Open Access Article
Open Access ArticleBis-indolylation of aldehydes and ketones using silica-supported FeCl3: molecular docking studies of bisindoles by targeting SARS-CoV-2 main protease binding sites†
Barnali Deb a,
Sudhan Debnath
a,
Sudhan Debnath *b,
Ankita Chakraborty‡
*b,
Ankita Chakraborty‡
 a and
Swapan Majumdar
a and
Swapan Majumdar *a
*a
aDepartment of Chemistry, Tripura University, Suryamaninagar, 799 022, India. E-mail: smajumdar@tripurauniv.ac.in; Fax: +91-381-237-4802; Tel: +91-381-237-9070
bDepartment of Chemistry, Netaji Subhash Mahavidalaya, Tripura 799114, India
First published on 16th September 2021
Abstract
We report herein an operationally simple, efficient and versatile procedure for the synthesis of bis-indolylmethanes via the reaction of indoles with aldehydes or ketones in the presence of silica-supported ferric chloride under grindstone conditions. The prepared supported catalyst was characterized by SEM and EDX spectroscopy. The present protocol has several advantages such as shorter reaction time, high yield, avoidance of using harmful organic solvents during the reaction and tolerance of a wide range of functional groups. Molecular docking studies targeted toward the binding site of SARS-CoV-2 main protease (3CLpro or Mpro) enzymes were investigated with the synthesized bis-indoles. Our study revealed that some of the synthesized compounds have potentiality to inhibit the SARS-CoV-2 Mpro enzyme by interacting with key amino acid residues of the active sites via hydrophobic as well as hydrogen bonding interactions.
Introduction
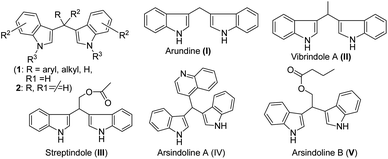
Giving high priority to social responsibility and public awareness to conserve our mother nature, the synthetic organic chemistry community designs any chemical process in such a way so that it can eliminate or reduce the use of harmful chemicals as much as possible during manufacturing and processing to keep the environment clean and sustainable for future generations. Therefore, the development of efficient and environmentally benign synthetic strategies is the prime task of organic chemists in academic and industrial settings.1 Thus, over the last decade, tremendous efforts have been devoted to the exploration of an eco-friendly methodology in both academia and industries using solvent-free techniques,2 ionic liquids,3 water media,4 phase transfer catalysts,5 solid-supported catalysts,6 microwave irradiation7 or grindstone/ball-milling processes.8 The use of grindstone chemistry is very fascinating for the synthesis of desired functional molecules via simple mixing of the precursors and catalysts with much faster reaction rates and selectivity under benign reaction conditions. In recent times, much attention has been paid to organic transformations promoted by solid-supported reagents or catalysts and till now they occupy a special position in the development of greener chemical processes due to their attractive properties such as high surface area, crystalline structure, less or no corrosion, high thermal stability, persistence in all organic solvents, no waste or disposal problems, and recyclability. Among various solid supports, silica gel is one of the extensively used surface material9 for different chemical transformations in organic chemistry, as it displays many significant advantages such as high surface area, thermal and chemical stability, easy availability, low cost and insolubility in organic solvents. Recently, silica-supported ferric chloride has been used as an efficient heterogeneous catalyst in many organic transformations by us10a and others10b–e in different occasions.The indole scaffold has been recognized as a prominent and privileged structural motif found in thousands of natural products, exhibiting a range of important biological activities and gaining technological interest.11 Many indole derivatives were found to possess anti-inflammatory activity along with analgesic and antipyretic properties. They inhibit the production of eicosanoids such as prostacyclin, thromboxanes and prostaglandins via inhibiting cyclooxygenase activity and thereby reduce edema. Indole analogues are the most important series of inhibitors with considerable anti-viral activities against hepatitis C virus (HCV), acting at a non-structural 5B RNA-dependent-RNA-polymerase (RdRp) thumb site I.12 Three compounds, namely, indigo, indirubin, and indicant containing an indole moiety isolated from the roots of the plant Isatis indigotica were tested against SARS-CoV 3CLpro, and they showed inhibitory activity on cell-free cleavage of SARS-CoV 3CLpro and their IC50 values were 37.3, 81.3 and 33.1 μg mL−1 respectively.13 Another indole derivative, 5-chloropyridin-3-yl-1H-indole-4-carboxylate showed excellent SARS-CoV 3CLpro inhibitory activity with an IC50 value of 30 nM.14 Arbidol, a small indole derivative, showed broad spectrum activity against influenza and other respiratory viral infections.15 It has a capacity to interact with both membrane and viral cellular proteins. The above-mentioned evidence indicates that indole derivatives act as anti-viral agents. Zhang et al. in their comprehensive overview on indole derivatives also highlighted the anti-viral activities of indole derivatives.16 They also promote the idea that indole macrocycles such as bis- and tris-indole may be exploited in future as novel lead compounds for use as anti-viral agents. Indole or its derivatives are also known as versatile intermediates in organic synthesis due to the feasibility of the 3-position of indole for electrophilic substitution.17 Consequently, 3-substituted indoles were carefully screened for components of drugs and are generally found to be of pharmaceutical interest in a variety of therapeutic areas. Bis(indolyl)methanes (BIMs) are a group of alkaloids with a basic skeleton of two indoles bridged by single methyl carbon; they are differentiated by the groups/substituents attached to the bridging methyl carbon (Fig. 1, 1).18 Several bis(indolyl)methanes and their derivatives have been isolated from terrestrial and marine natural sources, namely, parasitic bacteria, tunicates, and sponges, and some of these possess significant biological activities.19 The natural bis-indolylmethane (Fig. 1, I–V) and some of its analogues exhibit antibacterial, antifungal, anticancer and immunostimulatory properties including antipyretic, anti-inflammatory, anthelmintic, cardio-vascular, anticonvulsant, antimicrobial and selective COX-2 inhibitory activities. Bis(indol-3-yl)methanes (BIMs) and their oxidized derivatives are useful as dyes,20 colorimetric chemosensors,21 fluorescent chemosensors for Cu2+ cation22 and sensors of aspartate and glutamate.23
The acid-catalyzed reaction of electron-rich heterocyclic compounds such as indoles and pyrroles with p-dimethyl aminobenzaldehyde is known as the Ehrlich test.24 Generally, 3,3′-bis(indolyl)methanes are synthesized by an analogous reaction to the Ehrlich test, where indole reacts with aldehydes or ketones in the presence of an acid catalyst to produce an azafulvenium salt, which undergoes further addition reaction with a second indole molecule to produce bis(indolyl)methanes. The role of protic acid or Lewis acid is basically activation of carbonyl and the formation of azafuvenium ions. Except Ehrlich test-type condensation, other approaches for BIM synthesis, e.g. transition metal-catalysed oxidative coupling,25 Bartoli-type reaction26 and photoredox-catalysed ring opening functionalization of tetrahydroisoquinolines27 are known. However, due to the wide substrate and reagent scopes of Ehrlich test-type acid-catalysed electrophilic substitution reaction of indoles with aldehydes, it is one of the widely acceptable simple and straightforward approaches for the synthesis of bis-indolylmethanes. Consequently, several methods have been reported for the synthesis of these compounds. A variety of reagents such as protic acid,28 Lewis acids,29 transition and non-transitional metal salts,30 solid acidic catalysts31 and ionic liquids32 have been employed to accomplish this transformation. However, many of these methods suffer from disadvantages such as unsatisfactory yields, expensive catalysts, long reaction times, toxic organic solvents, laborious work-up procedures, requirement of special apparatus, and harsh reaction conditions. Thus, the development of simple, efficient, high-yielding, and eco-friendly methods using new recyclable catalysts for the synthesis of these compounds would be highly desirable. With the purpose to develop more efficient synthetic processes, reduce the number of separate reaction steps and minimize the by-products, herein we report a novel and efficient method for the preparation of bis-indolylmethane derivatives (BIM's) via multi-component assembly of indoles and aldehydes in the presence of silica-supported ferric chloride (SiO2-FeCl3) as an efficient, non-volatile, recyclable, easy-to-handle and eco-friendly catalyst (Scheme 1). Furthermore, molecular docking studies of the synthesized compounds were also performed to understand their binding affinities towards Mpro, a crucial life cycle enzyme of SARS-CoV-2.
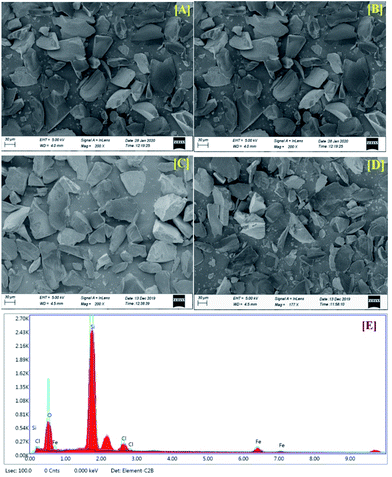
Silica-supported ferric chloride (FeCl3/SiO2) was first prepared by Mazur and Keinan10b who used it as a Lewis acid to dehydrative rearrangement of alcohols. We prepared the same with slight modifications in the procedure and characterized the supported catalyst by SEM analysis. Typically, to the slurry of silica gel (240–300 mesh, 40 g) in acetone (80 mL), anhydrous ferric chloride (5 g, 30.83 mmol) was added with vigorous stirring for 1 h. The excess acetone was removed under reduced pressure and then the mixture was dried under vacuum (∼1 mm Hg) for 24 h to obtain a free flowing solid. The catalyst was stored in a brown colour bottle at 4 °C for longer shelf life. SEM micrographs of the silica, and freshly prepared, six-month-old and recovered catalyst are shown in Fig. 2(A)–(D). An EDX spectrum of the catalyst is shown in Fig. 2(E). The stability of the catalyst was also judged by EDX spectra of the six-month-old catalyst and the recovered catalyst indicated that the quality of the catalyst do not deteriorate even after five cycles (ESI†).
Results and discussion
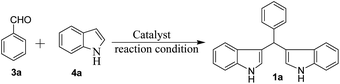
To optimize the reaction conditions for the synthesis of bis-indolylmethanes, we began with benzaldehyde (3a) and indole (4a) as model substrates using different amounts of catalysts under grindstone conditions. No product was obtained upon grinding of 3a and 4a in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar ratio in the absence of the catalyst (Table 1, entry 1), but at elevated temperatures, very little conversion was observed (Table 1, entry 2). However, grinding with chromatographic grade silica facilitated the reaction to yield 70% of 1a (Table 1, entry 3). Having in mind the Lewis acid character of FeCl3, we have employed 5 mol% of FeCl3 while grinding the mixture (Table 1, entry 4). Unfortunately, highly colored mixture of products (TLC) were obtained with full consumption of starting materials. We were pleased to observe that grinding the mixture with pre-prepared FeCl3-SiO2 (20 mg, containing 2 mol% of FeCl3) for 3 h provides clean transformation of the substrates to the desired product in 92% yield (Table 1, entry 5), but the reaction was much faster at 80 °C, which afforded 96% of the yield of 1a in 5 min (Table 1, entry 6). To evaluate the catalytic efficiency, we used different amounts of catalyst FeCl3-SiO2 in this condensation reaction (Table 1, entry 7 and 8). Increase in the amount of catalyst (4 mol%) did not improve further but decreases the amount of catalyst to 1 mol% and slightly decreases the yield of 1a. SiO2-FeCl3 in the presence of solvent CH2Cl2 and EtOH at room temperature gave desired product in 84% and 87% of yield respectively within 15 min and 90 min (Table 1, entries 9 and 10). However, under identical conditions (Table 1, entry 6), other silica-supported catalysts such as SiO2-KHSO4, SiO2-HClO4, and SiO2-H2SO4 have given bis(indolyl) product (1a) in 65%, 58%, and 78% of yield respectively (Table 1, entries 11–13). Furthermore, we also tested the catalytic activity of different nano catalysts such as Cu nanoparticles, nanoTitania Silica (TS 1) and resin-based catalyst Amberlite IR 120H+, which gave 90%, 50% and 90% of yield and the required time was 40–120 min (Table 1, entries 14–16). Amongst all, SiO2-FeCl3 was found to be an efficient catalyst for the synthesis of 1a. The present protocol is also applicable for large-scale synthesis (10 mmol) of bis(indolyl) methane (entry 17), which gave 95% yield of 1a in 15 min.
2 molar ratio in the absence of the catalyst (Table 1, entry 1), but at elevated temperatures, very little conversion was observed (Table 1, entry 2). However, grinding with chromatographic grade silica facilitated the reaction to yield 70% of 1a (Table 1, entry 3). Having in mind the Lewis acid character of FeCl3, we have employed 5 mol% of FeCl3 while grinding the mixture (Table 1, entry 4). Unfortunately, highly colored mixture of products (TLC) were obtained with full consumption of starting materials. We were pleased to observe that grinding the mixture with pre-prepared FeCl3-SiO2 (20 mg, containing 2 mol% of FeCl3) for 3 h provides clean transformation of the substrates to the desired product in 92% yield (Table 1, entry 5), but the reaction was much faster at 80 °C, which afforded 96% of the yield of 1a in 5 min (Table 1, entry 6). To evaluate the catalytic efficiency, we used different amounts of catalyst FeCl3-SiO2 in this condensation reaction (Table 1, entry 7 and 8). Increase in the amount of catalyst (4 mol%) did not improve further but decreases the amount of catalyst to 1 mol% and slightly decreases the yield of 1a. SiO2-FeCl3 in the presence of solvent CH2Cl2 and EtOH at room temperature gave desired product in 84% and 87% of yield respectively within 15 min and 90 min (Table 1, entries 9 and 10). However, under identical conditions (Table 1, entry 6), other silica-supported catalysts such as SiO2-KHSO4, SiO2-HClO4, and SiO2-H2SO4 have given bis(indolyl) product (1a) in 65%, 58%, and 78% of yield respectively (Table 1, entries 11–13). Furthermore, we also tested the catalytic activity of different nano catalysts such as Cu nanoparticles, nanoTitania Silica (TS 1) and resin-based catalyst Amberlite IR 120H+, which gave 90%, 50% and 90% of yield and the required time was 40–120 min (Table 1, entries 14–16). Amongst all, SiO2-FeCl3 was found to be an efficient catalyst for the synthesis of 1a. The present protocol is also applicable for large-scale synthesis (10 mmol) of bis(indolyl) methane (entry 17), which gave 95% yield of 1a in 15 min.
| Entry | Catalyst | Solvent | Temp (°C) | Time (min) | Yielda (%) |
|---|---|---|---|---|---|
| a Isolated yield.b Calculated based on the amount of FeCl3 used during preparation.c Reaction in 10 mmol scale. | |||||
| 1 | No | No | rt | 120 | NR |
| 2 | No | No | 80 | 15 | Mixture |
| 3 | SiO2 | No | 80 | 180 | 70 |
| 4 | FeCl3 | No | 80 | 30 | mixture |
| 5 | SiO2-FeCl3 (2 mol% FeCl3)b | No | rt | 180 | 92 |
| 6 | SiO2-FeCl3 (2 mol%) | No | 80 | 5 | 96 |
| 7 | SiO2-FeCl3 (4 mol%) | No | 80 | 5 | 96 |
| 8 | SiO2-FeCl3 (1 mol%) | No | 80 | 20 | 92 |
| 9 | SiO2-FeCl3 | CH2Cl2 | rt | 15 | 84 |
| 10 | SiO2-FeCl3 | EtOH | rt | 90 | 87 |
| 11 | SiO2-KHSO4 | No | 80 | 10 | 65 |
| 12 | SiO2-HClO4 | No | 80 | 15 | 58 |
| 13 | SiO2-H2SO4 | No | 80 | 10 | 78 |
| 14 | Cu nano | No | 80 | 90 | 90 |
| 15 | Nano TS 1 | No | 80 | 40 | 50 |
| 16 | Amberlite IR 120H+ | No | 80 | 120 | 90 |
| 17 | SiO2-FeCl3c | No | 80 | 15 | 95 |
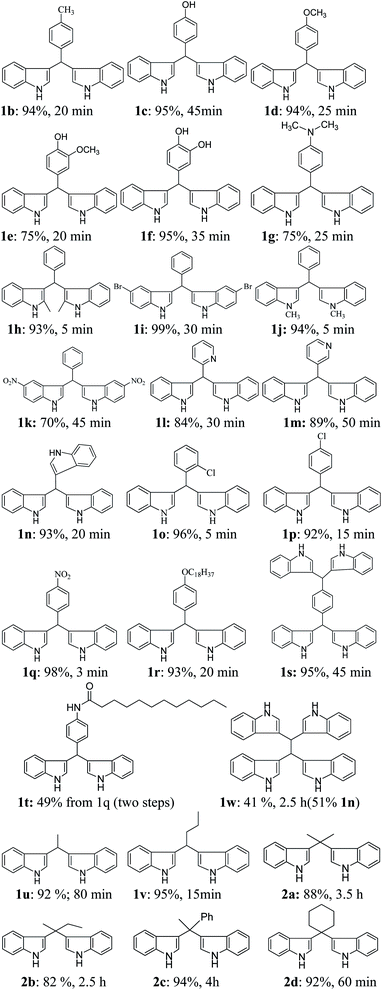
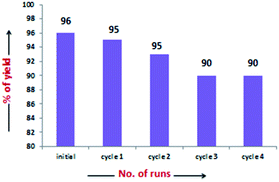
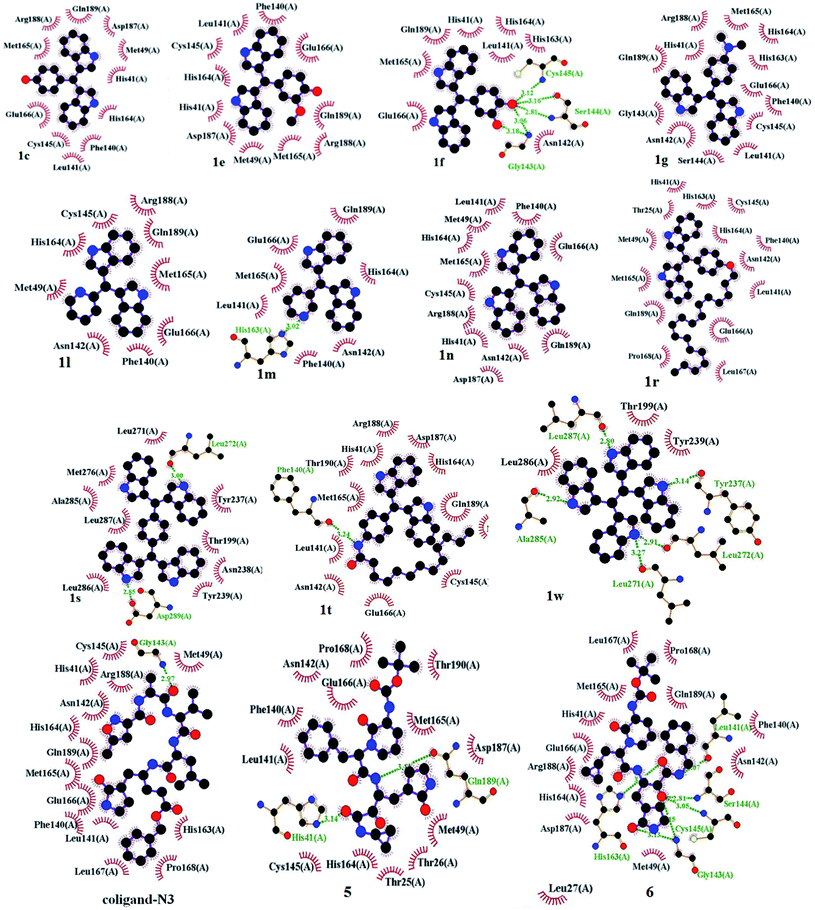
The scope of the current protocol was investigated by using a large variety of aldehydes and ketones with indoles. The aromatic aldehydes containing various substituents such as Me, OMe, OH, Cl, NMe2, and NO2 with simple or substituted indoles under the standard reaction condition (Table 1, entry 6) result in good to excellent yields of 1 (Fig. 3). From the results summarised in Fig. 3, it was observed that the required reaction time is little more in the case of electron-releasing substituents present in aromatic aldehyde (1b–1g) than in electron-withdrawing ones (1o–1q), which increases the reaction rate. In case of methyl substituents present in the indole ring (1h and1j), the reaction is completed within 5 min but –Br substituents present in position 5 of the indole moiety required little longer time, which may be due to the –I effect, but both gave excellent yields (1i). The presence of an electron-withdrawing nitro group in the indole scaffold took longer reaction times, which extend up to 45 min and yield of 1k was found only 70%. This present method is equally effective for the synthesis of hetero aromatic bis(indolyl) methane compounds (1l, 1m) and for long alkoxy chain containing bisindolyl methane derivatives (1r), in very good yield within 30–50 min reaction time. This reaction was further explored for the synthesis of tri-indolylmethane in high yields (1n) by the condensation of indol-3-carbaldehyde with two equivalents of indole under identical conditions. Interestingly, the catalyst was effectively used for the synthesis of 1,4-bis(di(1H-indol-3-yl)methyl)benzene (1s) and no mono product was detected under this reaction condition. The reaction of 4 equivalents of indole with 1 equivalent of terephthalaldehyde proceeded successfully to give the product (1s) in an excellent yield within 40–45 min. Next, we paid our attention towards the synthesis of bis-indolylmethane derivatives using aliphatic aldehydes and ketones. The aliphatic aldehydes such as acetaldehydes, butyraldehyde and glyoxal (40% aq) also reacted with indole under standardized reaction condition and yielded the corresponding bis-indoles in 92% (1u), 95% (1v) and 41% (1w) yields respectively. Unexpectedly, the low yield of 1w is due to the concomitant formation of 1n in 51% yield by C–C bond cleavage followed by indolylation of the so-generated indole-3-carboxaldehyde. Furthermore, reactions of ketones with indole were also proceeded satisfactorily, but they needed comparatively longer reaction times and afforded a little lower yield of products than that of aldehydes due to the lower reactivity of ketones. The reaction of ketones such as acetones, butanone, acetophenone and cyclohexanone was allowed individually with simple indole using 2 mol% of SiO2-FeCl3 under grindstone conditions, and the respective bisindolyl products were isolated in 88% (2a, 3.5 h), 82% (2b, 2.5 h), 94% (2c, 4 h) and 92% (2d, 1 h) respectively. We have also investigated the catalytic recyclability of the catalyst SiO2-FeCl3 from the reaction of indole and benzaldehyde. After completion of the reaction, the catalyst was filtered by dissolving the products in dichloromethane or ethyl acetate, washed several times, dried under vacuum and used for the next cycle. The results (96% initial, 95%, 93%, 90% and 90%) evidently showed that the very little loss of catalytic efficiency in each cycle (Fig. 4).
From the careful analysis of the results presented in Table 1, particularly entries 3–6, it is very much clear that the effect of SiO2 and FeCl3 is synergic. Thus, we presumed that, both Lewis acid behaviour of FeCl3 and surface of silica being used in the catalyst are responsible for the catalytic activity. The formation of polynuclear iron(III) complex by the coordination of aldehydic/ketonic carbonyl10a facilitates the nucleophilic attack to the carbonyl center by indole followed by the elimination of water, as silica is a water adsorbent, and then attacks the second molecule of indole reacting with the arylidene/alkylidene moiety so generated after the dehydration process to form bis-indolylmethanes via the Michael addition-type process.
Molecular docking study
The world is now facing a serious health crisis due to coronavirus disease (COVID-19) since December 2019. The causative agent for COVID-19 was severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).33 Two proteases that facilitate the processing of functional proteins of SARS-CoV-2 are the 3C-like protease (3CLpro) and the papain-like protease (PLpro). 3CLpro executes proteolytic cleavages at a maximum number of sites (11 sites) within the polyprotein, hence it is also termed the main protease (Mpro). This is a key enzyme of SARS-CoV-2, which plays a pivotal role in mediating viral replication and transcription that make it an attractive drug target to combat COVID-19.34 Mpro are located at the cleft of domains that comprised a His–Cys catalytic dyad (cysteine-145 and histidine-41 residues). Here, cysteine-145 acts as a common nucleophile and plays a major role in the proteolytic functioning of Mpro.35 Recently, Adhikari36 and Sarma37 independently reviewed the anti-SARS effect of some small molecules as 3CLpro inhibitors with their various binding modes of interactions to the target protein for potential remedy of corona virus. Structure-based drug discovery (SBDD) is becoming an essential tool in assisting fast and cost-efficient lead discovery and optimization. Due to the various drawbacks associated with the traditional wet lab screening, the application of the rational drug design approach is proven to be faster and less costly than the traditional high-throughput screening of drug discovery. After successful demonstration of silica-supported ferric chloride in the synthesis of various bis-indolylmethanes and having in mind the efficacy of various indole derivatives, we decided to perform molecular docking studies of the synthesized bis-indole compounds against Mpro. A set of compounds (1c, 1e–g, 1l–n, 1r, 1s and 1w) were selected keeping in mind their H-bond donor–acceptor ability, π–π interactions capacity and hydrophobicity. The compound 1r is appended with a long alkyl chain, which is expected to have hydrophobic interactions with the hydrophobic amino acid residues in the active site of the enzyme. However, an amide bond has special features due to the resonance contribution of lone pair of nitrogen to carbonyl group that restrict the C–N rotation by a substantial amount of double bond character, which ultimately provides a geometrically confined structure38 and plays a very important role in biology. Thus, apart from 1r, we have designed and synthesized another long alkyl chain appended compound 1t from bis-bisindolyl methane 1q on the basis of molecular docking analysis. The nitro group of 1q was successfully reduced using hydrazine hydrate-Pd/C (10%) under refluxing in ethanol followed by acylation with dodecanoyl chloride in the presence of Et3N in dichloromethane at room temperature to afford 1t in 49% yield (two steps). Molecular docking studies of the compounds were carried out using AutoDock Vina, and AutoDock 4.2.39 The X-ray crystal structures of SARS-CoV-2 life cycle protein Mpro (PDB ID: 6LU7, 2.16 Å) were retrieved from the RCSB protein data bank.40 PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC)41 was used for the visual analysis of protein–ligand complexes and LigPlot+ Version, v.2.2 was used to generate 2D protein–ligand interactions.41b The structures of the synthesized compounds were sketched using Chem Draw Professional 15.1 and were saved as sdf formatted files. Then the structures were imported in Pymol and converted into pdb file format. In the AutoDock Tools interface the pdf formatted files of all the compounds were imported followed by conversion into pdbqt format for compatibility with Autodock 4.2 and Autodock Vina using AutoDock Tools1.5.6.42The Mpro was imported in AutoDock Tools 1.5.6 and prepared by removal of water, removal of the bound ligand, addition of polar hydrogen atoms followed by computing Gasteiger and adding Kollman partial charges to the protein. Finally, the protein was saved in the pdbqt format. The amino acid residues of 6LU7 from the 5.0 Å distance of the centre of the co-ligand (N3) were LEU-27, HIS-41, HIS-42, LEU-141, GLY-143, GLU-166, PRO-168, ARG-188, THR-190, and GLN-192.
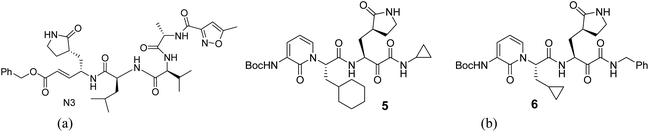
For the docking study, we considered known inhibitor—co-ligand (N3) and two other peptide-based inhibitors 5 and 6 as reference (Fig. 5). Co-ligand (N3) can specifically inhibit Mpro from multiple coronaviruses, including SARS-CoV and MERS-CoV, and inhibit SARS-CoV-2 replication in human Calu-3 lung cells.43 The active site amino acid residues were selected using PyMOL, and the residues are shown in Fig. 6. The grid centre dimension of Mpro was x = −10.883, y = 13.934, z = 68.209 and grid size was x = 58, y = 68, z = 70. Selecting these residues, a receptor grid was generated. The molecular docking was carried out using Windows 10, OS architecture 64 bit, Core (TM) 2 Due CPU machine.
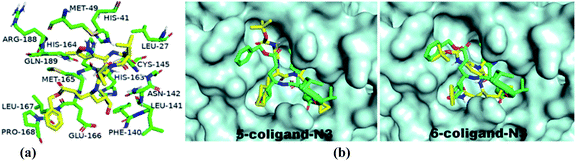 | ||
| Fig. 6 (a) 3D interactions of the co-ligand with active site amino acid residues of Mpro are depicted by PyMOL in the X-ray crystallographic protein-co-ligand complex of 6LU7. (b) Binding pose of known inhibitors 5 and 6 in the receptor-active site of SARS-CoV-2 Mpro is depicted in PyMOL [structures 5 and 6 (yellow) and co-ligand N3 (green)]. | ||
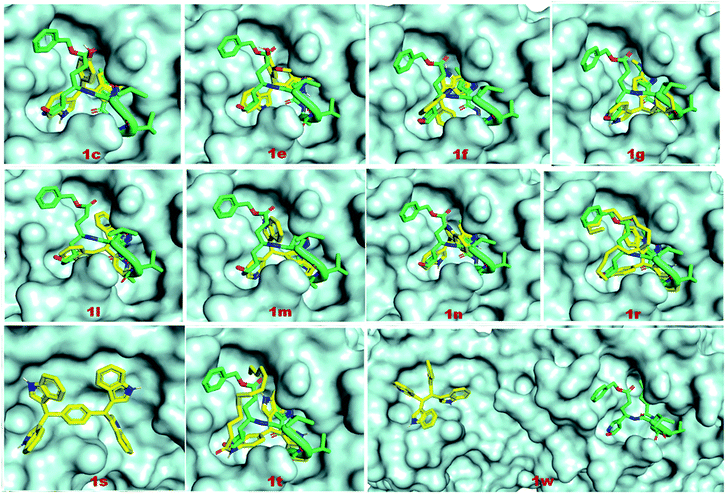
The binding affinities of coligand (N3) and two known SARS-CoV-2 Mpro inhibitors 5 and 6 were found to be −6.6, −7.7 and −7.9 respectively, as predicted using AutoDock Vina (Table 2). The binding affinities of the compounds 1c, 1e, 1f, 1g, 1l, 1m, 1n, 1r, 1s, 1t and 1w were −7.8, −7.6, −7.9, −7.3, −7.3, −6.7, −7.9, −5.8, −9.1, −6.0 and −7.7 kcal mol−1 respectively, as predicted using AutoDock Vina. The predicted binding affinities of compounds 1c, 1e, 1f, 1g, 1l, 1m, 1n, 1s and 1w were less than the binding affinity of co-ligands (N3). The binding affinities of 1c, 1f, 1n, 1s and 1w were less than or equal to the binding affinity of known inhibitor 5. The binding affinities of 1f, 1n and 1s were less than or equal to the binding affinities of another known inhibitor 6. The binding energies of coligand (N3) and two known SARS-CoV-2 3CLpro inhibitors 5 and 6 were −7.35, −9.71, and −8.43 kcal mol−1, respectively. The binding energies of the compounds 1c, 1e, 1f, 1g 1l, 1m, 1n, 1r, 1s, 1w and 1t were −9.05, −8.88, −8.58, −9.29, −8.32, −8.31, −9.13, −6.60, −12.37, −8.58 and −12.27 kcal mol−1 respectively, as predicted using AutoDock. The binding energies of ligands 1c, 1e, 1f, 1g, 1l, 1m, 1n, 1s, 1t and 1w were less than the binding energy of the co-ligand. The binding energies of 1s and 1w were less than the binding energy of known inhibitor 5. Similarly, the binding energies of 1c, 1e, 1f, 1g, 1n, 1s, 1t and 1w were less than the binding energy of another known inhibitor 6. The docking scores of the ligands 1s and 1w were comparatively very good but not bound to the active site of Mpro. Therefore, these two ligands are not considered as good inhibitors. In two docking algorithm, the compounds 1c, 1e, 1f and 1n showed good binding affinities towards SARS-CoV-2 Mpro. The docking score and interacting amino acid residues of all the compounds, known inhibitors and co-ligands are shown in Table 2. The compounds 1c, 1e, 1f, 1r, 1g, 1l, 1m, 1n, 1t, 5 and 6 bind in the active site region in a similar pose to the X-ray crystallographic bound co-ligand (N3). The binding pose of all the compounds, known inhibitors and co-ligands are shown in Fig. 7. The key amino acid residues that interact with known SARS-CoV-2 Mpro inhibitors identified by MD simulation are HIS-41, GLY-143, and GLU-166.44 The compounds 1c, 1e 1f, 1g, 1n, 1r and 1t exhibited hydrophobic interactions with at least two residues out of three key interacting residues. The other key interacting amino acid residues of SARS-CoV main protease is CYS-145.45 The compounds 1c, 1e 1f 1g, 1l, 1n, 1r and 1t also showed hydrophobic interactions with CYS-145. Therefore, except compounds 1s and 1w all other compounds showed interactions with key amino acid residues of SARS-CoV-2 Mpro. On the basis of docking scores, interacting amino acid residues and in comparison with co-ligand and known inhibitors, it is concluded that the compounds 1c, 1e, 1f and 1n may be the potential SARS-CoV-2 Mpro inhibitors (Fig. 8).
| Compounds | Binding affinity | Binding energy | Ki (nM) | Interacting amino acid residues |
|---|---|---|---|---|
| a Ki values expressed in μM. | ||||
| Co-ligand (N3) | −6.6 | −7.35 | 4.07a | Hydrophobic: HIS-41, MET-49, LEU-27, PHE-140, LEU-141, ASN-142, CYS-145, HIS-163, HIS-164, MET-165, GLU-166, LEU-167, PRO-168, ARG-188, GLN-189; H-bonding: GLY-143 (2.97 Å) |
| 5 | −7.7 | −9.71 | 76.45 | Hydrophobic: THR-25, THR-26, MET-49, PHE-140, LEU-141, ASN-142, CYS-145, HIS-164, MET-165, PRO-168, ASP-187, THR-190; H-bonding: HIS-41 (3.14 Å), GLN-189 (3.11 Å) |
| 6 | −7.9 | −8.43 | 664.7 | Hydrophobic: HIS-41, MET-49, PHE-140, ASN-142, HIS-164, MET-165, GLU-166, LEU-167, PRO-168, ASP-187, ARG-188, GLN-189; H-bonding: LEU-141 (3.07 Å), GLY-143 (3.13, 3.15 Å), SER-144 (2.81 Å), CYS-145 (3.05 Å), HIS-163 (3.00 Å) |
| 1c | −7.8 | −9.05 | 232.93 | Hydrophobic: HIS-41, MET-49, PHE-140, LEU-141, CYS-145, HIS-164, MET-165, GLU-166, ASP-187, ARG-188, GLN-189 |
| 1e | −7.6 | −8.88 | 307.65 | Hydrophobic: HIS-41, MET-49, PHE-140, LEU-141, CYS-145, HIS-164, MET-165, GLU-166, ASP-187, ARG-188, GLN-189 |
| 1f | −7.9 | −8.58 | 515.25 | Hydrophobic: HIS-41, HIS-164, HIS-163, LEU-141, ASN-142, GLU-166, MET-165, GLN-189. H-bonding: CYS-145 (3.12), SER-144 (3.16, 2.81), ASN-142 (3.06, 3.18 Å) |
| 1g | −7.3 | −9.29 | 154.54 | Hydrophobic: PHE-140, LEU-141, ASN-142, GLY-143, SER-144, CYS-145, HIS-163, HIS-164, MET-165, GLU-166, ARG-188, GLN-189 |
| 1l | −7.3 | −8.32 | 794.7 | Hydrophobic: MET-49, PHE-140, ASN-142, CYS-145, HIS-164, MET-165, GLU-166, ARG-188, GLN-189 |
| 1m | −6.7 | −8.31 | 813.9 | Hydrophobic: PHE-140, LEU-141, ASN-142, HIS-164, MET-165, GLU-166, GLN-189. H-bonding: HIS-163 (3.02 Å) |
| 1n | −7.9 | −9.13 | 203.59 | Hydrophobic: HIS-41, MET-49, PHE-140, LEU-141, ASN-142, CYS-145, HIS-164, MET-165, GLU-166, ASP-187, ARG-188, GLN-189 |
| 1r | −5.8 | −6.6 | 14.35a | Hydrophobic: THR-25, HIS-41, MET-49, PHE-140, LEU-141, ASN-142, CYS-145, HIS-163, HIS-164, MET-165, GLU-166, LEU-167, PRO-168, GLN-189 |
| 1s | −9.1 | −12.37 | 0.8513 | Hydrophobic: THR-199, TYR-237, ASN-238, TYR-239, LEU-271, MET-276, ALA-285, LEU-286, LEU-287; H-bonding: LEU-272 (3.00 Å), ASP-289 (2.85 Å) |
| 1t | −6.0 | −8.58 | 517.49 | Hydrophobic: HIS-41, MET-49, LEU-141, ASN-142, CYS-145, HIS-164, MET-165, GLU-166, ASP-187, ARG-188, GLN-189, THR-190; H-bonding: PHE-140 (3.24 Å) |
| 1w | −7.7 | −12.27 | 1.01 | Hydrophobic: TYR-239, LEU-286 THR-199; H-bonding: TYR-237 (3.14 Å), LEU-271 (3.27 Å), LEU-272 (2.91 Å), ALA-285 (2.92 Å), LEU-287 (2.80 Å) |
Conclusions
In conclusion, we have successfully demonstrated the utility of silica-supported ferric chloride in the synthesis of bisindolyl methane derivatives under grindstone conditions. The catalyst was characterized by SEM/EDX spectra. Our study revealed that the quality of this supported material is retained after six months as well as after recycling several times. The present protocol has several advantages like operational simplicity, shorter reaction time, high yield of the products and tolerance of a wide range of functional groups. We also investigated the molecular docking study of the synthesised compounds by targeting the binding site of SARS-CoV-2 main protease (3CLpro or Mpro) enzymes. Our study revealed that some of the synthesized compounds could be potential candidates as antiviral agents against SARS-CoV-2 Mpro enzyme as the computational study indicates that some of the synthesised BIM's interacting with key amino acids residues of the active sites by hydrophobic as well as hydrogen bonding interactions.Experimental section
General method
The starting materials indole or substituted indoles, various types of aldehydes (aromatic, aliphatic, heterocyclic etc.), anhydrous ferric chloride and silica gel 230–400 mesh were purchased either from Sigma Aldrich chemical Co., USA or Acros Organics or SRL India and used as received. All the solvents were distilled prior to use. 1H NMR and 13C NMR spectra were recorded at ambient temperature using a Bruker Ascend 400 MHz spectrometer (400 MHz for 1H and 100 MHz for 13C). Chemical shifts were reported in parts per million from the tetramethylsilane internal reference, and coupling constants were reported in Hertz. Proton multiplicities were represented as s (singlet), d (doublet), dd (double doublet), t (triplet), q (quartet), and m (multiplet). Infrared spectra were recorded using a Fourier transform infrared (FT-IR, Model: Spectrum 100) spectrometer KBr pellets or in thin films. The surface morphology of the silica gel-supported material was estimated using a Field Emission Scanning Electron Microscope Model-Sigma 300, Carl Zeiss instrument.General experimental procedure for the synthesis of 1–2
A mixture of indoles (2.0 mmol), corresponding aldehydes or ketones (1.0 mmol) and FeCl3-SiO2 (20 mg, 2 mol% FeCl3) were taken in a cone-shaped flask and mixed thoroughly using a wide glass rod or spatula. Then, the flask was immersed onto a preheated water bath of temperature 80 °C with continuous grinding of the mixture. After completion of the reaction (monitored by TLC), the catalyst was recovered by dissolving the product in a suitable solvent like ethyl acetate, dichloromethane or methanol. The residue (catalyst) was collected by filtration, dried under vacuum and recycled. The filtrate containing the product was concentrated by distillation under reduced pressure and the distillate was collected for recycling purposes. In most cases, the pure product was crystallized out form the residual filtrate. Only in few cases the product was purified by column chromatography (ethyl acetate–hexane). The synthesized products were characterized by melting point, IR data, NMR, and spectral analysis and compared with the reported compounds. Spectral data of all known compounds are available in ESI.†Experimental procedure for the synthesis of 1t from 1q
A mixture of 1q (367 mg, 1 mmol), Pd–C (10%, 40 mg) and hydrazine hydrate (1.5 mmol) in ethanol (10 mL) was refluxed for 2 h. After cooling, the mixture was filtered off and the residue was washed with ethyl acetate (2 × 10 mL). The combined filtrate was evaporated under reduced pressure and the residue was used for next step without purification. To the stirred solution of the residue in dichloromethane (5 mL), triethylamine (500 μL) and lauroyl chloride (270 mg, 1.2 mmol) were added successively and stirred for 4 h at room temperature. After completion of the reaction (TLC), water (10 mL) was added and extracted with dichloromethane (3 × 10 mL), dried over anhydrous Na2SO4 and evaporated to residue. The purification of the crude product by column chromatography (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, ethyl acetate–hexane) afforded the title compound 1t.
1, ethyl acetate–hexane) afforded the title compound 1t.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors also grateful to Department of Science and Technology (DST), Govt. of India for providing 400 MHz NMR facility under DST-FIST programme (No SR/FST/CSI-263/2015) and Central Instrumentation Centre (CIC) Tripura University from Instrumental facilities.Notes and references
-
(a) I. Eilks and F. Rauch, Chem. Educ. Res. Pract., 2012, 13, 57–58 RSC
; (b) C. J. Clarke, W.-C. Tu, O. Levers, A. Bröhl and J. P. Hallett, Chem. Rev., 2018, 118(2), 747–800 CrossRef CAS PubMed
; (c) S. A. Matlin, G. Mehta, H. Hopf and A. Krief, Nat. Chem., 2015, 7, 941–943 CrossRef CAS PubMed
; (d) R. A. Sheldon, Green Chem., 2005, 7, 267–278 RSC
.
-
(a) C. Reichardt, Org. Process Res. Dev., 2007, 11, 105–113 CrossRef CAS
; (b) A. Sarkar, S. Satra, S. K. Kundu, A. Hajra, G. V. Zyryanov, O. N. Chupakhin, V. N. Charushin and A. Majee, Green Chem., 2016, 18, 4475–4525 RSC
.
-
(a) R. R. Hawker and J. B. Harper, Adv. Phys. Org. Chem., 2018, 52, 49–85 CrossRef CAS
; (b) T. Itoh, Chem. Rev., 2017, 117, 10567–10607 CrossRef CAS PubMed
; (c) A. S. Amarasekara, Chem. Rev., 2016, 116, 6133–6183 CrossRef CAS PubMed
; (d) A. R. Hajipour and F. Refiee, Org. Prep. Proced. Int., 2015, 47, 249–308 CrossRef CAS
.
-
(a) B. H. Lipshutz, S. Ghorai and M. Cortes-Clerget, Chem.–Eur. J., 2018, 24, 6672–6695 CrossRef CAS PubMed
; (b) R. N. Butler and A. G. Coyne, Org. Biomol. Chem., 2016, 14, 9945–9960 RSC
; (c) D. K. Romney, F. H. Arnold, B. H. Lipshutz and C.-J. Li, J. Org. Chem., 2018, 83, 7319–7322 CrossRef CAS PubMed
.
-
(a) S. Liu, Y. Kumatabara and S. Shirakawa, Green Chem., 2016, 18, 331–341 RSC
; (b) L. Zong and C.-H. Tan, Acc. Chem. Res., 2017, 50, 842–856 CrossRef CAS PubMed
; (c) D. C. M. Albanese, F. Foschi and M. Penso, Org. Process Res. Dev., 2016, 20, 129–139 CrossRef CAS
; (d) J. Schörgenhumer, M. Tiffner and M. Waser, Beilstein J. Org. Chem., 2017, 13, 1753–1769 CrossRef PubMed
.
-
(a) P. Gracia-Garcıa, J. M. Moreno, U. Dıaz, M. Bruix and A. Corma, Nat. Commun., 2016, 7, 10835 CrossRef PubMed
; (b) M. Kaur, S. Sharma and P. M. S. Bedi, Chin. J. Catal., 2015, 36, 520–549 CrossRef CAS
; (c) S. An, D. Song, B. Lu, X. Yang and Y.-H. Guo, Chem.–Eur. J., 2015, 21, 10786–10798 CrossRef CAS PubMed
; (d) F. Su, L. Ma, D. Song, X. Zhang and Y. Guo, Green Chem., 2013, 15, 885–890 RSC
.
-
(a) R. B. NasirBaig and R. S. Varma, Chem. Soc. Rev., 2012, 41, 1559–1584 RSC
; (b) V. Polshettiwar and R. S. Varma, Acc. Chem. Res., 2008, 41, 629–639 CrossRef CAS PubMed
.
-
(a) A. Stolle, T. Szuppa, S. E. S. Leonhardt and B. Ondruschka, Chem. Soc. Rev., 2011, 40, 2317–2329 RSC
; (b) S. Santra, D. S. Kopchuk, I. S. Kovalev, G. V. Zyryanov, A. Majee, V. N. Charushin and O. N. Chupakhin, Green Chem., 2016, 18, 423–426 RSC
; (c) A. Khaskel, P. Gogoi, P. Barman and B. Bandyopadhyay, RSC Adv., 2014, 4, 35559–35567 RSC
.
- S. Onitsuka, Y. Z. Jin, A. C. Shaikh, H. Furuno and J. Inanaga, Molecules, 2012, 17, 11469–11483 CrossRef CAS PubMed
.
-
(a) S. Majumdar, A. Chakraborty, S. Bhattacharjee, S. Debnath and D. K. Maiti, Tetrahedron Lett., 2016, 57, 4595–4598 CrossRef CAS
; (b) E. Keinan and Y. Mazur, J. Org. Chem., 1978, 43, 1020–1022 CrossRef CAS
; (c) D. Habibi and M. Nasrollahzadeh, Synth. Commun., 2010, 40, 3159–3167 CrossRef CAS
; (d) A. Fadel and J. Salaün, Tetrahedron, 1985, 41, 413–420 CrossRef CAS
; (e) M. A. Chari, D. Shobha and K. Mukkanti, Catal. Commun., 2006, 7, 787–790 CrossRef CAS
.
-
(a) P. F. Rosales, G. S. Bordin, A. E. Gowar and S. Moura, Fitoterapia, 2020, 143, 104558 CrossRef CAS PubMed
; (b) T. P. Singh and O. M. Singh, Mini Rev. Med. Chem., 2018, 18, 9–25 CrossRef CAS PubMed
.
- F. Zhao, N. Liu, P. Zhan, X. Jiang and X. Liu, Eur. J. Med. Chem., 2016, 94, 218–228 CrossRef PubMed
.
- C.-W. Lin, F.-J. Tsai, C.-H. Tsai, C.-C. Lai, L. Wan, T.-Y. Ho, C.-C. Hsieh and P.-D. L. Chao, Antiviral Res., 2005, 68, 36–42 CrossRef CAS PubMed
.
- A. K. Ghosh, G. Gong, V. Grum-Tokars, D. C. Mulhearn, S. C. Baker, M. Coughlin, B. S. Prabhakar, K. Sleeman, M. E. Johnson and A. D. Mesecar, Bioorg. Med. Chem. Lett., 2008, 18, 5684–5688 CrossRef CAS PubMed
.
- J. Blaising, S. J. Polyak and E.-I. Pecheur, Antiviral Res., 2014, 107, 84–94 CrossRef CAS PubMed
.
- M.-Z. Zhang, Q. Chen and G.-F. Yang, Eur. J. Med. Chem., 2015, 89, 421–441 CrossRef CAS PubMed
.
- G. M. Ziarani, R. Moradi, T. Ahmadi and N. Lashgari, RSC Adv., 2018, 8, 12069–12103 RSC
.
-
(a) P. J. Praveena, P. S. Parameswaran and M. S. Majik, Synthesis, 2015, 47, 1827–1837 CrossRef
; (b) M. Karthik, A. K. Tripathi, N. M. Gupta, M. Palanichamy and V. Murugeson, Catal. Commun., 2004, 5, 371–375 CrossRef CAS
; (c) M. Chakraborty, N. Ghosh, R. Basak and Y. Harigaya, Tetrahedron Lett., 2002, 43, 4075–4078 CrossRef
; (d) B. P. Bandgar and K. A. Shaikh, Tetrahedron Lett., 2003, 44, 1959–1961 CrossRef CAS
; (e) A. V. Reddy, K. Ravinder, V. L. N. Reddy, T. V. Goud, V. Ravikanth and Y. Venkateswarlu, Synth. Commun., 2003, 33, 3687–3694 CrossRef
.
-
(a) M. Xia, S. Wang and W. B. Yuan, Synth. Commun., 2004, 34, 3175–3182 CrossRef CAS
; (b) M. Damodiran, D. Muralidharan and P. T. Perumal, Bioorg. Med. Chem. Lett., 2009, 19, 3611–3614 CrossRef CAS PubMed
; (c) G. W. Gribble, J. Chem. Soc., Perkin Trans. 1, 2000, 7, 1045–1075 RSC
; (d) A. J. K. Karamyan and M. T. Hamann, Chem. Rev., 2010, 110, 4489–4497 CrossRef PubMed
; (e) M. Mari, A. Tassoni, S. Lucarini, M. Fanelli, G. Piersanti and G. Spadoni, Eur. J. Org. Chem., 2014, 2014, 3822–3830 CrossRef CAS
; (f) T. Endo, M. Tsuda, J. Fromont and J. Kobayashi, J. Nat. Prod., 2007, 70, 423–424 CrossRef CAS PubMed
.
-
(a) R. M. F. Batista, S. P. G. Costa, R. M. P. Silva, N. E. M. Lima and M. M. M. Raposo, Dyes Pigments, 2014, 102, 293–300 CrossRef CAS
; (b) N. Kumari, S. Jha and S. Bhattacharya, Chem.–Asian J., 2012, 7, 2805–2812 CrossRef CAS PubMed
.
-
(a) X. He, S. Hu, K. Liu, Y. Guo, J. Xu and S. Shao, Org. Lett., 2006, 8, 333–336 CrossRef CAS PubMed
; (b) L. Wang, X. He, Y. Guo, J. Xu and S. Shao, Beilstein J. Org. Chem., 2011, 7, 218–221 CrossRef CAS PubMed
; (c) H. J. Kim, H. Lee, J. H. Lee, D. H. Choi, J. H. Jung and J. S. Kim, Chem. Commun., 2011, 47, 10918–10920 RSC
.
- R. Martinez, A. Espinosa, A. Tarraga and P. Molina, Tetrahedron, 2008, 64, 2184–2191 CrossRef CAS
.
- L. Wang, X. He, Y. Guo, J. Xu and S. Shao, Beilstein J. Org. Chem., 2011, 7, 218–221 CrossRef CAS PubMed
.
- A. C. Lamb, R. A. Federico-Prerez and Z.-L. Xue, Anal. Biochem., 2015, 484, 21–23 CrossRef CAS PubMed
.
-
(a) P. Saini, P. Kumari, S. Hazra and A. J. Elias, Chem.–Asian J., 2019, 14, 4154–4159 CrossRef CAS PubMed
; (b) D. Xia, Y. Wang, Z. Du, Q.-Y. Zheng and C. Wang, Org. Lett., 2012, 14, 588–591 CrossRef CAS PubMed
; (c) S. Karmakar, P. Das, D. Ray, S. Ghosh and S. K. Chattopadhyay, Org. Lett., 2016, 18, 5200 CrossRef CAS PubMed
; (d) R. R. Singh and R.-S. Liu, Chem. Commun., 2017, 53, 4593–4596 RSC
.
- T. Abe, S. Nakamura, R. Yanada, T. Choshi, S. Hibino and M. Ishikura, Org. Lett., 2013, 15, 3622–3625 CrossRef CAS PubMed
.
- C.-C. Chen, B.-C. Hong, W.-S. Li, T.-T. Chang and G.-H. Lee, Asian J. Org. Chem., 2017, 6, 426–431 CrossRef CAS
.
-
(a) B. V. Gregorovich, K. Liang, M. Clugston and S. Macdonald, Can. J. Chem., 1968, 46, 3291–3300 CrossRef CAS
; (b) M. Roomi and S. Macdonald, Can. J. Chem., 1970, 48, 139–143 CrossRef CAS
; (c) M. El-Sayeda, K. Mahmouda and A. Hilgerotha, Curr. Chem. Lett., 2014, 3, 7–14 CrossRef
; (d) T. Pillaiyar, M. Dawood, H. Irum and C. E. Müller, Arkivoc, 2018,(part iii), 1–19 Search PubMed
.
-
(a) A. Chatterjee, S. Manna, J. Benerji, C. Pascard, T. Prange and J. Shoolery, J. Chem. Soc., Perkin Trans. 1, 1980,(1), 553–555 RSC
; (b) M. Shiri, M. A. Zolfigol, H. G. Kruger and Z. C. Tanbakouchian, Chem. Rev., 2010, 110, 2250–2293 CrossRef CAS PubMed
; (c) L. Macha, D. Singh, H.-J. Rhee and H.-J. Ha, Org. Biomol. Chem., 2020, 18, 9473–9482 RSC
.
-
(a) M. M. Khodaei, I. Mohammadpoor, H. R. Memarian, A. R. Khosropour, K. Nikofar and P. Ghanbary, J. Heterocycl. Chem., 2008, 45, 1–5 CrossRef
; (b) K. Niknam, M. A. Zolfigol, T. Sadabadi and A. Nejati, J. Iran. Chem. Soc., 2006, 3, 31–32 Search PubMed
; (c) S. Mehrazma, N. Azizi and M. R. Saidi, Lett. Org. Chem., 2006, 3, 161–164 CrossRef CAS
; (d) M. Hosseini-Sarvari, Acta Chim. Slov., 2007, 54, 354–359 CAS
; (e) Z. H. Zhang, L. Yin and Y. M. Wang, Synthesis, 2005, 12, 1949–1954 Search PubMed
; (f) S. J. Ji, M. F. Zhou, D. G. Gu, S. Y. Wang and T. P. Loh, Synlett, 2003, 2077–2079 CrossRef CAS
; (g) X. Mi, S. Luo, J. He and J. P. Cheng, Tetrahedron Lett., 2004, 45, 4567–4570 CrossRef CAS
; (h) V. T. Kamble, B. P. Bandgar and S. N. Bavikar, Chin. J. Chem., 2007, 25, 13–15 CrossRef CAS
; (i) P. K. Pradhan, S. Dey, V. S. Giri and P. Jaisankar, Synthesis, 2005, 11, 1779–1782 Search PubMed
; (j) M. Xia, S. H. Wang and W. B. Yuan, Synth. Commun., 2004, 34, 3175–3182 CrossRef CAS
; (k) L. P. Mo, Z. C. Ma and Z. H. Zhang, Synth. Commun., 2005, 35, 1997–2004 CrossRef CAS
; (l) N.-K. Nguyen, M.-T. Ha, H. Y. Bui, Q. T. Trinh, B. N. Tran, V. T. Nguyen, T. Q. Hung, T. T. Dang and X. H. Vu, Catal. Commun., 2021, 149, 106240 CrossRef CAS
(7 pages)..
-
(a) M. Chakrabarty, N. Ghosh, R. Basak and Y. A. Harigaya, Synth. Commun., 2004, 34, 421–434 CrossRef CAS
; (b) M. L. Deb and P. J. Bhuyan, Tetrahedron Lett., 2005, 47, 1441–1443 CrossRef
; (c) M. Karthik, A. K. Tripathi, N. M. Gupta, M. Palanichamy and V. Murugesan, Catal. Commun., 2004, 5, 371–375 CrossRef CAS
; (d) G. Penieres-Carrillo, J. G. García-Estrada, J. L. Gutiérrez-Ramírez and C. Alvarez-Toledano, Green Chem., 2003, 5, 337–339 RSC
; (e) C. Ramesh, J. Banerjee, R. Pal and B. Das, Adv. Synth. Catal., 2003, 345, 557–559 CrossRef CAS
; (f) H. A. Soliman, A. Y. Mubarak and S. S. Elmorsy, Chin. Chem. Lett., 2016, 27, 353–358 CrossRef CAS
; (g) H. Firouzabadi, N. Iranpoor, M. Jafarpour and A. Ghaderi, J. Mol. Catal. Chem., 2006, 253, 249–251 CrossRef CAS
; (h) N. Azizi, L. Torkian and M. R. Saidi, J. Mol. Catal. Chem., 2007, 275, 109–112 CrossRef CAS
; (i) T. K. Khatab, A. M. Abdelghany and H. A. Soliman, Silicon, 2018, 10, 703–708 CrossRef CAS
.
-
(a) B. Wu, J. Wen, J. Zhang, J. Li, Y.-Z. Xiang and X.-Q. Yu, Synlett, 2009, 500–504 CAS
; (b) S. S. Shinde, B. S. Chi and D. Y. Lee, Org. Lett., 2008, 10, 733–735 CrossRef CAS PubMed
; (c) S. A. Siddiqui, U. C. Narkhede, S. S. Palimkar, T. Daniel, R. J. Lahoti and K. V. Srinivasan, Tetrahedron, 2005, 61, 3539–3546 CrossRef CAS
; (d) G. V. M. Sharma, J. J. Reddy, P. S. Lakshmi and P. R. A. Krishna, Tetrahedron Lett., 2004, 45, 7729–7732 CrossRef CAS
; (e) H. J. Park and J. C. Lee, Synlett, 2009, 79–80 CAS
; (f) R. S. Balaskar, B. B. Shingate, M. S. Shingare and D. V. Mane, Arab. J. Chem., 2016, 9, S120–S123 CrossRef CAS
; (g) J. Jin, Y. Li, S. Xiang, W. Fan, S. Guo and D. Huang, Org. Biomol. Chem., 2021, 19, 4076–4081 RSC
.
-
(a) C. Huang, Y. Wang, X. Li, L. Ren, J. Zhao, Y. Hu, L. Zhang, G. Fan, J. Xu, X. Gu, Z. Cheng, T. Yu, J. Xia, Y. Wei, W. Wu, X. Xie, W. Yin, H. Li, M. Liu, Y. Xiao, H. Gao, L. Guo, J. Xie, G. Wang, R. Jiang, Z. Gao, Q. Jin, J. Wang and B. Cao, Lancet, 2020, 395, 497–506 CrossRef CAS PubMed
; (b) A. E. Gorbalenya, S. C. Baker, R. S. Baric, R. J. de Groot, C. Drosten, A. A. Gulyaeva, B. L. Haagmans, C. Lauber, A. M. Leontovich, B. W. Neuman, D. Penzar, S. Perlman, L. L. M. Poon, D. V. Samborskiy, I. A. Sidorov, I. Sola and J. Ziebuhr, Nat. Microbiol., 2020, 5, 536–544 CrossRef PubMed
.
-
(a) K. Anand, J. Ziebuhr, P. Wadhwani, J. R. Mesters and R. Hilgenfeld, Science, 2003, 300, 1763–1767 CrossRef CAS PubMed
; (b) V. Grum-Tokars, K. Ratia, A. Begaye, S. C. Baker and A. D. Mesecar, Virus Res., 2008, 133, 63–73 CrossRef CAS PubMed
.
- A. K. Ghosh, K. Xi, V. Grum-Tokars, X. Xu, K. Ratia, W. Fu, K. V. Houser, S. C. Baker, M. E. Johnson and A. D. Mesecar, Bioorg. Med. Chem. Lett., 2007, 17, 5876–5880 CrossRef CAS PubMed
.
- B. Adhikari and N. Sahu, ChemistrySelect, 2021, 6, 2010–2028 CrossRef CAS PubMed
.
- M. Konwar and D. Sarma, Tetrahedron, 2021, 77, 131761 CrossRef CAS PubMed
(20 pages)..
- C. R. Kemnitz and M. J. Loewen, J. Am. Chem. Soc., 2007, 129, 2521–2528 CrossRef CAS PubMed
.
-
(a) G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 6, 2785–2791 CrossRef PubMed
; (b) O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CAS
.
- X. Liu, B. Zhang, Z. Jin, H. Yang and Z. Rao, RCSB PDB id: 6LU7, 2020, https://doi:10.2210/pdb6LU7/pdb Search PubMed.
-
(a) L. DeLano, The PyMOL molecular graphics system, DeLano Scientific, San Carlos, CA, 2002, http://www.pymol.org Search PubMed
; (b) R. A. Laskowski and M. B. Swindells, J. Chem. Inf. Model., 2011, 51, 2778–2786 CrossRef CAS PubMed
.
- M. F. Sanner, J. Mol. Graph. Model., 1999, 17, 57–61 CAS
.
- L. Zhang, D. Lin, X. Sun, U. Curth, C. Drosten, L. Sauerhering, S. Becker, K. Rox and R. Hilgenfeld, Science, 2020, 368, 409–412 CrossRef CAS PubMed
.
- R. Yoshino, N. Yasu and M. Sekijima, Sci. Rep., 2020, 10, 12493 CrossRef CAS PubMed
.
- T. Regnier, D. Sarma, K. Hidaka, U. Bacha, E. Freire, Y. Hayashi and Y. Kiso, Bioorg. Med. Chem. Lett., 2009, 19, 2722–2727 CrossRef CAS PubMed
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra05679d |
| ‡ Present address: Department of Chemistry, Faculty of Science and Technology, ICFAI UniversityTripura, Kamalghat, Mohanpur, Agartala, Tripura 799210. |
| This journal is © The Royal Society of Chemistry 2021 |