Open Access Article
Open Access ArticleRole of polyunsaturated phospholipids in liquid-ordered and liquid-disordered phases†
Jing Yang *a,
Jianyu Jina and
Shiben Li
*a,
Jianyu Jina and
Shiben Li *b
*b
aCollege of Education, Wenzhou University, Wenzhou, Zhejiang 325035, China. E-mail: yangjing@wzu.edu.cn
bDepartment of Physics, Wenzhou University, Wenzhou, Zhejiang 325035, China. E-mail: shibenli@wzu.edu.cn
First published on 9th August 2021
Abstract
Polyunsaturated phospholipids interact complexly with other membrane components. We have examined pair interactions among ternary lipid bilayers composed of saturated DPPC, polyunsaturated PLePC, and cholesterol in the liquid-ordered and the liquid-disordered phases by all-atom molecular dynamics simulations. The results show that PLePC exhibits strong repulsion with DPPC and cholesterol in the liquid-disordered phase. When the bilayer changes to the liquid-ordered phase, the repulsion of PLePC with DPPC and cholesterol reduces significantly. The phase state of the bilayer which affects the order of acyl tails as well as their density distributions along the bilayer normal is a key factor regulating the role of PLePC in lipid mixtures. Polyunsaturated phospholipids play a strong repulsive role in the liquid-disordered phase but a weak role in the liquid-ordered phase.
1 Introduction
Saturated phospholipids, unsaturated phospholipids, and cholesterol are key components of biological membranes.1 The presence of cholesterol in membranes can induce the liquid-disordered (Ld) and liquid-ordered (Lo) phases depending on its concentration.1,2 While the structural diversities of lipids make their mutual interactions complex, the influences of membrane phases are also critical.3 The diverse pair interactions among membrane components determine their mutual preferences and lateral distributions in the plane of membranes.4,5 Investigating lipid–lipid and cholesterol–lipid interactions is important for further understanding of lipid rafts,5,6 as well as phase separation7,8 and lateral heterogeneity9,10 of membranes.While several evidences have shown attractive interactions between saturated lipids and cholesterol, the interactions involving unsaturated lipids are repulsive due to the double bonds in acyl tails.11,12 Kulig et al. explored the miscibility of lipid mixtures composed of phosphatidic acids (PAs) and phosphatidylcholines (PCs) with various acyl chains, and demonstrated that the miscibility of membrane components strongly depends on the acyl chain unsaturation. They observed that interactions between PA and PC molecules vary from attractive for systems in which all lipid tails are saturated, to repulsive for systems containing lipids with saturated and unsaturated acyl tails.13 In coarse-grained molecular dynamics (MD) simulations of multi-component lipid systems, Lin et al. observed that liquid–liquid domain stability increases with the degree of lipid polyunsaturation, and showed that lipid polyunsaturation is a driving force for liquid–liquid phase separation.14 The mixing behavior and phase separation of membrane components are virtually governed by the lipid–lipid and cholesterol–lipid interactions. By utilizing the nearest-neighbor recognition (NNR) method, Regen et al. measured the interactions among exchangeable mimics of saturated 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), monounsaturated 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), and cholesterol. They further proposed a push–pull mechanism for lipid raft formation, in which cholesterol and saturated lipids are pulled together by favorable interactions in the Lo phase, leading to transient domains, and that both cholesterol and saturated lipids are repelled by unsaturated lipids in the Ld phase.11,15,16 Moreover, NNR measurements showed that unsaturated lipids with higher degrees of unsaturation exert stronger repulsive forces on cholesterol and saturated lipids in the Ld phase.17
The lipid–lipid and lipid–cholesterol interactions among membranes are influenced by both the lipid unsaturation degree and the phase state of the membrane. While the key role of polyunsaturated phospholipids in the Ld phase has been unveiled,17 our knowledge on their role in the Lo phase is still limited. In this paper, we have performed all-atom MD simulations on ternary mixtures composed of saturated PC, polyunsaturated PC, and cholesterol. The pair interactions are compared in different phases. The results are consistent with the push–pull mechanism derived from experiments and reveal that polyunsaturated phospholipids play a strong repulsive role in the Ld phase but a weak role in the Lo phase.
2 Methods
Polyunsaturated phospholipids, 1-palmitoyl-2-linolenoyl-sn-glycero-3-phosphocholine (PLePC), which have three double bonds in the acyl tails, together with saturated DPPC and cholesterol (CHOL) were used to construct ternary DPPC/PLePC/CHOL bilayers. The chemical structures of DPPC, PLePC, CHOL molecules are shown in Fig. 1. Two sets of DPPC/PLePC/CHOL bilayers in the Ld and the Lo phases were created by the CHARMM-GUI web-based tool.18,19 Each system has 128 lipid/cholesterol molecules and 5120 TIP3P water molecules,20 corresponding to a hydration number of 40 water per lipid. Ternary mixtures composed of saturated PC, unsaturated PC, and CHOL can yield rich phase behavior at various lipid ratios and different temperatures.21,22 Therefore, choosing appropriate lipid ratios is crucial for modeling ternary lipid bilayers in the Ld and the Lo phases. However, to the best of our knowledge, the ternary phase diagram for DPPC/PLePC/CHOL has not been reported. Recently, Carpenter et al. completed sweeping studies of the ternary phase diagram for DPPC/DOPC/CHOL with a refined Martini coarse-grained force field that improves simulation fidelity to experimental phase diagrams.23 Although DOPC (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1/18
1/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) has a lower unsaturation degree than PLePC (16
1) has a lower unsaturation degree than PLePC (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) adopted in our models, lipid ratios that reproduce the Ld and the Lo phases of DPPC/DOPC/CHOL in the simulations of Carpenter et al. can be considered as trials for modeling the DPPC/PLePC/CHOL mixtures. Given that our all-atom bilayer has only 128 PC/CHOL molecules, we set the number of CHOL to 12 and 56 for the Ld phase and the Lo phase respectively. The numbers of DPPC and PLePC were set equal to obtain adequate statistical data. For the bilayer in the Ld phase, the components are 58 DPPC, 58 PLePC, and 12 CHOL, which corresponds to a cholesterol concentration of 9%. For the bilayer in the Lo phase, the components are 36 DPPC, 36 PLePC, and 56 CHOL, corresponding to a cholesterol concentration of 44%.
3) adopted in our models, lipid ratios that reproduce the Ld and the Lo phases of DPPC/DOPC/CHOL in the simulations of Carpenter et al. can be considered as trials for modeling the DPPC/PLePC/CHOL mixtures. Given that our all-atom bilayer has only 128 PC/CHOL molecules, we set the number of CHOL to 12 and 56 for the Ld phase and the Lo phase respectively. The numbers of DPPC and PLePC were set equal to obtain adequate statistical data. For the bilayer in the Ld phase, the components are 58 DPPC, 58 PLePC, and 12 CHOL, which corresponds to a cholesterol concentration of 9%. For the bilayer in the Lo phase, the components are 36 DPPC, 36 PLePC, and 56 CHOL, corresponding to a cholesterol concentration of 44%.
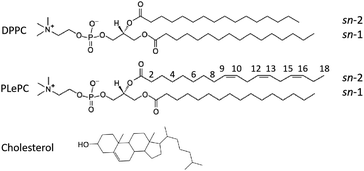 | ||
Fig. 1 Chemical structures of DPPC (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/16 0/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0), PLePC (16 0), PLePC (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18 0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3), and cholesterol molecules with the numbering of highlighted carbon sites considered in this work. 3), and cholesterol molecules with the numbering of highlighted carbon sites considered in this work. | ||
All-atom MD simulations were performed using NAMD 2.13 (ref. 24) and the CHARMM36 force field.25,26 The input files generated by CHARMM-GUI were used for minimization, equilibration, and production runs.19 Each system was equilibrated in the NPT ensemble at 1 atm and 318.15 K for 100 ns followed by a 400 ns NPT production run. A time step of 2 fs was used. Covalent bonds with hydrogen atoms of lipids were kept rigid using SHAKE,27 and water molecules were kept rigid using SETTLE.28 The particle mesh Ewald method was employed to compute long-range electrostatic interactions.29 The Lennard-Jones interactions were smoothed over the range of 10 to 12 Å by the force-based switching function.19,30 Pressure was controlled by means of the Langevin piston Nosé–Hoover method.31 Temperature was controlled by the Langevin dynamics with a damping coefficient of 1 ps−1.32
3 Results and discussion
To quantify the order of lipid acyl tails, we computed the deuterium order parameter,33 SCD, for DPPC and PLePC in the Ld and the Lo phases. The order parameter is defined as
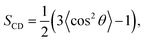 | (1) |
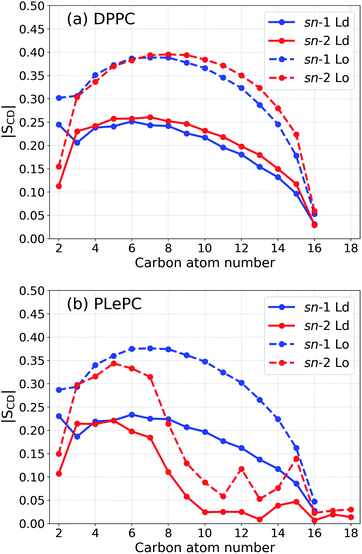 | ||
| Fig. 2 Order parameter |SCD| for the acyl tails of DPPC (a) and PLePC (b) in the Ld phase (with 9% cholesterol) and the Lo phase (with 44% cholesterol). | ||
The lateral packing of lipid–lipid and cholesterol–lipid pairs among ternary mixtures are evaluated by the two-dimensional lateral radial pair distribution function g(r), where r is the projected distance on the x–y plane between centers of mass of two molecules.37,38 The g(r) of various pairs among DPPC/PLePC/CHOL mixtures in the Ld and the Lo phases are shown in Fig. 3. When the bilayer changes from the Ld phase to the Lo phase, the packing of DPPC–DPPC becomes more ordered since the first peak of g(r) increases (Fig. 3a). The first peak of g(r) for PLePC–PLePC is almost not affected, indicating a weak effect of the phase state on PLePC–PLePC interactions (Fig. 3b). The packing of DPPC–PLePC also becomes more ordered in the Lo phase (Fig. 3c). In contrast, the packing of CHOL–DPPC becomes looser in the Lo phase since the first peak of g(r) decreases (Fig. 3d), which is accompanied by a significant increase of g(r) for CHOL–PLePC, revealing that the packing of CHOL–PLePC becomes much tighter in the Lo phase (Fig. 3e). Therefore, the phase of the DPPC/PLePC/CHOL mixture has more complicated effects on cholesterol–lipid interactions than on lipid–lipid ones. The packing of CHOL–CHOL becomes slightly tighter in the Lo phase since the position of the first peak of g(r) shifts slightly inward (Fig. 3f). The observed ordering effect of CHOL on the lateral structure of a lipid bilayer was also reported in the simulations of Andoh et al., where the lateral packing of lipids is more structured for normal cell membranes (with 42% CHOL) than for leukemic cell membranes (with 23% CHOL).38
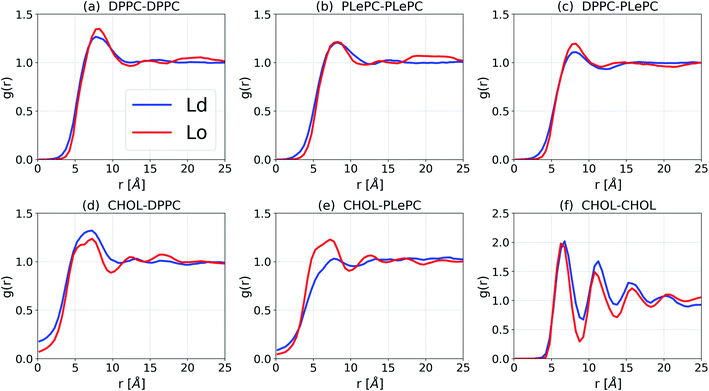 | ||
| Fig. 3 Lateral radial pair distribution function g(r) for DPPC/PLePC/CHOL mixtures in the Ld and the Lo phases. | ||
In order to compare lipid–lipid and cholesterol–lipid interactions systematically, we calculated the potential of mean force (PMF) for various pairs in a bilayer by
PMF(r) = −kBT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) g(r), g(r),
| (2) |
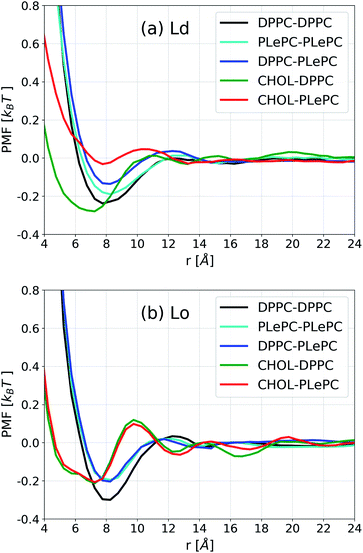 | ||
| Fig. 4 Potential of mean force (PMF) for lipid–lipid and cholesterol–lipid pairs in the Ld phase (a) and the Lo phase (b). | ||
In contrast to the striking differences among various pairs in the Ld phase, the PMF profiles of some pairs become indistinguishable in the Lo phase, as shown in Fig. 4b. When the bilayer changes to the Lo phase, the PMF profile for DPPC–PLePC overlaps with the PLePC–PLePC profile. Considering that the packing of PLePC–PLePC is almost not affected by the phases (Fig. 3b), the relatively lower DPPC–PLePC profile with respect to that in the Ld phase indicates that the repulsion between DPPC and PLePC is hindered in the Lo phase. The minimum for DPPC–DPPC is lower in the Lo phase due to more ordered packing. While the CHOL–DPPC and CHOL–PLePC profiles are significantly deviated in the Ld phase, they overlap in the Lo phase. Moreover, the minimum for CHOL–PLePC shifts inward to the same position as CHOL–DPPC at r ∼7.2 Å, indicating that the repulsion exerted by PLePC on CHOL vanishes in the Lo phase. Therefore, as the mixture changes to the Lo phase, the repulsive role of polyunsaturated PLePC is significantly eliminated, in terms of its interactions with saturated DPPC and CHOL. Compared to the CHOL–DPPC profile in the Ld phase, the minimum for CHOL–DPPC in the Lo phase increases to the same value as CHOL–PLePC, which suggests that DPPC's advantage in attracting CHOL vanishes in the Lo phase, as both DPPC and PLePC are in highly ordered states. The influence of local environment on lipid conformations was shown in a recent study of DSPC/DLiPC/CHOL mixtures, where the polyunsaturated DLiPC (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2/18
2/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) has a noticeably broader distribution of molecular conformations than that of saturated DSPC (18
2) has a noticeably broader distribution of molecular conformations than that of saturated DSPC (18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18
0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) in the Ld phase, but both PCs have restricted conformations in the Lo phase.45
0) in the Ld phase, but both PCs have restricted conformations in the Lo phase.45
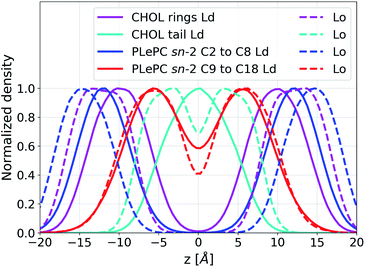
In addition to the lateral packing, the vertical structure of mixtures is also affected by the phases. The density distributions along the bilayer normal for CHOL, CHOL sterol rings, the CHOL hydrocarbon tail, the sn-1 chain of DPPC, the sn-1 and sn-2 chains of PLePC in the Ld phase are shown in Fig. 5a, where the origin of z is the center of mass of the bilayer. The density distributions of the saturated sn-1 chains of DPPC and PLePC are almost identical and flat in 4 Å < |z| < 10 Å, indicating an uniform distribution of saturated chains in each leaflet. In contrast, the density profile of the polyunsaturated sn-2 chain of PLePC has peaks at |z| ∼ 9 Å and much lower values at the center of the bilayer. Therefore, the vertical distribution of the PLePC sn-2 chain is nonuniform, and aggregates in the middle part of each leaflet, which leads to PLePC's repulsion with DPPC and CHOL. Moreover, the density profile of CHOL has peaks at |z| ∼ 9 Å, which overlap with the peaks of the PLePC sn-2 chain and further strengthen the repulsion between CHOL and PLePC. The density distributions of CHOL sterol rings and the CHOL hydrocarbon tail indicate that the peaks of the CHOL profile are contributed by sterol rings. Consequently, the repulsion between CHOL and PLePC is mainly due to interactions between the sterol rings of CHOL and the sn-2 chain of PLePC.
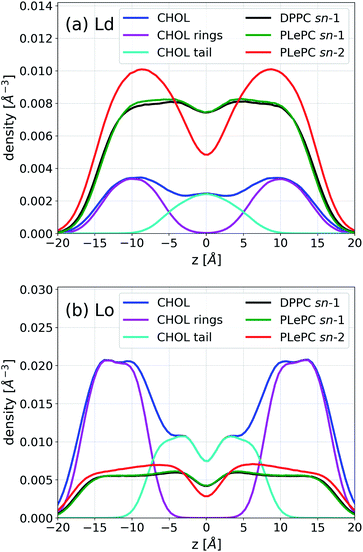 | ||
| Fig. 5 Density profiles of CHOL, CHOL sterol rings, the CHOL hydrocarbon tail, the sn-1 chain of DPPC, the sn-1 and sn-2 chains of PLePC in the Ld phase (a) and the Lo phase (b). | ||
Fig. 5b shows that the vertical density of PLePC sn-2 chain becomes much more uniform in the Lo phase, without the presence of clear peaks, which significantly weakens the repulsion of PLePC with DPPC and CHOL. The uniform distributions of both sn-1 and sn-2 chains extend to |z| ∼ 15 Å, indicating that the bilayer becomes thicker in the Lo phase. The density distribution of CHOL sterol rings also becomes more uniform in 10 Å < |z| < 14 Å. In contrast to the Ld phase, where the CHOL hydrocarbon tail aggregates in the central part of the bilayer, the profile has two peaks at |z| ∼ 3 Å in the Lo phase.
The maximum values of the density profiles for a given component are different in the Ld and the Lo phases according to its concentration in mixtures. To get rid of the effect of species concentrations, the normalized density distributions of CHOL sterol rings, the CHOL hydrocarbon tail, and the sn-2 chain of PLePC in different phases are compared in Fig. 6. Since the three double bonds of PLePC are located in the latter half of the sn-2 chain, the density distribution of the sn-2 chain is further divided into the saturated part considered from carbon number 2 to 8, and the polyunsaturated part considered from carbon number 9 to 18 (see Fig. 1). The results show that the normalized density distribution of CHOL sterol rings has a considerable overlap with the saturated and polyunsaturated parts of the PLePC sn-2 chain in the Ld phase, which means that CHOL interacts with the whole sn-2 chain in the Ld phase. When the bilayer changes to the Lo phase, the distributions of both CHOL sterol rings and the saturated part of the PLePC sn-2 chain move towards the surface of the bilayer, and still have a considerable overlap. The distribution of CHOL hydrocarbon tail also moves towards the surface as indicated by two peaks at |z| ∼ 3 Å in the Lo phase. In contrast, the polyunsaturated part almost remains in the same region of each leaflet, and its overlap with CHOL sterol rings reduces dramatically, which weakens its interaction with CHOL in the Lo phase. Moreover, as shown in Fig. 2b, the sn-2 chain in the Lo phase is more ordered, which further weakens its repulsion with CHOL. Therefore, the strong repulsion between CHOL and PLePC in the Ld phase can be attributed to the repulsive interaction between CHOL sterol rings and the polyunsaturated part of the sn-2 chain. When the bilayer changes to the Lo phase, CHOL moves towards the bilayer surface, and interacts less with the polyunsaturated part of the sn-2 chain, which vanishes the repulsion between them.
4 Conclusions
We have studied the pair interactions among ternary DPPC/PLePC/CHOL mixtures in the Ld and the Lo phases by all-atom MD simulations. The results show that the polyunsaturated PLePC has strong repulsion with DPPC and CHOL in the Ld phase, which is due to the nonuniform distribution of the PLePC sn-2 chain that aggregates in the middle part of each leaflet. When the mixture changes to the Lo phase, the distribution of the PLePC sn-2 chain becomes much more uniform, and CHOL moves towards the surface of the bilayer, which significantly weakens the repulsion of PLePC with DPPC and CHOL. The phase state of the bilayer which affects the order of acyl tails as well as their density distributions along the bilayer normal is a key factor regulating the role of PLePC in lipid mixtures. In general, polyunsaturated PLePC plays a strong repulsive role in the Ld phase but a weak role in the Lo phase. The results could be helpful for our understanding of the role of polyunsaturated phospholipids in lipid raft formation.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the National Natural Science Foundation of China (Grant No. 21973070) and the startup funding of Wenzhou University (Grant No. QD2021180).Notes and references
- T. Harayama and H. Riezman, Nat. Rev. Mol. Cell Biol., 2018, 19, 281–296 CrossRef CAS PubMed.
- E. Sezgin, I. Levental, S. Mayor and C. Eggeling, Nat. Rev. Mol. Cell Biol., 2017, 18, 361–374 CrossRef CAS.
- G. Enkavi, M. Javanainen, W. Kulig, T. Róg and I. Vattulainen, Chem. Rev., 2019, 119, 5607–5774 CrossRef CAS PubMed.
- P. F. F. Almeida, Biochim. Biophys. Acta, Biomembr., 2009, 1788, 72–85 CrossRef CAS.
- I. Levental, K. R. Levental and F. A. Heberle, Trends Cell Biol., 2020, 30, 341–353 CrossRef CAS PubMed.
- D. Bolmatov, D. Soloviov, M. Zhernenkov, D. Zav'Yalov, E. Mamontov, A. Suvorov, Y. Q. Cai and J. Katsaras, Langmuir, 2020, 36, 4887–4896 CrossRef CAS PubMed.
- R. X. Gu, S. Baoukina and D. Peter Tieleman, J. Am. Chem. Soc., 2020, 142, 2844–2856 CrossRef CAS.
- D. G. Ackerman and G. W. Feigenson, J. Phys. Chem. B, 2015, 119, 4240–4250 CrossRef CAS.
- M. Cebecauer, M. Amaro, P. Jurkiewicz, M. J. Sarmento, R. Šachl, L. Cwiklik and M. Hof, Chem. Rev., 2018, 118, 11259–11297 CrossRef CAS PubMed.
- D. Soloviov, Y. Q. Cai, D. Bolmatov, A. Suvorov, K. Zhernenkov, D. Zav'yalov, A. Bosak, H. Uchiyama and M. Zhernenkov, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 4749–4757 CrossRef CAS.
- S. L. Regen, Biochemistry, 2020, 59, 4617–4621 CrossRef CAS.
- P. F. Almeida, Langmuir, 2018, 35, 21–40 CrossRef PubMed.
- W. Kulig, H. Korolainen, M. Zatorska, U. Kwolek, P. Wydro, M. Kepczynski and T. Róg, Langmuir, 2019, 35, 5944–5956 CrossRef CAS.
- X. Lin, J. H. Lorent, A. D. Skinkle, K. R. Levental, M. N. Waxham, A. A. Gorfe and I. Levental, J. Phys. Chem. B, 2016, 120, 11930–11941 CrossRef CAS PubMed.
- M. R. Krause, T. a. Daly, P. F. Almeida and S. L. Regen, Langmuir, 2014, 30, 3285–3289 CrossRef CAS.
- C. Wang, M. R. Krause and S. L. Regen, J. Am. Chem. Soc., 2015, 137, 664–666 CrossRef CAS.
- C. Wang, Y. Yu and S. L. Regen, Angew. Chem., Int. Ed., 2017, 56, 1639–1642 CrossRef CAS PubMed.
- S. Jo, T. Kim, V. G. Iyer and W. Im, J. Comput. Chem., 2008, 29, 1859–1865 CrossRef CAS.
- J. Lee, X. Cheng, J. M. Swails, M. S. Yeom, P. K. Eastman, J. A. Lemkul, S. Wei, J. Buckner, J. C. Jeong, Y. Qi, S. Jo, V. S. Pande, D. A. Case, C. L. Brooks, A. D. MacKerell, J. B. Klauda and W. Im, J. Chem. Theory Comput., 2016, 12, 405–413 CrossRef CAS PubMed.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, J. Chem. Phys., 1983, 79, 926–935 CrossRef CAS.
- S. L. Veatch and S. L. Keller, Biochim. Biophys. Acta, Mol. Cell Res., 2005, 1746, 172–185 CrossRef CAS PubMed.
- G. W. Feigenson, Biochim. Biophys. Acta, Biomembr., 2009, 1788, 47–52 CrossRef CAS PubMed.
- T. S. Carpenter, C. A. López, C. Neale, C. Montour, H. I. Ingólfsson, F. Di Natale, F. C. Lightstone and S. Gnanakaran, J. Chem. Theory Comput., 2018, 14, 6050–6062 CrossRef CAS PubMed.
- J. C. Phillips, R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R. D. Skeel, L. Kalé and K. Schulten, J. Comput. Chem., 2005, 26, 1781–1802 CrossRef CAS PubMed.
- J. B. Klauda, R. M. Venable, J. A. Freites, J. W. O'Connor, D. J. Tobias, C. Mondragon-Ramirez, I. Vorobyov, A. D. MacKerell and R. W. Pastor, J. Phys. Chem. B, 2010, 114, 7830–7843 CrossRef CAS.
- J. B. Lim, B. Rogaski and J. B. Klauda, J. Phys. Chem. B, 2012, 116, 203–210 CrossRef CAS.
- J.-P. Ryckaert, G. Ciccotti and H. J. Berendsen, J. Comput. Phys., 1977, 23, 327–341 CrossRef CAS.
- S. Miyamoto and P. A. Kollman, J. Comput. Chem., 1992, 13, 952–962 CrossRef CAS.
- U. Essmann, L. Perera, M. L. Berkowitz, T. Darden, H. Lee and L. G. Pedersen, J. Chem. Phys., 1995, 103, 8577 CrossRef CAS.
- P. J. Steinbach and B. R. Brooks, J. Comput. Chem., 1994, 15, 667–683 CrossRef CAS.
- W. Hoover, Phys. Rev. A, 1985, 31, 1695–1697 CrossRef.
- S. E. Feller, Y. Zhang, R. W. Pastor and B. R. Brooks, J. Chem. Phys., 1995, 103, 4613 CrossRef CAS.
- L. S. Vermeer, B. L. De Groot, V. Réat, A. Milon and J. Czaplicki, Eur. Biophys. J., 2007, 36, 919–931 CrossRef CAS PubMed.
- C. Hofsäss, E. Lindahl and O. Edholm, Biophys. J., 2003, 84, 2192–2206 CrossRef.
- a. M. Smondyrev and M. L. Berkowitz, Biophys. J., 1999, 77, 2075–2089 CrossRef CAS.
- T. Róg and M. Pasenkiewicz-Gierula, Biochimie, 2006, 88, 449–460 CrossRef.
- C. Hong, D. P. Tieleman and Y. Wang, Langmuir, 2014, 30, 11993–12001 CrossRef CAS PubMed.
- Y. Andoh, S. Okazaki and R. Ueoka, Biochim. Biophys. Acta, Biomembr., 2013, 1828, 1259–1270 CrossRef CAS.
- J. Timko, A. De Castro and S. Kuyucak, J. Chem. Phys., 2011, 134, 204510 CrossRef PubMed.
- A. J. Sodt, R. W. Pastor and E. Lyman, Biophys. J., 2015, 109, 948–955 CrossRef CAS PubMed.
- D. Hakobyan and A. Heuer, J. Phys. Chem. B, 2013, 117, 3841–3851 CrossRef CAS.
- S. L. Niu and B. J. Litman, Biophys. J., 2002, 83, 3408–3415 CrossRef CAS PubMed.
- O. Engberg, V. Hautala, T. Yasuda, H. Dehio, M. Murata, J. P. Slotte and T. K. Nyholm, Biophys. J., 2016, 111, 546–556 CrossRef CAS.
- T. K. Nyholm, S. Jaikishan, O. Engberg, V. Hautala and J. P. Slotte, Biophys. J., 2019, 116, 296–307 CrossRef CAS PubMed.
- W. F. Bennett, J. E. Shea and D. P. Tieleman, Biophys. J., 2018, 114, 2595–2605 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra02692e |
| This journal is © The Royal Society of Chemistry 2021 |