Open Access Article
Open Access ArticleA carbon dot-based Co-nanozyme with alkaline phosphatase – mechanism and application†
Haiyan Fan a,
Kanat Dukenbayev‡
a,
Kanat Dukenbayev‡
 b,
Qinglei Sunc,
Medina Khamijana,
Akhrorbek Turdaliyeva,
Alibek Ysmaiyla,
Aigerim Tassanbiyevaa,
Cuiping Ma
b,
Qinglei Sunc,
Medina Khamijana,
Akhrorbek Turdaliyeva,
Alibek Ysmaiyla,
Aigerim Tassanbiyevaa,
Cuiping Ma d and
Yingqiu Xie
d and
Yingqiu Xie *a
*a
aSchool of Sciences and Humanities, Nazarbayev University, Nur-Sultan, Republic of Kazakhstan. E-mail: xieautumnus@yahoo.com; yingqiu.xie@nu.edu.kz
bSchool of Engineering and Digital Sciences, Nazarbayev University, Nur-Sultan, Republic of Kazakhstan
cKey Laboratory for Applied Technology of Sophisticated Analytical Instrument of Shandong Province, Shandong Analysis and Test Center, Qilu University of Technology (Shandong Academy of Sciences), Jinan, China
dShandong Provincial Key Laboratory of Biochemical Engineering, Qingdao Nucleic Acid Rapid Detection Engineering Research Center, College of Marine Science and Biological Engineering, Qingdao University of Science and Technology, Qingdao, 266042, China
First published on 21st October 2021
Abstract
Elevated levels of alkaline phosphatase (ALP) are associated with bone metastasis, liver cancer, prostate cancer, breast cancer, and many other diseases or stem cell marker. It is therefore of great significance to quantitatively detect the ALP levels by a rapid, highly sensitive, and easy-to-use strip paper test. In the present work, we discovered an enhancement of ALP activity upon the addition of cauliflower-derived carbon dots (CFCDs), which can be applied as a sensor for ALP. The mixed ALP and CFCDs exhibited a typical Michaelis Menten mechanism with increased Vmax and reduced Km compared to ALP alone. High-Resolution Atomic Force Microscopy (HR-AFM) reveals the dimensions of ALP, the CFCDs, and the phosphatase substrate para-nitrophenyl phosphate (pNPP), as well as the potential interactions among them. The role of the CFCDs was identified as the addition of reaction centers to ALP; in other words, a competitive activator. Besides the improved kinetics, the yield of dephosphorylated product was also increased by at least twice upon the addition of CFCDs. Taking advantage of this effect, a portable CFCD-based paper strip assay was developed to achieve sensitive detection of abnormally elevated ALP levels and visualization of cancer stem cells or proteins by phosphatase-conjugated antibodies. Our findings show great promise for disease diagnosis and bioassays related to ALP enhancement that may be used for protein or cell detection.
1 Introduction
The concept of an artificial enzyme has exhibited some breakthroughs from the perspectives of both the conventional definition and related mechanism of natural enzymes ever since Ronald Breslow proposed it.1 Materials such as metal-based and carbon-based nanomaterials, functionalized quantum dots, polymers, and biomolecular-based materials have been developed to fulfill hydrolase, peroxidase, superoxide dismutase, oxidase and other enzymatic functions.2–12 In order to lower the cost, enhance the biocompatibility, simplify the synthesis, and take advantage of their environmental friendliness, carbon nanodots (CDs) have been introduced to the family of artificial enzymes. So far, the majority of CD-based nanozymes have mainly demonstrated peroxidase and oxidase activities.13–17 In our previous work, the attention was shifted to the impact of soybean-derived CDs on the protein tyrosine phosphatase.18Metal-based or carbon-based, the nanozyme generally catalyzes various reactions in a unique mechanism from that of the natural enzyme. For instance, using Au nanoparticles shelled with a single layer of 1-sulfanyloctane, followed by azacrown functionalization, Houillon et al. successfully loaded Zn2+ on the surface of the nano-assemblies through coordination between Zn2+ and the amine groups. The obtained nanozyme exhibited superior hydrolase activity toward a 2-hydroxypropyl p-nitrophenyl phosphate (HPNP) substrate.19 The authors attributed the extraordinary enzymatic effect to the more effective usage of the substrates, because more hydrolase centers, with coordinated Zn2+, were formed with the addition of Zn2+ to the particular assembly.
As mentioned before, CDs have only been utilized so far as peroxidases. Xia et al. investigated the enzymatic mechanism of the CDs developed from glucose and cyclodextrin.14 Based on quantum mechanical calculations using density function theory, the authors pointed out that the peroxidase activity exhibited by CDs was not initiated by the production of ˙OH like most other peroxidases, but through the activation of a H2O2 molecule by its noncovalent binding to the CDs on one hand and to the peroxidase substrate 3,3′,5,5′-tetramethylbenzidine (TMB) on the other hand.
CDs and their chemical and/or biological functions are somewhat mysterious, as they do not have a specific chemical composition, or a well-defined structure. All we have learned so far is that CDs mostly appear as semispherical shaped particles with the individual size usually smaller than 10 nm.20 While there are generally some functional groups such as C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, O–H and N–H distributed on the surface, their optical properties, in particular, their photoluminescence, mostly depend on the particle size rather than the functional groups.21–24 Using o-phenylenediamine and citric acid as the starting material, Lu et al. synthesized CDs emitting 7 different colors from violet to red by tuning the pH and the temperature and duration of the hydrothermal reaction, and for the first time, the authors proposed that the emitting color of CDs is correlated with the distribution of sp2 and sp3 hybridization in the carbon core.25–27
O, O–H and N–H distributed on the surface, their optical properties, in particular, their photoluminescence, mostly depend on the particle size rather than the functional groups.21–24 Using o-phenylenediamine and citric acid as the starting material, Lu et al. synthesized CDs emitting 7 different colors from violet to red by tuning the pH and the temperature and duration of the hydrothermal reaction, and for the first time, the authors proposed that the emitting color of CDs is correlated with the distribution of sp2 and sp3 hybridization in the carbon core.25–27
Within the general enzymatic mechanism, Michaelis–Menten, for instance, the formation of the enzyme–substrate complex is critical in the determination of the overall reaction rate. For the natural protein enzyme, there are only a few activation sites that allow binding with the substrate. In ALP, for instance, the activation sites are commonly known as two Zn2+ distanced by 4.6 Å in the protein.28–30 A perfect fit between the shape of the activation site and that of the substrate is necessary to render a strong binding between them. Therefore, the conformation and associated folding structure of the protein enzyme are vital for protein enzymes. With nanozymes, on the other hand, the relevant functional groups are most likely exposed and ready to bind with the substrates. Hence, nanozymes may provide broader and more flexible binding sites to the substrates.
Based on our knowledge from previous work, there is strong binding between the representative functional group, a protonated amide group, on the soybean derived CDs, and the protein tyrosine phosphate group.18 In the present work, we attempt to combine CFCDs and ALP, as the functional groups on the surface of CFCDs can potentially form strong intermolecular bonding (hydrogen bonds, for instance) with ALP. We expect that the combination of CFCDs and ALP may enhance the number of activation sites and therefore improve the kinetics and sensitivity of phosphatase.
Phosphatase has been widely studied in cell signaling and diagnosis. Elevated levels of alkaline phosphatase (ALP) in the blood or tissue fluid are associated with bone metastasis, liver cancer, prostate cancer, breast cancer, and many other diseases or stem cell characters.31–35 It is therefore of considerable significance to quantitatively detect the ALP levels in body fluid, in a rapid, highly sensitive, and easy-to-use manner. However, the current test assays designed for ALP detection are insensitive because of their limited activity for colorimetric detection during substrate catalytic reaction.36,37 In the present work, we examined the phosphatase activator activity of CFCDs and explored its application in a paper strip test for ALP detection. We discovered that CFCDs, when combined with ALP, dramatically accelerated the phosphatase reaction, and greatly enhanced the yield of phosphatase products; thus, an improved sensitivity was achieved. As a result, a highly sensitive paper strip-based portable test of ALP concentration in the medium was developed, and cancer stem-like cells were labeled. The present study holds enormous potential in the diagnosis of cancer, and may be applicable in clinical practice.
2 Experimental section
2.1 Synthesis of CFCDs
The CFCDs were synthesized using hydrothermal treatment of a mixture of dried cauliflower powder and ethylenediamine in 60 mL of water at 200 °C for five hours, as described previously with other CDs. The liquid phase was then collected, followed by centrifugation and oven drying at 60 °C.2.2 FT-IR, scanning electron microscopy (SEM) and high-resolution atomic-force microscopy (HR-AFM)
The FT-IR spectrum was recorded for the dried CFCDs on a FT-IR spectrometer (Thermo Scientific Nicolet iS5) at the resolution of 2 cm−1. For the SEM assay, the samples were dried on foil paper and subjected to scanning by a Carl Zeiss SUPRA 55 Scanning Electron Microscope.HR-AFM was performed by applying the AC-Mode of tapping mode in air by an AFM Smart SPM 1000 (AIST-NT, Russia). For the protein-based binding assay, Calf Intestinal Alkaline Phosphatase (CIAP, New England Biolabs) as the ALP was mixed with CFCDs or vehicle as a control.
2.3 Statistical analysis
For data analysis and comparison of different treatments, Student's t-test was used, with p values not more than 0.05–0.01 thought of as statistically significant.2.4 The para-nitrophenyl phosphate (pNPP) and 5-bromo-4-chloro-3-indolyl phosphate-nitro blue tetrazolium liquid substrate system (BCIP-NBT)-based phosphatase assays and paper strip assay
Phosphatase assays were performed using pNPP and BCIP-NBT (Molecular probe, Life Technologies). Using pNPP and NBT/BCIP as substrates, the UV-visible absorbance was measured at 410 nm, and 590 nm, respectively. para-Nitrophenol (pNP) (Biorad) was used as the standard to measure the product concentration and kinetics.For the rapid paper strip assay, BCIP-NBT as a substrate was applied to filter paper with color and CIAP Phosphatase (New England Biolabs) was used as an ALP standard. The standard curve was made by measuring absorbance at 590 nm using a spectrophotometer. For the cell line assay, medium from cell culture was used for testing using white filter paper by applying BCIP-NBT with or without CFCDs, or alternatively with β-mercaptoethanol (β-ME).
2.5 BCIP-NBT based phosphatase staining of a single-cell formed colony
MDAMB231 cells were applied to form a colony by the standard protocol of a soft agar assay. The colonies formed, confirmed as potential stem-like cells, were stained with the control of BCIP-NBT only, or BCIP-NBT solution mixed with cauliflower CDs and β-ME. After 24 h, the cells were investigated and photographed by an inverted microscope on soft agar plates.2.6 BCIP-NBT based phosphatase-conjugated immunocytochemistry (ICC) assay
The cells were cultured in a CO2 incubator and subjected to an ICC assay by fixing according to the Cell Signaling manufacture protocol (https://www.cellsignal.com/contents/resources-protocols/immunofluorescence-general-protocol/if). The YAP antibody was used as the primary antibody (Santa Cruz) and a phosphatase-conjugated antibody was used as a secondary antibody. Finally, BCIP-NBT was used for staining of the antibodies, and images were taken for further analysis.3 Results and discussion
3.1 Characterization of CFCDs
The SEM image of the CFCDs in Fig. 1A indicates that at the working concentration, the CFCDs stay in the aggregated form. The morphology for these aggregations is a cylindrical shaped tablet with an average diameter in the range from 20 nm to 120 nm and thickness of 1–3 nm suggested by the AFM image. The Fourier transform infrared (FT-IR) spectrum shows that the dominant functional group distributed on the surface of the CFCDs is an amide, exhibiting peaks at 1640 cm−1, 1560 cm−1, 1300 cm−1, and 3320 cm−1, representing C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, N–H, C–N, and N–H stretching vibrations, respectively (Fig. 1B). These IR absorption bands are coincident with the ones shown in the FT-IR spectrum of soybean-derived CDs (SBCDs),18 suggesting that both CDs, though prepared with different precursors, contain the same chemical components, and the effective surface functional group is protonated amide. The aqueous solution of the CFCDs exhibits a light-yellow color, and its absorption spectrum shows three main peaks at 340 nm, 390 nm and 540 nm, wherein, excitation at 540 nm does not emit fluorescence (Fig. 1C). The fluorescence spectra exhibit peaks at 436 nm and 470 nm corresponding to the excitation wavelengths of 340 nm and 390 nm, respectively.
O, N–H, C–N, and N–H stretching vibrations, respectively (Fig. 1B). These IR absorption bands are coincident with the ones shown in the FT-IR spectrum of soybean-derived CDs (SBCDs),18 suggesting that both CDs, though prepared with different precursors, contain the same chemical components, and the effective surface functional group is protonated amide. The aqueous solution of the CFCDs exhibits a light-yellow color, and its absorption spectrum shows three main peaks at 340 nm, 390 nm and 540 nm, wherein, excitation at 540 nm does not emit fluorescence (Fig. 1C). The fluorescence spectra exhibit peaks at 436 nm and 470 nm corresponding to the excitation wavelengths of 340 nm and 390 nm, respectively.
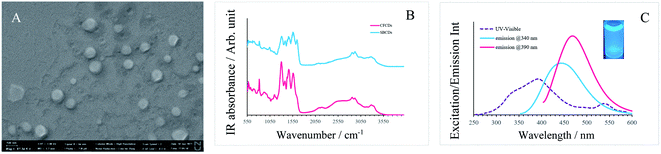 | ||
| Fig. 1 (A) SEM image of CFCDs at the working concentration. (B) FT-IR spectrum of the CFCDs and SBCDs.18 (C) UV-visible (purple dashed line) and fluorescence (solid blue at the excitation wavelength of 340 nm, solid red at the excitation wavelength of 390 nm) spectra of the aqueous solution of CFCDs. | ||
3.2 Enzymatic kinetics for the CFCDs, ALP and ALP-CFCDs
As indicated in Fig. 2A, the dependence of the reaction rate on the concentration of the substrate pNPP demonstrates a typical Michaelis–Menten mechanism for both ALP and ALP-CFCDs, whereas no significant phosphatase activity was observed for CFCDs alone. The values of Vmax and Km obtained from the linear fitting in Fig. 2B are summarized in Table 1. It is evident that the addition of CFCDs to ALP not only enhances the maximum reaction rate, but also greatly enhances the binding affinity with the substrate. The reaction with CFCDs only, although it exhibits nearly a hundred times slower rate, showed the strongest binding affinity with the substrate among the three systems.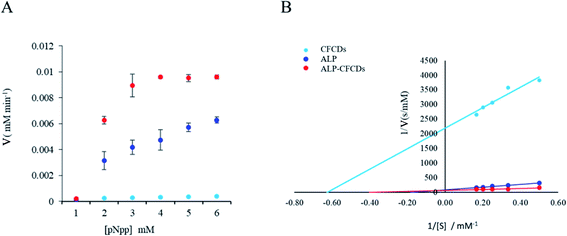 | ||
| Fig. 2 ALP enzyme kinetics upon CFCD coenzyme addition. (A) The plot of the initial reaction rate (V) versus the pNPP substrate (S) concentration at 25 °C and pH 7. (B) The plot of 1/V versus 1/[S]. | ||
| System | Vmax (mM min−1) | Km |
|---|---|---|
| CFCDs | 0.000456 | 1.59 |
| ALP | 0.0117 | 5.44 |
| ALP-CFCDs | 0.0147 | 2.48 |
Based on the enzymatic kinetics, CFCDs do not show evident phosphatase activity by themselves. They instead act as a competitive promoter of ALP. Based on the morphology of CFCDs at the working concentration, it's quite likely that ALP is distributed on the surface of particles of CFCDs, and thus more active sites are provided per unit surface area for the substrates.
3.3 Revealing the binding between ALP and CFCDs using HR-AFM imaging
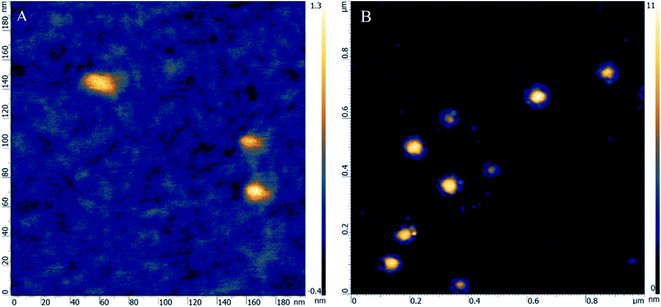
The HR-AFM technique was utilized to collect more evidence for the binding between ALP and CFCDs from the perspective of dimension change. As depicted in Fig. 3A, the HR-AFM image of ALP exhibits a structure with dimensions of 20 nm × 10 nm × 1.3 nm, which is consistent with the dimensions described for the general E. coli ALP reported.38 The individual unit on the image may be considered as a single protein. Upon addition of CFCDs, the dimension enlarges to ∼100 nm × 100 nm × 10 nm, wherein individual ALP proteins are found attaching to the surface of the tablet of CFCDs with a diameter of ∼100 nm, as indicated in Fig. 3B. Considering the structure of ALP, it is most likely that the phosphate group surrounding the Zn2+ on ALP interacts with the protonated amide group on the CFCDs.18 Therefore, the coordination between CFCDs and ALP creates new phosphatase centers along the interface between the two.3.4 Beyond the Michaelis–Menten mechanism
The HR-AFM images indicate that there is strong binding between the CFCDs and ALP. On the other hand, the enzymatic kinetic results suggest that the binding with the substrate is enhanced after loading ALP on the surface of the CFCDs. However, the CFCDs themselves do not show obvious phosphatase activity. A question arises in terms of the role of CFCDs in the enhancement of phosphatase activity. No matter whether it is the alkaline phosphatase or protein tyrosine phosphatase (PTP), a nuclear affiliation group is necessary to attack the phosphorous atom within the enzyme–substrate (ES) complex. In ALP, it takes Zn2+ to bind with the substrate, and a hydroxyl group originally coordinated with Zn2+ acts as the nuclear affiliation group,39 whereas in PTP, the C–S− group on cysteine binds with the substrate and acts as the nuclear affiliation group.40 There is no significant amount of Zn2+ nor C–S− groups in CFCDs; nevertheless, the promote the ALP activity and increase the yield of the dephosphorylated product. It is most likely that the effect of CFCDs is initiated at the onset of the dissociation of ES. As we pointed out earlier, the phosphatase center is on the interface between the CFCDs and ALP. We are reminded here that the actual enzymatic reaction should be expressed using the equation below.
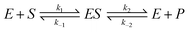 | (1) |
The Michaelis–Menten mechanism takes an approximation to simplify the problem. Regardless, the dissociation of ES to the final product is a reversible reaction. We hypothesize in the present work that CFCDs promote the phosphatase mainly through the interaction with the dephosphorylated product, para-nitro-phenoxide. Such interaction favors the dissociation of ES. As the dissociation of ES is the rate-determining step, the CFCDs, therefore, exhibit a promoting effect on phosphatase.
3.5 CFCD-based paper strip assay for ALP detection
Taking advantage of the fact that CFCDs enhance not only the enzymatic kinetics but also the yield of the dephosphorylated product, a CFCD-based paper strip assay was developed for sensitive detection of ALP. By using ALP standard 0.00001, 0.001, 0.01 and 0.1 U μL−1, we tested the colorimetric response of a paper strip that was pretreated with BCIP and NBT w/o CFCDs. As shown in Fig. 4, an observable color change occurs at the ALP concentration of 0.1 U μL−1 after 12 min without the addition of CFCDs. An obvious color change is observed at the ALP level of 0.01 U μL−1 within 2 min upon the addition of CFCDs. In general, a level of blood ALP concentration above 0.01 U μL−1 is usually associated with a risk of bone diseases, neurological diseases, and cancer.36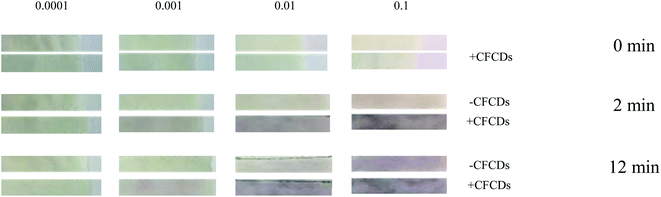 | ||
| Fig. 4 CFCDs enhance the sensitivity of the paper strip test assay for ALP detection based on the NBT-BCIP substrate. All the paper tests were performed at 25 °C and pH = 7. | ||
In a separate experiment, we applied the paper as mentioned above to the strip test for samples derived from the culture medium of cancer cells with different cell lines. As expected, the MDAMB231 cell sample with a relatively higher level of ALP than other cancer cells used in the present work shows an observable color change within 5 minutes in the presence of CFCDs (Fig. S1†). Meanwhile, the actual quantity of ALP in the MDAMB231 cell sample was able to be determined using the calibration curves established with or without CFCDs. Both obtained standard curves have the same trend, and the actual level of ALP was identified as 4.53 U mL−1 without CFCDs, and 5.3 U mL−1 with CFCDs (Fig. S2†).
Overall, the addition of CFCDs greatly enhanced the detection sensitivity and accuracy of ALP. For a proper paper test to detect an abnormally elevated ALP level, high sensitivity toward the level of ALP is not desired, instead, the color change is expected to occur at the abnormally high level. With the addition of CFCDs, the observable color change occurs at the ALP concentration of 0.01 U μL−1, which makes the abnormally high level of ALP visualizable on the paper test, whereas with the average level of ALP, no color change is observed. The paper strip test developed in the present work is sufficient to detect ALP at higher than normal levels as the estimated clinical diagnosis of a normal range of ALP in adult is 44–147 U μL−1 (https://www.urmc.rochester.edu/encyclopedia/content.aspx?contenttypeid=167%26contentid=alkaline_phosphatase). This is of significance for a portable monitor of potential bone metastasis of cancer. Given that elevation of the ALP level occurs along with many diseases and stem cells,31–35 the as-prepared paper strip may be potentially applied to rapid cancer diagnosis.
3.6 CFCDs enhance the visualization of cell staining of phosphatase expressed cancer stem-like cells
Given that ALP activity is also elevated in stem cells, we applied CFCDs to a live stain of stem cells. Using MDAMB231 cells as a model of cancer stem-like cells and NBT/BCIP as a substrate, we stained a single cell-derived colony in soft agar for the detection of ALP activity. After 24 h, the stain showed an intense dark blue color, as shown in ESI Fig. S3,† compared to the control. Our data suggest that the addition of CFCDs may achieve signaling amplification, hence visualizing the stem-like cells directly in vivo. The rapid identification of cancer stem-like cells was then fulfilled. On the other hand, the elevation of the ALP level is also associated with cancer stem cells, and the as-prepared paper strip may be applied to stem cell tracking or sorting instead of the antibody-based markers in cancer stem cell or spheres as we used before in prostate cancer spheres.3.7 The CFCD enhances the visualization of phosphatase-conjugated immunocytochemistry assay in cells
In a further test, CFCDs were applied to phosphatase-conjugated immunoassays, wherein YAP primary antibody and phosphatase-conjugated secondary antibody were used to stain cells to investigate the YAP protein levels in PC3 cells. As shown in Fig. 5, the YAP antibody staining intensity and the cell image resolution were greatly enhanced upon the addition of CFCDs without altering the expression pattern. In detail, YAP expression was found in both the nucleus and cytosol, and in CFCD-treated cells, the localization of YAP remained the same. Thus, the CFCDs may be used for bioassays such as ICC.4 Conclusions
Cauliflower-derived carbon dots (CFCDs) exhibit a novel mechanism of co-enzymatic alkaline phosphatase. Combined with the enzymatic kinetic characterization and HR-AFM imaging, the co-enzymatic effect led by CFCDs was proposed. The intermolecular interaction between CFCDs and para-nitro-phenoxide was believed to play an essential role in the co-enzymatic effect. The CFCD-integrated testing of ALP achieved 10–100 times more sensitive results under reasonable conditions compared with the clinical test. The present study sets a milestone for the potential of a regular paper strip to test the ALP level and visualize cancer stem-like cells.List of abbreviations
| CND, c-dots | Carbon dots |
| HR-AFM | High-resolution atomic force microscopy |
| ALP | Alkaline phosphatase |
Funding
This work was supported by the Faculty-Development Competitive Research grants program with grant ID: 110119FD4531 i.e. ID: 16797152 (ID: 15798117) to YX; and ID: 110119FD4542 i.e. ID: 16796808 (ID: 15874919) to HF and YX.Data availability
The raw/processed data required to reproduce these findings cannot be shared at this time as the data also forms part of an ongoing study or patent.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We would like to thank the Nazarbayev University for the research funding of the Faculty-Development Competitive Research grants program (ID: 16797152 to YX; and ID: 16796808 to HF and YX). We also would like to thank all lab members or students of Dr Xie's lab for kind help on this project. We especially thank Sultan Makhmetov.References
- R. Breslow and L. E. Overman, Artificial enzyme combing a metal catalytic group and a hydrophobic binding cavity, J. Am. Chem. Soc., 1970, 92, 1075–1077 CrossRef CAS PubMed.
- I. Nath, J. Chakraborty and F. Verpoort, Chem. Soc. Rev., 2016, 45, 4127–4170 RSC.
- D. Jiang, D. Ni, Z. T. Rosenkrans, P. Huang, X. Yan and W. Cai, Nanozyme: new horizons for responsive biomedical applications, Chem. Soc. Rev., 2019, 48, 3683 RSC.
- M. Diez-Castellnou, F. Mancin and P. Scrimin, J. Am. Chem. Soc., 2014, 136, 1158–1161 CrossRef CAS PubMed.
- L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang, N. Gu, T. Wang, J. Feng, J. Yang, D. Yang, S. Perrett and X. Yan, Nat. Nanotechnol., 2007, 2, 577–583 CrossRef CAS PubMed.
- J. Chen, S. Patil, S. Seal and J. F. McGinnis, Nat. Nanotechnol., 2006, 1, 142–150 CrossRef CAS PubMed.
- B. C. M. Martindale, G. A. M. Hutton, C. A. Caputo and E. Reisner, Solar hydrogen production using carbon quantum dots and a molecular nickel catalyst, J. Am. Chem. Soc., 2015, 137(18), 6018–6025 CrossRef CAS PubMed.
- D. Mosconi, D. Mazzier, S. Silvestrini, A. Privitera, C. Marega, L. Franco and A. Moretto, Synthesis and photochemical applications of processable polymers enclosing photoluminescent carbon quantum dots, ACS Nano, 2015, 9(4), 4156–4164 CrossRef CAS PubMed.
- A. Zammataro, C. M. A. Gangemi, A. Pappalardo, R. M. Toscano, R. Puglisi, G. Nicotra, M. E. Fragalà, N. Tuccitto and G. T. Sfrazzetto, Covalently functionalized carbon nanoparticles with a chiral Mn–Salen: a new nanocatalyst for enantioselective epoxidation of alkenes, Chem. Commun., 2019, 55(36), 5255–5258 RSC.
- H. Wei and E. Wang, Chem. Soc. Rev., 2013, 42, 6060–6093 RSC.
- C. Korsvik, S. Patil, S. Seal and W. T. Self, Chem. Commun., 2007, 1056–1058 RSC.
- A. Asati, S. Santra, C. Kaittanis, S. Nath and J. M. Perez, Angew. Chem., Int. Ed., 2009, 48, 2308–2312 CrossRef CAS PubMed.
- T. Liu, Z. Cui, J. Zhou, Y. Wang and Z. Zou, Synthesis of pyridine-rich N, S, X0-doped carbon quantum dots as effective enzyme mimics, Nanoscale Res. Lett., 2017, 12, 375 CrossRef PubMed.
- Y. Lv, M. Ma, Y. Huang and Y. Xia, Carbon dots nanozymes: how to be close to natural enzymes, Chem.–Eur. J., 2019, 25, 954–960 CAS.
- B. Garg and T. Bisht, Carbon nanodots as peroxidase nanozymes for biosensing, Molecules, 2016, 21, 1653 CrossRef CAS PubMed.
- S. Liu, J. Tian, L. Wang, Y. Luo and X. Sun, A general strategy for the production of photoluminescent carbon nitride dots from organic amines and their application as novel peroxidase-like catalysts for colorimetric detection of H2O2 and glucose, RSC Adv., 2012, 2, 411–413 RSC.
- P. Zhang, D. Sun, A. Cho, A. Weon, S. Lee, J. Lee, J. W. Han, D. P. Kim and W. Choi, Modified carbon nitride nanozyme as bifunctional glucose oxidase-peroxidase for metal-free bioinspired cascade photocatalysis, Nat. Commun., 2019, 10, 940–954 CrossRef PubMed.
- Y. Xie, H. Fan, W. Lu, Q. Yang, A. Nurkesh, T. Yeleusizov, A. Maipas, J. Lu, L. Manarbek, Z. Chen and E. Benassi, Nuclear MET requires ARF and is inhibited by carbon nanodots through binding to phosphor-tyrosine in prostate cancer, Oncogene, 2019, 38(16), 2967–2983, DOI:10.1038/s41388-018-0608-2.
- F. Manea, F. B. Houillon, L. Pasquato and P. Scrimin, Angew. Chem., Int. Ed., 2004, 43, 6165–6169 CrossRef CAS PubMed.
- X. Xu, R. Ray, Y. Gu, H. J. Ploehn, L. Gearheart, K. Raker and W. A. Sorivens, Electrophoretic analysis and purification of fluorescent single-walled carbon nanotube fragments, J. Am. Chem. Soc., 2004, 126, 12736–12737 CrossRef CAS PubMed.
- H. Li, et al., Water-soluble fluorescent carbon quantum dots and photocatalyst design, Angew. Chem., Int. Ed., 2010, 49, 4430–4434 CrossRef CAS PubMed.
- H. Li, X. He, Z. Kang, H. Huang, Y. Liu, J. Liu, S. Lian, C. H. Tsang, X. Yang and S. T. Lee, Water-soluble fluorescent carbon quantum dots and photocatalyst design, Angew. Chem., Int. Ed., 2010, 49, 4430–4434 CrossRef CAS PubMed.
- Z. Gan, X. Wu, G. Zhou, J. Shen and P. K. Chu, Is there real upconversion photoluminescence from graphene quantum dots?, Adv. Opt. Mater., 2013, 1, 554–558 CrossRef.
- J. Liu, K. Zhang, M. Yang, H. Sun and B. Yang, One-step hydrothermal synthesis of nitrogendoped conjugated carbonized polymer dots with 31% efficient red emission for in vivo imaging, Small, 2018, 14, e1703919 CrossRef PubMed.
- B. Wang, J. Yu, L. Sui, S. Zhu, Z. Tang, B. Yang and S. Lu, Rational design of multi-color-emissive carbon dots in a single reaction system by hydrothermal, Adv. Sci., 2021, 8, 2001453 CrossRef CAS PubMed.
- Y. Ru, L. Sui, H. Song, X. Liu, Z. Tang, S.-Q. Zhang, B. Yang and S. Lu, Rational design of multicolor-emitting chiral carbonized polymer dots for full-color and white circularly polarized luminescence, Angew. Chem., Int. Ed., 2021, 60, 14091–14099 CrossRef CAS PubMed.
- X. Yang, L. Sui, Y. Zhang, Z. Tang, B. Yang and S. Lu, Red-emitting, self-oxidizing carbon dots for the preparation of white LEDs with super-high color rendering index, Sci. China: Chem., 2021, 64, 1–7 Search PubMed.
- K. M. Holtz and E. R. Kantrowitz, The mechanism of the alkaline phosphatase reaction: insights from NMR, crystallography and site-specific mutagenesis, FEBS Lett., 1999, 462, 7–11 CrossRef CAS PubMed.
- J. G. Reinhild and G. A. Kfoury, Zinc-dependent enzymes in zinc-depleted rats; intestinal alkaline phosphatase, Am. J. Clin. Nutr., 1969, 22, 1250–1263 CrossRef PubMed.
- U. Sharma, D. Pal and R. Prasad, Alkaline Phosphatase: an overview, Clin. Biochem., 2014, 29, 269–278 CAS.
- S. Damera, K. L. Raphael, B. C. Baird, A. K. Cheung, T. Greene, S. Beddhu, S. Damera, K. L. Raphael, B. C. Baird, A. K. Cheung, T. Greene and S. Beddhu, Serum alkaline phosphatase levels associate with elevated serum C-reactive protein in chronic kidney disease, Kidney Int., 2011, 79(2), 228–233 CrossRef CAS PubMed.
- D. W. Moss, Perspectives in alkaline phosphatase research, Clin. Chem., 1992, 38(12), 2486–2492 CrossRef CAS.
- F. Nayeem, K. E. Anderson, M. Nagamani, J. J. Grady and L. J. Lu, Alkaline phosphatase and percentage body fat predict circulating C-reactive protein in premenopausal women, Biomarkers, 2010, 15(8), 663–670 Search PubMed.
- C. T. Lee, Y. C. Tsai, H. Y. Ng, Y. Su, W. C. Lee, L. C. Lee, T. T. Chiou, S. C. Liao and K. T. Hsu, Association between C-reactive protein and biomarkers of bone and mineral metabolism in chronic hemodialysis patients: a cross-sectional study, J. Renal Nutr., 2009, 19, 220–227 Search PubMed.
- B. M. Cheung, K. L. Ong, R. V. Cheung, L. Y. Wong, N. M. Wat, S. Tam, G. M. Leung, C. H. Cheng, J. Woo, E. D. Janus, C. P. Lau, T. H. Lam and K. S. Lam, Association between plasma alkaline phosphatase and C-reactive protein in Hong Kong Chinese, Clin. Chem. Lab. Med., 2008, 46(4), 523–527 CAS.
- C. Payne and R. A. Wilbey, Alkaline phosphatase activity in pasteurized milk: a quantitative comparison of fluorophos and colourimetric procedures, Int. J. Dairy Technol., 2009, 62, 308–314 CrossRef CAS.
- L. Zhang, J. Nie, H. Wang, J. Yang, B. Wang and Y. Zhang, Instrument-free quantitative detection of alkaline phosphatase using paper-based devices, Anal. Methods, 2017, 9, 3375–3379 RSC.
- K. M. Holtz and E. R. Kantrowitz, FEBS Lett., 1999, 462, 7–11 CrossRef CAS PubMed.
- J. E. Coleman, Structure and mechanism of alkaline phosphatase, Annu. Rev. Biophys. Biomol. Struct., 1992, 21, 1441–1483 CrossRef PubMed.
- D. Barford, Molecular mechanisms of the protein serine/threonine phosphatase, Trends Biochem. Sci., 1996, 21, 407–412 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra04483d |
| ‡ Co-first authors. |
| This journal is © The Royal Society of Chemistry 2021 |