Open Access Article
Open Access ArticleDifferent phosphorylation and farnesylation patterns tune Rnd3–14-3-3 interaction in distinct mechanisms†
Jun
Hu
,
Xue-Meng
Sun
,
Jing-Yun
Su
,
Yu-Fen
Zhao
and
Yong-Xiang
Chen
 *
*
Key Laboratory of Bioorganic Phosphorus Chemistry and Chemical Biology (Ministry of Education), Department of Chemistry, Tsinghua University, Beijing 100084, China. E-mail: chen-yx@mail.tsinghua.edu.cn
First published on 26th January 2021
Abstract
Protein posttranslational modifications (PTMs) are often involved in the mediation or inhibition of protein–protein interactions (PPIs) within many cellular signaling pathways. Uncovering the molecular mechanism of PTM-induced multivalent PPIs is vital to understand the regulatory factors to promote inhibitor development. Herein, Rnd3 peptides with different PTM patterns as the binding epitopes and 14-3-3ζ protein were used as models to elucidate the influences of phosphorylation and farnesylation on binding thermodynamics and kinetics and their molecular mechanism. The quantitative thermodynamic results indicate that phosphorylated residues S210 and S218 (pS210 and pS218) and farnesylated C241 (fC241) enhance Rnd3–14-3-3ζ interactions in the presence of the essential pS240. However, distinct PTM patterns greatly affect the binding process. Initial association of pS240 with the phosphate-binding pocket of one monomer of the 14-3-3ζ dimer triggers the binding of pS210 or pS218 to another monomer, whereas the binding of fC241 to the hydrophobic groove on one 14-3-3ζ monomer induces the subsequent binding of pS240 to the adjacent pocket on the same monomer. Based on the experimental and molecular simulation results, we estimate that pS210/pS218 and pS240 mediate the multivalent interaction through an additive mechanism, whereas fC241 and pS240 follow an induced fit mechanism, in which the cooperativity of these two adjacent PTMs is reflected by the index ε described in our established thermodynamic binding model. Besides, these proposed binding models have been further used for describing the interaction between 14-3-3ζ and other substrates containing adjacent phosphorylation and lipidation groups, indicating their potential in general applications. These mechanistic insights are significant for understanding the regulatory factors and the design of PPI modulators.
Introduction
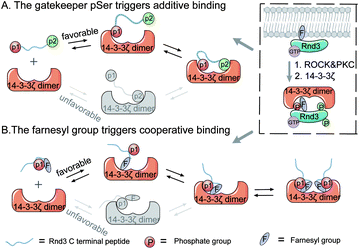
Protein–protein interactions (PPIs) are essential for almost all biological processes,1 including cellular signal transduction, in which multivalent effects are pervasive to commonly achieve strong yet reversible interactions.2,3 Multivalent PPIs often occur on protein surfaces and especially involve the intrinsically disordered regions (IDRs) of proteins.4–6 Diverse protein post-translational modifications (PTMs), such as phosphorylation, glycosylation, lipidation and methylation, that provide extra functionalities on protein surfaces play pivotal roles in mediating or inhibiting many important PPIs through electrostatic forces, hydrogen bonding, or hydrophobic effects.5 Particularly, recent advances in the field of liquid–liquid phase separation demonstrate that PTM-mediated multivalent PPIs play essential roles in some important liquid phase biomolecular condensates.7 Since PPIs in signaling pathways are often associated with the pathogenesis of many human diseases, they have become promising drug targets and have attracted intense attention from both academia and the pharmaceutical industry.8–11 Thus, the fundamental understanding of multivalent PPIs on the molecular scale, including those with diverse PTMs, is of great significance for further modulator development.14-3-3 proteins are ubiquitously expressed in all eukaryotic organisms and often work as homodimers. By interacting with a multitude of phosphorylated partner proteins with diverse functions, 14-3-3 proteins regulate a very large number of physiological processes.12 In addition to the general phosphorylated substrates, 14-3-3 can recognize some proteins that have been modified with other types of PTMs, such as glycosyl moieties,13 lipids14 and even multiple PTMs.15 The Rnd3 protein, a member of the Rho-GTPase family with well-known functions in the regulation of actin cytoskeleton dynamics,16 cell-cycle progression, cell polarity, differentiation, apoptosis and survival,17,18 is a newly characterized interaction partner of 14-3-3. Rnd3 has a flexible C-terminal tail that contains multiple phosphorylation sites19 and ends with a farnesyl group and a methylated cysteine.20 Previous reports have revealed that the 14-3-3ζ protein can stabilize Rnd3 in the cytoplasm by recognizing phosphorylated S240 (pS240) and the adjacent farnesylated C241 (fC241) on the Rnd3 protein.14,21,22 In addition, Rnd3 phosphorylation at either S210 or S218 in combination with pS240 is required for the optimal binding of Rnd3 to 14-3-3ζ in cells.14 However, the molecular mechanism of the contribution of multiple phosphorylated residues to binding as well as the cooperative effects of phosphorylation and farnesylation in this multivalent PPI remain unclear.
Physiological multivalent PPIs appear in a variety of cellular processes, such as signal transduction and pathogen-host cell recognition.23,24 Binding-enhancing multivalent interactions often adopt two kinds of molecular mechanisms. One is the positive cooperative (synergistic) mechanism, which sometimes displays an induced fit effect that has been thoroughly studied in the field of enzymology,25 illuminating the process by which binding at one site induces a series of conformational changes in the protein to facilitate binding more tightly.26 Another mechanism is the noncooperative (additive) mechanism, which displays the multisite interaction combinations, which often involve distal binding sites27 and has been systematically studied in host–guest systems.3,28,29 This second mechanism is well-reflected in the binding pattern between 14-3-3 dimers and its multiphosphorylated protein partners.30,31 Moreover, a thermodynamic model for this kind of multivalent PPI was established based on ditopic host–guest systems featuring effective molarity.32 However, the effects of multiphosphorylation on the binding kinetics of 14-3-3 and partner proteins such as Rnd3 remain elusive. In addition, the cooperative effects of adjacent phosphorylation and farnesylation epitopes on the Rnd3–14-3-3ζ interaction have not been well elucidated. Thus, it is necessary to address these issues.
Here, we used the C-terminal domain of Rnd3 with different PTM patterns and 14-3-3ζ as a model to elucidate the regulatory mechanisms of phosphorylation and farnesylation toward multivalent interactions (Fig. 1). First, the pS240 epitope was verified to be indispensable in the binding of the multiphosphorylated Rnd3 peptide to the 14-3-3ζ protein. By using thermodynamic assays, we further determined that the other phosphorylated sites, pS210 and pS218, as well as farnesylated C241, can enhance Rnd3–14-3-3ζ binding, although alone, they hardly mediate stable binding. Moreover, kinetic studies determined that these auxiliary PTM epitopes play different roles in the binding process. The initial association of pS240 with the phosphate binding pocket on one monomer of the 14-3-3ζ dimer triggers the binding of pS210 or pS218 to the other monomer, whereas the initial binding of fC241 to the hydrophobic groove on one 14-3-3ζ monomer induces the subsequent binding of pS240 to the adjacent pocket on the same monomer. Moreover, based on the experimental data and molecular simulation results, we estimate that pS210/pS218 and pS240 mediate a multivalent interaction through the additive mechanism, whereas fC241 and pS240 follow the induced-fit mechanism to mediate binding, in which the cooperativity of these two adjacent PTMs is reflected by the index ε described in our established thermodynamic binding model. In addition, the farnesyl group of the Rnd3 peptide acts as a kinetic stabilizer33 for the interaction, which may guide the development of new modulators (inhibitors or stabilizers) for PPIs.9,34 Besides, these proposed models have been further used for describing the interaction between 14-3-3ζ and other substrates containing adjacent phosphorylation and lipidation groups, indicating their potential in general applications.
Results and discussion
Generation of Rnd3 peptides with various PTM patterns and their interactions with the 14-3-3ζ protein
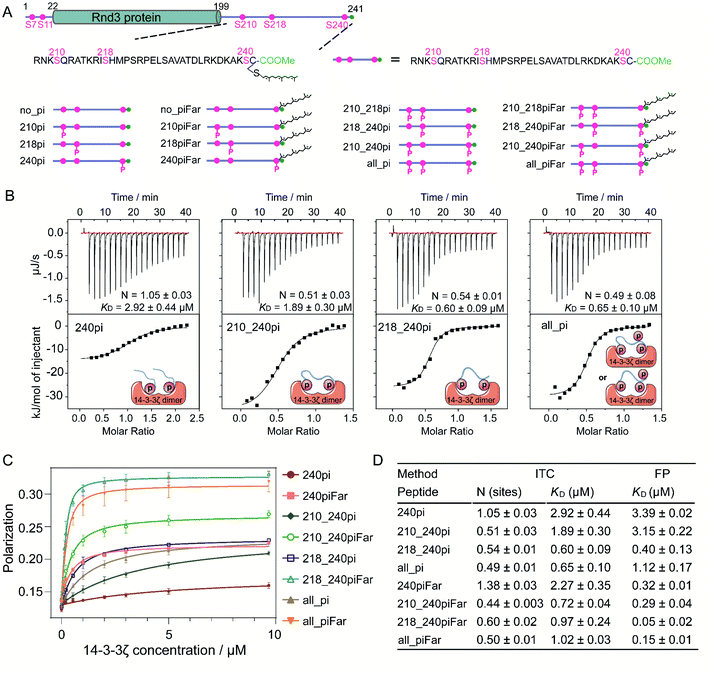
Sixteen 35-mer Rnd3 C-terminal peptides (207–241) with different PTM patterns at S210, S218, S240 and C241 were synthesized following the strategy previously developed35,36 (Fig. 2A). In addition, sixteen fluorescent Rnd3 peptides bearing a fluorescein (FAM) moiety at the N-terminus for fluorescence polarization assays were synthesized (Fig. S4–S6 and Table S1†). The 14-3-3ζ protein fused to the intein–CBD tag was expressed in E. coli cells, purified by using chitin beads and finally cleaved from the tag through thiolysis.37,38 14-3-3 proteins tend to form homodimers or heterodimers.39,40 In a 14-3-3 dimer, the two identical phosphate-binding sites on each monomer are approximately 34 Å apart,41 indicating that the 14-3-3 dimer might interact with one protein bearing two active phosphorylated sites or two proteins each bearing one active phosphorylated site. Accordingly, our native PAGE and matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) results from the generated 14-3-3ζ as well as the SDS-PAGE results of its photocrosslinked product confirmed the dimer state of 14-3-3ζ under physiological conditions (Fig. S10†).Phosphorylation at S210 or S218 enhances the Rnd3–14-3-3 interaction by binding to one pS recognition pocket that is distinct from the gatekeeper residue pS240
To understand the specific roles of multiphosphorylation and farnesylation in the Rnd3–14-3-3ζ interaction at the molecular level, we measured the binding thermodynamics between sixteen Rnd3 C-terminal peptides with different PTM patterns (Fig. 2A) and 14-3-3ζ by using both isothermal titration calorimetry (ITC) and fluorescence polarization (FP) assays. As indicated by the ITC and FP results, among all the phosphorylated Rnd3 peptides (240pi, 210_240pi, 218_240pi and all_pi) as well as the unmodified peptide (no_pi), only peptides containing pS240 displayed measurable binding affinities to 14-3-3ζ (Fig. 2B–D, S11 and S12†), demonstrating that pS240 plays a dominant role in the PPI. Although pS210 and pS218 alone could not mediate stable Rnd3–14-3-3ζ binding, both of these PTMs (Fig. S11 and S12†), particularly pS218, could enhance the interaction to some extent in the presence of pS240, as reflected by the improved binding affinities to 14-3-3ζ of peptides 210_240pi, 218_240pi and all_pi compared to that of 240pi (Fig. 2B–D). These biophysical quantitative data confirmed previously published cellular results regarding different contributions from pS240, pS210 and pS218 to the Rnd3–14-3-3ζ interaction.14Next, we examined the binding modes of the multiphosphorylated Rnd3 peptide with 14-3-3ζ according to the binding stoichiometry measured by ITC (Fig. 2B and D). The singly phosphorylated peptide 240pi binds to 14-3-3ζ with a stoichiometry of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, indicating that each monomer of the 14-3-3ζ dimer interacts with one Rnd3 C-terminal tail bearing the singly phosphorylated S240 residue, which is in accordance with a previous report using a short 10-mer Rnd3 peptide. In contrast, we found for the first time that the doubly phosphorylated peptides 210_240pi and 218_240pi, as well as the triply phosphorylated peptide all_pi, bind to 14-3-3ζ in a stoichiometry of 1
1, indicating that each monomer of the 14-3-3ζ dimer interacts with one Rnd3 C-terminal tail bearing the singly phosphorylated S240 residue, which is in accordance with a previous report using a short 10-mer Rnd3 peptide. In contrast, we found for the first time that the doubly phosphorylated peptides 210_240pi and 218_240pi, as well as the triply phosphorylated peptide all_pi, bind to 14-3-3ζ in a stoichiometry of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (Fig. 2B). These results reveal that one Rnd3 C-terminal tail bearing two or three phosphorylated serines including pS240 interacts with one 14-3-3ζ dimer, as shown in Fig. 2B. As mentioned above, the distance between two phosphate-binding sites on the 14-3-3ζ dimer is approximately 34 Å, which fits the distance between pS210 or pS218 and pS240 on Rnd3 (>34 Å). Moreover, because the two phosphate binding sites on the 14-3-3ζ dimer are far away from each other, we estimated that the binding of the multiphosphorylated Rnd3 C-terminal tail to 14-3-3ζ might follow the additive mechanism, by which these pS epitopes act jointly to mediate binding with functional specificity.27 pS240 functions as a gatekeeper42,43 in this multivalent interaction.
2 (Fig. 2B). These results reveal that one Rnd3 C-terminal tail bearing two or three phosphorylated serines including pS240 interacts with one 14-3-3ζ dimer, as shown in Fig. 2B. As mentioned above, the distance between two phosphate-binding sites on the 14-3-3ζ dimer is approximately 34 Å, which fits the distance between pS210 or pS218 and pS240 on Rnd3 (>34 Å). Moreover, because the two phosphate binding sites on the 14-3-3ζ dimer are far away from each other, we estimated that the binding of the multiphosphorylated Rnd3 C-terminal tail to 14-3-3ζ might follow the additive mechanism, by which these pS epitopes act jointly to mediate binding with functional specificity.27 pS240 functions as a gatekeeper42,43 in this multivalent interaction.
In addition, a complete thermodynamic picture, including the enthalpic and entropic changes due to the interaction, was drawn according to the ITC results.32 The binding of peptide 240pi to 14-3-3ζ is an exothermic and entropy-increasing process reflected by the ΔH and TΔS data (Table 1). However, the changes in enthalpy and entropy upon the binding of peptides 210_240pi, 218_240pi and all_pi to 14-3-3ζ were obviously lower than those of 240pi, indicating that phosphorylation at S210 and S218 induces the binding system to release more heat and become more orderly, which fits the general enthalpic and entropic laws of electrostatic interactions.
| Rnd3 peptide | ΔH (kJ mol−1) | TΔS (kJ mol−1) |
|---|---|---|
| no_piFar | 2.94 ± 0.17 | 28.83 |
| 240pi | −14.79 ± 0.52 | 16.82 |
| 210_240pi | −36.66 ± 1.60 | −3.99 |
| 218_240pi | −26.04 ± 0.61 | 9.48 |
| all_pi | −29.92 ± 0.71 | 5.42 |
| 240piFar | −14.91 ± 0.41 | 17.29 |
| 210_240piFar | −30.76 ± 0.30 | 4.32 |
| 218_240piFar | −23.35 ± 1.24 | 11.00 |
| all_piFar | −26.20 ± 0.11 | 7.28 |
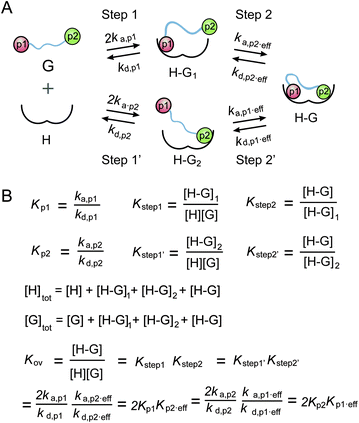
A thermodynamic model was established to describe the multivalent interaction between multiply phosphorylated partner proteins and 14-3-3,32 which was utilized to analyze the multivalent interactions between multiply phosphorylated Rnd3 and 14-3-3ζ (Fig. 3). However, due to the very weak binding affinity to 14-3-3ζ of the pS210 and pS218 epitopes on Rnd3, which could not be detected by either ITC or FP assays, the corresponding affinity constants in the previous equations of the thermodynamic model were lacking. Thus, we accordingly modified the equations by introducing two new parameters for the second binding step as follows: the real effective association rate constant ka,p2,eff and the actual effective affinity constant Kp2,eff. By comparing the calculated Kp2,eff values of 210_240pi and 218_240pi, we found different contributions of pS210 and pS218 to Rnd3–14-3-3ζ binding. K210,eff and K218,eff were determined to be 0.54 and 4.24, respectively. Because K210,eff is much smaller than K218,eff, we deduced that the pS218 site has a greater enhancement effect on the overall binding than the pS210 site.
Taken together, these quantitative thermodynamic results prove that phosphorylation at S210 and S218 within the Rnd3 C-terminal tail can enhance the interaction between Rnd3 bearing the gatekeeper pS240 and 14-3-3ζ. Moreover, pS218 displays a greater enhancement effect than pS210, as reflected by the actual effective affinity constant Kp2,eff in our binding thermodynamic model modified equation. In addition, we determined the binding mode in which one doubly or triply phosphorylated Rnd3 tail containing the essential pS240 binds to a 14-3-3ζ dimer via two remote phosphate binding sites, which might act through a multivalent additive mechanism.
Phosphorylation at S218 induces the deceleration of the dissociation of Rnd3 from 14-3-3ζ
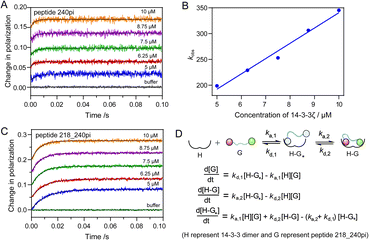
The binding mode of one multiply phosphorylated Rnd3 to a 14-3-3ζ dimer was determined by the thermodynamic studies in the previous section, but the kinetics of the binding process, particularly the recognition of different pS epitopes, remain unclear. Thus, we measured the association rate constant, kon, and dissociation rate constant, koff, of representative Rnd3 peptides (240pi, 218pi, and 218_240pi) labeled with the fluorophore FAM to 14-3-3ζ by using real-time FP detection in a stopped-flow apparatus. The polarization value of the fluorescent peptides increased upon rapid mixing with 14-3-3ζ and reached a plateau after a certain period of time. The time-resolved FP curves of Rnd3 peptide 240pi and 218_240pi with increasing concentrations of 14-3-3ζ are shown in Fig. 4A and C.The real-time FP signal change of peptide 218pi containing pS218 upon the addition of 14-3-3ζ could not be observed, probably due to their weak binding. For peptide 240pi containing only one phosphorylation epitope pS240, we assume that the binding process is a one-step reversible interaction. Each fitting curve yielded the constant kobs (kobs = kon × [14-3-3ζ] + koff) according to pseudo first-order conditions, where [FAM-240pi] ≪ [14-3-3ζ]. The constants kon and koff were found to be 29.63 ± 2.31 μM−1 s−1 and 44.25 ± 17.81 s−1, respectively (Fig. 4A and B and Table 2). The calculated KD (1.49 μM) 240pi based on these rate constants is in reasonable agreement with the KD (3.39 μM) from steady-state titration.
| Rate constants | 240pi | 218_240pi | 240piFar |
|---|---|---|---|
| k a,1 (μM−1 s−1) | 29.63 | 57.46 | 39.21 |
| k d,1 (s−1) | 44.25 | 35.20 | 19.94 |
| k a,2 (s−1) | — | 62.20 | 25.59 |
| k d,2 (s−1) | — | 38.89 | 54.53 |
| K D (μM) | 1.49 | 0.23 | 0.37 |
In addition, the dissociation process can also be detected by a competition assay, in which an excess of unlabeled Rnd3 peptides were used to release the fluorescent Rnd3 peptides bound to 14-3-3ζ. The FP value decreased over time after rapid mixing and reached a steady state (Fig. S15†). The data were fitted to a single exponential equation to give the dissociation rate constant koff. The koff of 240pi obtained from the competitive dissociation assay is 63.92 s−1, which is in reasonable agreement with the koff from the fitting of association curves as mentioned above.
For the peptide 218_240pi containing two remote phosphorylation epitopes, we assumed that the binding process was a two-step reversible interaction. The group of association curves of 218_240pi with different concentrations of 14-3-3ζ (Fig. 4C) was fitted using numerical integration of a set of differential equations describing a two-step binding mechanism (Fig. 4D). First, we should determine which phosphorylation epitope (pS240 or pS218) initiated the binding. We estimate that these two epitopes have less influence on each other in the first binding step and last dissociation step, which thereby displays similar binding thermodynamics and kinetics with 240pi or 218pi to 14-3-3ζ. However, due to the very weak binding affinity of pS218 to 14-3-3ζ, we couldn't measure the binding kinetics data of 218pi for further comparison with the data of 240pi. Thus, we tried two rounds of fitting to obtain the binding kinetic data, in which the association of pS218 or pS240 with 14-3-3ζ was respectively set as the first binding step. The phosphate randomly associates with either of the phosphate binding sites of the 14-3-3 dimer, providing the association rate constant of step one to be ka,1 = 2 × kon (kon,240pi or kon,218pi).
Assuming that pS218 initiates the binding: ka,218pi > ka,240pi, together with KD,218pi ≫ KD,240pi, it can be deduced that: kd,218pi > kd,240pi, which means that the koff of 240pi binding to 14-3-3ζ is the kd,1 in this two-step binding equation (Fig. 4D). The group of association curves of 218_240pi with 14-3-3ζ were then fitted to obtain the corresponding constants: ka,1 = 16.70 μM−1 s−1, kd,1 = 53.76 s−1, ka,2 = 29.84 s−1, and kd,2 = 8.64 s−1 using numerical integration of a set of differential equations as shown in Fig. 4D. However, the calculated ka,1 (16.70 μM−1 s−1), which equals the 2 × kon of 218pi, is much smaller than the 2 × kon of 240pi (59.26 μM−1 s−1), which is contradictory to the prerequisite that pS218 initiates the binding of 218_240pi to 14-3-3ζ.
Next, assuming that pS240 initiates the binding, the 2 × kon of 240pi binding to 14-3-3ζ is the ka,1 in this two-step binding equation. The group of association curves of 218_240pi with 14-3-3ζ was then fitted to obtain the corresponding rate constants using numerical integration of a set of differential equations as shown in Fig. 4D. The fitting curves matched well with the recording curves, giving the rate constants as: ka,1 = 57.46 μM−1 s−1, kd,1 = 35.20 s−1, ka,2 = 66.20 s−1, and kd,2 = 38.89 s−1 (Table 2). The calculated KD and koff for 218_240pi based on these four rate constants are respectively 0.23 μM and 9.69 s−1, which are in reasonable agreement with the previously determined KD (0.40 μM) from steady-state titration and koff (8.42 s−1) from the competitive dissociation assays.
These results demonstrate that phosphorylation at S218 does not accelerate the association but decelerates the dissociation of Rnd3 peptides embedded with the gatekeeper pS240 from 14-3-3ζ, resulting in the enhancement of the binding affinity. We therefore estimated that one monomer of the 14-3-3ζ dimer first recognizes the essential pS240 on Rnd3 in the rate-determining step, which can then facilitate the subsequent association of another monomer with the pS218 or pS210 site because of the proximity effect.44 Furthermore, the second phosphate-involved recognition in the multivalent interaction retards the dissociation of the Rnd3–14-3-3ζ complex, which is in agreement with the properties of electrostatic interactions.
Farnesylation at C241 enhances the binding between Rnd3 and 14-3-3ζ in a cooperative manner with the essential pS240
In addition to multiphosphorylation, Rnd3 contains a constitutive hydrophobic farnesyl group at C241, which not only plays a key role in the association of Rnd3 with the membrane but also largely enhances the interaction between Rnd3 containing pS240 and 14-3-3ζ, as described in previously published work.14 Farnesylation at C241 has been reported to largely improve the binding affinity of a 10-mer Rnd3 peptide containing pS240 to 14-3-3ζ, as indicated by an ITC-determined KD of 69 nM over 1.06 μM. Moreover, structural studies have revealed that a hydrophobic groove close to the phosphate-binding pocket on 14-3-3ζ can accommodate the farnesyl group of the Rnd3 peptide. However, the molecular mechanism by which farnesylation and phosphorylation act cooperatively in the multivalent Rnd3–14-3-3ζ interaction is still not clear. We therefore determined the thermodynamics and kinetics of the binding with adjacent pS240 and fC241.First, all synthesized 35-mer Rnd3 peptides containing a farnesyl group were investigated by ITC and FP assays to obtain the thermodynamic data of binding to 14-3-3ζ. Farnesylated peptide no_piFar without any phosphate modifications displayed a very weak interaction with 14-3-3ζ (Fig. S11 and S12†). In contrast, 240piFar containing both fC241 and pS240 displayed a much higher binding affinity to 14-3-3ζ than no_piFar, confirming the essential role of pS240 in the multivalent PPI involving fC241. Moreover, the binding stoichiometry of 240piFar to 14-3-3ζ is close to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 as was also observed for 240pi, indicating that farnesylation shows little change in the binding mode of a pS240-containing Rnd3 peptide to one 14-3-3ζ monomer via its phosphate-binding pocket. Interestingly, for farnesylated Rnd3 peptides, the KD values determined by the FP assay were smaller than those measured by the ITC experiment. Particularly for 240piFar and 218_240piFar, the difference between the two sets of KD values was close to an order of magnitude. However, for the corresponding peptides without farnesylation, the two sets of KD values were close. Thus, we selected only one set of KD values determined by the same technique for binding affinity comparison. We found that the farnesyl group can largely improve the binding affinities of all the Rnd3 peptides containing pS240, as indicated by the FP KD in Fig. 2C and D and S12.†
1 as was also observed for 240pi, indicating that farnesylation shows little change in the binding mode of a pS240-containing Rnd3 peptide to one 14-3-3ζ monomer via its phosphate-binding pocket. Interestingly, for farnesylated Rnd3 peptides, the KD values determined by the FP assay were smaller than those measured by the ITC experiment. Particularly for 240piFar and 218_240piFar, the difference between the two sets of KD values was close to an order of magnitude. However, for the corresponding peptides without farnesylation, the two sets of KD values were close. Thus, we selected only one set of KD values determined by the same technique for binding affinity comparison. We found that the farnesyl group can largely improve the binding affinities of all the Rnd3 peptides containing pS240, as indicated by the FP KD in Fig. 2C and D and S12.†
Next, we examined the thermodynamic binding modes of Rnd3 peptides containing both farnesylation and double or triple phosphorylation epitopes with 14-3-3ζ. All of the binding stoichiometries of the peptides 210_240piFar, 218_240piFar and all_piFar to 14-3-3ζ were approximately 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, showing the same result as the corresponding peptides without the farnesylation epitope. These results indicate that one 14-3-3ζ dimer can accommodate one Rnd3 peptide containing fC241, pS240 and either pS210 or pS218 via two phosphate-binding sites and one farnesyl binding site (Fig. S11†). In addition, after comparing the ΔH and TΔS data of all the farnesylated peptides with the data of the corresponding nonfarnesylated peptides (Table 1), we found that farnesylation leads to a decrease in ΔH except for with the peptide 240piFar, which displays less of a change, and an increase in TΔS, which is in contrast to the effects of phosphorylation at S210 or S218 on ΔH and TΔS described earlier. These data indicate the occurrence of hydrophobic interactions between Rnd3 and 14-3-3ζ. The hydrophobic surface was surrounded by water molecules, which formed an ice-like cage structure called a clathrate cage.45 The surface of the cage is covered with ordered hydrogen bonds, which were broken by the interaction between the farnesyl group and the hydrophobic groove on 14-3-3ζ. The resulting release of free water molecules leads to an increase in the entropy of the system.46
2, showing the same result as the corresponding peptides without the farnesylation epitope. These results indicate that one 14-3-3ζ dimer can accommodate one Rnd3 peptide containing fC241, pS240 and either pS210 or pS218 via two phosphate-binding sites and one farnesyl binding site (Fig. S11†). In addition, after comparing the ΔH and TΔS data of all the farnesylated peptides with the data of the corresponding nonfarnesylated peptides (Table 1), we found that farnesylation leads to a decrease in ΔH except for with the peptide 240piFar, which displays less of a change, and an increase in TΔS, which is in contrast to the effects of phosphorylation at S210 or S218 on ΔH and TΔS described earlier. These data indicate the occurrence of hydrophobic interactions between Rnd3 and 14-3-3ζ. The hydrophobic surface was surrounded by water molecules, which formed an ice-like cage structure called a clathrate cage.45 The surface of the cage is covered with ordered hydrogen bonds, which were broken by the interaction between the farnesyl group and the hydrophobic groove on 14-3-3ζ. The resulting release of free water molecules leads to an increase in the entropy of the system.46
Thus, we have revealed that the hydrophobic farnesyl group on Rnd3, which displays weak binding to 14-3-3ζ, largely enhances the binding of Rnd3 to 14-3-3ζ at a stoichiometry of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in combination with the adjacent essential pS240. Therefore, we estimated the occurrence of great cooperativity between fC241 and pS240 in this multivalent PPI. To address this issue, we tried to establish a thermodynamic model for the Rnd3–14-3-3ζ interaction mediated by the potential cooperativity of fC241 and pS240. As mentioned earlier, we built a modified thermodynamic binding equation to describe the PPI mediated by the two ditopic pS residues. Herein, we took the multivalent PPI mainly mediated by adjacent fC241 and pS240 as a host–guest system but introduced the effective factor coefficient eff and the index of cooperativity ε, which is similar to the effective molarity term (EM),32 as shown in Fig. 5. effa,xx represents the degree to which the association process in the second binding step is affected by the association in the first binding step and effd,xx represents the degree to which the dissociation process in the second binding step is affected by the first binding event. According to the equation Kmonomer = εKp1KF in Fig. 5, where Kmonomer is the association constants of 240piFar (K240piFar = 3.13 μM−1), Kp1 is the association constants of 240pi (K240pi = 0.29 μM−1), and KF is the association constants of no_piFar (K240Far = 0.13 μM−1) (Fig. 6A, FP data), we calculated the ε value of this binding as 83.02 μM, which is much larger than 1, indicating great cooperativity between these two adjacent modifications in the multivalent PPI.
1 in combination with the adjacent essential pS240. Therefore, we estimated the occurrence of great cooperativity between fC241 and pS240 in this multivalent PPI. To address this issue, we tried to establish a thermodynamic model for the Rnd3–14-3-3ζ interaction mediated by the potential cooperativity of fC241 and pS240. As mentioned earlier, we built a modified thermodynamic binding equation to describe the PPI mediated by the two ditopic pS residues. Herein, we took the multivalent PPI mainly mediated by adjacent fC241 and pS240 as a host–guest system but introduced the effective factor coefficient eff and the index of cooperativity ε, which is similar to the effective molarity term (EM),32 as shown in Fig. 5. effa,xx represents the degree to which the association process in the second binding step is affected by the association in the first binding step and effd,xx represents the degree to which the dissociation process in the second binding step is affected by the first binding event. According to the equation Kmonomer = εKp1KF in Fig. 5, where Kmonomer is the association constants of 240piFar (K240piFar = 3.13 μM−1), Kp1 is the association constants of 240pi (K240pi = 0.29 μM−1), and KF is the association constants of no_piFar (K240Far = 0.13 μM−1) (Fig. 6A, FP data), we calculated the ε value of this binding as 83.02 μM, which is much larger than 1, indicating great cooperativity between these two adjacent modifications in the multivalent PPI.
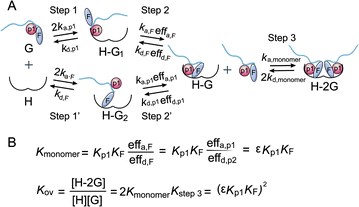 | ||
| Fig. 5 (A) Cooperative binding scheme of phosphorylated and farnesylated Rnd3 peptides with 14-3-3ζ. The binding process is divided into two steps. Guest (G) represents the Rnd3 peptides and host (H) represents 14-3-3ζ. ka,p1 is the association rate constant of the peptide bearing the phosphorylation epitope at S240 with 14-3-3ζ. kd,p1 is the dissociation rate constant of the peptide bearing the phosphorylation epitope at S240 with 14-3-3ζ. effa,xx is the degree to which the association process in the second binding step is affected by the association in the first binding step and effd,xx is the degree to which the dissociation process in the second binding step is affected by the first binding event. p1 in orange represents pS240 in the Rnd3 peptide and F in gray represents the farnesyl group in the Rnd3 peptide. (B) Mass action laws and mass balance equations for the interaction. K is the affinity constant. The kinetic parameters and equation derivation of the first two steps are shown in Fig. S16.† | ||
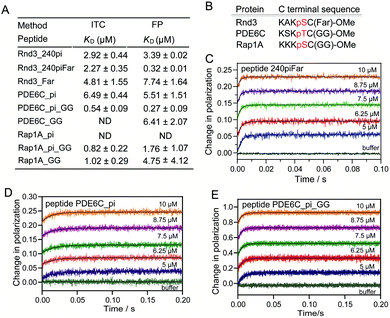
Besides, we explored the interaction of 14-3-3ζ with its other potential substrates from the C-terminus of Rap1A and PDE6C, which have a similar sequence and modifications to the C-terminus of Rnd3 (-K-X-K-pS/pT-fC/gC-OMe) as shown in Fig. 6B and Table S1.† Both Rap1A and PDE6C have geranylgeranylation epitopes instead of farnesylation epitopes on the C-terminal Cys. These Rap1A and PDE6C peptides with different phosphorylation and lipidation patterns were further applied in the measurements of their binding affinities to 14-3-3ζ by ITC and FP assays. The results indicate that Rap1A peptide bearing both phosphate and lipid (Rap1A_pi_GG) display a relatively weaker binding affinity (KD = 1.76 μM) to 14-3-3ζ while PDE6C_pi_GG shows a stronger binding affinity (KD = 0.27 μM) as shown in Fig. 6A. We estimate that the local sequence anchored with the phosphate epitope might lead to their different binding affinities. The ITC and FP results of a series of Rap1A and PDE6C peptides with different PTM patterns show that the lipid group greatly enhances the binding affinities of phosphorylated PDE6C peptides to 14-3-3ζ. According to the equation Kmonomer = εKp1KF in Fig. 5, where Kmonomer is the association constants of PDE6C_pi_GG (3.70 μM−1), Kp1 is the association constants of PDE6C_pi (0.18 μM−1), and KF is the association constants of PDE6C_GG (0.16 μM−1), we calculated the ε value of this binding as 128.47 μM. These results indicate great cooperativity between phosphorylation and geranylgeranylation in the multivalent PPI of PDE6C and 14-3-3ζ, which is in agreement with the Rnd3–14-3-3ζ interaction.
In brief, these quantitative thermodynamic results prove that farnesylation at C241 of the Rnd3 C-terminal tail can enhance the interaction between 14-3-3ζ and Rnd3 mediated by phosphorylated S240. The binding mode is that one Rnd3 peptide containing fC241, pS240, and pS210 or pS218 binds a 14-3-3ζ dimer via two separate phosphate-binding sites and one farnesyl binding groove. Moreover, fC241 mainly introduces hydrophobic interactions and acts cooperatively with the adjacent essential pS240 for the multivalent PPI. Moreover, we found that 14-3-3ζ can also bind to the C-terminus of Rap1A and PDE6C bearing similar lipid and phosphate modifications with Rnd3 via a cooperative mechanism.
Farnesylation at C241 greatly increases the association rate and reduces the dissociation rate of Rnd3 bearing pS240 to 14-3-3ζ
In the multiphosphorylation-involved Rnd3–14-3-3ζ interaction, the pS240-binding pocket on one 14-3-3ζ monomer and the remote pS210- or p218-binding pocket on the other monomer are far apart and probably hardly mutually influence. Indeed, pS210 and p218 enhance the binding affinities of Rnd3 to 14-3-3ζ mainly by decelerating the dissociation rate while maintaining the association rate. In contrast, fC241 is adjacent to pS240, which provides the possibility for these two kinds of modifications to mutually affect Rnd3 binding to 14-3-3ζ. Thus, in the following work, we first determined the binding kinetics of farnesyl-involved Rnd3–14-3-3ζ multivalent PPIs to further reveal the role of farnesylation in the binding process. Following a similar method mentioned earlier, different farnesylated and phosphorylated Rnd3 peptides with or without the fluorophore FAM were used to determine their rate constants of binding to 14-3-3ζ by using real-time FP detection in a stopped-flow apparatus.For Rnd3 peptide 240piFar containing adjacent phosphorylation and farnesylation epitopes, we assumed that the binding process was a two-step reversible interaction, too. The group of association curves of 240piFar with different concentrations of 14-3-3ζ (Fig. 6C) was fitted using numerical integration of a set of differential equations describing a two-step binding mechanism (Fig. S17†). Different from 218_240pi on which two remote phosphorylation epitopes respectively bind to each monomer of a 14-3-3ζ dimer, these two adjacent epitopes bind to the same 14-3-3ζ monomer.
Due to the relatively weak binding affinity of fC241 to 14-3-3ζ, we couldn't measure the binding kinetics data of no_piFar for direct comparison with the data of 240_pi to deduce which epitope initiates the binding process. Thus, we tried two rounds of fitting to obtain the binding kinetic data, in which the association of p240 or fC241 with 14-3-3ζ was respectively set as the first binding step.
Assuming that p240 initiates the binding (kon,240pi > kon,Far), we used the kon of 240pi binding to 14-3-3ζ as the ka,1 in this two-step binding equation. The group of association curves of 240piFar with 14-3-3ζ was then fitted to obtain the corresponding rate constants kd,1, ka,2, and kd,2 using numerical integration of a set of differential equations as shown in Fig. S17.† The fitted curves matched well with the recorded curves and gave the rate constants as: ka,1 = 31.07 μM−1 s−1, kd,1 = 12.97 s−1, ka,2 = 60.17 s−1, and kd,2 = 344.64 s−1. kd,2 is much larger than the koff of 240pi (44.25 s−1), indicating that kd,2 represents the koff of no_piFar. However, KD,240pi is slightly smaller than KD,Far (Fig. 6A), according to the equation KD = koff/kon, then we calculated kon,Far > kon,240pi, which is contradictory to the prerequisite that pS240 initiates the binding of 240piFar to 14-3-3ζ. Next, we assumed that fC241 initiated the binding. Since the kon of no_piFar binding to 14-3-3ζ cannot be determined, we set ka,1, kd,1, ka,2, and kd,2 as unknown constants. The fitted curves matched well with the recorded curves. The rate constants were determined: ka,1 = 39.21 μM−1 s−1, kd,1 = 19.94 s−1, ka,2 = 25.59 s−1, and kd,2 = 54.53 s−1 (Fig. 6C and Table 2). The calculated KD and koff for 240piFar based on these four rate constants are respectively 0.37 μM and 10.87 s−1, which are in reasonable agreement with the previously determined KD (0.32 μM) from steady-state titration and koff (8.02 s−1) from the competitive dissociation assays.
Based on these results, we conclude that farnesylation at C241 accelerates the association of Rnd3 peptides embedded with the pS240 with 14-3-3ζ and impeded the release of the Rnd3 peptide from the 14-3-3ζ. However, as mentioned above, the farnesylated Rnd3 peptides without the essential pS240 displayed weak interactions with 14-3-3ζ. We estimate that the farnesyl group at C241 and the phosphate group at S240 might mutually affect the conformation of two adjacent binding sites to achieve optimal binding, which is further explored and is described in the next section.
Besides, we measured the binding kinetics of PDE6C peptides with different modification patterns (Fig. 6D and E) by using a stopped-flow apparatus coupled with an FP detector. The fitting of the association curves PDE6C_pi with increasing concentration of 14-3-3ζ followed the one-step binding model, resulting in kon and koff of 4.06 ± 0.22 μM−1 s−1 and 33.29 ± 1.70 s−1. For the peptide PDE6C_pi_GG, we assumed that the binding process was a two-step reversible interaction and we also tried two rounds of fitting to obtain the binding kinetic data, in which the association of the phosphate group or geranylgeranyl group with 14-3-3ζ was respectively set as the first binding step. Assuming that the phosphate group initiated the binding (kon,PDE6C_pi > kon,PDE6C_GG), we used the kon of PDE6C_pi binding to 14-3-3ζ as the ka,1 in this two-step binding equations. The fitting result gave unreasonable parameters r1 ≈ 1, r0 ≈ 0, r2 ≈ 0 (eqn (S8)†), so we estimated that it was not the phosphate of PDE6C which initiates the binding. Next, we assumed that the geranylgeranyl group initiated the binding. Since the kon of PDEC6_GG binding to 14-3-3ζ cannot be determined, we set ka,1, kd,1, ka,2, and kd,2 as unknown constants. The fitted curves matched well with the recorded curves and the rate constants were determined: ka,1 = 23.71 μM−1 s−1, kd,1 = 15.29 s−1, ka,2 = 63.13 s−1, and kd,2 = 51.49 s−1. The calculated KD for PDE6C_pi_GG based on these four rate constants are 0.50 μM, which is in reasonable agreement with the previously determined KD (0.27 μM) from steady-state titration. These kinetic results demonstrate that the established binding kinetic models are suitable for evaluation of the interaction of 14-3-3ζ with other substrates bearing lipid and phosphate modifications.
The cooperativity between adjacent farnesylation and phosphorylation might result from induced fit effects
In earlier work, we revealed the influence of farnesylation on the binding thermodynamics and kinetics of the Rnd3–14-3-3ζ multivalent interaction. To further explore the molecular mechanism by which farnesylation at C241 and the adjacent essential phosphorylation at S240 cooperatively mediate the tight multivalent PPI, the interactions of the Rnd3 peptides no_piFar, 240pi and 240piFar with 14-3-3ζ were investigated by molecular dynamics (MD) simulations using GROMACS47 based on the reported crystallographic structure of 14-3-3ζ bound to a 10-mer Rnd3 peptide with a free carboxylic acid end from the Protein Data Bank (PDB code: 4bg6) and the structures of three Rnd3 peptides with the native carboxylic methyl ester that were built in PyMOL. The MD simulation time of each complex was set to 100 ns. We collected the most frequently occurring structures for further analysis. The structure of the complex of 240piFar and 14-3-3ζ from our MD simulation results was in general agreement with the previously reported crystallographic structure of a similar complex14 (Fig. S18†), proving the plausibility of the MD simulation results.We evaluated the binding affinity of each complex by analyzing their binding free energies from the simulation results (Fig. S19†), which were in accordance with the experimental results that the 240piFar complex displays the strongest binding while the no_piFar complex is the most unstable. Then, we analyzed the changes in the root-mean-square deviation (RMSD) of the structure of 14-3-3ζ in each complex and the root-mean-square-fluctuation (RMSF) of its residues. The overall RMSD of 14-3-3ζ in complexation with 240piFar displays the slightest changes among the three complexes (Fig. S20†). Moreover, the RMSF data show that the residues on 14-3-3ζ in complexation with 240piFar display minimal fluctuations (Fig. S21†), which is consistent with the RMSD results, indicating that the 240piFar complex bound to 14-3-3ζ displays the most stable structure among the three complexes. In addition, we analyzed the structural changes in different Rnd3 peptides in each complex. The results indicated that peptide no_piFar in the complex hardly achieved a stable structure within the MD simulations. Altogether, these MD results demonstrate that both farnesylation at C241 and phosphorylation at S240 are beneficial to the binding affinity.
To uncover the molecular mechanism of cooperativity between adjacent farnesylation and phosphorylation in the multivalent interaction, we further analyzed the conformational changes in the Rnd3–14-3-3ζ binding surfaces in each complex.
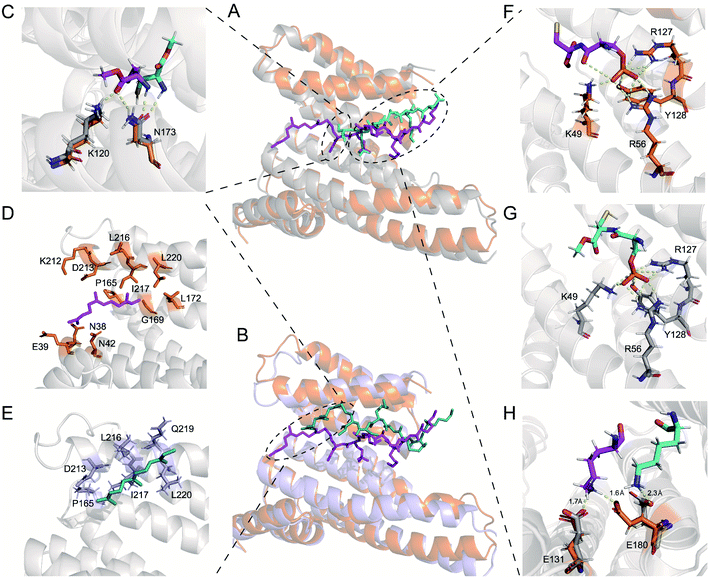
First, we compared specific movements of the key residues of 14-3-3ζ due to its association with 240pi or 240piFar. In the phosphate-binding pocket of 14-3-3ζ, the orientation of some residues changed slightly (Fig. 7A, F and G). For example, the amino group of the side chain of K49, which interacts with the oxygen atom of the phosphate group of 240pi, moves slightly away from the phosphate group of 240piFar. In addition, the phosphate group in 240piFar displays a polar interaction with the amide bond within its own mainchain (this interaction is replaced in the crystallographic structure by the binding of phosphate to the C-terminal free carboxyl group), while there is no such internal interaction for 240pi in complexation with 14-3-3ζ. Therefore, we further compared the conformational changes of the Rnd3 peptides in each complex. K235 in 240pi exhibits a polar interaction with D231 of 14-3-3ζ, while K235 in 240piFar interacts with both the C-terminal carboxyl ester and the side chain carboxyl group of D231, allowing D231 to stretch toward the cavity (Fig. S18†). D236 in the 240piFar peptide has a strong polar interaction with R60 of 14-3-3ζ at a distance of 1.7 Å, whereas D236 of 240pi faces the other side of 14-3-3ζ, displaying a distance of 8.3 Å to R60 (Fig. S18†). K237 in 240piFar interacts with both E131 and E180 of 14-3-3ζ at respective distances of 1.7 Å and 1.6 Å, while K237 of 240pi interacts only with E180 at a distance of 2.3 Å (Fig. 7H). In addition, the orientation of C241 in 240piFar was also flipped, with its methyl ester facing K120 and N173 of 14-3-3ζ, resulting in the formation of hydrogen bonds, while the methyl ester of C241on 240pi is far from K120 and N173 (Fig. 7C). These results strongly supported the fact that farnesylation induces conformational changes in the binding surfaces between Rnd3 and pS240 embedded and 14-3-3ζ to enhance their interaction.
Next, we explored the effects of S240 phosphorylation on the farnesyl binding surface by comparing the structures of the no_piFar complex and 240piFar. Whereas no_piFar constantly changed its binding state to 14-3-3ζ during the simulation and hardly reached a stable conformation, which was probably due to the lack of the essential pS240 on no_piFar, a weak interaction with 14-3-3ζ was observed (Fig. S25†). Therefore, we took out the structure that occurred with the highest frequency in the simulation for analysis (Fig. 7B). The results showed that the farnesyl group still binds to the hydrophobic groove on 14-3-3ζ but shifts away from the locking sites (Fig. 7D and E). These results demonstrated that phosphorylation at pS240 also affects the conformation of the binding surface between farnesylated Rnd3 and 14-3-3ζ and helps to lock the farnesyl group in the right binding position to 14-3-3ζ.
Altogether, the MD simulation results corroborate that farnesylation can enhance the binding between 14-3-3ζ and Rnd3 bearing pS240. Moreover, the conformational analysis demonstrated the cooperativity of the adjacent farnesylation and phosphorylation PTMs in the multivalent interaction, probably through the induced fit mode. The farnesyl group in Rnd3 peptides here acts as the kinetic stabilizer48,49 through inducing conformational changes of the Rnd3–14-3-3ζ binding surfaces to reach the optimal fitting mode.
Conclusions
In this article, we illustrated the mechanistic effects of different phosphorylation and farnesylation patterns on the binding thermodynamics and kinetics of Rnd3 to 14-3-3ζ by using the C-terminal domain of Rnd3 with different PTM patterns and the 14-3-3ζ protein. pS240 was verified to act as the gatekeeper for the binding between Rnd3–14-3-3ζ. Moreover, the critical auxiliary epitopes pS210, pS218 and fC241 are proved to enhance this multivalent binding in the presence of pS240, while the mechanisms of affinity enhancement by additional phosphorylation and farnesylation modifications are greatly different. For the binding of multiphosphorylated Rnd3 peptides to 14-3-3ζ, pS240-mediated association is the first and rate-determining step, while remote pS210 or pS218 does not accelerate the binding process but does slow dissociation to improve the affinity. However, for Rnd3 peptides modified by adjacent phosphorylation and farnesylation groups, fC241 initiates the first step of binding, functioning as a kinetic stabilizer, which stimulates the subsequent association of pS240 with 14-3-3ζ via pulling it into the adjacent phosphate-binding cavity, leading to great enhancement of affinity. Moreover, the MD stimulation results demonstrated the cooperativity of adjacent farnesylation and phosphorylation groups in the multivalent Rnd3–14-3-3ζ interaction through mutually affecting the conformation of two binding sites. According to the mechanisms of these binding processes, we described two binding models for the Rnd3–14-3-3ζ PPIs mediated by distinct multi-PTM patterns: gatekeeper-triggered additive binding and farnesyl group triggered “induce-fit” cooperative binding. In these models, the parameters ε and kp,eff were introduced. By comparing these parameters in the same binding model, we can reveal the degree to which the different binding epitopes participate in the binding process. Moreover, we explored both binding thermodynamics and kinetics of 14-3-3ζ to its other substrates from Rap1A and PDE6C driven by adjacent phosphorylation and lipidation, which follow the cooperative mechanism, too.Altogether, this work not only provides a new insight into the binding mechanism of 14-3-3ζ to Rnd3 as well as other substrates bearing lipid and phosphate modifications, but also establishes general models for exploring the binding thermodynamics and particularly kinetics of other multivalent PPIs mediated by remote or adjacent PTM epitopes, even providing a reference for analyzing other types of multivalent host–guest interactions. Besides, uncovering the mechanism of multivalent PPIs is of critical importance for further developing thermodynamic and kinetic inhibitors or stabilizers of PPIs to regulate binding processes.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (91753122, 21672125) and the National Key R&D Program of China (2018YFA0507600). We thank Luo SZ and Chen JL from Beijing University of Chemical Technology for help in MD simulation.Notes and references
- D. E. Scott, A. R. Bayly, C. Abell and J. Skidmore, Nat. Rev. Drug Discovery, 2016, 15, 533–550 CrossRef CAS.
- M. Mammen, S.-K. Choi and G. M. Whitesides, Angew. Chem., Int. Ed., 1998, 37, 2754–2794 CrossRef.
- C. Fasting, C. A. Schalley, M. Weber, O. Seitz, S. Hecht, B. Koksch, J. Dernedde, C. Graf, E. W. Knapp and R. Haag, Angew. Chem., Int. Ed. Engl., 2012, 51, 10472–10498 CrossRef CAS.
- P. E. Wright and H. J. Dyson, Nat. Rev. Mol. Cell Biol., 2015, 16, 18–29 CrossRef CAS.
- A. Bah and J. D. Forman-Kay, J. Biol. Chem., 2016, 291, 6696–6705 CrossRef CAS.
- J. Weng and W. Wang, Curr. Opin. Struct. Biol., 2020, 62, 9–13 CrossRef CAS.
- S. F. Banani, H. O. Lee, A. A. Hyman and M. K. Rosen, Nat. Rev. Mol. Cell Biol., 2017, 18, 285–298 CrossRef CAS.
- L. G. Milroy, T. N. Grossmann, S. Hennig, L. Brunsveld and C. Ottmann, Chem. Rev., 2014, 114, 4695–4748 CrossRef CAS.
- J. Bosch, Drug Discovery Today: Technol., 2017, 24, 3–9 CrossRef.
- J. Ohkanda, Chem. Rec., 2013, 13, 561–575 CrossRef CAS.
- H. Lu, Q. Zhou, J. He, Z. Jiang, C. Peng, R. Tong and J. Shi, Signal Transduction Targeted Ther., 2020, 5, 213–235 CrossRef.
- Y. Cau, D. Valensin, M. Mori, S. Draghi and M. Botta, Curr. Med. Chem., 2018, 25, 5–21 CrossRef CAS.
- C. A. Toleman, M. A. Schumacher, S. H. Yu, W. Zeng, N. J. Cox, T. J. Smith, E. J. Soderblom, A. M. Wands, J. J. Kohler and M. Boyce, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 5956–5961 CrossRef CAS.
- P. Riou, S. Kjaer, R. Garg, A. Purkiss, R. George, R. J. Cain, G. Bineva, N. Reymond, B. McColl, A. J. Thompson, N. O'Reilly, N. Q. McDonald, P. J. Parker and A. J. Ridley, Cell, 2013, 153, 640–653 CrossRef CAS.
- T. Obsil and V. Obsilova, Semin. Cell Dev. Biol., 2011, 22, 663–672 CrossRef CAS.
- K. Aoki, F. Maeda, T. Nagasako, Y. Mochizuki, S. Uchida and J. Ikenouchi, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 1863–1871 CrossRef.
- R. G. Hodge and A. J. Ridley, Nat. Rev. Mol. Cell Biol., 2016, 17, 496–510 CrossRef CAS.
- L. Paysan, L. Piquet, F. Saltel and V. Moreau, Mol. Cancer Res., 2016, 14, 1033–1044 CrossRef CAS.
- P. Riou, P. Villalonga and A. J. Ridley, Bioessays, 2010, 32, 986–992 CrossRef CAS.
- P. J. Roberts, N. Mitin, P. J. Keller, E. J. Chenette, J. P. Madigan, R. O. Currin, A. D. Cox, O. Wilson, P. Kirschmeier and C. J. Der, J. Biol. Chem., 2008, 283, 25150–25163 CrossRef CAS.
- K. Riento, N. Totty, P. Villalonga, R. Garg, R. Guasch and A. J. Ridley, EMBO J., 2005, 24, 1170–1180 CrossRef CAS.
- J. P. Madigan, B. O. Bodemann, D. C. Brady, B. J. Dewar, P. J. Keller, M. Leitges, M. R. Philips, A. J. Ridley, C. J. Der and A. D. Cox, Biochem. J., 2009, 424, 153–161 CrossRef CAS.
- L. B. Case, X. Zhang, J. A. Ditlev and M. K. Rosen, Science, 2019, 363, 1093–1097 CrossRef CAS.
- G. Lasso, S. V. Mayer, E. R. Winkelmann, T. Chu, O. Elliot, J. A. Patino-Galindo, K. Park, R. Rabadan, B. Honig and S. D. Shapira, Cell, 2019, 178, 1526–1541 CrossRef CAS.
- D. E. Koshland Jr., Angew. Chem., Int. Ed. Engl., 1995, 33, 2375–2378 CrossRef.
- C. M. Hong, D. M. Kaphan, R. G. Bergman, K. N. Raymond and F. D. Toste, J. Am. Chem. Soc., 2017, 139, 8013–8021 CrossRef CAS.
- A. Gao, K. Shrinivas, P. Lepeudry, H. I. Suzuki, P. A. Sharp and A. K. Chakraborty, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E11053–E11060 CrossRef CAS.
- J. D. Badjic, A. Nelson, S. J. Cantrill, W. B. Turnbull and J. F. Stoddart, Acc. Chem. Res., 2005, 38, 723–732 CrossRef CAS.
- J. Huskens, A. Mulder, T. Auletta, C. A. Nijhuis, M. J. Ludden and D. N. Reinhoudt, J. Am. Chem. Soc., 2004, 126, 6784–6797 CrossRef CAS.
- L. M. Stevers, C. V. Lam, S. F. R. Leysen, F. A. Meijer, D. S. van Scheppingen, R. M. J. M. de Vries, G. W. Carlile, L. G. Milroy, D. Y. Thomas, L. Brunsveld and C. Ottmann, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 1152–1161 CrossRef.
- D. J. Kast and R. Dominguez, Nat. Commun., 2019, 10, 483 CrossRef CAS.
- L. M. Stevers, P. J. de Vink, C. Ottmann, J. Huskens and L. Brunsveld, J. Am. Chem. Soc., 2018, 140, 14498–14510 CrossRef CAS.
- S. Connelly, S. Choi, S. M. Johnson, J. W. Kelly and I. A. Wilson, Curr. Opin. Struct. Biol., 2010, 20, 54–62 CrossRef CAS.
- A. Ballone, F. Centorrino and C. Ottmann, Molecules, 2018, 23, 1386–1399 CrossRef.
- V. Diaz-Rodriguez, D. G. Mullen, E. Ganusova, J. M. Becker and M. D. Distefano, Org. Lett., 2012, 14, 5648–5651 CrossRef CAS.
- F.-Y. Li, Z.-F. Zhang, S. Voss, Y.-W. Wu, Y.-F. Zhao, Y.-M. Li and Y.-X. Chen, Chem. Sci., 2020, 11, 826–832 RSC.
- Z. P. Gates, J. R. Stephan, D. J. Lee and S. B. Kent, Chem. Commun., 2013, 49, 786–788 RSC.
- S. Batjargal, C. R. Walters and E. J. Petersson, J. Am. Chem. Soc., 2015, 137, 1734–1737 CrossRef CAS.
- Y. M. Gu, Y. H. Jin, J. K. Choi, K. H. Baek, C. Y. Yeo and K. Y. Lee, FEBS Lett., 2006, 580, 305–310 CrossRef CAS.
- X. Yang, W. H. Lee, F. Sobott, E. Papagrigoriou, C. V. Robinson, J. G. Grossmann, M. Sundstrom, D. A. Doyle and J. M. Elkins, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 17237–17242 CrossRef CAS.
- C. Johnson, S. Crowther, M. J. Stafford, D. G. Campbell, R. Toth and C. MacKintosh, Biochem. J., 2010, 427, 69–78 CrossRef CAS.
- M. B. Yaffe, FEBS Lett., 2002, 513, 53–57 CrossRef CAS.
- B. Kostelecky, A. T. Saurin, A. Purkiss, P. J. Parker and N. Q. McDonald, EMBO Rep., 2009, 10, 983–989 CrossRef CAS.
- W. J. Errington, B. Bruncsics and C. A. Sarkar, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 25659–25667 CrossRef CAS.
- K. A. Dill, T. M. Truskett, V. Vlachy and B. Hribar-Lee, Annu. Rev. Biophys. Biomol. Struct., 2005, 34, 173–199 CrossRef CAS.
- P. Atkins and J. De Paula, Physical Chemistry, Oxford University Press, 2011 Search PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, SoftwareX, 2015, 1–2, 19–25 CrossRef.
- P. J. de Vink, S. A. Andrei, Y. Higuchi, C. Ottmann, L. G. Milroy and L. Brunsveld, Chem. Sci., 2019, 10, 2869–2874 RSC.
- E. Sijbesma, K. K. Hallenbeck, S. Leysen, P. J. de Vink, L. Skora, W. Jahnke, L. Brunsveld, M. R. Arkin and C. Ottmann, J. Am. Chem. Soc., 2019, 141, 3524–3531 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: LC-MS of peptides, experimental details and figures. See DOI: 10.1039/d0sc05838f |
| This journal is © The Royal Society of Chemistry 2021 |