Open Access Article
Open Access ArticleDiversified strategy for the synthesis of DNA-encoded oxindole libraries†
Xuan
Wang
ab,
Jiaxiang
Liu
a,
Ziqin
Yan
a,
Xiaohong
Liu
cde,
Sixiu
Liu
a,
Yanrui
Suo
a,
Weiwei
Lu
a,
Jinfeng
Yue
a,
Kaixian
Chen
cde,
Hualiang
Jiang
cde,
Yujun
Zhao
 *a,
Mingyue
Zheng
*a,
Mingyue
Zheng
 *de,
Dongcheng
Dai
*de,
Dongcheng
Dai
 *b and
Xiaojie
Lu
*b and
Xiaojie
Lu
 *a
*a
aState Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 501 Haike Road, Zhang Jiang Hi-Tech Park, Pudong, Shanghai 201203, P. R. China. E-mail: xjlu@simm.ac.cn
bAmgen Asia R&D Center, Amgen Biopharmaceutical R&D (Shanghai) Co., Ltd., 4560 Jinke Road, Building No. 2, 13th Floor, Pudong, Shanghai 201210, P. R. China
cShanghai Institute for Advanced Immunochemical Studies, School of Life Science and Technology, ShanghaiTech University, Pudong, Shanghai 201210, P. R. China
dDrug Discovery and Design Center, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 555 Zuchongzhi Road, Shanghai 201203, P. R. China
eUniversity of Chinese Academy of Sciences, No. 19A Yuquan Road, Beijing 100049, P. R. China
First published on 4th January 2021
Abstract
DNA-encoded library technology (DELT) employs DNA as a barcode to track the sequence of chemical reactions and enables the design and synthesis of libraries with billions of small molecules through combinatorial expansion. This powerful technology platform has been successfully demonstrated for hit identification and target validation for many types of diseases. As a highly integrated technology platform, DEL is capable of accelerating the translation of synthetic chemistry by using on-DNA compatible reactions or off-DNA scaffold synthesis. Herein, we report the development of a series of novel on-DNA transformations based on oxindole scaffolds for the design and synthesis of diversity-oriented DNA-encoded libraries for screening. Specifically, we have developed 1,3-dipolar cyclizations, cyclopropanations, ring-opening of reactions of aziridines and Claisen–Schmidt condensations to construct diverse oxindole derivatives. The majority of these transformations enable a diversity-oriented synthesis of DNA-encoded oxindole libraries which have been used in the successful hit identification for three protein targets. We have demonstrated that a diversified strategy for DEL synthesis could accelerate the application of synthetic chemistry for drug discovery.
Introduction
The advances in molecular biology, in particular genomic sciences, has had a deep impact on the identification of potential disease-related protein targets in drug discovery.1 Medicinal chemistry programs mostly rely on screening compound collections populated by a range of molecules derived from a set of known and robust chemical reactions. Based on the comprehensive analysis conducted by Dean G. Brown, high-throughput screening dominates early drug discovery, in conjunction with fragment-based screening and knowledge directed screens.2 These lead generation strategies have successfully identified potential hit compounds, however the limited chemical space3 and high operation cost4 restricts the hit discovery for multiple protein targets and limits the scope of application in small biotech startups and university laboratories due to prohibitive costs. It was through continued innovation by chemists that led to the introduction of DNA-encoded library technology pioneered in 1992 by Brenner and Lerner5 and has become a vital tool in the current investigational repertoire.As an interdisciplinary technology, the integration of combinatorial chemistry, molecular biology, next generation DNA-sequencing and informatic analysis shaped the scene of the DNA-encoded library technology platform and brought it to a new level with larger compound collections, reduced operation costs and screening time. Barcoded small molecules translated from phage display6 with a small molecule warhead (phenotype) individually coupled to encoding DNA tags (genotype) serve to retrace the synthetic history. The encoded compound libraries were constructed using a combinatorial strategy followed by successive iterative steps of both chemical transformation and oligonucleotide ligation.7 DNA barcodes have facilitated affinity selection8 to interrogate vast numbers of compounds in a single pool, exceeding by several orders of magnitude the capacity of the traditional “one-compound per well” screening approach. The approach has dramatically decreased the cost and cycle times, and will become a general technology broadly applied to industry and academia alike.8,9
Nevertheless, every new technology has strengths and weaknesses and DELT is no exception. The encoded compound collections are predominantly composed of limited scaffolds, focused on peptides and aromatic rings.10 One reason for this limitation is that the scope of chemical reactions applicable in solution-phase DEL libraries are constrained to robust reactions that preserve the integrity of the oligonucleotides; few new synthetic methodologies can be applied to DEL library construction.11 To preserve the integrity of the DNA barcodes, chemists have continued to expand the scope of mild chemical transformations and DNA-protecting strategies (solid-phase DEL,12 reversible absorption of DNA onto resin,13 micellar catalyst-promoted reactions14), which provides new opportunities for DEL library synthesis.
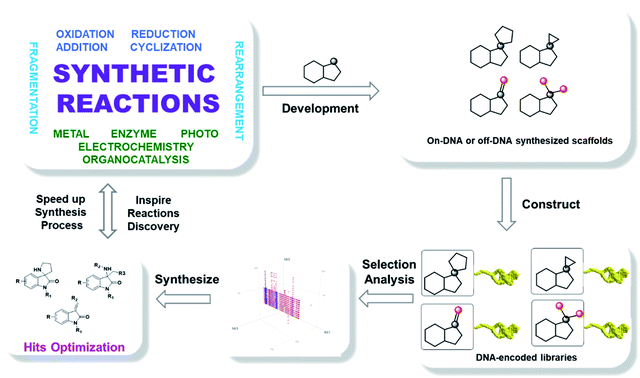
In addition to the directed DNA protection strategy, diversified library construction could be enabled by the identification of mild chemical transformations. As illustrated in Fig. 1, a multitude of innovative chemistries were developed in academia to synthesize drug-like scaffolds, which could be introduced to the encoded library to expand chemical space through innovative designs. This route enables an enhancement of the crosstalk between synthetic and medicinal chemists in industry and academia and an opportunity to impact the future of drug discovery.15
In DELT library construction, an ideal scaffold provides mature synthetic methods, enabling synthesis both on-DNA or off-DNA and possessing multiple functional groups and appendage points for DNA-linking along with an abundance of readily available building blocks. Based on an extensive literature analysis, the oxindole core16 was selected to validate our initial proposal. Firstly, the C3 position of the oxindole could be converted into a number of diverse structures through well-established transformations; secondly, at least two functional groups are appended on these scaffolds; thirdly, oxindole-type compounds are prevalent in nature and have the potential to be transformed into drugs.
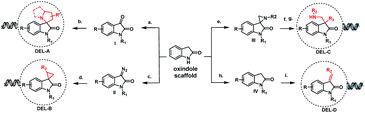
As illustrated in Fig. 2, four types of DELs were constructed from oxindoles to validate the concept: DEL-A, the spiropyrrolizidine centered DEL was built on-DNA using a three-component cyclization reaction from available isatins I. DEL-B, the spirocyclopropyl centered DEL was synthesized by a palladium-promoted on-DNA cycloaddition reaction from diazoisatin derivatives II. DEL-C, spiroaziridine derivatives III were converted from I using an aza-Corey–Chaykovsky reaction, which then underwent a ring-opening reaction. DEL-D, unsaturated centered DEL was synthesized from oxindole IV by an on-DNA aldol reaction. These DELs arising from a simple oxindole core, spanned spiro, disubstituted and unsaturated structures, elegantly fit the diversity-oriented synthesis (DOS) strategy, which was recently demonstrated by Schreiber, et al.17
Results and discussion
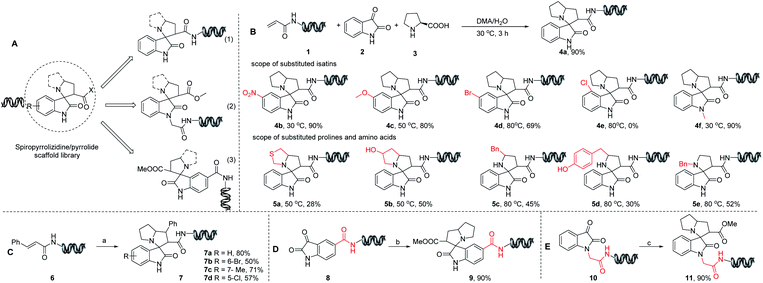
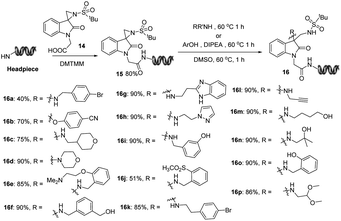
Our efforts commenced with the investigation of on-DNA three-component reaction that can form highly functionalized spiro-pyrrolizidine/pyrrolidine oxindoles.18 DNA barcodes could be conjugated to the scaffold via three attachment sites: pyrrolizidine ring (1), nitrogen of oxindole (2) and aromatic ring (3), which could attenuate the steric hindrance caused by oligonucleotide tags and facilitate target–binder interactions by presenting multiple orientations (Fig. 3A).As shown in Fig. 3B, DNA-linked acryl functionalized compound 1 was obtained in good conversion with 100 equiv. of isatin and 100 equiv. of proline via a three-component cyclization reaction at 30 °C. We observed that higher reaction temperatures were required for converting the starting material 1 to the desired products with some of the substituted isatins (Fig. 3B, 4c and 4d). The lack of reactivity in 4e at 80 °C was presumably a consequence of the steric hindrance of the chloride substitution. Elevated temperatures also caused the formation of side products and DNA decomposition. A representative set of experimental results showing the interplay between temperature and isatin derivatives is found in the ESI S2.† 50 °C was chosen as the preferred temperature to balance product formation and minimize side product formation.
Further diversity was explored with cyclic/acyclic α-amino acids (Fig. 3B). Cyclic amino acids such as thiazolidine-4-carboxylic acid and 4-hydroxypyrrolidine-2-carboxylic acid provided moderate conversions under the optimized reaction conditions. Only benzyl-functionalized acyclic amino acid variants provided the desired products at 80 °C, likely due to the decreased stability of the intermediate. DNA-appended acrylamide 6via on-DNA Heck coupling reaction developed previously were also tested in this annulation reaction (Fig. 3C) and moderate conversions were obtained, proving the possibility of two on-DNA methodologies in sequence. Next, we turned to isatin-functionalized DNA headpieces covalently linked through amide bond formation (Fig. 3D and E). We observed formation of the desired product along with 50% of byproduct, which had a molecular weight of desired product plus 18 Da (ESI S5†). Several amide bond forming reaction conditions were tested with no improvement in the product ratio. Fortunately, these byproducts didn't impact the subsequent on-DNA spiro-annulation reaction under the optimized reaction conditions. In addition, we also carried out the proof-of-concept synthesis to apply this new on-DNA annulation reaction via Heck cross-coupling reaction,19 Suzuki cross-coupling,20 nitro reduction, reductive amination and amide bond formation. These transformations could be found in the ESI S6.†
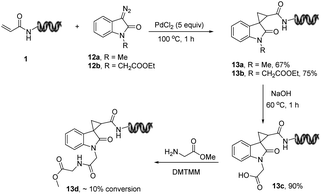
Spirocyclopropyl oxindoles are useful building blocks with a broad spectrum of biological and pharmaceutical applications such as HIV-1 NNRT1 inhibitors and arginine vasopressin inhibitors.21 Various approaches to access spirocyclopropyl fragments have been established between diazo compounds and electron-deficient olefins using a variety of metal catalysts. In particular, InCl3 has been demonstrated to promote reaction with 3-diazoindole and electro-deficient olefins in the presence of water.21 This was the preferred starting point to develop the corresponding DNA-compatible cycloaddition reactions. A range of promoters including Cu, Ag, Pd, Pt, Au, Ru, Ga, Mg, Fe and Zr were tested and PdCl2 provided 67% of the desired on-DNA spirocyclopropyl oxindole 13a. Unreacted DNA-conjugated acryl represented the mass balance at 100 °C, and low DNA recovery was found upon LC-MS analysis (ESI S7,†Fig. 4). Unfortunately, lower reaction temperatures also significantly decreased the conversions. Meanwhile, a proof-of-concept library synthesis using ester functionalized diazoisatin 12b as a bifunctional chemical linker was performed, providing the desired product 13b. After hydrolysis, the resulting carboxylic acid 13c enabled further diversity via acylation but with low conversion (13d, 10% conversion). Although this on-DNA set of reaction conditions was not robust enough for constructing spirocyclopropyl centered DNA-encoded libraries due to the low DNA recovery and suboptimal conversions to the desired products, it opens the door for more future exploration of this type of on-DNA transformation.
Highly strained spiroaziridine oxindoles and 3,3′-disubstituted oxindoles are attracting attention for their interesting bioactivities.22 We proposed to synthesize the spiroaziridine oxindole scaffolds first followed by DNA barcoding conjugation. DEL-C was constructed via DNA compatible ring-opening reactions to introduce aliphatic amines and phenol building blocks into the library. As illustrated in Fig. 5, the starting point for this exploration was the synthesis of carboxylic acid functionalized spiroaziridine oxindoles. From the methodologies established over past decades, we selected an aza-Corey–Chaykovsky reaction23 to access the aziridine unit via the addition of sulfur ylides to isatin derived N-tert-butanesulfinyl ketimines. Following oxidation and ester hydrolysis, the desired scaffold 14 was covalently linked to DNA barcoding via DMTMM promoted amide formation. We next turned to investigate the aminolysis and hydrolysis of the aziridine motif through a DNA-compatible ring-opening reaction. Both aliphatic amines and phenols provided moderate to excellent conversions. Weakly nucleophilic aromatic amines showed a lack of reactivity. Meanwhile, we also observed that DNA-conjugated 3,3′-dihydroxyloxindoles were formed as byproducts at elevated temperatures under basic conditions, possibly due to the residual base in the final step. Therefore, a spin filter procedure was required to remove the remaining basic reagents and to maintain neutral conditions. The corresponding DEL construction and validation are described in the next section.
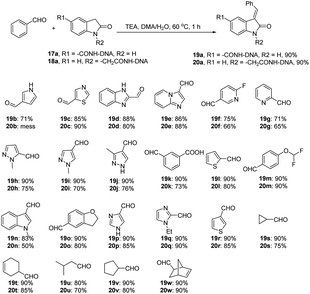
2-Indolinone is a well-known scaffold and present in drugs for cancer treatment.16e,24 ‘Two tagging points’ were employed to construct 3-alkenyl oxindole DNA-encoded libraries via a Claisen–Schmidt condensation reaction of substituted indoline-2-ones with various aldehydes in the presence of TEA (Fig. 6). Both aliphatic aldehydes and aromatic aldehydes delivered modest to excellent conversions. However, ketones gave negligible product formation even under forcing basic conditions, possibly due to relatively low nucleophilicity. Depending on the targets to be screened, such libraries may provide potential starting points for covalent drugs. The corresponding DEL construction and validation are described in the next section.
Encoded libraries construction
DNA-encoded libraries A, C and D were constructed by the newly developed DNA-compatible reactions with readily accessible building blocks such as amino acids, amines, borates and aldehydes. The size of the combined libraries was approximately 1.05 million (Fig. 7A). To determine the physicochemical properties of the compounds that compose the three libraries, 6000 compounds were randomly selected to summarize the physicochemical properties as shown in Fig. 7B. The full library molecule (tri-synthon) have a slightly higher molecule weight (ca. 650 Da on average) than Lipinski rule-of-5 compliant compounds, while the other properties meet the range of the typical hit compound profiles. Comparing the chemical space of DELs with 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 752 compounds from DrugBank, we applied the Principle Component Analysis (PCA) for both pool and found the overlap between them was more than 90%.
752 compounds from DrugBank, we applied the Principle Component Analysis (PCA) for both pool and found the overlap between them was more than 90%.
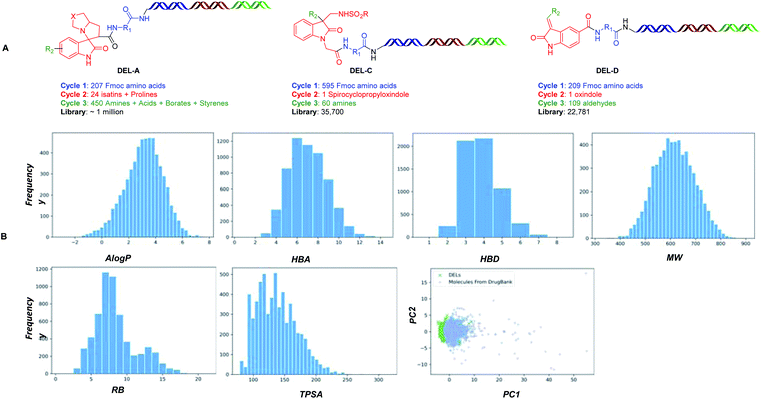 | ||
| Fig. 7 (A) The structures of DNA-encoded libraries. DNA-encoded library A, which was divided into five sub-libraries: DEL-A1, a di-synthon library; DEL-A2, a tri-synthon library contained a Heck coupling reaction; DEL-A3, a tri-synthon library contained a Suzuki coupling reaction; DEL-A4, a tri-synthon library contained an amidation reaction, reductive amination reaction and urea formation; DEL-A5, a tri-synthon library contained an amidation reaction, these reaction schemes are listed in S6 of ESI;† DNA-encoded library C based on DNA-compatible ring-opening methodology; DNA-encoded library D based on DNA compatible Claisen condensation methodology. (B) Physicochemical property distribution of DELs, including AlogP, HBA, HBD, MW, RB, TPSA and PCA. For PCA, PC means principal component, and it's a linear combination of variables (e.g. MW, AlogP, etc.). Each PC value accounts for the percentage of the total variance around the PCs. When PC1 & PC2 accounts for the majority of the variation (>70%), we could use a 2D graph to represent the chemical space. | ||
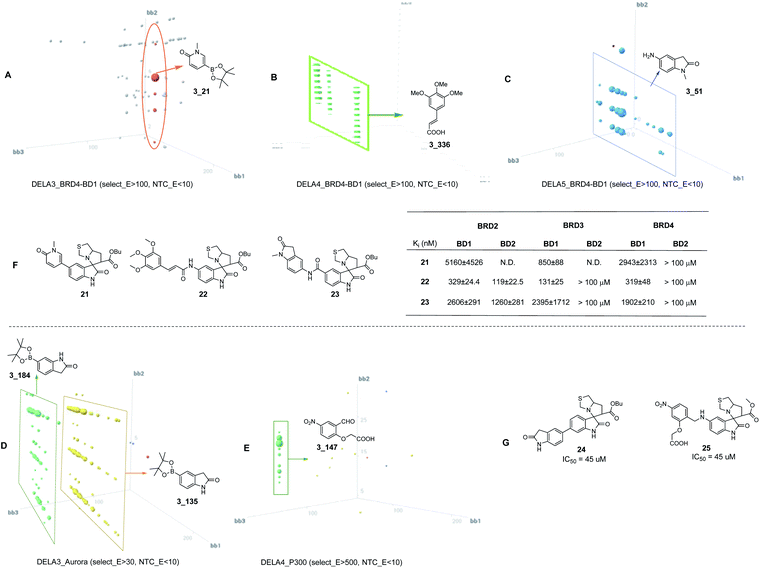
Not surprisingly, the properties of the di-synthon with the oxindole core were more attractive for hit-to-lead optimization. To evaluate whether the divergent isatin based DEL also worked for the screening of biological targets, we performed affinity screenings for three protein targets BRD4-BD1 (Fig. 8A–C), Aurora A (Fig. 8D) and p300 (Fig. 8E). DEL-A library pool was screened against BRD4-BD1 protein using the known inhibitor JQ1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 as a positive control. After the affinity screening, the samples were subjected to NGS and the generated data was translated into the corresponding copy number for the data analysis. In order to fully analyze the data among five different isatin libraries (DEL A1–A5), we used the enrichment for the normalization of different DELs26 (see the ESI† for the enrichment calculation). Fig. 8A–E illustrated the three-dimensional view of the corresponding selection outcomes for each individual library. From the data analysis, we found that DEL-A3–5 had obvious features for the BRD4-BD1, and the competition experiment with JQ1 further confirmed that the features are competitive binders likely within the similar binding pocket with JQ1 (ESI S8†). As illustrated in the three cubes (Fig. 8A–C), nascent SAR has been identified for DEL A3–5. From the detailed analysis, we have synthesized the truncated compounds 21–23 for activity confirmation with BRD2, BRD3 and BRD4. All synthesized compounds have moderate Ki values which demonstrated our rationale for the design and synthesis of the diversified oxindole libraries.
25 as a positive control. After the affinity screening, the samples were subjected to NGS and the generated data was translated into the corresponding copy number for the data analysis. In order to fully analyze the data among five different isatin libraries (DEL A1–A5), we used the enrichment for the normalization of different DELs26 (see the ESI† for the enrichment calculation). Fig. 8A–E illustrated the three-dimensional view of the corresponding selection outcomes for each individual library. From the data analysis, we found that DEL-A3–5 had obvious features for the BRD4-BD1, and the competition experiment with JQ1 further confirmed that the features are competitive binders likely within the similar binding pocket with JQ1 (ESI S8†). As illustrated in the three cubes (Fig. 8A–C), nascent SAR has been identified for DEL A3–5. From the detailed analysis, we have synthesized the truncated compounds 21–23 for activity confirmation with BRD2, BRD3 and BRD4. All synthesized compounds have moderate Ki values which demonstrated our rationale for the design and synthesis of the diversified oxindole libraries.
For Aurora A, we found the libraries had features with moderate enrichment (<100) in “target” and low in the “no target” control. However, obvious SAR has been identified for the cycle 3 BBs (Fig. 8D). 3_184 and 3_135 have similar structures and the latter was selected for synthesis off-DNA (24). 24 showed weak activity against Aurora A, demonstrating that moderate enrichment also could correlate with binding affinity. For the p300, a potential feature was found in DEL-A4 with good enrichment and the cycle 3 BB 3_147 was selected (Fig. 8E). Truncated structure 25 was synthesized off-DNA and validated in a biochemical assay. 25 showed weak activity against the target, but demonstrated the potential for multi-target selection with our designed DELs.
Conclusion
In summary, based on the oxindole scaffolds, we have developed a series of on-DNA synthetic transformations including 1,3-dipolar cyclization, cyclopropanation, ring-opening of aziridines and Claisen–Schmidt condensations to construct diverse oxindole derivatives. Among these reactions, the cyclopropanation and ring-opening reaction of aziridines are the first reported examples of on-DNA reactions, which opens the door for future exploration and applications. The majority of these transformations enable the diversity-oriented synthesis of DNA-encoded oxindole libraries which have been demonstrated with the design and synthesis of spiropyrrolizidine-oxindole DEL A libraries. Three protein targets have been screened and active hit compounds were identified and confirmed with moderate to good activities. Expanding the scope of synthetic transformations compatible for DEL library synthesis, particularly based on the oxindole scaffolds, more active hits could be identified as starting points for medicinal chemistry programs. In addition, oxindole libraries based on the novel on-DNA reactions are currently under construction and additional results from DEL selections will be reported in the due course.Experimental procedures
Full details of synthesis and LC-MS/MS analysis are provided in the ESI.†Author contributions
Jiaxiang Liu, Ziqin Yan, and Xiaohong Liu contributed equally to this article.Conflicts of interest
The authors declare no competing interests.Acknowledgements
We gratefully acknowledge financial support from the National Natural Science Foundation of China (81773634 to M. Z., 21877117 and 91953203 to X. L., 21907103 to X. W., 81673295 to Y. Z.), National Science & Technology Major Project “Key New Drug Creation and Manufacturing Program”, China (Number: 2018ZX09711002 to H. J., No. 2018ZX09711002-005 to X. L.), and “Personalized Medicines—Molecular Signature-based Drug Discovery and Development”, Strategic Priority Research Program of the Chinese Academy of Sciences (XDA12050201 to M. Z.; XDA12040330 and XDA12050405 to X. L.), Shanghai Commission of Science and Technology (18431907100 to X. L.). We also thank Dr Ryan Wurz of Amgen for critical reading.References
- (a) M. Schenone, V. Dancik, B. K. Wagner and P. A. Clemons, Nat. Chem. Biol., 2013, 9, 232–240 CrossRef CAS; (b) C. Finan, A. Gaulton, F. A. Kruger, R. T. Lumbers, T. Shah, J. Engmann, L. Galver, R. Kelley, A. Karlsson, R. Santos, J. P. Overington, A. D. Hingorani and J. P. Casas, Sci. Transl. Med., 2017, 9, eaag1166 CrossRef; (c) M. Luo, J. Jiao and R. Wang, BMC Bioinf., 2019, 20, 198 CrossRef.
- D. G. Brown and J. Bostrom, J. Med. Chem., 2018, 61, 9442–9468 CrossRef CAS.
- (a) J. L. Reymond and M. Awale, ACS Chem. Neurosci., 2012, 3, 649–657 CrossRef CAS; (b) M. González-Medina, J. J. Naveja, N. Sánchez-Cruz and J. L. Medina-Franco, RSC Adv., 2017, 7, 54153–54163 RSC.
- S. Morgan, P. Grootendorst, J. Lexchin, C. Cunningham and D. Greyson, Health Policy, 2011, 100, 4–17 CrossRef.
- S. Brenner and R. A. Lerner, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 5381–5383 CrossRef CAS.
- G. P. Smith and V. A. Petrenko, Chem. Rev., 1997, 97, 391–410 CrossRef CAS.
- M. A. Clark, R. A. Acharya, C. C. Arico-Muendel, S. L. Belyanskaya, D. R. Benjamin, N. R. Carlson, P. A. Centrella, C. H. Chiu, S. P. Creaser, J. W. Cuozzo, C. P. Davie, Y. Ding, G. J. Franklin, K. D. Franzen, M. L. Gefter, S. P. Hale, N. J. Hansen, D. I. Israel, J. Jiang, M. J. Kavarana, M. S. Kelley, C. S. Kollmann, F. Li, K. Lind, S. Mataruse, P. F. Medeiros, J. A. Messer, P. Myers, H. O'Keefe, M. C. Oliff, C. E. Rise, A. L. Satz, S. R. Skinner, J. L. Svendsen, L. Tang, K. van Vloten, R. W. Wagner, G. Yao, B. Zhao and B. A. Morgan, Nat. Chem. Biol., 2009, 5, 647–654 CrossRef CAS.
- D. Neri and R. A. Lerner, Annu. Rev. Biochem., 2018, 87, 479–502 CrossRef CAS.
- (a) F. Buller, L. Mannocci, J. Scheuermann and D. Neri, Bioconjugate Chem., 2010, 21, 1571–1580 CrossRef CAS; (b) M. A. Clark, Curr. Opin. Chem. Biol., 2010, 14, 396–403 CrossRef CAS; (c) R. E. Kleiner, C. E. Dumelin and D. R. Liu, Chem. Soc. Rev., 2011, 40, 5707–5717 RSC; (d) R. M. Franzini, D. Neri and J. Scheuermann, Acc. Chem. Res., 2014, 47, 1247–1255 CrossRef CAS; (e) H. Salamon, M. Klika Skopic, K. Jung, O. Bugain and A. Brunschweiger, ACS Chem. Biol., 2016, 11, 296–307 CrossRef CAS; (f) S. L. Belyanskaya, Y. Ding, J. F. Callahan, A. L. Lazaar and D. I. Israel, ChemBioChem, 2017, 18, 837–842 CrossRef CAS; (g) R. A. Goodnow Jr, C. E. Dumelin and A. D. Keefe, Nat. Rev. Drug Discovery, 2017, 16, 131–147 CrossRef; (h) N. Favalli, G. Bassi, J. Scheuermann and D. Neri, FEBS Lett., 2018, 592, 2168–2180 CrossRef CAS; (i) Y. Zhou, C. Li, J. Peng, L. Xie, L. Meng, Q. Li, J. Zhang, X. D. Li, X. Li, X. Huang and X. Li, J. Am. Chem. Soc., 2018, 140, 15859–15867 CrossRef CAS; (j) P. Dickson and T. Kodadek, Org. Biomol. Chem., 2019, 17, 4676–4688 RSC; (k) J. Ottl, L. Leder, J. V. Schaefer and C. E. Dumelin, Molecules, 2019, 24, 1629 CrossRef CAS.
- R. M. Franzini and C. Randolph, J. Med. Chem., 2016, 59, 6629–6644 CrossRef CAS.
- (a) Z. J. Gartner, M. W. Kanan and D. R. Liu, Angew. Chem., Int. Ed., 2002, 41, 1796–1800 CrossRef CAS; (b) A. L. Satz, J. Cai, Y. Chen, R. Goodnow, F. Gruber, A. Kowalczyk, A. Petersen, G. Naderi-Oboodi, L. Orzechowski and Q. Strebel, Bioconjugate Chem., 2015, 26, 1623–1632 CrossRef CAS; (c) M. L. Malone and B. M. Paegel, ACS Comb. Sci., 2016, 18, 182–187 CrossRef CAS; (d) K. Götte, S. Chines and A. Brunschweiger, Tetrahedron Lett., 2020, 151889 CrossRef.
- M. Potowski, F. Losch, E. Wunnemann, J. K. Dahmen, S. Chines and A. Brunschweiger, Chem. Sci., 2019, 10, 10481–10492 RSC.
- (a) D. T. Flood, S. Asai, X. Zhang, J. Wang, L. Yoon, Z. C. Adams, B. C. Dillingham, B. B. Sanchez, J. C. Vantourout, M. E. Flanagan, D. W. Piotrowski, P. Richardson, S. A. Green, R. A. Shenvi, J. S. Chen, P. S. Baran and P. E. Dawson, J. Am. Chem. Soc., 2019, 141, 9998–10006 CrossRef CAS; (b) Y. Ruff, R. Martinez, X. Pelle, P. Nimsgern, P. Fille, M. Ratnikov and F. Berst, ACS Comb. Sci., 2020, 22, 120–128 CrossRef CAS.
- (a) M. K. Skopic, K. Gotte, C. Gramse, M. Dieter, S. Pospich, S. Raunser, R. Weberskirch and A. Brunschweiger, J. Am. Chem. Soc., 2019, 141, 10546–10555 CrossRef CAS; (b) J. H. Hunter, L. Prendergast, L. F. Valente, A. Madin, G. Pairaudeau and M. J. Waring, Bioconjugate Chem., 2020, 31, 149–155 CrossRef CAS.
- D. P. Rotella, ACS Chem. Neurosci., 2016, 7, 1315–1316 CrossRef CAS.
- (a) N. R. Ball-Jones, J. J. Badillo and A. K. Franz, Org. Biomol. Chem., 2012, 10, 5165–5181 RSC; (b) L. Hong and R. Wang, Adv. Synth. Catal., 2013, 355, 1023–1052 CrossRef CAS; (c) M. A. Borad, M. N. Bhoi, N. P. Prajapati and H. D. Patel, Synth. Commun., 2014, 44, 1043–1057 CrossRef CAS; (d) M. Xia and R.-Z. Ma, J. Heterocycl. Chem., 2014, 51, 539–554 CrossRef CAS; (e) A. Leoni, A. Locatelli, R. Morigi and M. Rambaldi, Expert Opin. Ther. Pat., 2016, 26, 149–173 CrossRef CAS; (f) G. Muller, T. Berkenbosch, J. C. Benningshof, D. Stumpfe and J. Bajorath, Chemistry, 2017, 23, 703–710 CrossRef; (g) G. J. Mei and F. Shi, Chem. Commun., 2018, 54, 6607–6621 RSC.
- C. J. Gerry, M. J. Wawer, P. A. Clemons and S. L. Schreiber, J. Am. Chem. Soc., 2019, 141, 10225–10235 CrossRef CAS.
- (a) S. Rehn, J. Bergman and B. Stensland, Eur. J. Org. Chem., 2004, 2004, 413–418 CrossRef; (b) T. L. Pavlovskaya, F. G. Yaremenko, V. V. Lipson, S. V. Shishkina, O. V. Shishkin, V. I. Musatov and A. S. Karpenko, Beilstein J. Org. Chem., 2014, 10, 117–126 CrossRef.
- X. Wang, H. Sun, J. Liu, W. Zhong, M. Zhang, H. Zhou, D. Dai and X. Lu, Org. Lett., 2019, 21, 719–723 CrossRef CAS.
- Y. Ding and M. A. Clark, ACS Comb. Sci., 2015, 17, 1–4 CrossRef CAS.
- S. Muthusamy and R. Ramkumar, Synlett, 2015, 26, 2156–2160 CrossRef CAS.
- (a) F. M. Ismail, D. O. Levitsky and V. M. Dembitsky, Eur. J. Med. Chem., 2009, 44, 3373–3387 CrossRef CAS; (b) J. Kaur, S. S. Chimni, S. Mahajan and A. Kumar, RSC Adv., 2015, 5, 52481–52496 RSC; (c) G. Zhu, S. Liu, S. Wu, L. Peng, J. Qu and B. Wang, J. Org. Chem., 2017, 82, 4317–4327 CrossRef CAS; (d) S. Hajra, S. Singha Roy, A. Biswas and S. A. Saleh, J. Org. Chem., 2018, 83, 3633–3644 CrossRef CAS.
- (a) E. J. Corey and M. Chaykovsky, J. Am. Chem. Soc., 1965, 87, 1353–1364 CrossRef CAS; (b) S. Hajra, S. M. Aziz, B. Jana, P. Mahish and D. Das, Org. Lett., 2016, 18, 532–535 CrossRef CAS.
- A. Andreani, Eur. J. Med. Chem., 1990, 25, 187–190 CrossRef CAS.
- P. Filippakopoulos, J. Qi, S. Picaud, Y. Shen, W. B. Smith, O. Fedorov, E. M. Morse, T. Keates, T. T. Hickman, I. Felletar, M. Philpott, S. Munro, M. R. McKeown, Y. Wang, A. L. Christie, N. West, M. J. Cameron, B. Schwartz, T. D. Heightman, N. La Thangue, C. A. French, O. Wiest, A. L. Kung, S. Knapp and J. E. Bradner, Nature, 2010, 468, 1067–1073 CrossRef CAS.
- R. M. Franzini, T. Ekblad, N. Zhong, M. Wichert, W. Decurtins, A. Nauer, M. Zimmermann, F. Samain, J. Scheuermann, P. J. Brown, J. Hall, S. Graslund, H. Schuler and D. Neri, Angew. Chem., Int. Ed., 2015, 54, 3927–3931 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Supplemental experimental procedures and supplemental data. See DOI: 10.1039/d0sc06696f |
| This journal is © The Royal Society of Chemistry 2021 |