Open Access Article
Open Access ArticleRapid structural discrimination of IgG antibodies by multicharge-state collision-induced unfolding†
Zhibin Yina,
Mingyi Duab,
Dong Chenab,
Wenyang Zhanga,
Wenjie Huanga,
Xinzhou Wu *b and
Shijuan Yan
*b and
Shijuan Yan *a
*a
aGuangdong Key Laboratory for Crop Germplasm Resources Preservation and Utilization, Agro-biological Gene Research Center, Guangdong Academy of Agricultural Sciences, Guangzhou, 510640, China. E-mail: shijuan@agrogene.ac.cn
bState Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, Key Laboratory of Natural Pesticide and Chemical Biology of the Ministry of Education, South China Agricultural University, Guangzhou, 510642, China. E-mail: wuxz@scau.edu.cn
First published on 12th November 2021
Abstract
Immunoglobulin G (IgG) antibodies are an important class of biotherapeutics that target various diseases, such as cancers, neurodegenerative disorders, and autoimmune diseases, yet rapid discrimination of IgG antibodies remains a great challenge due to heterogeneity, flexibility, and large size. Herein, we demonstrate a simplified multicharge-state collision-induced unfolding (CIU) method for rapid differentiation of four IgG isotypes that differ in terms of the numbers and patterns of disulfide bonds, bypassing tedious single charge-state selection in advance. The results presented herein reveal that gas-phase unfolding behaviors have a strong dependence on charge states outside IgG surfaces; therefore, multicharge-state CIU analysis of IgG subtypes could offer a great opportunity to gain deeper insights into their gas-phase structural differentiation. The full discrimination of IgG antibody isoforms that possess different disulfide bond numbers and even subtle disulfide bonding patterns can be achieved based on their charge-dependent gas-phase unfolding behaviors and root-mean square deviation in CIU difference spectra. Taken together, the incorporation of all charge states observed in a native ion mobility-mass spectrometry (IM-MS) experiment to CIU analysis could make this strategy sensitive to more subtle structural discrepancies, facilitating the rapid discrimination and evaluation of innovative structurally similar biotherapeutic candidates with unexplored functions.
1. Introduction
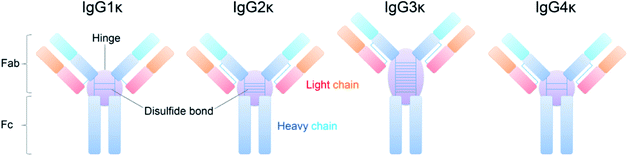
As highly active and specific antibodies produced by the immune system, immunoglobulins (Igs) play a vital role in mediating an immunological response network.1 Recent decades have witnessed an increasing interest in developing Igs as biotherapeutics, which benefit from higher efficiency, higher specificity, and fewer side effects than conventional small-molecule-based therapeutics. Antibodies have proven to be a clinically and commercially successful drug class with regard to clinical treatment for various diseases, such as cancer, cardiovascular and neurodegenerative disorders, and other autoimmune diseases.2–4 Among them, as the most abundant of the five classes of Igs, Ig gamma (IgG) comprises three-quarters of all human antibodies in serum. To this end, all marked antibody drugs approved for clinical use are IgG-based therapeutic design templates, which have been widely exploited, including antibody–drug conjugates5,6 and bispecific antibodies.7 Despite the tremendous market for IgG antibodies, their structural and compositional complexity normally compromises their pharmaceutical benefits, potentially posing challenges in the discovery and development of antibody-based therapeutics.8,9 Thus, the clinical significance of these therapeutic proteins accompanied by high structural complexity has stimulated the development of analytical methods for rapid and comprehensive characterization of therapeutic antibodies.The architecture of IgG antibodies is relatively conserved as a characteristic Y-shaped topology through evolution, which is composed of two heavy chains and two light chains united by disulfide bonds. The number of disulfide bonds within the hinge region and connection type between heavy and light chains contribute to four subclasses of human IgGs (i.e., IgG1κ, IgG2κ, IgG3κ, and IgG4κ). More specifically, IgG1κ, IgG2κ, and IgG4κ afford similar molecular weights (∼150 kDa), topologies, and overall hinge lengths, whereas IgG3κ possesses up to 11 interchain disulfide bonds in the hinge region, resulting in a longer hinge and less structural flexibility.10 Additionally, IgG antibodies normally exhibit a high degree of heterogeneity because of variable posttranslational modifications (PTMs), such as glycosylation, oxidation, and disulfide linkage variations within therapeutic antibodies, which might decrease drug efficacy and even produce drug safety issues.11,12 These subtle differences resulting from intrinsic structural features could affect their binding-site specificity with antigens and cognate receptors.13
Currently, conventional biophysical methods, including NMR, X-ray crystallography, and cryo-EM, have attracted increasing attention by providing high-resolution crystal structures in structural biology. Although these techniques have generated impressive results, it remains a challenging issue to acquire structural characterization of full-length IgG antibodies because of their heterogeneity, flexibility, and large size.14 To date, very few intact IgGs can be obtained in the protein data bank (PDB),15 and most available IgG structures are only Fab or Fc domains. Alternatively, a conventional liquid chromatography-tandem mass spectrometry (LC-MS/MS)-based bottom-up strategy has been widely adopted for structural sequencing and PTM characterization.16–18 Unfortunately, sequence–structure correlations can be missing in the laborious enzymatic digestion process. To circumvent this limitation, middle-down and top-down approaches have recently been demonstrated for enhanced characterization of antibodies.19,20 Notably, Cotham et al. utilized restricted enzymatic digestion and 193 nm ultraviolet photodissociation (UVPD) for middle-down proteomics, allowing comprehensive sequencing of large peptides ranging from 4–12 kDa for unambiguous IgG identification.21 Fornelli et al. combined the IdeS enzyme and electron transfer dissociation (ETD) fragmentation for sequencing and characterization of IgGs.22 Mao et al. used top-down electron capture dissociation (ECD) with a 9.4 T Fourier transform ion cyclotron resonance (FTICR) mass spectrometry (MS) for structural characterization of intact IgG1κ with extensive sequence coverage.23 Recently, Burgt et al. used matrix-assisted laser desorption/ionization (MALDI) FTICR MS coupled with in-source decay fragmentation for top- and middle-down characterization of antibodies.24 As a highly complementary method, native MS has recently emerged as a promising method for producing intact gas-phase protein complexes and even membrane proteins, providing valuable stoichiometry and topology information.25,26 Gratifyingly, native MS can be hyphenated with ion mobility-mass spectrometry (IM-MS), which has been widely acknowledged in studying intact protein assemblies, such as dynamics,13,27 thermal stability,28,29 protein refolding,26,30 and antibody–drug conjugates.31,32 Despite these successful applications, great challenges remain because the available IM-MS normally has limited spectral resolution, and therefore, fails to produce sufficient information for defining macromolecular structures in detail.
Inspired by the pioneering efforts of Ruotolo's group, these challenges motivated the development of native IM-MS-based collision-induced unfolding (CIU), which has gained rapidly increasing attention owing to its ability to rapidly interrogate subtle tertiary structure differences and chemical modifications in the protein isoforms based on their unfolding patterns and gas-phase stabilities.9,33,34 Upon stepwise collisional activation of protein assemblies, a range of partially unfolded intermediates can occur in the gas phase, which provides a multitude of comparative and diagnostic information of protein complexes detected by IM-MS.35,36 Unfortunately, only protein ions with a single charge state can be quadrupole-selected prior to CIU analysis, resulting in iterative data collection process or a loss of complementary information acquired by multiple charge states. Furthermore, the CIU patterns of protein assemblies are strongly dependent on charge-state selection, but to what extent charge states affect their gas-phase structures is still in an early stage of exploration.
In this manuscript, we demonstrate a simplified multicharge-state CIU method for rapid differentiation of four IgG isotypes without the need for charge-state selection in advance. IgG antibodies are yielded in native IM-MS, allowing this multicharge-state CIU strategy to reflect their gas-phase unfolding behaviors in a rapid, simple, and sensitive manner. Benefiting from the integrated advantages of reduced data acquisition time and complementary structural information of protein assemblies with multiple charge states, this method affords the ability to rapidly characterize subtle tertiary structure differences in terms of different disulfide bonding numbers and patterns. Additionally, the effect of charge states on unfolding behaviors and gas-phase stabilities of IgG antibodies was investigated, exhibiting a strong correlation between CIU fingerprints and charge states. Furthermore, we used this method to discriminate four IgG isoforms that possess different disulfide bond numbers and even subtle disulfide bonding patterns based on their charge-dependent gas-phase unfolding behaviors and root-mean square deviation (RMSD) in CIU difference spectra. Altogether, these results demonstrate that this simplified multicharge-state CIU method offers great potential for the characterization of intact IgG antibodies, facilitating rapid screening and evaluation of candidate antibodies for innovator biotherapeutics.
2. Experimental
2.1 Chemicals and materials
Four homologous monoclonal antibodies from human myeloma, including IgG1κ, IgG2κ, IgG3κ, and IgG4κ, were purchased from Sigma Aldrich (St. Louis, MO, USA). Ammonium acetate (AA) and cesium iodide (CsI) were purchased from Sigma Aldrich (St. Louis, MO, USA). Borosilicate capillaries with inner and outer diameters of 0.58 mm and 1.0 mm, respectively, were purchased from Sutter Instruments (Novato, CA, USA). Amicon centrifugal filters with a 100 kDa cutoff were purchased from Merck Millipore (Darmstadt, Germany).2.2 Sample preparation of IgG antibodies
Individual stock solutions of four protein complexes were initially dissolved in Milli-Q water at a concentration of 50 µM. To achieve a better desalting effect, the stock solutions were buffer-exchanged with 200 mM AA solution using centrifugal filters with a cutoff of 100 kDa. Afterwards, the buffer-exchanged protein solution was diluted with 200 mM AA to a concentration of 5 µM for native MS analysis.2.3 IM-MS experiments
Before native MS analysis, borosilicate capillaries were first pulled to a tip with an ∼1 µm inner diameter using a P-97 puller (Sutter Instruments, Novato, CA, USA). Four protein aliquots (∼3 µL) were introduced to the nanoESI emitter and analyzed in positive ion mode using a quadrupole ion mobility time-of-flight mass spectrometer (Synapt G2-Si HDMS, Waters, Milford, MA, UK). Given that the tip was easy to plug under high electrospray voltage, protein ions were generated and maintained by applying a potential as low as 0.8–1.0 kV for the optimal sensitivity and stable spray. Before data acquisition, the IM-MS instrument was mass-calibrated using 50 mg mL−1 CsI solution. To avoid conformational disturbance, the dry gas temperature was maintained at as low as 30 °C. MassLynx 4.2 (Waters, Milford, MA, UK) was used for IM-MS data analysis, and mass spectra were obtained using Origin 9.0 software (Origin Lab, North Hampton, MA, USA). The detailed instrumental parameters are listed in Table S1.†2.4 CIU experiments
To achieve rapid discrimination of intact antibodies with multiple charge states, no ions of interest were mass-selected prior to the IMS measurement. The collision voltage in the trap cell was gradually increased from 4 V to 200 V with a 2 V stepwise increment to acquire CIU fingerprints with different charge states. It should be noted that a 4 V dropout voltage was the minimum voltage for ion transmission from the trap to the IMS cell; thus, it was chosen as the starting collision energy. In theory, a larger dropout voltage can accelerate the data acquisition process; however, a lower imaging resolution of CIU fingerprints can be foreseen due to the smaller pixels. There is a trade-off in balancing desirable imaging resolution and reasonable acquisition time. Thus, a 2 V stepwise increment was utilized to provide more structural differences. The CIU fingerprints and feature analysis for four IgG subtypes were achieved by a software package termed CIUSuite2 developed by Ruotolo's group.34 The “Compare” function in CIUSuite2 was adopted to visualize the significant difference between two CIU fingerprints. Moreover, RMSD calculations can be adopted as an indicator to summarize and quantify CIU differences. Specifically, the calculated RMSD values turned out to be larger if the difference between two CIU fingerprints became more significant. Additionally, the “Average” function in CIUSuite2 software was adopted to produce the averaged CIU fingerprints from multiple duplicate trials. Given the reproducibility of multi-charge-state CIU data is indispensable to produce reliable comparative results, all the multi-charge-state CIU plots were averaged by triplicates unless otherwise noted.3. Results and discussion
3.1 Native MS of IgG subtypes
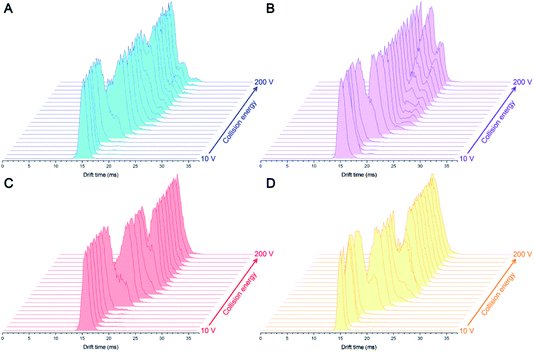
Four IgG subtypes differing in terms of the number and pattern of disulfide bonds were chosen as model systems, and a detailed schematic diagram with regard to IgG subtype structures is shown in Fig. 1. Among them, IgG1κ, IgG2κ, and IgG4κ have identical domain structures and similar molecular weights of ∼150 kDa because they share more than 90% sequence identity. The numbers and patterns of interchain disulfide bonds contribute to their main differences. Specifically, IgG1κ and IgG4κ afford 4 interchain disulfide bonds, whereas IgG2κ possesses 6 interchain disulfide bonds. In sharp contrast, IgG3κ has a higher molecular weight of ∼170 kDa due to repeated amino acid sequences, which results in a total of 13 interchain disulfide bonds.10 Prior to the CIU analysis of four IgG subtypes, native mass spectra of IgG1κ, IgG2κ, IgG3κ, and IgG4κ were first obtained. As shown in Fig. 2, the most abundant charge states of these four antibody ions range from 23+ to 24+, which is in accordance with previous reports.13,37 This strongly suggests that their native-like structures can be well preserved due to the moderate nanoESI source. Thompson et al. reported that highly charged ion signals ranging from 32+ to 56+ can be observed under denaturing conditions.8 The mass spectra were dominated by a relatively narrow peak width for the distinct charge states of the four IgG ions because nano-ESI could be more tolerant to salt contamination, avoiding undesired salt adduction effects. Exceptionally, the peak width of IgG3κ can be slightly broadened due to the presence of higher heterogeneity.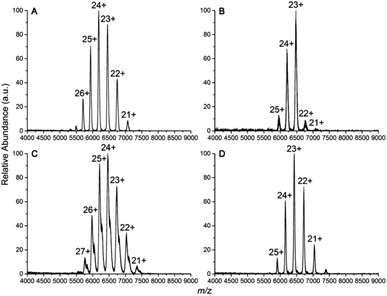 | ||
| Fig. 2 Native mass spectra of (A) IgG1κ, (B) IgG2κ, (C) IgG3κ, and (D) IgG4κ in 200 mM ammonium acetate buffer. | ||
3.2 IM-MS drift-time distributions of IgG subtypes
Precise measurement of collision cross section (CCS) values for biomolecule ions is key to structural biology.38 However, the limited IM resolution and dynamic conformational changes cause a dilemma in the identification of structurally similar IgG with high heterogeneity. Thus, these challenges motivated the development of the CIU method, which adopts stepwise collisional heating in combination with native IM-MS to visualize the unfolding features of protein ions in the gas phase.34 As shown in Fig. 3, the drift time (DT) for the dominant charge state (23+) of four IgG ions was acquired at different collision voltages ranging from 10 V to 200 V. Under the lower energy (10 V) condition, four 23+ protein ions afford a relatively narrow DT distribution at ∼14.93 ms, ∼14.77 ms, ∼15.32 ms, and ∼14.76 ms, which correspond to compact structures. A larger DT value of IgG3κ can be ascribed to the fact that IgG3κ affords a higher molecular weight and less structural flexibility. Notably, when the collision energy was increased to 60 V, the dominant peaks were split and tailed by the small peaks with larger DTs, indicating that some partial unfolding occurs for their gas-phase conformations. Upon further increases in the collision voltage, the native peak was fully converted into the unfolding conformation at larger DTs. This observation was not entirely unexpected. Zhong et al. demonstrated that the CIU behaviors of protein ions are sensitive to their domain structure.39 Under high-energy collision, gas-phase protein ions tend to unfold in the CIU analysis, which makes another stable conformation occur with larger CCS.35,39 Similar phenomena are observed for different charge states except that different collision voltages are required to initiate the folding process (Fig. S1–S4†). As previously reported, the ions of interest with the specific charge state should be separated in the trap cell prior to IM separation, which demands a tedious and lengthy data collection time decided by the required charge numbers. Moreover, submicrometer nanoESI emitters easily clog if a prolonged electrospray process is adopted.40,41 Herein, all IgG ions with different charge states were simultaneously injected for CIU analysis without charge separation, thereby avoiding the time-consuming acquisition process. Altogether, this strategy will expand the horizons of the IM-MS platform in the rapid differentiation of protein complexes more than IgG with different charge states and sufficiently high resolution (2 V increments) in a single experiment.3.3 Differential CIU fingerprints of IgG subtypes as a function of charge state
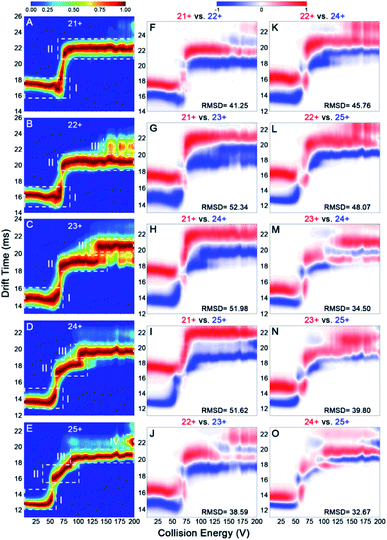
Previous reports have revealed that the dissociation/unfolding pathways of protein complexes are highly dependent upon their initial charge states.42,43 Hall et al. demonstrated that atypical CID dissociation behavior of protein complexes, such as loss of folded monomeric subunits, can occur for higher charge states of precursor ions.42 Thus, simultaneous acquisition of CIU behaviors for various IgGs with all charge states observed in native mass spectra could offer a great opportunity to gain deeper insights into gas-phase structural differentiation of IgGs. Fig. 4 shows the CIU fingerprints of IgG1κ at different charge states ranging from 21+ to 25+. Notably, two main CIU features for 21+ IgG1κ can be observed, including the initial compact state (state I) and the unfolded state (state II), over the increased acceleration voltages (Fig. 4A). As expected, the lowest charge state (21+) requires higher voltages to unfold and generally accesses fewer intermediate structures along the unfolding pathway than higher charge states. As the charge state increases (i.e., 22+ and 23+), another unfolded state (state III) occurs at higher energies. Additionally, the first transition voltage from the initial compact state (state I) to the first unfolded state (state II) gradually decreases from 73.8 V to 54.4 V for the lowest and highest charge state. Many studies have demonstrated that manipulating the charge number carried on the protein surface in a controllable manner can alter protein structures from compact to extended conformations.44,45 As shown in Fig. 4B–E, lower collision voltages are required for the transition from state II to state III when the charge state is increased, suggesting that IgG1κ ions with higher charge states are prone to unfolding. The third unfolded state (state IV) even be observed for 25+ IgG1κ ions. Intriguingly, similar results can also be obtained for the other three IgG ions (Fig. S5–S7†), and the required voltages among each transition state for these four IgG ions are listed in Table S2.† Thus, although all charge states ranging from 21+ to 25+ are observed in the native mass spectrum (Fig. 2A), which corresponds to the compact and native-like conformations of IgG1κ, the gas-phase unfolding behaviors can be completely different.To visualize the effect of charge states on CIU fingerprints of IgG ions, we further investigated the significant differences among various charge states ranging from 21+ to 25+ using the “Compare” function of CIUSuite2. Specifically, RMSD calculations were used to summarize CIU differences for quantification. Recently, Wu et al. used CIU difference fingerprints and RMSD to rapidly discriminate monosaccharide derivate isomers from binary mixtures.46 Although native MS normally affords a native-like conformation with low charge states, narrow charge-state distributions, and minimized Coulomb repulsions, subtle surface-charge variation could still affect their gas-phase structures and CIU behaviors. As shown in Fig. 4F–O, the CIU difference plots and the corresponding RMSD values of IgG1κ ions were obtained to exhibit the absolute difference among multifarious charge states. Interestingly, the results revealed that the structural discrepancy between one charge difference (i.e., 22+ vs. 23+) is smaller than that between two or three charge differences (i.e., 22+ vs. 24+ or 22+ vs. 25+). This is foreseeable because the redundant charges might contribute to the slight structural changes derived from unfavorable coulombic repulsions. Additionally, IgG structures can be divided into antigen-binding fragments (Fabs) and crystallizable segments (Fcs) by a central hinge, which provides relative flexibility, such as Fab–Fab and Fab–Fc movements.47 Hansen et al. demonstrated that IgG ions might undergo collapse in the gas phase due to their intrinsic structural features (i.e., Fab arm flexibility and hinge length); therefore, higher charges might drive the structural variation.13 Additionally, the RMSD value (32.67) between 24+ and 25+ IgG1κ was lower than that (41.25) between 21+ and 22+ IgG1κ, indicating that the conformational difference was reduced for high-charge-state IgG ions. Similar phenomena are observed for the other three IgG ions (Fig. S5–S7†). Taken together, the data presented in Fig. 4 strongly support the hypothesis that more charge states on the protein surface can result in more extended conformations, which significantly alters the gas-phase unfolding behaviors of IgG ions.
3.4 Rapid discrimination of intact IgG subtypes using multicharge-state CIU
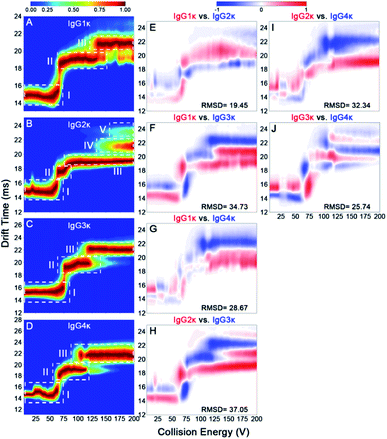
According to the mentioned-above results, some subtle structural differences might be missing using a single charge state only. To this end, we investigated the effect of disulfide bonding patterns within IgG ions on their charge-dependent CIU behaviors. IgG1κ and IgG4κ differ by one amino acid (i.e., a proline residue in IgG1 and a serine residue in IgG4) in the Fab region, resulting in different connection types of disulfide bonds between the light chain and heavy chain (Fig. 1).10 Considering that both IgG1κ and IgG4κ possess the same interchain disulfide bonds, similar CIU patterns of IgG1κ and IgG4κ at higher charge states ranging from 23+ to 25+ can be obtained (Fig. 5A and D), which is also in agreement with the reported results by Ruotolo's groups.37 However, the greater differences correlating to distinct patterns of disulfide bonds can be clearly distinguished at low charge states (i.e., 21+ and 22+), likely due to their distinct disulfide bonding patterns. Specifically, 21+ IgG4κ has one more intermediate unfolding state (state III) than 21+ IgG1κ, whereas 22+ IgG1κ has longer state II and shorter state III than 22+ IgG4κ. Moreover, the mean Fab–Fab angles of IgG1κ and IgG4κ are 117 ± 43° and 128 ± 39°, respectively, which might account for why the second unfolding state (state III) is more stable for IgG4κ.27 This result further supports that CIU is highly sensitive to disulfide bonding patterns within IgG antibodies. One issue to note is that although the 23+ or 24+ charge state affords the highest signal-to-noise in the native mass spectrum (Fig. 2), it is not the optimal charge state to provide differentiated CIU data for these two antibodies with high sequence identity and structural similarity. Thus, simultaneous acquisition of multicharge-state CIU data observed in a native MS experiment will offer superior potential in discriminating subtle differences in terms of disulfide bonding patterns.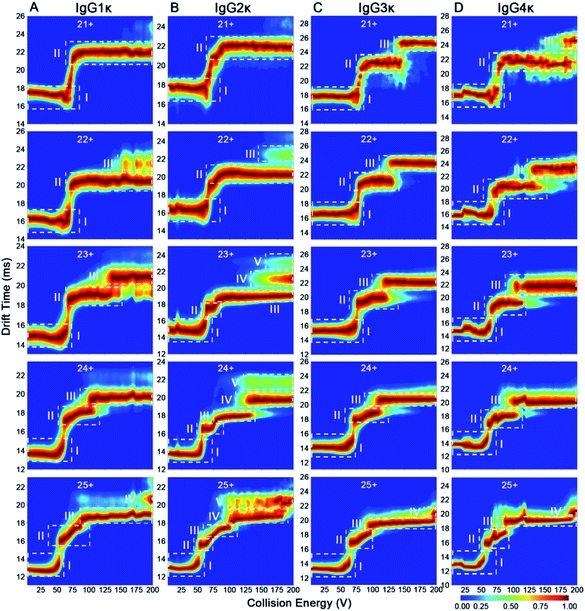 | ||
| Fig. 5 CIU fingerprints of (A) IgG1κ, (B) IgG2κ, (C) IgG3κ, and (D) IgG4κ at different charge states ranging from 21+ to 25+. | ||
In addition to disulfide bonding patterns, CIU fingerprints of IgG antibodies can be strongly dependent on their disulfide bond numbers, which are the main differences in these IgG subtypes. In detail, there are 4 interchain disulfide bonds in both IgG1κ and IgG4κ but 6 and 13 interchain disulfide bonds in IgG2κ and IgG3κ, respectively. Considering that IgG2κ and IgG4κ afford the same disulfide bonding patterns within Fab arms but different numbers of disulfide bonds, significantly different CIU behaviors can be observed for both IgG2κ and IgG4κ. More specifically, only state I and state II occupy the CIU fingerprints of 21+ IgG2κ, but one more unfolding pattern (state III) occupies the CIU fingerprints of 21+ IgG4κ ions (Fig. 5B and D). Additionally, as the charge states increase (e.g., 23+), a total of five conformational families occupy the CIU feature of IgG2κ, whereas only three conformational families exist in that of IgG4κ. On the other hand, given that IgG3κ affords a total of 13 disulfide bonds that are nearly twofold greater than the other three IgG ions, IgG3κ exhibits relatively “stable” CIU behaviors due to a more compact hinge region. Specifically, CIU fingerprints of IgG3κ are dominated by the three principle IM features at the lowest charge state (21+). As the charge state increases, state II and state III in the CIU fingerprint of IgG3κ are relatively stable, suggesting that more disulfide bonds within IgG3κ contribute to its higher gas-phase stability. In sharp contrast, although only state I and state II occupy the CIU fingerprints of 21+ IgG2κ, a rapid transition from state II to state V in the CIU fingerprint of IgG2κ occurs over the collision voltage range. Similar phenomena can be observed in the CIU behaviors of IgG1κ and IgG4κ, in which the first unfolding state (state II) can be gradually transformed into the second unfolding state (state III) over the collision voltage range. Taken together, the results presented herein indicate that gas-phase CIU behaviors can be sensitive not only to disulfide bond patterns between light and heavy chains but also to overall disulfide bond numbers for four IgG subtypes. More importantly, the subtle structural discrepancy that is difficult to detect by CIU patterns at the single charge state can be clearly discriminated using multicharge-state CIU analysis.
To better quantitatively visualize the gas-phase structural differences of these CIU fingerprints in detail, the RMSD parameters among the four IgG subtypes were further acquired. Previous observations have revealed that a low RMSD value below ∼6% corresponds to technical replicates of CIU analysis, which indicates that a gas-phase structural discrepancy among IgG subtypes exists if an RMSD value higher than the threshold value.37,46 Additionally, a higher RMSD value reflects that there is more gas-phase structural discrepancy among IgG subtypes. Fig. 6 and S8–S11† show the CIU difference plots, and the corresponding RMSD values of four IgG subtypes with various charge states ranging from 21+ to 25+ are listed in Table S3.† It should be mentioned that the effect of disulfide bonding patterns and numbers on their gas-phase CIU behaviors can be strongly dependent on charge states. Specifically, given that IgG1κ and IgG4κ differ only slightly in disulfide bond patterns within heavy and light chains, their structural difference can be significantly enhanced under a low charge state (ranging from 21+ to 23+), whereas it is changed into the smallest difference under a high charge state (i.e., 24+ and 25+) compared to other IgG subtype pairs (Table S3†). Notably, this result suggests that the effect of disulfide bonding patterns on gas-phase IgG structures can be weakened for extended conformations with more charges residing on the protein surface. In contrast, the structural difference between IgG1κ and IgG2κ can be gradually enlarged with an increase in charge states from 21+ to 25+. Notably, the CIU method is not only sensitive to the structural differences among various IgG subtypes but within one IgG isoform. Hernandez-Alba et al., have demonstrated that although both wt-IgG4s (natalizumab and reslizumab) share the same number and network of disulfide bonds in the hinge region and very similar primary sequences, clear CIU differences can be observed between both 22+ wt-IgG4 ions at higher collision voltages.48 This observation strongly supports that the CIU method enables the in-depth differentiation of structural differences within IgG subtypes or one IgG isoform.
4. Conclusions
In summary, this work demonstrates a multicharge-state CIU method for rapid differentiation of four IgG isotypes that differ in the numbers and patterns of disulfide bonds in a single native IM-MS experiment without the need for charge-state selection in advance. Given that IgG antibodies are yielded in their native forms, the correlation between surface charge states and gas-phase unfolding patterns of IgG antibodies was investigated. The results indicate that CIU fingerprints are strongly dependent on their charge states on the protein surface, supporting the notion that redundant surface charge states can alter protein structures from compact to more extended conformations, and therefore, result in unique gas-phase unfolding behaviors. In addition to charge state, CIU fingerprints are highly sensitive to disulfide bonding numbers and patterns among IgG antibodies. Rapid and comprehensive discrimination of IgG isoforms can be achieved based on charge-dependent gas-phase unfolding behaviors and RMSD in CIU difference spectra. Furthermore, numerous possibilities remain in the future to investigate the effects of charge states on gas-phase structures of proteins and large protein complexes. Using an optimized workflow and automated data processing, this method holds great potential in high-throughput screening protein–ligand interactions and biotherapeutic characterization. Overall, incorporating all charge states observed in a native IM-MS experiment into CIU analysis will offer higher sensitivity in rapid screening and discriminating biotherapeutic candidates with more subtle structural differences, thereby stimulating more research studies on large protein complexes.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by the National Natural Science Foundation of China (Grant No. 21904142) and the Key Realm R&D Program of Guangdong Province (Grant No. 2020B0202090005). This work was also supported by the Special Fund for Scientific Innovation Strategy-Construction of High-level Academy of Agriculture Science (Grant No. R2021YJ-QG004 and R2020PY-JX019).References
- P. C. Taylor, A. C. Adams, M. M. Hufford, I. de la Torre, K. Winthrop and R. L. Gottlieb, Nat. Rev. Immunol., 2021, 21, 382–393 CrossRef CAS PubMed.
- P. J. Carter and G. A. Lazar, Nat. Rev. Drug Discovery, 2018, 17, 197–223 CrossRef CAS PubMed.
- E. Jabbour, S. Paul and H. Kantarjian, Nat. Rev. Clin. Oncol., 2021, 18, 418–433 CrossRef CAS PubMed.
- K. Rajasekhar and T. Govindaraju, RSC Adv., 2018, 8, 23780–23804 RSC.
- A. Beck, L. Goetsch, C. Dumontet and N. Corvaïa, Nat. Rev. Drug Discovery, 2017, 16, 315–337 CrossRef CAS PubMed.
- E. Robinson, J. P. M. Nunes, V. Vassileva, A. Maruani, J. C. F. Nogueira, M. E. B. Smith, R. B. Pedley, S. Caddick, J. R. Baker and V. Chudasama, RSC Adv., 2017, 7, 9073–9077 RSC.
- A. F. Labrijn, M. L. Janmaat, J. M. Reichert and P. W. H. I. Parren, Nat. Rev. Drug Discovery, 2019, 18, 585–608 CrossRef CAS PubMed.
- N. J. Thompson, S. Rosati, R. J. Rose and A. J. R. Heck, Chem. Commun., 2013, 49, 538–548 RSC.
- Y. Tian and B. T. Ruotolo, Analyst, 2018, 143, 2459–2468 RSC.
- H. Liu and K. May, mAbs, 2012, 4, 17–23 CrossRef PubMed.
- J. Wypych, M. Li, A. Guo, Z. Zhang, T. Martinez, M. J. Allen, S. Fodor, D. N. Kelner, G. C. Flynn, Y. D. Liu, P. V. Bondarenko, M. S. Ricci, T. M. Dillon and A. Balland, J. Biol. Chem., 2008, 283, 16194–16205 CrossRef CAS PubMed.
- A. Bertolotti-Ciarlet, W. Wang, R. Lownes, P. Pristatsky, Y. Fang, T. McKelvey, Y. Li, Y. Li, J. Drummond, T. Prueksaritanont and J. Vlasak, Mol. Immunol., 2009, 46, 1878–1882 CrossRef CAS PubMed.
- K. Hansen, A. M. Lau, K. Giles, J. M. McDonnell, W. B. Struwe, B. J. Sutton and A. Politis, Angew. Chem., Int. Ed., 2018, 57, 17194–17199 CrossRef CAS PubMed.
- L. Bongini, D. Fanelli, F. Piazza, P. De Los Rios, S. Sandin and U. Skoglund, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 6466–6471 CrossRef CAS PubMed.
- E. O. Saphire, P. W. H. I. Parren, R. Pantophlet, M. B. Zwick, G. M. Morris, P. M. Rudd, R. A. Dwek, R. L. Stanfield, D. R. Burton and I. A. Wilson, Science, 2001, 293, 1155–1159 CrossRef CAS PubMed.
- A. Beck, E. Wagner-Rousset, D. Ayoub, A. Van Dorsselaer and S. Sanglier-Cianférani, Anal. Chem., 2013, 85, 715–736 CrossRef CAS PubMed.
- W. Li, L. Lin, D. Yan, Y. Jin, Y. Xu, Y. Li, M. Ma and Z. Wu, Anal. Chem., 2020, 92, 3237–3245 CrossRef PubMed.
- L. Lin, Q. Yu, J. Zheng, Z. Cai and R. Tian, Clin. Proteomics, 2018, 15, 42 CrossRef PubMed.
- P. B. Pandeswari and V. Sabareesh, RSC Adv., 2019, 9, 313–344 RSC.
- J. Camperi, A. Goyon, D. Guillarme, K. Zhang and C. Stella, Analyst, 2021, 146, 747–769 RSC.
- V. C. Cotham, A. P. Horton, J. Lee, G. Georgiou and J. S. Brodbelt, Anal. Chem., 2017, 89, 6498–6504 CrossRef CAS PubMed.
- L. Fornelli, D. Ayoub, K. Aizikov, A. Beck and Y. O. Tsybin, Anal. Chem., 2014, 86, 3005–3012 CrossRef CAS PubMed.
- Y. Mao, S. G. Valeja, J. C. Rouse, C. L. Hendrickson and A. G. Marshall, Anal. Chem., 2013, 85, 4239–4246 CrossRef CAS PubMed.
- Y. E. M. van der Burgt, D. P. A. Kilgour, Y. O. Tsybin, K. Srzentić, L. Fornelli, A. Beck, M. Wuhrer and S. Nicolardi, Anal. Chem., 2019, 91, 2079–2085 CrossRef CAS PubMed.
- A. C. Leney and A. J. R. Heck, J. Am. Soc. Mass Spectrom., 2017, 28, 5–13 CrossRef CAS PubMed.
- B. Yan and J. Bunch, J. Am. Soc. Mass Spectrom., 2021, 32, 690–699 CrossRef CAS PubMed.
- K. J. Pacholarz, M. Porrini, R. A. Garlish, R. J. Burnley, R. J. Taylor, A. J. Henry and P. E. Barran, Angew. Chem., Int. Ed., 2014, 53, 7765–7769 CrossRef CAS PubMed.
- K. J. Pacholarz, S. J. Peters, R. A. Garlish, A. J. Henry, R. J. Taylor, D. P. Humphreys and P. E. Barran, ChemBioChem, 2016, 17, 46–51 CrossRef CAS PubMed.
- M. J. Edgeworth, J. J. Phillips, D. C. Lowe, A. D. Kippen, D. R. Higazi and J. H. Scrivens, Angew. Chem., Int. Ed., 2015, 54, 15156–15159 CrossRef CAS PubMed.
- B. Yan, A. J. Taylor and J. Bunch, J. Am. Soc. Mass Spectrom., 2019, 30, 1179–1189 CrossRef CAS PubMed.
- E. J. Larson, D. S. Roberts, J. A. Melby, K. M. Buck, Y. Zhu, S. Zhou, L. Han, Q. Zhang and Y. Ge, Anal. Chem., 2021, 93, 10013–10021 CrossRef CAS PubMed.
- A. Ehkirch, V. D'Atri, F. Rouviere, O. Hernandez-Alba, A. Goyon, O. Colas, M. Sarrut, A. Beck, D. Guillarme, S. Heinisch and S. Cianferani, Anal. Chem., 2018, 90, 1578–1586 CrossRef CAS PubMed.
- D. A. Polasky, S. M. Dixit, D. D. Vallejo, K. D. Kulju and B. T. Ruotolo, Anal. Chem., 2019, 91, 10407–10412 CrossRef CAS PubMed.
- D. A. Polasky, S. M. Dixit, S. M. Fantin and B. T. Ruotolo, Anal. Chem., 2019, 91, 3147–3155 CrossRef CAS PubMed.
- J. D. Eschweiler, R. M. Martini and B. T. Ruotolo, J. Am. Chem. Soc., 2017, 139, 534–540 CrossRef CAS PubMed.
- K. Pisupati, Y. Tian, S. Okbazghi, A. Benet, R. Ackermann, M. Ford, S. Saveliev, C. M. Hosfield, M. Urh, E. Carlson, C. Becker, T. J. Tolbert, S. P. Schwendeman, B. T. Ruotolo and A. Schwendeman, Anal. Chem., 2017, 89, 4838–4846 CrossRef CAS PubMed.
- Y. Tian, L. Han, A. C. Buckner and B. T. Ruotolo, Anal. Chem., 2015, 87, 11509–11515 CrossRef CAS PubMed.
- P. Lössl, M. van de Waterbeemd and A. J. Heck, EMBO J., 2016, 35, 2634–2657 CrossRef PubMed.
- Y. Zhong, L. Han and B. T. Ruotolo, Angew. Chem., Int. Ed., 2014, 53, 9209–9212 CrossRef CAS PubMed.
- D. N. Mortensen and E. R. Williams, Analyst, 2016, 141, 5598–5606 RSC.
- J. Hu, Q.-Y. Guan, J. Wang, X.-X. Jiang, Z.-Q. Wu, X.-H. Xia, J.-J. Xu and H.-Y. Chen, Anal. Chem., 2017, 89, 1838–1845 CrossRef CAS PubMed.
- Z. Hall, A. Politis, M. F. Bush, L. J. Smith and C. V. Robinson, J. Am. Chem. Soc., 2012, 134, 3429–3438 CrossRef CAS PubMed.
- Z. Hall, H. Hernández, J. A. Marsh, S. A. Teichmann and C. V. Robinson, Structure, 2013, 21, 1325–1337 CrossRef CAS PubMed.
- C. C. Going and E. R. Williams, Anal. Chem., 2015, 87, 3973–3980 CrossRef CAS PubMed.
- M. A. Zenaidee, M. G. Leeming, F. Zhang, T. T. Funston and W. A. Donald, Angew. Chem., Int. Ed., 2017, 56, 8522–8526 CrossRef CAS PubMed.
- X. Wu, Y. Zhang, R. Qin, P. Li, Y. Wen, Z. Yin, Z. Zhang and H. Xu, Talanta, 2021, 224, 121901 CrossRef CAS PubMed.
- Z. Zhang, H. Pan and X. Chen, Mass Spectrom. Rev., 2009, 28, 147–176 CrossRef CAS PubMed.
- O. Hernandez-Alba, E. Wagner-Rousset, A. Beck and S. Cianférani, Anal. Chem., 2018, 90, 8865–8872 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra06486j |
| This journal is © The Royal Society of Chemistry 2021 |