | Tube/bulb | Spherical; 10 nm | E. coli, P. aeruginosa, B. subtilis, F. oxysporium, F. oxysporum | 296 |
| 19 | Sunroot tuber (Helianthus tuberosus) | Tube/bulb | Spherical; 10–70 nm | R. solanacearum, X. axonopodis | 297 |
| 20 | Dioscorea alata | Tube/bulb | Spherical; 10–25 nm | E. coli, S. auricularis | 298 |
| 21 | Crocus haussknechtii Bois | Tube/bulb | Spherical;; 10–25 nm | S. aureus, P. aeruginosa | 299 |
| 22 | Allium sativum (garlic) | Tube/bulb | Spherical; 50–70 nm | P. aeruginosa, B. licheniformis | 300 |
| 23 | Sargassum muticum | Whole plant | Spherical; 43–79 nm | B. subtilis, K. pneumoniae, S. typhi | 301 |
| 24 | Brassica oleracea L. (Broccoli) | Whole plant | Spherical; 30–45 nm | E. coli, B. subtilis and S. aureus, Aspergillus sp., Pneumocystis sp. | 302 |
| 25 | Vernonia cinerea L. | Whole plant | Spherical; 40–75 nm | C. albicans, Penicillium spp | 303 |
| 26 | Artemisia marschalliana | Whole plant | Spherical; 5–50 nm | S. aureus, B. cereus, A. baumannii, P. aeruginosa | 304 |
| 27 | Linum usitatissimum L. | Whole plant | Spherical; 49–54 nm, 19–24 nm respectively | E. coli, K. pneumoniae, S. aureus | 305 |
| 28 | Elaeagnus umbellata | Whole plant | Spherical; 20–100 nm | E. coli, S. aureus | 306 |
| 29 | Sida cordifolia | Whole plant | Spherical; 3–8 nm | B. subtilis, S. aureus, E. coli, K. pneumonia, A. hydrophila, P. fluorescence, F. branchiophilum, E. tarda, Y. ruckeri | 307 |
| 30 | Sida acuta | Whole plant | Spherical; 14.9 nm | E. coli, S. aureus, S. faecalis | 308 |
| 31 | Rheum ribes | Whole plant | Spherical; 3.32 ± 0.58 nm | S. pyogenes, S. aureus, S. typhimurium, E. coli | 309 |
| 32 | Blumea eriantha | Whole plant | Spherical; 50 nm | B. cereus, B. subtilis, S. aureus, E. coli | 310 |
| 33 | Arnicae anthodium | Whole plant | Irregular; 90–118 nm | S. aureus, E. coli, P. aeruginosa, C. albicans | 311 |
| 34 | Salacia chinensis | Whole plant | Irregular; 20–80 nm | S. aureus, P. aeruginosa, E. coli, S. typhi | 312 |
| 35 | Ferocactus echidne | Whole plant | Elliptical; 20–60 nm | E. coli, S. aureus, C. Albicans | 313 |
| 36 | Rosa indica | Petals | Spherical; 23.52–60.83 nm | S. mutans, E. coli, K. pneumoniae, E. faecalis | 43 |
| 37 | Hibiscus rosa-sinensis | Petals | Cube; 76.25 ± 0.17 nm | V. cholerae, E. coli, K. pneumoniae, S. aureus | 314 |
| 38 | Clitoria ternatea L. | Petals | Spherical and flat plate; 35–80 nm | S. aureus, Shigella sp. | 39 |
| 39 | Euphorbia antiquorum L. | Latex | Spherical; 10–50 nm | E. coli, K. pneumoniae, P. mirabilis, V. cholerae, E. faecalis | 315 |
| 40 | Calotropis gigantea L. | Latex | Spherical; 5–30 nm | Bacillus, Enterococci, Shigella, P. aeruginosa, K. pneumonia, Staphylococcus, E. coli | 316 |
| 41 | Cocoa | Pod | Spherical; 4–32 nm | E. coli, K. pneumoniae, S. pyogenes, S. aureus P. aeruginosa, A. flavus, A. fumigatus and A. niger | 317 |
| 42 | Cola nitida | Pod | Spherical; 12–80 nm | K. granulomatis, P. aeruginosa, E. coli, S. aureus, A. niger, A. flavus, A. fumigatus | 318 |
| 43 | Taxus yunnanensis | Callus | Spherical; 6.4–27.2 nm | E. coli, S. aureus, S. paratyphi, B. subtilis | 319 |
| 44 | Chlorophytum borivilianum L. | Callus | Spherical; 52.0 nm | P. aeruginosa, B. subtilis, Methicillin-resistant Escherichia coli, S. aureus, C. albicans | 320 |
3.10 From peels
Recently, utilization of peel extract as a reducing agent for the green synthesis of nanoparticles with selective size gaining immense attention. To date, various peel extract was used for the biosynthesis of AgNPs (Table 4, entries 9–17). Peel extract of Cavendish banana,45 banana (Musa paradisiaca).288 Carica papaya289 and Citrus sinensis290 were utilized for the green synthesis of AgNPs. The synthesized AgNPs were tested against various bacterial strains to investigate its bactericidal activity and found that the AgNPs obtained from various peel extract displayed great cell disruption. The influence of various factors on the bio-reduction of AgNO3 was investigated and found that 1.75 mM AgNO3, 20.4 mg dry banana peel 4.5 pH and 72 h incubation time. The authors reported that the synthesized AgNPs displayed synergistic effects with the antibiotic levofloxacin.288 Citrus maxima peel extract was utilized as both reducing and capping agent for the green synthesis of AgNPs with size ranging from 4–11 nm. Bactericidal activity of the synthesized AgNPs was tested against E. coli and S. aureus and found that the Citrus maxima peel extract mediated AgNPs showed great inhibitory action against both the bacterial strain. Furthermore, bactericidal activity was also examined against plant pathogens such as F. oxysporum and V. dahlia, showed excellent inhibitory action against both the pathogens.291 Besides, Punica granatum,292 Citrus × clementine,293 and Solanum melongena L.294 peel extracts were exploited for the green synthesis of AgNPs and tested its bactericidal activity against various bacterial strains. The results revealed that the synthesized AgNPs displayed great cell wall damage of both Gram-positive and Gram-negative bacteria. Interestingly, AgNPs derived from Punica granatum peel extract showed high ZOI against S. aureus (16.5 mm) compared to E. coli (15.5 mm).292 Microwave irradiation technique has been utilized for the green synthesis of AgNPs from Solanum melongena L. peel extract to increase the rate of bioreduction process of AgNO3. Moreover, the size and the shape of the nanoparticles generated via this process is 92.4 nm and spherical.294 In another work, Dutta et al.295 have reported the green synthesis of AgNPs using Citrus limetta peel extract as both reducing and capping agent. Investigation of bactericidal activity of the synthesized AgNPs against various pathogens such as C. albicans, C. glabrata, C. parapsilosis, C tropicalis, M. luteus, S. mutans, S. epidermidis, S. aureus, E. coli revealed that the Citrus limetta peel extract mediated AgNPs have cell disruption potential and hence can be used in pharmaceutical industries. Furthermore, the antifungal activity test of the synthesized AgNPs against Candida species revealed that the nanoparticle has the ability of cell membrane distortion. The effect of AgNPs on micro-morphological changes of C. albicans was clearly visible and found that AgNPs induces the cell blebs and a thick exudate deposition around the cell that demonstrate the leakage of intercellular components. From the results, the authors reported that Citrus limetta peel extract mediated AgNPs have excellent antifungal activity.3.11 From tube/bulb
A diverse tube/bulb extract of plants are reported for the green synthesis of AgNPs (Table 4, entries 18–22). Recently, onion (Allium cepa) extract was employed as a reducing as well as a capping agent for the biosynthesis of spherical AgNPs with size ranging from 10–23 nm. The authors reported that the synthesized AgNPs have excellent antimicrobial activity against B. subtilis, B. cereus, B. licheniformis, S. aureus, S. mutans, E. coli, K. pneumoniae, S. typhimurium, P. aeruginosa, P. vulgaris, S. marcescens, C. albicans.296 A wide variety of tube/bulb extract such as Sunroot tuber,297 Dioscorea alata298 and Crocus haussknechtii Bois299 were utilized for the green synthesis of AgNPs and examined the bactericidal activity of the synthesized nanoparticles against both Gram-positive and Gram-negative bacterial strains. It is reported that, spherical AgNPs with size range 10–25 nm was obtained using 20 mM AgNO3, 0.5 mL Crocus haussknechtii Bois extract at pH 7 and temperature of 75 °C.299 An obvious result was obtained in case of Dioscorea alata mediated AgNPs, where it is observed that the ZOI for E. coli is greater than S. aureus.298 In another work, aqueous extract of Allium sativum was employed as both reducing and capping agent for the synthesis of AgNPs. To investigate the antimicrobial activity, the authors have applied the synthesized NPs to the pathogenic bacteria such as P. aeruginosa and B. licheniformis and found that the nanoparticle has the cell permeable ability and hence can be used in biomedical applications to make antimicrobial drug.3003.12 From the whole plant
The exploitation of plant extract in the biosynthesis of AgNPs is an important field in nanobiotechnology. To date, numerous literatures are available for the green synthesis of AgNPs using plant extract (Table 4, entries 23–35). Sargassum muticum,301 Brassica oleracea L. (Broccoli),302 Vernonia cinerea L.,303 Artemisia marschalliana304 and Linum usitatissimum L.305 plant extracts were exploited for the green synthesis of AgNPs. The resulted nanoparticles were utilized against both Gram-negative and Gram-positive bacterial strains to investigate its bactericidal activity, and it is observed that the plant extract mediated AgNPs are capable of bacterial cell damage at a very low concentration. Different concentration of AgNPs derived from Artemisia marschalliana plant extract was used against S. aureus, B. cereus, A. baumannii, and P. aeruginosa and found that the ZOI is highest for S. aureus unlike other plant extract mediated AgNPs. Therefore, the authors claimed that the photosynthesized AgNPs from Artemisia marschalliana plant extract can compete with the commercial antibiotics.304 In comparison of the bactericidal activity of AgNPs derived from callus extract and whole plant extract revealed that callus extract mediated AgNPs are smaller in size and thus displayed high bactericidal activity.305 Ali et al.306 have reported shape/size-selective green synthesis of AgNPs using Elaeagnus umbellate extract and treated against various bacterial pathogens such as E. coli and S. aureus to examine its bactericidal activity. The results showed that the Elaeagnus umbellate extract mediated AgNPs can effectively damage the cell membranes as well as releases cellular matrix and hence can be used in pharmaceutics. To further investigate the morphological changes of the bacteria S. aureus and E. coli, SEM analysis was performed, where the images displayed that before AgNPs treatment the cell membranes of the two bacteria remain intake and have a regular morphology. However, after the treatment of AgNPs, no definite cell wall was observed, and membrane disruption occurs. Besides, antimicrobial activity of AgNPs derived from plant extract of Sida cordifolia,307 Sida acut,308 Rheum ribes,309 Blumea eriantha310 and Arnicae anthodium311 were examined against various bacterial strains. The small size of AgNPs (50 nm) derived from Blumea eriantha plant extract displayed excellent growth inhibition of bacterial cell as it provides a high surface area to the pathogens and thus effects more compared to the larger AgNPs.310 Similarly, Salacia chinensis extract was employed for the green synthesis of AgNPs and examine their antimicrobial activity against S. aureus, P. aeruginosa, E. coli, S. typhi. The antimicrobial activity testing results revealed that the Salacia chinensis extract mediated AgNPs showed high inhibition activity against S. aureus and P. aeruginosa. However, it showed minimum inhibition activity against E. coli and S. typhi.312 Plant extract of Ferocactus echidne was utilized for the green synthesis of AgNPs. The synthesized nanoparticles were utilized against various human pathogens and found that the nanoparticle is active against both Gram-positive and Gram-negative bacteria.3133.13 From petals, latex, pod and callus
Different parts of plants such as petals (Table 4, entries 36–38), latex (entries 39 and 40), pod (entries 41 and 42) and callus (entries 43 and 44) in the form of their aqueous/alcoholic extract have been utilized for the green synthesis of AgNPs. It is reported that Rosa indica43 and Hibiscus rosa-sinensis314 petal extract were utilized for the green synthesis of spherical AgNPs. Both the petal extract mediated AgNPs displayed good bactericidal activity against Gram-negative bacteria compared to Gram-positive bacteria.43,314 The size of the synthesized nanoparticle plays a vital role in the bactericidal activity test. Smaller the size of the nanoparticle greater is the surface available to adhere to the microorganisms, which led to the change in the Physico-chemical properties of the bacterial cell and finally led to bacterial cell damage.314 Vanaraj et al.39 reported the green synthesis of AgNPs by using Clitoria ternatea L. extract as a bioreducing agent and examined their antimicrobial activity against S. aureus and Shigella sp. and found that the synthesized AgNPs can effectively disrupt the cell membranes of both the bacterial pathogens.Latex extract of Euphorbia antiquorum L. was employed for the green synthesis of AgNPs with size ranging from 10–50 nm. Antimicrobial activity of the synthesized AgNPs was tested against various human pathogens such as E. coli, K. pneumoniae, P. mirabilis, V. cholera and E. faecalis and showed mild inhibition activity against all mentioned pathogens.315 Similarly, antimicrobial activity of spherical AgNPs derived from Calotropis gigantea L. against various human pathogens has been investigated and displayed remarkable activity against both Gram-positive and Gram-negative bacteria.316 Pod extract of Cocoa was utilized for the biosynthesis of AgNPs. The synthesized nanoparticles showed great inhibition against E. coli and K. pneumonia. Moreover, the nanoparticle improves the activity of cefuroxime and ampicillin synergistically.317 In addition, Lateef et al.318 have reported the green synthesis of AgNPs using pod extract of Cola nitida as a reducing as well as capping and stabilizing agent. Antimicrobial activity of the synthesized AgNPs revealed that at different AgNPs concentration ranging from 50–150 μg mL−1 showed great inhibition activity against K. granulomatis, P. aeruginosa, and E. coli. Besides, incorporation of 5 μg mL−1 of pod extract of Cola nitida mediated AgNPs into the paint completely inhibits the growth of bacteria such as S. aureus, E. coli, P. aeruginosa, A. niger, A. flavus and A. fumigatus and hence can be utilized in paint manufacture industries and biomedical.
Recently, callus extract of Taxus yunnanensis has been employed as a reducing and stabilizing agent for the green synthesis of AgNPs and examined their bactericidal activity against both Gram-positive and Gram-negative bacteria. The bactericidal activity test of the synthesized AgNPs revealed that the inhibition effect is more pronounced in case of Gram-positive compared to Gram-negative bacteria. Therefore, callus extract of Taxus yunnanensis mediated AgNPs can be used in antibiotic therapeutics, an alternative to the antibacterial drug.319 Spherical and well-dispersed AgNPs were also prepared from callus extract of Chlorophytum borivilianum L. It is reported that the synthesized nanoparticle can effectively inhibit almost all kinds of human pathogens.320
4. Mechanism of antibacterial inhibition by bioinspired AgNPs
The actual mode and reactive species, whether AgNPs321–323 or the released Ag+,80 in the bactericidal activity of AgNPs is not well established to date and is still a topic of hot debate. However, most of the recent studies revealed that released Ag+, not the actual AgNPs, is possible the antimicrobial agent that causes cell damage and consequent death.11,324,325 Several pathways have been that proposed for the bactericidal activity of AgNPs which include the generation of reactive oxygen species,326 free radicals derived from the surface of AgNPs,327 silver ion stress,328 coating agents,329 interactions with the bacterial cell that leads to depletion of intracellular ATP level322 and damage in respiratory enzymes.330The possible mechanism for the antibiotic activity of AgNPs is displayed in Fig. 9. It is reported that the smaller the size of AgNPs greater is the bactericidal activity as it provides a greater surface to the bacterial membrane. The interaction between the positively charged Ag ion with the negatively charged cell membranes led to the disruption of the cell morphology and hence cell leakage occurred, resulting in cell death. Besides, AgNPs bind strongly with phosphorus and sulfur of the extracellular and intracellular membrane proteins, thus affects the cell replication, respiration and finally, the lifetime of the cell. Apart from that, AgNPs can also bind with the thiol and amino groups of membrane protein and led to the formation of reactive oxygen species (ROS), which inhibits the cell respiration. The excellent bactericidal activity of AgNPs can be attributed to the interaction with the plasma membrane and peptidoglycan cell wall of the bacterial strain.331 It has also been suggested that the interaction of AgNPs with cell wall increases the membrane permeability by forming pores or pits and thereby causing the death of bacteria.332,333
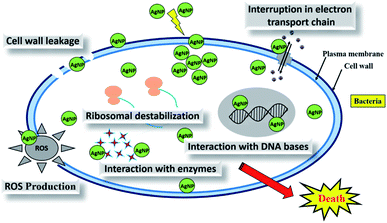 | ||
| Fig. 9 Possible mechanism for the bactericidal activity of AgNPs. This figure has been reproduced from ref. 331 with permission from Elsevier, copyright 2004. | ||
5. Conclusion and future outlook
Taking into account the many benefits of green synthesis of AgNPs using plant extracts and their excellent antimicrobial activities as bare or in conjugation with antibiotic drugs, there is no doubt that this research field will continue to attract much interest in recent years. Here different biogenic methods for the synthesis of AgNPs using phytochemicals, nontoxic, inexpensive, and eco-friendly route has been comprehensively reviewed. The antimicrobial susceptibility of the produced AgNPs against several pathogenic microbes has also been highlighted. Although the rapid and green synthetic methods using plant extracts have shown great potential in AgNPs, understanding the mechanism by which phytochemicals of these plants are involved in the synthesis and the mode of antimicrobial inhibition are still not fully understood. In addition, controlling the shape of biosynthesized AgNPs, which have many positive effects on its activities, remained largely unanswered till today although chemical methods are already well-known for shape-controlled synthesis. This problem is potentially due to the large number of different phytochemicals present in the plant extract, making it difficult for a systematic control of the interaction with the produced AgNPs. Hence, better understanding of each phytochemical, quantities and their interaction will pave the way for shape-selective synthesis of biogenic NPs. In general, the smaller the NPs better its antimicrobial activities due to the increase in surface are that are in contact with the microbial cell.118,141 Of the same size range, antimicrobial activities of AgNPs are in the order; triangular > pentagonal, hexagonal, cubic, nano-rod > spherical. Triangular one showed the highest activity mainly due to better edge fitting due to sharp edge and predominant stable (1 1 1) facet.109,112,113 Hexagonal, cubic, nano-rod have bend edge, which might have reduce their efficacy towards microbes as compared to triangular shape NPs.334 whereas spherical shape NPs with no sharp edge and predominantly (1 0 0) facets showed least antimicrobial effects.113Several authors revealed that Gram-positive bacteria (e.g. S. aureus), due to their thick cell wall of peptidoglycan layer (∼20–80 nm thick), are less susceptible to AgNPs than Gram-negative bacteria (e.g. E. coli) with cell wall consisting of lipopolysaccharides at the exterior, followed underneath by layer of peptidoglycan (∼7–8 nm).75,102,103,107,108,118,130,166,236 However, this is not the case everywhere.108,154,194,228,230,280 In the light of this, one must look into the role of lipopolysaccharides in Gram-negative that might have acted as a shield against some AgNPs and also the synergetic effect of AgNPs and biomolecules that act as a capping that might have alter the mode of interaction of NPs with the cell wall. Hence understanding the underlying mechanism of the interaction is still a challenge.
The antimicrobial efficacy of AgNPs can be greatly enhance by its synergistic interaction with many well-known antibiotic drugs.10,11,77,80,104,127,183,261,288,317 This opens a new and exciting opportunity in combating numerous newly evolved highly infectious multi drug-resistant microbes. Hence, this research field has become a ‘hot’ topic in recent years although it is in its infancy. To have a better insight, understanding the mechanism of interaction of the AgNPs with drugs and the alteration in the mode of attack due to the synergetic interaction towards the microbes needs to be well understood and validated experimentally.
The successful green synthesis of AgNPs and evaluation, understanding the antimicrobial activities is a complex process till today although this research field has been explored several decades. However, looking at the literature we can draw several assumptions which potentially provide us AgNPs with high antimicrobial activities. Hence, knowing the complexity of the research on the green synthesis and antimicrobial activity of AgNPs, the below points are worth considered during AgNPs synthesis:
(1) Chemical composition of the plant extract
It is believed that the oxidation of different biomolecules such as flavonoids, ketones, aldehydes, tannins, carboxylic acids, phenolic and the protein of the plant are mainly responsible for the reduction of Ag+ to Ag0. In addition, the stability and size of the produced AgNPs depends on the biomolecules acting as a capping agent.138 Hence, one must first investigate the biomolecules present in the plant extract and its capping efficacy for successful synthesis of AgNPs. In general, greater the capping activity greater the stability and lesser is the average particle size of NPs.335,336 However, this is not true in some case.337,338 Hence, specific interaction of the biomolecules and the formed NPs also need attention on case to case basis.(2) Concentration of the plant extract
The shape and size of the synthesized AgNPs depend on the concentration of the plant extract used. At dilute concentration, formation of the NPs may not even take places; hence one need to investigate carefully. Formation of AgNPs is usually accompanied by colour change and prominent UV-Vis absorption at around 430 cm−1. Increase in extract concentration leads to formation a large number of NPs to a certain level.76 However, while desired concentration of extract can afford a highly disperse AgNPs with high antimicrobial activity, high concentration of extract often leads to agglomeration and large NPs, as excess reducing agents potential caused secondary reduction process on the surface of the preformed nuclei.339(3) Concentration of AgNO3
The number of AgNPs increased with the increase in AgNO3 concentration up to the level where all the AgNO3 salt are consumed i.e. all Ag+ are reduced to Ag0, which can be easily monitored by increasing intensities in UV-Vis spectroscopy. Once all the AgNO3 are consumed, an equilibrium will be reach. Hence one need to see the balance between the AgNO3 and the amount of reducing agent present in the extract.76,235,261(4) Extraction solvent
Different biochemicals in the plant has different level of solubility in solvents; hence the successful extraction of the desired biochemicals for synthesis of AgNPs largely depends on the extraction solvent used. Phenolic compounds are known to be highly soluble in ethanol, methanol as well as their mixture with water (ethanol–water or methanol–water).43,185,199,200,204 Hence, these are a solvent of choice for extraction along with pure water which is most used.(5) Extraction time and temperature
Another important factor to consider for the successful synthesis of biogenic AgNPs is the extraction temperature. It is well known that solubility of biochemicals increased with increase in extraction temperature and time. Hence, more chemical will be extracted at a higher temperature which will make it a strong reducing agent. But there is a possibility of extracting a non-reactive biochemicals or decomposition of biochemicals at a long time at higher temperature.185,187(6) pH
pH can change the electrical charges of biomolecules in the plant extract that might have affect the nature of their capping and stabilizing affinity and subsequently the growth of NPs. Increased in pH usually resulted in the increase rate of formation as well as promotes homogeneous distribution of size of NPs.149,340 However, under acidic condition slow formation and agglomeration took place resulting in larger NPs.151,152,341 At the same time, high pH can lead to precipitation of AgOH which is undesirable.150 Hence, neutral pH (7) is highly recommended if an external buffer is used.(7) Reaction time
The size of the NPs are reported to increases with time74,111,163 as indicated by a red-shift in UV-Vis. Spectrometer data. Hence vigilant monitoring of the reaction to get a stable small size NPs is critical.(8) Reaction temperature
High temperature is usually required to achieve complete reduction of AgNO3 to AgNPs using chemical route,337,342 although from economic and green chemistry prospective, RT reaction is the best choice. However, when it comes to green NPs synthesis, RT process, despite there are some exceptions, usually afford spherical shape NPs which are less susceptible to microbes as mentioned earlier. In the meantime, synthesis of different shapes of NPs for specific purpose is highly desirable. Literature review revealed that formation of cubic,334,343 pentagonal, hexagonal,111 triangular, rod-shape nanowire343 AgNPs happen usually above RT, although some other parameters such as capping agents and stabilizers concentration needs to be taken care. Hence, in addition to increasing the speed of reaction and decreasing the size of NPs with temperature, one must consider the reaction temperature to produce NPs with different shape for a specific purpose, particularly as a potent antimicrobial.Conflicts of interest
None to declare.References
- M. Ayelén Vélez, M. Cristina Perotti, L. Santiago, A. María Gennaro and E. Hynes, Bioactive compounds delivery using nanotechnology: design and applications in dairy food, Elsevier Inc., 2017 Search PubMed.
- A. Bera and H. Belhaj, J. Nat. Gas Sci. Eng., 2016, 34, 1284–1309 CrossRef CAS.
- L. J. Frewer, N. Gupta, S. George, A. R. H. Fischer, E. L. Giles and D. Coles, Trends Food Sci. Technol., 2014, 40, 211–225 CrossRef CAS.
- V. J. Mohanraj and Y. Chen, Trop. J. Pharm. Res., 2007, 5, 561–573 Search PubMed.
- L. Stadler, M. Homafar, A. Hartl, S. Najafishirtari, R. Zbo, M. Petr, M. B. Gawande, J. Zhi and O. Reiser, ACS Sustainable Chem. Eng., 2019, 7, 2388–2399 CrossRef CAS.
- M. D. Purkayastha and A. K. Manhar, Nanosci. Food Agri., 2016, 2, 59–128 Search PubMed.
- S. Chatterjee, Dhanurdhar and L. Rokhum, Renewable Sustainable Energy Rev., 2017, 72, 560–564 CrossRef CAS.
- S. Bagheri and N. M. Julkapli, J. Magn. Magn. Mater., 2016, 416, 117–133 CrossRef CAS.
- A. P. Ingle, A. Biswas, C. Vanlalveni, R. Lalfakzuala, I. Gupta, P. Ingle, L. Rokhum and M. Rai, Microb. Bionanotechnol., 2020, 135–161 Search PubMed.
- C. Medina, M. J. Santos-Martinez, A. Radomski, O. I. Corrigan and M. W. Radomski, Br. J. Pharmacol., 2007, 150, 552–558 CrossRef CAS.
- H. Deng, D. McShan, Y. Zhang, S. S. Sinha, Z. Arslan, P. C. Ray and H. Yu, Environ. Sci. Technol., 2016, 50, 8840–8848 CrossRef CAS.
- C. Xu, O. U. Akakuru, J. Zheng and A. Wu, Front. Bioeng. Biotechnol., 2019, 7, 141 CrossRef.
- S. Bagheri, M. Yasemi, E. Safaie-Qamsari, J. Rashidiani, M. Abkar, M. Hassani, S. A. Mirhosseini and H. Kooshki, Artif. Cells, Nanomed., Biotechnol., 2018, 46, 462–471 CrossRef CAS.
- A. Muthuraman, N. Rishitha and S. Mehdi, in Design of Nanostructures for Theranostics Applications, 2018, pp. 529–562 Search PubMed.
- S. Tortorella and T. C. Karagiannis, in Molecular Mechanisms and Physiology of Disease: Implications for Epigenetics and Health, 2014 Search PubMed.
- S. Ahmed, M. Ahmad, B. L. Swami and S. Ikram, J. Adv. Res., 2016, 7, 17–28 CrossRef CAS.
- S. R. Vijayan, P. Santhiyagu, R. Ramasamy, P. Arivalagan, G. Kumar, K. Ethiraj and B. R. Ramaswamy, Enzyme Microb. Technol., 2016, 95, 45–57 CrossRef CAS.
- S. Ahmed, Annu, S. Ikram and S. Yudha, J. Photochem. Photobiol., B, 2016, 161, 141–153 CrossRef CAS.
- P. Mohanpuria, N. K. Rana and S. K. Yadav, J. Nanopart. Res., 2008, 10, 507–517 CrossRef CAS.
- G. Pathak, K. Rajkumari and L. Rokhum, Nanoscale Adv., 2019, 1, 1013–1020 RSC.
- S. A. Saiqa Ikram, J. Nanomed. Nanotechnol., 2015, 6, 1000309 CrossRef.
- S. Ahmed and S. Ikram, Nano Res. Appl., 2015, 1, 1–6 Search PubMed.
- K. Rajkumari, D. Das, G. Pathak and L. Rokhum, New J. Chem., 2019, 43, 2134–2140 RSC.
- B. Changmai, I. B. Laskar and L. Rokhum, J. Taiwan Inst. Chem. Eng., 2019, 102, 276–282 CrossRef CAS.
- B. Changmai, P. Sudarsanam and L. Rokhum, Ind. Crops Prod., 2020, 145, 111911 CrossRef CAS.
- B. Nath, B. Das, P. Kalita and S. Basumatary, J. Cleaner Prod., 2019, 239, 118112 CrossRef CAS.
- S. Nour, N. Baheiraei, R. Imani, M. Khodaei, A. Alizadeh, N. Rabiee and S. M. Moazzeni, J. Mater. Sci.: Mater. Med., 2019, 30, 120 CrossRef.
- O. Bondarenko, K. Juganson, A. Ivask, K. Kasemets, M. Mortimer and A. Kahru, Arch. Toxicol., 2013, 87, 1181–1200 CrossRef CAS.
- A. Pal, S. Shah and S. Devi, Mater. Chem. Phys., 2009, 114, 530–532 CrossRef CAS.
- T. Wu, H. Shen, L. Sun, B. Cheng, B. Liu and J. Shen, ACS Appl. Mater. Interfaces, 2012, 4, 2041–2047 CrossRef CAS.
- X. Zhang, H. Sun, S. Tan, J. Gao, Y. Fu and Z. Liu, Inorg. Chem. Commun., 2019, 100, 44–50 CrossRef CAS.
- V. R. Remya, V. K. Abitha, P. S. Rajput, A. V. Rane and A. Dutta, Chem. Int., 2019, 3, 165–171 Search PubMed.
- M. Rafique, I. Sadaf, M. S. Rafique and M. B. Tahir, Artif. Cells, Nanomed., Biotechnol., 2017, 45, 1272–1291 CrossRef CAS.
- C. Vanlalveni, K. Rajkumari, A. Biswas, P. P. Adhikari, R. Lalfakzuala and L. Rokhum, Bionanoscience, 2018, 8, 624–631 CrossRef.
- N. Durán, P. D. Marcato, G. I. H. De Souza, O. L. Alves and E. Esposito, J. Biomed. Nanotechnol., 2007, 3, 203–208 CrossRef.
- A. J. Kora, R. B. Sashidhar and J. Arunachalam, Carbohydr. Polym., 2010, 82, 670–679 CrossRef CAS.
- A. Ahmad, Y. Wei, F. Syed, K. Tahir, A. U. Rehman, A. Khan, S. Ullah and Q. Yuan, Microb. Pathog., 2017, 102, 133–142 CrossRef CAS.
- S. Sumitha, S. Vasanthi, S. Shalini, S. V. Chinni, S. C. B. Gopinath, P. Anbu, M. B. Bahari, R. Harish, S. Kathiresan and V. Ravichandran, Molecules, 2018, 23, 3311 CrossRef.
- S. Vanaraj, B. B. Keerthana and K. Preethi, J. Inorg. Organomet. Polym. Mater., 2017, 27, 1412–1422 CrossRef CAS.
- S. M. Ali, N. M. H. Yousef and N. A. Nafady, J. Nanomater., 2015, 1–10 Search PubMed.
- V. Dhand, L. Soumya, S. Bharadwaj, S. Chakra, D. Bhatt and B. Sreedhar, Mater. Sci. Eng., C, 2016, 58, 36–43 CrossRef CAS.
- B. Ajitha, Y. Ashok Kumar Reddy, S. Shameer, K. M. Rajesh, Y. Suneetha and P. Sreedhara Reddy, J. Photochem. Photobiol., B, 2015, 194, 84–92 CrossRef.
- R. Manikandan, B. Manikandan, T. Raman, K. Arunagirinathan, N. M. Prabhu, M. Jothi Basu, M. Perumal, S. Palanisamy and A. Munusamy, Spectrochim. Acta, Part A, 2015, 138, 120–129 CrossRef CAS.
- A. Biswas, C. Vanlalveni, P. P. Adhikari, R. Lalfakzuala and L. Rokhum, IET Nanobiotechnol., 2018, 12, 933–938 CrossRef.
- T. Kokila, P. S. Ramesh and D. Geetha, Appl. Nanosci., 2015, 5, 911–920 CrossRef CAS.
- V. Kathiravan, S. Ravi, S. Ashokkumar, S. Velmurugan, K. Elumalai and C. P. Khatiwada, Spectrochim. Acta, Part A, 2015, 139, 200–205 CrossRef CAS.
- A. K. Jha, K. Prasad and A. R. Kulkarni, Colloids Surf., B, 2009, 71, 226–229 CrossRef CAS.
- N. Vigneshwaran, N. M. Ashtaputre, P. V. Varadarajan, R. P. Nachane, K. M. Paralikar and R. H. Balasubramanya, Mater. Lett., 2007, 61, 1413–1418 CrossRef CAS.
- S. Shivaji, S. Madhu and S. Singh, Process Biochem., 2011, 46, 1800–1807 CrossRef CAS.
- M. J. Ahmed, G. Murtaza, A. Mehmood and T. M. Bhatti, Mater. Lett., 2015, 153, 10–13 CrossRef CAS.
- A. Miri, M. Sarani, M. Rezazade Bazaz and M. Darroudi, Spectrochim. Acta, Part A, 2015, 141, 287–291 CrossRef CAS.
- S. Medda, A. Hajra, U. Dey, P. Bose and N. K. Mondal, Appl. Nanosci., 2014, 5, 875–880 CrossRef.
- P. Premasudha, M. Venkataramana, M. Abirami, P. Vanathi, K. Krishna and R. Rajendran, Bull. Mater. Sci., 2015, 38, 965–973 CrossRef CAS.
- B. Ajitha, Y. A. K. Reddy and P. S. Reddy, J. Photochem. Photobiol., B, 2015, 146, 1–9 CrossRef CAS.
- M. Kumara Swamy, K. M. Sudipta, K. Jayanta and S. Balasubramanya, Appl. Nanosci., 2014, 5, 73–81 CrossRef.
- S. R. Goswami, T. Sahareen, M. Singh and S. Kumar, J. Ind. Eng. Chem., 2015, 26, 73–80 CrossRef CAS.
- M. Harshiny, M. Matheswaran, G. Arthanareeswaran, S. Kumaran and S. Rajasree, Ecotoxicol. Environ. Saf., 2015, 121, 135–141 CrossRef CAS.
- N. Krithiga, A. Rajalakshmi and A. Jayachitra, J. Nanosci., 2015, 1, 128204 Search PubMed.
- S. S. Sana, V. R. Badineni, S. K. Arla and V. K. Naidu Boya, Mater. Lett., 2015, 145, 347–350 CrossRef CAS.
- P. Velmurugan, M. Cho, S. S. Lim, S. K. Seo, H. Myung, K. S. Bang, S. Sivakumar, K. M. Cho and B. T. Oh, Mater. Lett., 2015, 138, 272–275 CrossRef CAS.
- D. Bose and S. Chatterjee, Indian J. Microbiol., 2015, 55, 163–167 CrossRef CAS.
- G. Marslin, R. K. Selvakesavan, G. Franklin, B. Sarmento and A. C. P. Dias, Int. J. Nanomed., 2015, 10, 5955–5963 CAS.
- B. Sadeghi, A. Rostami and S. S. Momeni, Spectrochim. Acta, Part A, 2015, 134, 326–332 CrossRef CAS.
- A. Devadiga, K. V. Shetty and M. B. Saidutta, Int. Nano Lett., 2015, 5, 205–214 CrossRef CAS.
- R. K. Salar, P. Sharma and N. Kumar, Resour.-Effic. Technol., 2015, 1, 106–115 Search PubMed.
- A. Verma and M. S. Mehata, J. Radiat. Res. Appl. Sci., 2016, 9, 109–115 CrossRef CAS.
- V. Ravichandran, S. Vasanthi, S. Shalini, S. Adnan and A. Shah, Mater. Lett., 2016, 180, 264–267 CrossRef CAS.
- S. Ahmed, A. K. Manzoor and S. Ikram, J. Bionanosci., 2016, 10, 282–287 CrossRef CAS.
- B. Sundararajan, G. Mahendran, R. Thamaraiselvi and B. D. Ranjitha Kumari, Bull. Mater. Sci., 2016, 39, 423–431 CrossRef CAS.
- D. Bose and S. Chatterjee, Appl. Nanosci., 2016, 6, 895–901 CrossRef CAS.
- Y. K. Mohanta, S. K. Panda, K. Biswas, A. Tamang, J. Bandyopadhyay, D. De, D. Mohanta and A. K. Bastia, IET Nanobiotechnol., 2016, 10, 438–444 CrossRef.
- C. S. Espenti, K. S. V. K. Rao and K. M. Rao, Mater. Lett., 2016, 147, 129–133 CrossRef.
- K. Anandalakshmi, J. Venugobal and V. Ramasamy, Appl. Nanosci., 2016, 6, 399–408 CrossRef CAS.
- S. Ahmed, Saifullah, M. Ahmad, B. L. Swami and S. Ikram, J. Radiat. Res. Appl. Sci., 2016, 9, 1–7 CrossRef.
- K. Khanra, S. Panja, I. Choudhuri, A. Chakraborty and N. Bhattacharyya, Nanomed. J., 2015, 7, 128–133 CAS.
- J. L. López-Miranda, M. Vázquez, N. Fletes, R. Esparza and G. Rosas, Mater. Lett., 2016, 176, 285–289 CrossRef.
- K. Jyoti, M. Baunthiyal and A. Singh, J. Radiat. Res. Appl. Sci., 2016, 9, 217–227 CrossRef CAS.
- S. Soman and J. G. Ray, J. Photochem. Photobiol., B, 2016, 163, 391–402 CrossRef CAS.
- B. Ajitha, Y. A. K. Reddy, P. S. Reddy, Y. Suneetha, H. J. Jeon and C. W. Ahn, J. Mol. Liq., 2016, 219, 474–481 CrossRef CAS.
- D. McShan, Y. Zhang, H. Deng, P. C. Ray and H. Yu, J. Environ. Sci. Health, Part C: Environ. Carcinog. Ecotoxicol. Rev., 2015, 33, 369–384 CrossRef CAS.
- V. P. Manjamadha and K. Muthukumar, Bioprocess Biosyst. Eng., 2016, 39, 401–411 CrossRef CAS.
- N. Chauhan, A. K. Tyagi, P. Kumar and A. Malik, Front. Microbiol., 2016, 7, 1748 Search PubMed.
- D. K. Verma, S. H. Hasan and R. M. Banik, J. Photochem. Photobiol., B, 2016, 155, 51–59 CrossRef CAS.
- B. Ajitha, Y. Ashok Kumar Reddy, K. M. Rajesh and P. Sreedhara Reddy, Mater. Today: Proc., 2016, 3, 1977–1984 Search PubMed.
- G. Kuppurangan, B. Karuppasamy, K. Nagarajan, R. Krishnasamy Sekar, N. Viswaprakash and T. Ramasamy, Appl. Nanosci., 2015, 6, 973–982 CrossRef.
- A. R. Allafchian, S. Z. Mirahmadi-Zare, S. A. H. Jalali, S. S. Hashemi and M. R. Vahabi, J. Nanostruct. Chem., 2016, 6, 129–135 CrossRef CAS.
- R. Kumari, G. Brahma, S. Rajak, M. Singh and S. Kumar, Orient. Pharm. Exp. Med., 2016, 16, 195–201 CrossRef.
- O. Azizian-Shermeh, A. Einali and A. Ghasemi, Adv. Powder Technol., 2017, 28, 3167–3171 Search PubMed.
- B. Bhuyan, A. Paul, B. Paul, S. S. Dhar and P. Dutta, J. Photochem. Photobiol., B, 2017, 173, 210–215 CrossRef CAS.
- S. Mahadevan, S. Vijayakumar and P. Arulmozhi, Microb. Pathog., 2017, 113, 445–450 CrossRef CAS.
- E. E. Elemike, D. C. Onwudiwe, O. E. Fayemi, A. C. Ekennia, E. E. Ebenso and L. R. Tiedt, J. Cluster Sci., 2016, 28, 309–330 CrossRef.
- N. Soni and R. C. Dhiman, Chin. Herb. Med., 2017, 9, 289–294 CrossRef.
- Y. K. Mohanta, S. K. Panda, R. Jayabalan, N. Sharma, A. K. Bastia and T. K. Mohanta, Front. Mol. Biosci., 2017, 4, 14 Search PubMed.
- E. E. Elemike, D. C. Onwudiwe, A. C. Ekennia, R. C. Ehiri and N. J. Nnaji, Mater. Sci. Eng., C, 2017, 75, 980–989 CrossRef CAS.
- I. Ocsoy, A. Demirbas, E. S. McLamore, B. Altinsoy, N. Ildiz and A. Baldemir, J. Mol. Liq., 2017, 238, 263–269 CrossRef CAS.
- H. S. A. Al-Shmgani, W. H. Mohammed, G. M. Sulaiman and A. H. Saadoon, Artif. Cells, Nanomed., Biotechnol., 2016, 45, 1234–1240, DOI:10.1080/21691401.2016.1220950.
- K. Sahayaraj and S. Rajesh, Sci. against Microb. Pathog. Communicating Current Research and Technological Advances. 2011, vol. 23, pp. 228–244 Search PubMed.
- E. E. Elemike, D. C. Onwudiwe, A. C. Ekennia and L. Katata-Seru, Res. Chem. Intermed., 2016, 43, 1383–1394 CrossRef.
- S. V. Otari, S. H. Pawar, S. K. S. Patel, R. K. Singh, S. Y. Kim, J. H. Lee, L. Zhang and J. K. Lee, J. Microbiol. Biotechnol., 2017, 27, 731–738 CrossRef CAS.
- T. Rasheed, M. Bilal, H. M. N. Iqbal and C. Li, Colloids Surf., B, 2017, 158, 408–415 CrossRef CAS.
- P. Thatoi, R. G. Kerry, S. Gouda, G. Das, K. Pramanik, H. Thatoi and J. K. Patra, J. Photochem. Photobiol., B, 2016, 163, 311–318 CrossRef CAS.
- L. Wang, Y. Wu, J. Xie, S. Wu and Z. Wu, Mater. Sci. Eng., C, 2018, 86, 1–8 CrossRef CAS.
- R. Geethalakshmi and D. V. L. Sarada, Ind. Crops Prod., 2013, 51, 107–115 CrossRef.
- R. G. Saratale, G. Benelli, G. Kumar, D. S. Kim and G. D. Saratale, Environ. Sci. Pollut. Res., 2017, 25, 10392–10406 CrossRef.
- A. Lateef, B. I. Folarin, S. M. Oladejo, P. O. Akinola, L. S. Beukes and E. B. Gueguim-Kana, Prep. Biochem. Biotechnol., 2018, 1–7, 1479864 Search PubMed.
- R. Vijayan, S. Joseph and B. Mathew, Part. Sci. Technol., 2018, 1–11, 1450312 Search PubMed.
- D. Kavaz, H. Umar and S. Shehu, Artif. Cells, Nanomed., Biotechnol., 2019, 1–11, 1536060 Search PubMed.
- F. Erci, R. Cakir-Koc and I. Isildak, Artif. Cells, Nanomed., Biotechnol., 2017, 1–9, 1415917 Search PubMed.
- S. Pal, Y. K. Tak and J. M. Song, Appl. Environ. Microbiol., 2007, 73, 1712–1720 CrossRef CAS.
- R. Vijayan, S. Joseph and B. Mathew, Artif. Cells, Nanomed., Biotechnol., 2017, 46, 861–871 CrossRef.
- M. Kumari, S. Pandey, V. P. Giri, A. Bhattacharya, R. Shukla, A. Mishra and C. S. Nautiyal, Microb. Pathog., 2017, 105, 346–355 CrossRef CAS.
- P. Kanmani and S. T. Lim, Process Biochem., 2013, 48, 1099–1106 CrossRef CAS.
- V. K. Sharma, R. A. Yngard and Y. Lin, Adv. Colloid Interface Sci., 2009, 145, 83–96 CrossRef CAS.
- A. Biswas and L. Rokhum, Int. Conf. Syst. Process. Physics, Chem. Biol., 2018, pp. 1–7 Search PubMed.
- M. Baghayeri, B. Mahdavi, Z. Hosseinpor-Mohsen Abadi and S. Farhadi, Appl. Organomet. Chem., 2017, 32, e4057 Search PubMed.
- S. Ghotekar, A. Savale and S. Pansambal, J. Water Environ. Nanotechnol., 2018, 3, 95–105 CAS.
- A. Biswas, L. Chawngthu, C. Vanlalveni, R. Hnamte, R. Lalfakzuala and L. Rokhum, J. Bionanosci., 2018, 12, 227–232 CrossRef CAS.
- N. Manosalva, G. Tortella, M. Cristina Diez, H. Schalchli, A. B. Seabra, N. Durán and O. Rubilar, World J. Microbiol. Biotechnol., 2019, 35, 1–9 CrossRef CAS.
- S. Onitsuka, T. Hamada and H. Okamura, Colloids Surf., B, 2019, 173, 242–248 CrossRef CAS.
- E. Bernardo-Mazariegos, B. Valdez-Salas, D. González-Mendoza, A. Abdelmoteleb, O. Tzintzun Camacho, C. Ceceña Duran and F. Gutiérrez-Miceli, Rev. Argent. Microbiol., 2019, 51, 103–109 Search PubMed.
- K. Kanagamani, P. Muthukrishnan, K. Shankar, A. Kathiresan, H. Barabadi and M. Saravanan, J. Cluster Sci., 2019, 30, 1415–1424 CrossRef CAS.
- S. Paosen, S. Jindapol, R. Soontarach and S. P. Voravuthikunchai, APMIS, 2019, 127, 764–778 CrossRef CAS.
- E. H. Ibrahim, M. Kilany, H. A. Ghramh, K. A. Khan and S. ul Islam, Saudi J. Biol. Sci., 2019, 26, 1689–1694 CrossRef CAS.
- M. Nilavukkarasi, S. Vijayakumar and S. Prathip Kumar, Mater. Sci. Energy Technol., 2020, 3, 371–376 Search PubMed.
- Z. Shunying, Y. Yang, Y. Huaidong, Y. Yue and Z. Guolin, J. Ethnopharmacol., 2005, 96, 151–158 CrossRef.
- P. Moteriya and S. Chanda, J. Inorg. Organomet. Polym. Mater., 2020, 30, 3920–3932 CrossRef CAS.
- W. Huang, M. Yan, H. Duan, Y. Bi, X. Cheng and H. Yu, J. Nanomater., 2020, 1–7, 9535432 Search PubMed.
- F. Ö. Küp, S. Çoşkunçay and F. Duman, Mater. Sci. Eng., C, 2020, 107, 110207 CrossRef.
- M. A. Ramadan, A. E. Shawkey, M. A. Rabeh and A. O. Abdellatif, J. Herb. Med., 2020, 20, 100289 CrossRef.
- S. Javan bakht Dalir, H. Djahaniani, F. Nabati and M. Hekmati, Heliyon, 2020, 6, e03624 CrossRef.
- R. K. Chahande, B. A. Mehere, P. K. Pantawane, P. B. Chouke and S. R. Murai, Mater. Today: Proc., 2020, 29, 923–928 CAS.
- S. A. Khan, S. Shahid and C. S. Lee, Biomolecules, 2020, 10, 835 CrossRef CAS.
- A. O. Nyabola, P. G. Kareru, E. S. Madivoli, S. I. Wanakai and E. G. Maina, J. Inorg. Organomet. Polym. Mater., 2020, 30, 3493–3501 CrossRef CAS.
- S. Jebril, R. Khanfir Ben Jenana and C. Dridi, Mater. Chem. Phys., 2020, 248, 122898 CrossRef CAS.
- R. Parvataneni, Drug Chem. Toxicol., 2020, 43, 307–321 CrossRef CAS.
- E. S. Madivoli, P. G. Kareru, A. N. Gachanja, S. M. Mugo, D. S. Makhanu, S. I. Wanakai and Y. Gavamukulya, J. Inorg. Organomet. Polym. Mater., 2020, 30, 2842–2850 CrossRef CAS.
- A. Biswas, C. Vanlalveni, P. P. Adhikari, R. Lalfakzuala and L. Rokhum, Micro Nano Lett., 2019, 14, 799–803 CrossRef CAS.
- C. Vijilvani, M. R. Bindhu, F. C. Frincy, M. S. AlSalhi, S. Sabitha, K. Saravanakumar, S. Devanesan, M. Umadevi, M. J. Aljaafreh and M. Atif, J. Photochem. Photobiol., B, 2020, 202, 111713 CrossRef CAS.
- M. Maghimaa and S. Ali, J. Photochem. Photobiol., B, 2020, 204, 111806 CrossRef CAS.
- M. A. Asghar, E. Zahir, M. A. Asghar, J. Iqbal and A. A. Rehman, PLoS One, 2020, 15, e0234964 CrossRef CAS.
- B. Reidy, A. Haase, A. Luch, K. A. Dawson and I. Lynch, Materials, 2013, 6, 2295–2350 CrossRef CAS.
- A. A. Lourthuraj, M. M. Selvam, M. S. Hussain, A. W. A. Abdel-Warith, E. M. I. Younis and N. A. Al-Asgah, Saudi J. Biol. Sci., 2020, 27, 1753–1759 CrossRef CAS.
- A. K. Keshari, R. Srivastava, P. Singh, V. B. Yadav and G. Nath, J. Ayurveda Integr. Med., 2020, 11, 37–44 CrossRef.
- A. J. Kora, J. Mounika and R. Jagadeeshwar, Fungal Biol., 2020, 124, 671–681 CrossRef CAS.
- A. M. Elgorban, A. E. R. M. El-Samawaty, M. A. Yassin, S. R. Sayed, S. F. Adil, K. M. Elhindi, M. Bakri and M. Khan, Biotechnol. Biotechnol. Equip., 2015, 30, 56–62 CrossRef.
- A. Nouri, M. Tavakkoli Yaraki, A. Lajevardi, Z. Rezaei, M. Ghorbanpour and M. Tanzifi, Colloids Interface Sci. Commun., 2020, 35, 100252, DOI:10.1016/j.colcom.2020.100252.
- M. Ghaedi, M. Yousefinejad, M. Safarpoor, H. Z. Khafri and M. K. Purkait, J. Ind. Eng. Chem., 2015, 31, 167–172 CrossRef CAS.
- S. Muthukrishnan, S. Bhakya, T. Senthil Kumar and M. V. Rao, Ind. Crops Prod., 2015, 63, 119–124 CrossRef CAS.
- N. L. Gavade, A. N. Kadam, M. B. Suwarnkar, V. P. Ghodake and K. M. Garadkar, Spectrochim. Acta, Part A, 2015, 136, 953–960 CrossRef CAS.
- T. J. I. Edison and M. G. Sethuraman, Process Biochem., 2012, 47, 1351–1357 CrossRef CAS.
- R. Vivek, R. Thangam, K. Muthuchelian, P. Gunasekaran, K. Kaveri and S. Kannan, Process Biochem., 2012, 47, 2405–2410 CrossRef CAS.
- Y. Sun, Y. Yin, B. T. Mayers, T. Herricks and Y. Xia, Chem. Mater., 2002, 14, 4736–4745 CrossRef CAS.
- R. M. Gengan, K. Anand, A. Phulukdaree and A. Chuturgoon, Colloids Surf., B, 2013, 105, 87–91 CrossRef CAS.
- P. Logeswari, S. Silambarasan and J. Abraham, J. Saudi Chem. Soc., 2015, 19, 311–317 CrossRef.
- H. Kolya, P. Maiti, A. Pandey and T. Tripathy, J. Anal. Sci. Technol., 2015, 6, 33 CrossRef.
- S. N. Sinha and D. Paul, Spectrosc. Lett., 2015, 48, 600–604 CrossRef CAS.
- X. Yao, M. Jericho, D. Pink and T. Beveridge, J. Bacteriol., 1999, 181, 6865–6875 CrossRef CAS.
- K. Elangovan, D. Elumalai, S. Anupriya, R. Shenbhagaraman, P. K. Kaleena and K. Murugesan, J. Photochem. Photobiol., B, 2015, 151, 118–124 CrossRef CAS.
- S. B. Ulaeto, G. M. Mathew, J. K. Pancrecious, J. B. Nair, T. P. D. Rajan, K. K. Maiti and B. C. Pai, ACS Biomater. Sci. Eng., 2020, 6, 235–245 CrossRef CAS.
- B. Ajitha, Y. A. K. Reddy, H. J. Jeon and C. W. Ahn, Adv. Powder Technol., 2018, 29, 86–93 CrossRef CAS.
- M. Khatami, S. Pourseyedi, M. Khatami, H. Hamidi, M. Zaeifi and L. Soltani, Bioresour. Bioprocess., 2015, 2, 19 CrossRef.
- H. Xu, L. Wang, H. Su, L. Gu, T. Han, F. Meng and C. Liu, Food Biophys., 2014, 10, 12–18 CrossRef.
- M. S. Alsalhi, S. Devanesan, A. A. Alfuraydi, R. Vishnubalaji, M. A. Munusamy, K. Murugan, M. Nicoletti and G. Benelli, Int. J. Nanomed., 2016, 11, 4439–4449 CrossRef CAS.
- A. Lateef, M. A. Akande, M. A. Azeez, S. A. Ojo, B. I. Folarin, E. B. Gueguim-Kana and L. S. Beukes, Nanotechnol. Rev., 2016, 5, 507–520 CAS.
- M. K. Choudhary, J. Kataria, S. S. Cameotra and J. Singh, Appl. Nanosci., 2016, 6, 105–111 CrossRef CAS.
- Z. H. Pak, H. Abbaspour, N. Karimi and A. Fattahi, Appl. Sci., 2016, 6, 69 CrossRef.
- M. Khatami, M. S. Nejad, S. Salari and P. G. N. Almani, IET Nanobiotechnol., 2016, 10, 237–243 CrossRef.
- M. Khatami, R. Mehnipor, M. H. S. Poor and G. S. Jouzani, J. Cluster Sci., 2016, 27, 1601–1612 CrossRef CAS.
- A. Chahardoli, N. Karimi and A. Fattahi, Iran. J. Pharm. Res., 2017, 16, 1167–1175 Search PubMed.
- M. T. Haseeb, M. A. Hussain, K. Abbas, B. G. M. Youssif, S. Bashir, S. H. Yuk and S. N. A. Bukhari, Int. J. Nanomed., 2017, 12, 2845–2855 CrossRef CAS.
- M. Dhayalan, M. I. J. Denison, L. Anitha Jegadeeshwari, K. Krishnan and N. Nagendra Gandhi, Nat. Prod. Res., 2016, 31, 465–468 CrossRef.
- S. Pirtarighat, M. Ghannadnia and S. Baghshahi, Nanomed. J., 2017, 4, 184–190 CAS.
- R. Kumar, P. Sharma, A. Bamal, S. Negi and S. Chaudhary, Green Process. Synth., 2017, 6, 0146 Search PubMed.
- Y. He, F. Wei, Z. Ma, H. Zhang, Q. Yang, B. Yao, Z. Huang, J. Li, C. Zeng and Q. Zhang, RSC Adv., 2017, 7, 39842–39851 RSC.
- S. Balakrishnan, I. Sivaji, S. Kandasamy, S. Duraisamy, N. S. Kumar and G. Gurusubramanian, Environ. Sci. Pollut. Res., 2017, 24, 14758–14769 CrossRef CAS.
- A. Qidwai, R. Kumar and A. Dikshit, Green Chem. Lett. Rev., 2018, 11, 176–188 CrossRef CAS.
- M. A. Ansari and M. A. Alzohairy, J. Evidence-Based Complementary Altern. Med., 2018, 1–9, 1860280 Search PubMed.
- A. Rautela, J. Rani and M. Debnath, J. Anal. Sci. Technol., 2019, 10, 1–10 CrossRef.
- N. G. Girón-Vázquez, C. M. Gómez-Gutiérrez, C. A. Soto-Robles, O. Nava, E. Lugo-Medina, V. H. Castrejón-Sánchez, A. R. Vilchis-Nestor and P. A. Luque, Results Phys., 2019, 13, 102142 CrossRef.
- L. Hernández-Morales, H. Espinoza-Gómez, L. Z. Flores-López, E. L. Sotelo-Barrera, A. Núñez-Rivera, R. D. Cadena-Nava, G. Alonso-Núñez and K. A. Espinoza, Appl. Surf. Sci., 2019, 489, 952–961 CrossRef.
- R. Varghese, M. A. Almalki, S. Ilavenil, J. Rebecca and K. C. Choi, Saudi J. Biol. Sci., 2019, 26, 148–154 CrossRef CAS.
- S. Arokiyaraj, S. H. Choi, Y. Lee, R. Bharanidharan, V. I. Hairul-Islam, B. Vijayakumar, Y. K. Oh, V. Dinesh-Kumar, S. Vincent and K. H. Kim, Molecules, 2015, 20, 384–395 CrossRef.
- M. Adnan, M. Obyedul Kalam Azad, A. Madhusudhan, K. Saravanakumar, X. Hu, M. H. Wang and C. D. Ha, Nanotechnology, 2020, 31, 26 CrossRef.
- F. A. Qais, A. Shafiq, I. Ahmad, F. M. Husain, R. A. Khan and I. Hassan, Microb. Pathog., 2020, 144, 104172 CrossRef.
- M. F. Zayed, R. A. Mahfoze, S. M. El-kousy and E. A. Al-Ashkar, Colloids Surf., A, 2020, 585, 124167 CrossRef CAS.
- J. H. Kim, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2017, 1063, 196–203 CrossRef CAS.
- T. Belwal, P. Dhyani, I. D. Bhatt, R. S. Rawal and V. Pande, Food Chem., 2016, 207, 115–124 CrossRef.
- H. Padalia, P. Moteriya and S. Chanda, Arabian J. Chem., 2015, 8, 732–741 CrossRef CAS.
- N. Gogoi, P. J. Babu, C. Mahanta and U. Bora, Mater. Sci. Eng., C, 2015, 46, 463–469 CrossRef CAS.
- P. Moteriya and S. Chanda, Artif. Cells, Nanomed., Biotechnol., 2016, 45, 1556–1567 CrossRef.
- A. Ebrahiminezhad, Y. Barzegar, Y. Ghasemi and A. Berenjian, Chem. Ind. Chem. Eng. Q., 2017, 232, 31–37 CrossRef.
- N. Chandrasekhar and S. P. Vinay, Appl. Nanosci., 2017, 7, 851–861 CrossRef CAS.
- M. Hariram, S. Vivekanandhan, V. Ganesan, S. Muthuramkumar, A. Rodriguez-uribe, A. K. Mohanty and M. Misra, Bioresour. Technol. Rep., 2019, 7, 100298 CrossRef.
- M. R. Bindhu, M. Umadevi, G. A. Esmail, N. A. Al-Dhabi and M. V. Arasu, J. Photochem. Photobiol., B, 2020, 205, 111836 CrossRef CAS.
- B. Ajitha, Y. A. K. Reddy, Y. Lee, M. J. Kim and C. W. Ahn, Appl. Organomet. Chem., 2019, 33, e4867 CrossRef.
- A. K. Mittal, D. Tripathy, A. Choudhary, P. K. Aili, A. Chatterjee, I. P. Singh and U. C. Banerjee, Mater. Sci. Eng., C, 2015, 53, 120–127 CrossRef CAS.
- S. Pugazhendhi, E. Kirubha, P. K. Palanisamy and R. Gopalakrishnan, Appl. Surf. Sci., 2015, 357, 1801–1808 CrossRef CAS.
- P. R. Rathi Sre, M. Reka, R. Poovazhagi, M. Arul Kumar and K. Murugesan, Spectrochim. Acta, Part A, 2015, 135, 1137–1144 CrossRef CAS.
- N. H. Rao, N. Lakshmidevi, S. V. N. Pammi, P. Kollu, S. Ganapaty and P. Lakshmi, Mater. Sci. Eng., C, 2016, 62, 553–557 CrossRef CAS.
- L. Pethakamsetty, K. Kothapenta, H. R. Nammi, L. K. Ruddaraju, P. Kollu, S. G. Yoon and S. V. N. Pammi, J. Environ. Sci., 2017, 55, 157–163 CrossRef CAS.
- K. M. Ezealisiji, X. S. Noundou and S. E. Ukwueze, Appl. Nanosci., 2017, 7, 905–911 CrossRef CAS.
- D. Wang, J. Markus, C. Wang, Y. J. Kim, R. Mathiyalagan, V. C. Aceituno, S. Ahn and D. C. Yang, Artif. Cells, Nanomed., Biotechnol., 2016, 45, 1548–1555 CrossRef.
- S. Kantipudi, L. Pethakamsetty, S. M. Kollana, J. R. Sunkara, P. Kollu, N. R. Parine, M. Rallabhandi and S. V. N. Pammi, IET Nanobiotechnol., 2018, 12, 133–137 CrossRef.
- G. Şeker Karatoprak, G. Aydin, B. Altinsoy, C. Altinkaynak, M. Koşar and I. Ocsoy, Enzyme Microb. Technol., 2017, 97, 21–26 CrossRef.
- S. Arokiyaraj, S. Vincent, M. Saravanan, Y. Lee, Y. K. Oh and K. H. Kim, Artif. Cells, Nanomed., Biotechnol., 2016, 45, 372–379 CrossRef.
- E. F. P. Henie, H. Zaiton and M. Suhaila, Int. Food Res. J., 2009, 16, 297–311 Search PubMed.
- F. Benakashani, A. Allafchian and S. A. H. Jalali, Green Chem. Lett. Rev., 2017, 10, 324–330 CrossRef CAS.
- J. Markus, D. Wang, Y. J. Kim, S. Ahn, R. Mathiyalagan, C. Wang and D. C. Yang, Nanoscale Res. Lett., 2017, 12, 46 CrossRef.
- M. Oves, M. Aslam, M. A. Rauf, S. Qayyum, H. A. Qari, M. S. Khan, M. Z. Alam, S. Tabrez, A. Pugazhendhi and I. M. I. Ismail, Mater. Sci. Eng., C, 2018, 89, 429–443 CrossRef CAS.
- T. T. N. Nguyen, T. T. Vo, B. N. H. Nguyen, D. T. Nguyen, V. S. Dang, C. H. Dang and T. D. Nguyen, Environ. Sci. Pollut. Res., 2018, 25, 34247–34261 CrossRef CAS.
- P. P. N. Vijay Kumar, R. L. Kalyani, S. C. Veerla, P. Kollu, U. Shameem and S. V. N. Pammi, Mater. Res. Express, 2019, 6, 10 Search PubMed.
- D. Garibo, H. A. Borbón-Nuñez, J. N. D. de León, E. García Mendoza, I. Estrada, Y. Toledano-Magaña, H. Tiznado, M. Ovalle-Marroquin, A. G. Soto-Ramos, A. Blanco, J. A. Rodríguez, O. A. Romo, L. A. Chávez-Almazán and A. Susarrey-Arce, Sci. Rep., 2020, 10, 12805 CrossRef CAS.
- M. N. Khan, T. A. Khan, Z. Khan and S. A. AL-Thabaiti, Bioprocess Biosyst. Eng., 2015, 38, 2397–2416 CrossRef CAS.
- A. A. Alfuraydi, S. Devanesan, M. Al-Ansari, M. S. AlSalhi and A. J. Ranjitsingh, J. Photochem. Photobiol., B, 2019, 192, 83–89 CrossRef CAS.
- P. S. Ramesh, T. Kokila and D. Geetha, Spectrochim. Acta, Part A, 2015, 142, 339–343 CrossRef CAS.
- S. Lokina, A. Stephen, V. Kaviyarasan, C. Arulvasu and V. Narayanan, Synth. React. Inorg., Met.-Org., Nano-Met. Chem., 2013, 45, 37–41 Search PubMed.
- S. J. Mane Gavade, G. H. Nikam, R. S. Dhabbe, S. R. Sabale, B. V. Tamhankar and G. N. Mulik, Adv. Nat. Sci.: Nanosci. Nanotechnol., 2015, 6, 045015 Search PubMed.
- N. Mapara, M. Sharma, V. Shriram, R. Bharadwaj, K. C. Mohite and V. Kumar, Appl. Microbiol. Biotechnol., 2015, 99, 10655–10667 CrossRef CAS.
- M. Ramar, B. Manikandan, P. N. Marimuthu, T. Raman, A. Mahalingam, P. Subramanian, S. Karthick and A. Munusamy, Spectrochim. Acta, Part A, 2015, 140, 223–228 CrossRef CAS.
- S. A. A. L. Rahisuddin, Z. Khan and N. Manzoor, Bioprocess Biosyst. Eng., 2015, 38, 1773–1781 CrossRef CAS.
- P. Yugandhar and N. Savithramma, Appl. Nanosci., 2015, 6, 223–233 CrossRef.
- G. Mahendran and B. D. Ranjitha Kumari, Food Sci. Hum. Well., 2016, 5, 207–218 CrossRef.
- P. Mosae Selvakumar, C. A. Antonyraj, R. Babu, A. Dakhsinamurthy, N. Manikandan and A. Palanivel, Synth. React. Inorg., Met.-Org., Nano-Met. Chem., 2015, 46, 291–294 CrossRef.
- Z. A. Ali, R. Yahya, S. D. Sekaran and R. Puteh, Adv. Mater. Sci. Eng., 2016, 2016, 4102196 Search PubMed.
- C. M. K. Kumar, P. Yugandhar and N. Savithramma, J. Intercult. Ethnopharmacol., 2017, 6, 296–310 CrossRef.
- M. M. O. Rashid, K. N. Akhter, J. A. Chowdhury, F. Hossen, M. S. Hussain and M. T. Hossain, BMC Complementary Altern. Med., 2017, 17, 336 CrossRef.
- N. Jayaprakash, J. J. Vijaya, K. Kaviyarasu, K. Kombaiah, L. J. Kennedy, R. J. Ramalingam, M. A. Munusamy and H. A. Al-Lohedan, J. Photochem. Photobiol., B, 2017, 169, 178–185 CrossRef CAS.
- S. Farhadi, B. Ajerloo and A. Mohammadi, Acta Chim. Slov., 2017, 67, 1 Search PubMed.
- S. A. Umoren, A. M. Nzila, S. Sankaran, M. M. Solomon and P. S. Umoren, Pol. J. Chem. Technol., 2017, 19, 128–136 CAS.
- Z. E. Jiménez Pérez, R. Mathiyalagan, J. Markus, Y. J. Kim, H. M. Kang, R. Abbai, K. H. Seo, D. Wang, V. Soshnikova and D. C. Yang, Int. J. Nanomed., 2017, 12, 709–723 CrossRef.
- B. A. Providence, A. A. Chinyere, A. A. Ayi, O. O. Charles, T. A. Elijah and H. L. Ayomide, Int. J. Phys. Sci., 2018, 13, 24–32 CrossRef.
- K. H. Oh, V. Soshnikova, J. Markus, Y. J. Kim, S. C. Lee, P. Singh, V. Castro-Aceituno, S. Ahn, D. H. Kim, Y. J. Shim, Y. J. Kim and D. C. Yang, Artif. Cells, Nanomed., Biotechnol., 2017, 46, 599–606 CrossRef.
- T. Sowmyya and G. Vijaya Lakshmi, Bionanoscience, 2017, 8, 179–195 Search PubMed.
- R. Dobrucka, M. Kaczmarek and J. Dlugaszewska, Adv. Nat. Sci.: Nanosci. Nanotechnol., 2018, 9, 025015 Search PubMed.
- G. M. Sangaonkar and K. D. Pawar, Colloids Surf., B, 2018, 164, 210–217 CrossRef CAS.
- N. Joshi, N. Jain, A. Pathak, J. Singh, R. Prasad and C. P. Upadhyaya, J. Sol-Gel Sci. Technol., 2018, 86, 682–689 CrossRef CAS.
- S. Batool, Z. Hussain, M. B. K. Niazi, U. Liaqat and M. Afzal, J. Drug Delivery Sci. Technol., 2019, 52, 403–414 CrossRef CAS.
- S. Andra, S. Balu, R. Ramoorthy, M. Muthalagu and V. S. Manisha, Mater. Today: Proc., 2019, 9, 639–644 CAS.
- F. K. Saidu, A. Mathew, A. Parveen, V. Valiyathra and G. V. Thomas, SN Appl. Sci., 2019, 1, 1368 CrossRef.
- M. I. Masum, M. M. Siddiqa, K. A. Ali, Y. Zhang, Y. Abdallah, E. Ibrahim, W. Qiu, C. Yan and B. Li, Front. Microbiol., 2019, 10, 820 CrossRef.
- J. Du, Z. Hu, Z. Yu, H. Li, J. Pan, D. Zhao and Y. Bai, Mater. Sci. Eng., C, 2019, 102, 247–253 CrossRef CAS.
- F. Gulbagca, S. Ozdemir, M. Gulcan and F. Sen, Heliyon, 2019, 5, e02980 CrossRef.
- C. Vishwasrao, B. Momin and L. Ananthanarayan, Waste Biomass Valorization, 2018, 10, 8 Search PubMed.
- M. M. R. Mollick, D. Rana, S. K. Dash, S. Chattopadhyay, B. Bhowmick, D. Maity, D. Mondal, S. Pattanayak, S. Roy, M. Chakraborty and D. Chattopadhyay, Arabian J. Chem., 2019, 12, 2572–2584, DOI:10.1016/j.arabjc.2015.04.033.
- R. Renuka, K. R. Devi, M. Sivakami, T. Thilagavathi, R. Uthrakumar and K. Kaviyarasu, Biocatal. Agric. Biotechnol., 2020, 24, 101567 CrossRef.
- M. Devi, S. Devi, V. Sharma, N. Rana, R. K. Bhatia and A. K. Bhatt, J. Tradit. Complement. Med., 2020, 10, 158–165 CrossRef.
- M. A. Odeniyi, V. C. Okumah, B. C. Adebayo-Tayo and O. A. Odeniyi, Sustainable Chem. Pharm., 2020, 15, 100197 CrossRef.
- D. Sasidharan, T. R. Namitha, S. P. Johnson, V. Jose and P. Mathew, Sustainable Chem. Pharm., 2020, 16, 100255 CrossRef.
- T. Shankar, P. Karthiga, K. Swarnalatha and K. Rajkumar, Resour.-Effic. Technol., 2017, 3, 303–308 Search PubMed.
- S. P. Vinay and N. Chandrasekhar, Mater. Today: Proc., 2019, 9, 499–505 CAS.
- P. Velusamy, J. Das, R. Pachaiappan, B. Vaseeharan and K. Pandian, Ind. Crops Prod., 2015, 66, 103–109 CrossRef CAS.
- I. Murali Krishna, G. Bhagavanth Reddy, G. Veerabhadram and A. Madhusudhan, Appl. Nanosci., 2015, 6, 681–689 CrossRef.
- J. Du, H. Singh and T. H. Yi, Bioprocess Biosyst. Eng., 2016, 39, 1923–1931 CrossRef CAS.
- A. C. d. J. Oliveira, A. R. de Araújo, P. V. Quelemes, D. Nadvorny, J. L. Soares-Sobrinho, J. R. S. d. A. Leite, E. C. da Silva-Filho and D. A. da Silva, Carbohydr. Polym., 2019, 213, 176–183 CrossRef CAS.
- A. V. Samrot, J. L. A. Angalene, S. M. Roshini, P. Raji, S. M. Stefi, R. Preethi, A. J. Selvarani and A. Madankumar, J. Cluster Sci., 2019, 30, 1599–1610 CrossRef CAS.
- M. Z. Siddiqui, A. R. Chowdhury, B. R. Singh, S. Maurya and N. Prasad, Natl. Acad. Sci. Lett., 2020 DOI:10.1007/s40009-020-00982-4.
- A. Mumtaz, H. Munir, M. T. Zubair and M. H. Arif, Mater. Res. Express, 2019, 6, 105308 CrossRef CAS.
- C. A. Eric, V. Benjamín, C. Monica, A. C. Mario, D. M. Francisco, A. R. Rogelio, R. Navor and M. B. Jose, Afr. J. Biotechnol., 2017, 16, 400–407 CrossRef.
- M. Khatami, I. Sharifi, M. A. L. Nobre, N. Zafarnia and M. R. Aflatoonian, Green Chem. Lett. Rev., 2018, 11, 125–134 CrossRef CAS.
- V. Ahluwalia, S. Elumalai, V. Kumar, S. Kumar and R. S. Sangwan, Microb. Pathog., 2018, 114, 402–408 CrossRef CAS.
- P. Moteriya and S. Chanda, J. Genet. Eng. Biotechnol., 2018, 16, 105–113 CrossRef.
- P. Karthiga, Biotechnol. Res. Innov., 2018, 2, 30–36 CrossRef.
- F. Zandpour, A. R. Allafchian, M. R. Vahabi and S. A. H. Jalali, IET Nanobiotechnol., 2018, 12, 491–495 CrossRef.
- M. Cakić, S. Glišić, D. Cvetković, M. Cvetinov, L. Stanojević, B. Danilović and K. Cakić, Colloid J., 2018, 80, 803–813 CrossRef.
- S. Dehghanizade, J. Arasteh and A. Mirzaie, Artif. Cells, Nanomed., Biotechnol., 2017, 46, 160–168 CrossRef.
- M. Moyo, M. Gomba and T. Nharingo, Int. J. Ind. Chem., 2015, 6, 329–338 CrossRef CAS.
- P. Yugandhar, R. Haribabu and N. Savithramma, 3 Biotech, 2015, 5, 1031–1039 CrossRef.
- A. Sasikala, M. Linga Rao, N. Savithramma and T. N. V. K. V. Prasad, Appl. Nanosci., 2014, 5, 827–835 CrossRef.
- D. Nayak, S. Ashe, P. R. Rauta, M. Kumari and B. Nayak, Mater. Sci. Eng., C, 2016, 58, 44–52 CrossRef CAS.
- S. Priya Velammal, T. A. Devi and T. P. Amaladhas, J. Nanostruct. Chem., 2016, 6, 247–260 CrossRef.
- S. Bhakya, S. Muthukrishnan, M. Sukumaran, M. Grijalva, L. Cumbal, J. F. Benjamin and M. V. Rao, RSC Adv., 2016, 6, 81436–81446 RSC.
- Q. Ahmed, N. Gupta, A. Kumar and S. Nimesh, Artif. Cells, Nanomed., Biotechnol., 2016, 45, 1192–1200 CrossRef.
- S. Pattanayak, M. Rahaman, D. Maity, S. Chakraborty, S. Kumar and S. Chattopadhyay, J. Saudi Chem. Soc., 2017, 21, 673–684 CrossRef CAS.
- G. Arya, R. M. Kumari, N. Gupta, A. Kumar and S. Nimesh, Artif. Cells, Nanomed., Biotechnol., 2017, 1–9 Search PubMed.
- P. Karthiga, S. Rajeshkumar and G. Annadurai, J. Cluster Sci., 2018, 29, 1233–1241 CrossRef CAS.
- S. Ramanathan, S. C. B. Gopinath, P. Anbu, T. Lakshmipriya, F. H. Kasim and C. G. Lee, J. Mol. Struct., 2018, 1160, 80–91 CrossRef CAS.
- M. Das and S. S. Smita, Appl. Nanosci., 2018, 8, 1059–1067 CrossRef CAS.
- E. C. Sekhar, K. S. V. K. Rao, K. M. S. Rao and S. B. Alisha, J. Appl. Pharm. Sci., 2018, 8, 1 Search PubMed.
- D. Bharathi, M. Diviya Josebin, S. Vasantharaj and V. Bhuvaneshwari, J. Nanostruct. Chem., 2018, 8, 83–92 CrossRef CAS.
- R. P. Illanes Tormena, E. V. Rosa, B. d. F. Oliveira Mota, J. A. Chaker, C. W. Fagg, D. O. Freire, P. M. Martins, I. C. Rodrigues da Silva and M. H. Sousa, RSC Adv., 2020, 10, 20676–20681 RSC.
- A.-R. Phull, Q. Abbas, A. Ali, H. Raza, S. J. kim, M. Zia and I. Haq, Future J. Pharm. Sci., 2016, 2, 31–36 CrossRef.
- J. H. Lee, J. M. Lim, P. Velmurugan, Y. J. Park, Y. J. Park, K. S. Bang and B. T. Oh, J. Photochem. Photobiol., B, 2016, 162, 93–99 CrossRef CAS.
- G. Sharma, J. S. Nam, A. R. Sharma and S. S. Lee, Molecules, 2018, 23, 2268 CrossRef.
- F. K. Alsammarraie, W. Wang, P. Zhou, A. Mustapha and M. Lin, Colloids Surf., B, 2018, 171, 398–405 CrossRef CAS.
- N. T. Selvi, R. Navamathavan, H. Y. Kim and R. Nirmala, Macromol. Res., 2019, 27, 1155–1160 CrossRef CAS.
- S. Devanesan, K. Ponmurugan, M. S. AlSalhi and N. A. Al-Dhabi, Int. J. Nanomed., 2020, 15, 4351–4362 CrossRef CAS.
- Y. Nan, L. I. Fuyan, J. Tiancai, L. I. U. Chongchong, S. U. N. Hushan, W. Lei and X. U. Hui, Acta Oceanol. Sin., 2017, 36, 95–100 Search PubMed.
- H. M. M. Ibrahim, J. Radiat. Res. Appl. Sci., 2015, 8, 265–275 CrossRef.
- J. Balavijayalakshmi and V. Ramalakshmi, J. Appl. Res. Technol., 2017, 15, 413–422 CrossRef.
- C. H. N. de Barros, G. C. F. Cruz, W. Mayrink and L. Tasic, Nanotechnol., Sci. Appl., 2018, 11, 1–14 CrossRef CAS.
- C. Huo, M. Khoshnamvand, P. Liu, C. G. Yuan and W. Cao, Mater. Res. Express, 2018, 6, 1 Search PubMed.
- M. Annu, S. Ahmed, G. Kaur, P. Sharma, S. Singh and S. Ikram, Toxicol. Res., 2018, 7, 923–930 CrossRef.
- R. G. Saratale, H. S. Shin, G. Kumar, G. Benelli, G. S. Ghodake, Y. Y. Jiang, D. S. Kim and G. D. Saratale, Environ. Sci. Pollut. Res., 2017, 25, 10250–10263 CrossRef.
- R. K. Das and D. Bhuyan, Nanotechnol. Environ. Eng., 2019, 4, 1 CrossRef.
- T. Dutta, N. N. Ghosh, M. Das, R. Adhikary, V. Mandal and A. P. Chattopadhyay, J. Environ. Chem. Eng., 2020, 8, 104019 CrossRef CAS.
- E. Z. Gomaa, J. Genet. Eng. Biotechnol., 2017, 15, 49–57 CrossRef.
- A. Aravinthan, M. Govarthanan, K. Selvam, L. Praburaman, T. Selvankumar, R. Balamurugan, S. Kamala-Kannan and J. H. Kim, Int. J. Nanomed., 2015, 10, 1977–1983 CAS.
- S. Pugazhendhi, P. Sathya, P. K. Palanisamy and R. Gopalakrishnan, J. Photochem. Photobiol., B, 2016, 159, 155–160 CrossRef CAS.
- M. Mosaviniya, T. Kikhavani, M. Tanzifi, M. Tavakkoli Yaraki, P. Tajbakhsh and A. Lajevardi, Colloids Interface Sci. Commun., 2019, 33, 100211 CrossRef CAS.
- M. Saha and P. K. Bandyopadhyay, Proc. Zool. Soc., 2019, 72, 180–186 CrossRef.
- S. Rajesh, V. Dharanishanthi and A. V. Kanna, J. Exp. Nanosci., 2014, 10, 1143–1152 CrossRef.
- P. Kuppusamy, S. J. A. Ichwan, N. R. Parine, M. M. Yusoff, G. P. Maniam and N. Govindan, J. Environ. Sci., 2015, 29, 151–157 CrossRef CAS.
- U. Ramaswamy, D. Mukundan, A. Sreekumar and V. Mani, Mater. Today: Proc., 2015, 2, 4600–4608 Search PubMed.
- S. Salehi, S. A. S. Shandiz, F. Ghanbar, M. R. Darvish, M. S. Ardestani, A. Mirzaie and M. Jafari, Int. J. Nanomed., 2016, 11, 1835–1846 CAS.
- S. Anjum and B. H. Abbasi, Int. J. Nanomed., 2016, 11, 715–728 CrossRef CAS.
- S. Ali, S. Perveen, M. Ali, T. Jiao, A. S. Sharma, H. Hassan, S. Devaraj, H. Li and Q. Chen, Mater. Sci. Eng., C, 2020, 108, 110421 CrossRef CAS.
- P. N. V. K. Pallela, S. Ummey, L. K. Ruddaraju, S. V. N. Pammi and S. G. Yoon, Microb. Pathog., 2018, 124, 63–69 CrossRef CAS.
- M. Idrees, S. Batool, T. Kalsoom, S. Raina, H. M. A. Sharif and S. Yasmeen, Environ. Technol., 2019, 40, 1071–1078 CrossRef CAS.
- A. Aygün, F. Gülbağça, M. S. Nas, M. H. Alma, M. H. Çalımlı, B. Ustaoglu, Y. C. Altunoglu, M. C. Baloğlu, K. Cellat and F. Şen, J. Pharm. Biomed. Anal., 2020, 179, 113012 CrossRef.
- R. R. Chavan, S. D. Bhinge, M. A. Bhutkar, D. S. Randive, G. H. Wadkar, S. S. Todkar and M. N. Urade, Mater. Today Commun., 2020, 24, 101320 CrossRef CAS.
- R. Dobrucka and J. Długaszewska, Indian J. Microbiol., 2015, 55, 168–174 CrossRef CAS.
- K. Jadhav, D. Dhamecha, B. Dalvi and M. Patil, Part. Sci. Technol., 2015, 33, 445–455 CrossRef CAS.
- A. T. Shah, M. I. Din, S. Bashir, M. A. Qadir and F. Rashid, Anal. Lett., 2015, 48, 1180–1189 CrossRef CAS.
- D. Nayak, S. Ashe, P. R. Rauta and B. Nayak, IET Nanobiotechnol., 2015, 9, 288–293 CrossRef.
- C. Rajkuberan, S. Prabukumar, G. Sathishkumar, A. Wilson, K. Ravindran and S. Sivaramakrishnan, J. Saudi Chem. Soc., 2017, 21, 911–919 CrossRef CAS.
- C. Rajkuberan, K. Sudha, G. Sathishkumar and S. Sivaramakrishnan, Spectrochim. Acta, Part A, 2015, 136, 924–930 CrossRef CAS.
- A. Lateef, M. A. Azeez, T. B. Asafa, T. A. Yekeen, A. Akinboro, I. C. Oladipo, L. Azeez, S. A. Ojo, E. B. Gueguim-Kana and L. S. Beukes, J. Nanostruct. Chem., 2016, 6, 159–169 CrossRef CAS.
- A. Lateef, M. A. Azeez, T. B. Asafa, T. A. Yekeen, A. Akinboro, I. C. Oladipo, L. Azeez, S. E. Ajibade, S. A. Ojo, E. B. Gueguim-kana and L. S. Beukes, J. Taibah Univ. Sci., 2015, 10, 551–562 CrossRef.
- Q. H. Xia, Y. J. Ma and J. W. Wang, Nanomaterials, 2016, 6, 160 CrossRef.
- F. Huang, Y. Long, Q. Liang, B. Purushotham, M. K. Swamy and Y. Duan, J. Nanomater., 2019, 2019, 2418785 Search PubMed.
- C. N. Lok, C. M. Ho, R. Chen, Q. Y. He, W. Y. Yu, H. Sun, P. K. H. Tam, J. F. Chiu and C. M. Che, J. Proteome Res., 2006, 5, 916–924 CrossRef CAS.
- M. Rai, A. Yadav and A. Gade, Biotechnol. Adv., 2009, 27, 76–83 CrossRef CAS.
- L. S. Dorobantu, C. Fallone, A. J. Noble, J. Veinot, G. Ma, G. G. Goss and R. E. Burrell, J. Nanopart. Res., 2015, 17, 172 CrossRef.
- F. Kang, P. J. Alvarez and D. Zhu, Environ. Sci. Technol., 2013, 48, 316–322 CrossRef.
- H. Xu, F. Qu, H. Xu, W. Lai, Y. A. Wang, Z. P. Aguilar and H. Wei, BioMetals, 2011, 25, 45–53 CrossRef.
- J. S. Kim, E. Kuk, K. N. Yu, J. H. Kim, S. J. Park, H. J. Lee, S. H. Kim, Y. K. Park, Y. H. Park, C. Y. Hwang, Y. K. Kim, Y. S. Lee, D. H. Jeong and M. H. Cho, Nanomedicine, 2007, 3, 91–101 Search PubMed.
- J. S. McQuillan, H. Groenaga Infante, E. Stokes and A. M. Shaw, Nanotoxicology, 2011, 6, 857–866 CrossRef.
- C. M. Zhao and W. X. Wang, Nanotoxicology, 2011, 6, 361–370 CrossRef.
- W. R. Li, X. B. Xie, Q. S. Shi, H. Y. Zeng, Y. S. Ou-Yang and Y. Ben Chen, Appl. Microbiol. Biotechnol., 2009, 85, 1115–1122 CrossRef.
- M. P. Patil and G. Kim, Appl. Microbiol. Biotechnol., 2017, 101, 79–92 CrossRef CAS.
- I. Sondi and B. Salopek-Sondi, J. Colloid Interface Sci., 2004, 275, 177–182 CrossRef CAS.
- J. R. Morones, J. L. Elechiguerra, A. Camacho, K. Holt, J. B. Kouri, J. T. Ramírez and M. J. Yacaman, Nanotechnology, 2005, 16, 2346–2353 CrossRef CAS.
- D. Yu and V. W. W. Yam, J. Am. Chem. Soc., 2004, 126, 13200–13201 CrossRef CAS.
- M. G. Bawendi, P. J. Carroll, W. L. Wilson and L. E. Brus, J. Chem. Phys., 1992, 96, 946–954 CrossRef CAS.
- N. Herron, Y. Wang and H. Eckert, J. Am. Chem. Soc., 1990, 112, 1322–1326 CrossRef CAS.
- Z. S. Pillai and P. V. Kamat, J. Phys. Chem. B, 2004, 108, 945–951 CrossRef CAS.
- A. Henglein and M. Giersig, J. Phys. Chem. B, 1999, 103, 9533–9539 CrossRef CAS.
- J. Y. Song and B. S. Kim, Bioprocess Biosyst. Eng., 2008, 32, 79–84 CrossRef.
- M. M. H. Khalil, E. H. Ismail, K. Z. El-Baghdady and D. Mohamed, Arabian J. Chem., 2014, 7, 1131–1139 CrossRef CAS.
- K. Chitra and G. Annadurai, BioMed Res. Int., 2014, 1–6 Search PubMed.
- S. S. Sana and L. K. Dogiparthi, Mater. Lett., 2018, 226, 47–51 CrossRef CAS.
- A. Khan, U. Farooq, T. Ahmad, R. Sarwar, J. Shafiq, Y. Raza, A. Ahmed, S. Ullah, N. Ur Rehman and A. Al-Harrasi, Int. J. Nanomed., 2019, 14, 3983–3993 CrossRef.
- Y. Sun and Y. Xia, Science, 2002, 298, 5176–5179 Search PubMed.
| This journal is © The Royal Society of Chemistry 2021 |