Open Access Article
Open Access ArticleDiscovery of four modified classes of triterpenoids delineated a metabolic cascade: compound characterization and biomimetic synthesis†
Bin
Zhou‡
 a,
Xin-Hua
Gao‡
a,
Xin-Hua
Gao‡
 a,
Min-Min
Zhang
b,
Cheng-Yu
Zheng
a,
Hong-Chun
Liu
b and
Jian-Min
Yue
a,
Min-Min
Zhang
b,
Cheng-Yu
Zheng
a,
Hong-Chun
Liu
b and
Jian-Min
Yue
 *a
*a
aState Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 555 Zuchongzhi Road, Shanghai 201203, People's Republic of China. E-mail: jmyue@simm.ac.cn
bDivision of Anti-tumor Pharmacology, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 555 Zuchongzhi Road, Shanghai 201203, People's Republic of China
First published on 18th June 2021
Abstract
Chemical studies on Dichapetalum gelonioides have afforded 18 highly modified complex triterpenoids belonging to four compound classes as defined by the newly adapted functional motifs associated with the A ring of the molecules. Their structures were determined by solid data acquired by diverse methods. The biosynthetic pathway for the four compound classes was rationalized via cascade modifications involving diverse chemical events. The subsequent biomimetic syntheses afforded all the desired products, including compounds 16 and 19 that were not obtained in our purification, which validated the proposed biosynthetic pathway. Besides, some compounds exhibited strong cytotoxic activities, especially 2 and 4 showed nanomolar potency against the NAMALWA tumor cell line, and a gross structure–activity relationship (SAR) of these compounds against the tested tumor cell lines was delineated.
Introduction
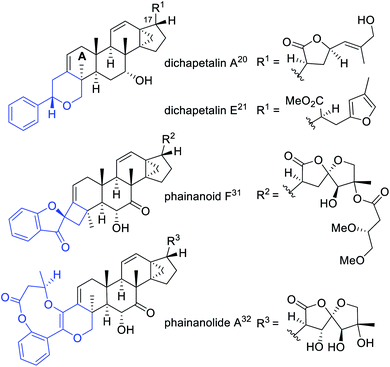
Biomimetic syntheses of natural products have always fascinated chemists, as the enzyme catalyzed reactions within organisms are especially atom-economic and efficient, as well as being highly regio- and stereoselective.1,2 Cascade reactions based on the biosynthetic mechanism have been recognized as a crucial synthetic strategy enabling consecutive chemical transformations.3–7 In addition to discovering structurally interesting and biologically significant natural molecules,8–15 we have been committed to the biomimetic and/or bioinspired syntheses of important natural compounds identified by our research group as well, which aimed to validate the proposed biosynthetic pathways from the chemical view.16–19Dichapetalin-type triterpenoids, a rare category of natural products featuring the modified skeletons by condensing a C6–C2 unit to the A ring of 13,30-cyclodammarane triterpenoids, were found as the characteristic components of Dichapetalum and Phyllanthus genera.20–32 The typical dichapetalin-type triterpenoids possess a common 2-phenylpyran moiety fused to the A ring of 13,30-cyclodammarane triterpenoids, with the major differences being the C-17 side chain and oxidation patterns.21,24–26 The first dichapetalin-type triterpenoid, dichapetalin A,20 was reported from D. madagascariense in 1995. Afterward, 50 more dichapetalin analogues with major modifications either in the newly added C6–C2 units or in the C-17 side chains were continuously identified from the Dichapetalum and Phyllanthus genera (Chart 1 and S1 in the ESI†).21–32 Some dichapetalin-type triterpenoids were found to exhibit distinct cytotoxic acitivities.20,23–27,29,30 Notably, instead of the ordinary dichapetalins with a 2-phenylpyran moiety, our previous work led to the identification of two types of skeletal new dichapetalins with potent immunosuppressive activities from the Phyllanthus genus for the first time, each with an unprecedented 4,5-spirocyclic and a 6/9/6 heterotricyclic system fused to the A ring of 13,30-cyclodammarane triterpenoids (Chart 1 and S1†).31,32 The interesting structures and significant biological activities of dichapetalin-type triterpenoids have attracted our great attention to conduct an in-depth chemical study on the species D. gelonioides, with the hope to discover more diverse modified structures of this biologically significant compound category, and further gain insights into their biosynthetic connections. In the current study, 18 highly modified triterpenoids, belonging to four compound classes as defined by the newly adapted functional motifs fused with the A ring of the molecules, including phenyl-butadiene (I), phenyl-endoperoxide (II), phenyl-furan (III), and phenyl-enedione (IV), were isolated and identified. The biosynthetic pathway for the four compound classes was rationalized involving diverse chemical events in a sequential mode. Inspired by the efficient biosynthetic initiatives, we thus designed a cascade chemical transformation strategy, which included singlet oxygen involved Diels–Alder [4 + 2] cycloaddition, base catalytic rearrangement, as well as dye-sensitized photooxygenation followed by acid induced rearrangement in turn. These cascade chemical reactions succeeded in biomimetic transformation of the four types of isolates effectively, which also afforded two related analogues 16 and 19 that were missed in our purification. Herein, we present a full account of the isolation, structural elucidation, chemical transformation, and bioactive evaluation of the four highly modified triterpenoid classes.
Results and discussion
Isolation and structural elucidation
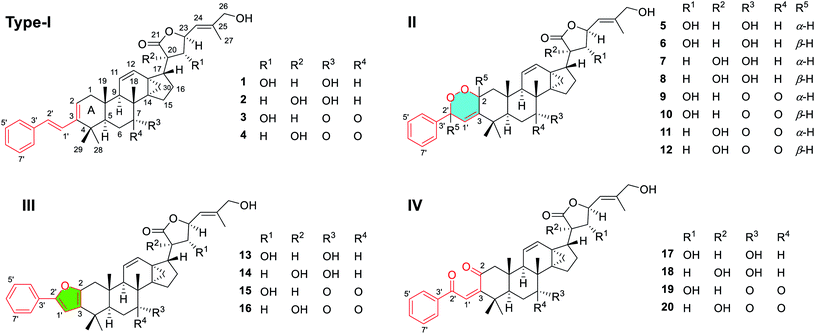
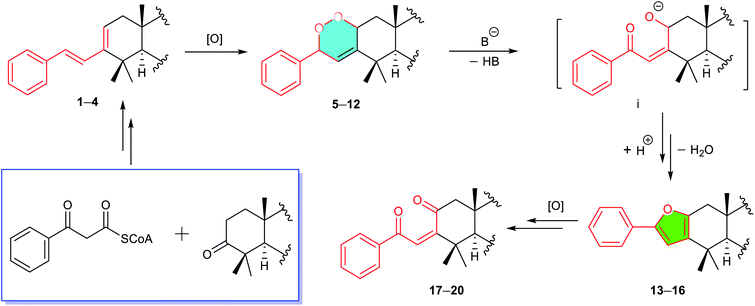
Chemical investigation of D. gelonioides has afforded 18 highly modified triterpenoids of four main compound classes as defined by the newly adapted functional motifs fused with the A ring of the molecules, including dichapegenins A–D (1–4, type-I) with a phenyl-butadiene group, dichapegenins E–L (5–12, type-II) with a phenyl-endoperoxide moiety, dichapegenins M–O (13–15, type-III) with a phenyl-furan functionality, and dichapegenins Q, R, and T (17, 18, and 20, type-IV) with a phenyl-enedione motif (Fig. 1).Compound 1 was obtained as colorless crystals. Its molecular formula was deduced by the HRMS (ESI) and 13C NMR data to be C38H48O5. Analysis of the 1H and 13C NMR data (Table S1†) revealed that the structure of 1 is highly related to dichapetalin-type triterpenoids, which are the characteristic components of the Dichapetalum species.25,26 An observation of five aromatic signals for a mono-substituted aromatic ring, in conjunction with the signals resonating at δH 6.94 and 7.09 (both J = 15.9 Hz) for a trans-oriented double bond, is indicative of a styrene functionality. The styrene group was linked to C-3 based on the HMBC cross-peak of H-2/C-1′ (Fig. S1A†). Compound 1 was thus identified as a highly modified dichapetalin-type triterpenoid with a unique phenyl-butadiene functionality.
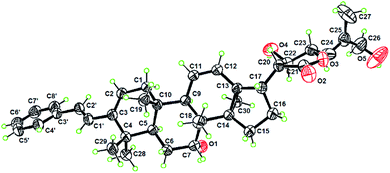
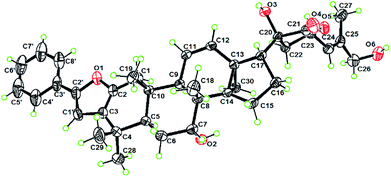
The planar structure of 1 was assigned by the COSY and HMBC data. Six spin coupling systems were established by the 1H–1H COSY correlations (Fig. S1A†). The HMBC correlations connected the six spin systems with the quaternary carbons and oxygen atoms, forming a 22,23,26-trihydroxydammar-24-en-21-oic acid-21,23-lactone scaffold with a tricyclo[4.3.1.01,6]decane and a styrene moiety for 1. The γ-lactone moiety as recognized by the key chemical shifts of C-21 (δC 175.3) and CH-23 (δH 4.90 and δC 79.3) was attached to C-17 by the HMBC cross-peak of H-22/C-17 and the COSY correlation of H-17/H-20. The relative configurations of all the stereogenic centers in 1 except for the C-22 are identical to their counterparts of dichapetalin-type triterpenoids,22,26 based on the NOESY data (Fig. S1B†). The single crystal X-ray diffraction study by using Cu Kα radiation allowed a final establishment of the absolute configuration for 1 (5R, 7R, 8R, 9R, 10S, 13S, 14S, 17S, 20R, 22R, 23S) [absolute structure parameter: 0.04 (13)] (Fig. 2, CCDC 2010767, see demonstration in the ESI†).
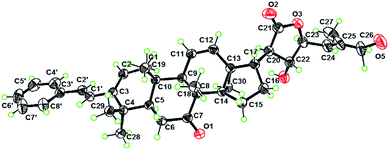
Analysis of the NMR data (Table S1†) of compounds 2–4 showed signals for the presence of a styrene functionality, indicating that 2–4 belong to the same compound class as 1. The 2D structures of 2–4 were elucidated by the NMR and MS data (the detailed structural elucidation of 2–4 is included in the ESI†), which differed from the structure of 1 in the oxidation patterns at C-7, C-20, and C-22. Compound 2 was verified to have a HO-20 group by the HMBC correlations (Fig. S29†) from H2-22 to C-20 and C-21. The similar negative Cotton effect observed at ca. 230 nm in the ECD spectrum caused by a γ-lactone n → π* transition suggested a 23R configuration for 2.26,33,34 The downfield shifted H-23 (ΔδH 0.29) of 2 compared to that of dichapetalin A21 due to the steric hindrance from HO-20 indicated that the HO-20 is α-oriented.33 The relative configurations for the other stereogenic centers in compounds 2–4 were assigned the same as those in compound 1, as supported by their similar NMR and NOESY data (Fig. S30, S39, and S48†). The absolute configuration (5R, 8R, 9R, 10S, 13S, 14S, 17S, 20R, 22R, 23S) of 3 was determined by X-ray crystallography using Cu Kα radiation [Fig. 3, CCDC 2082769, Flack parameter = 0.13 (12)]. Subsequently, the absolute configurations of 2 (5R, 7R, 8R, 9R, 10S, 13S, 14S, 17R, 20R, 23R) and 4 (5R, 8R, 9R, 10S, 13S, 14S, 17R, 20R, 23R) were assigned by the compatible ECD data with those of compounds 1 and 3 (see data under Section 2.4 in the ESI†), which are consistent with the biosynthetic consideration.
Compounds 5 and 6 share a molecular formula C38H48O7 as gleaned from the HRMS (ESI) and 13C NMR data, which has two more oxygen atoms than that of 1, requiring the presence of 15 degrees of unsaturation and an extra ring in their structures. The aforementioned data, as well as the presence of a trisubstituted double bond as judged by the NMR data in 5 and 6 instead of the conjugated double bond for the styrene motif in 1, suggested that 5 and 6 are the endoperoxide derivatives of 1. Compounds 5 and 6, sharing a common 2D structure with a novel phenyl-endoperoxide functionality, were thus established by their HMBC and COSY correlations (Fig. S2A†). The relative configurations of H-2 and H-2′ in 5 and 6 were determined by the NOESY correlations (Fig. 4). Both the H-2 and H-2′ in 5 are α-configured, as verified by the NOESY cross-peaks (Fig. 4A) of H-4′ (H-8′)/H-1β and H3-19, and of H-2/H-5 and H3-28. In contrast, both the H-2 and H-2′ in 6 are β-configured based on the NOESY correlations (Fig. 4B) between H-4′ (H-8′) with H-1α and H3-28, and between H-2 with H3-19 and H3-29. The relative configurations of the remaining stereocenters in 5 and 6 are identical to their counterparts in 1 based on the similar NMR patterns, as well as the NOESY data (Fig. S57 and S66†).
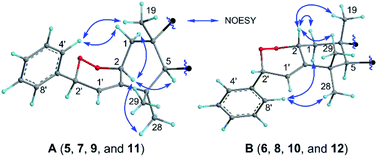 | ||
| Fig. 4 Selected NOESY correlations of the shared partial structures of (A) compounds 5, 7, 9, and 11 with α-oriented H-2 and H-2′, and (B) compounds 6, 8, 10, and 12 with β-oriented H-2 and H-2′. | ||
Similar to the case of 5/6, compounds 7/8, 9/10, and 11/12 were assigned as another three pairs of the endoperoxide derivatives of the corresponding phenyl-butadiene precursors 2, 3, and 4, respectively, based on their NMR evidence and the molecular formulae. The deductions were corroborated by their 2D NMR data (Fig. S2A†). Compounds with an odd number (5, 7, 9 and 11) and an even number (6, 8, 10 and 12) were thus assigned to have 2S, 2′S and 2R, 2′R absolute configurations, respectively, as the absolute configurations of the other stereocenters were readily established the same as their counterparts of 1–4 based on their comparable negative Cotton effects at ca. 230 nm in the ECD spectra (see data under Section 2.4 in the ESI†), which are consistent with the biosynthetic consideration.
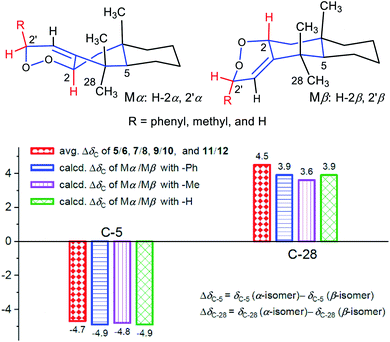
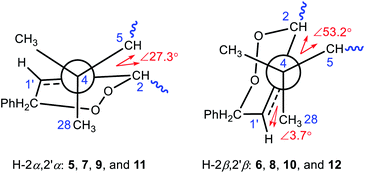
Comparison of the 13C NMR data (Tables S2 and S3†) showed distinct differential carbon chemical shifts (ΔδC) of C-5 and C-28 between the two isomeric endoperoxides. In order to verify whether the significant changes of ΔδC-5 and/or ΔδC-28 were caused by the effects of the 2′-phenyl group in different directions, three corresponding pairs of model molecules were thus defined as Mα/Mβ with the R groups as phenyl, methyl, and hydrogen, respectively (Fig. 5). The NMR calculations by using the time dependent density functional theory (TDDFT) gauge independent atomic orbital (GIAO) method (see demonstration in the ESI†)35,36 verified that the phenyl group does not account for the observed ΔδC, in which the calculated ΔδC-5 and ΔδC-28 for the three isomeric pairs of the model molecules were roughly consistent with the measured average ΔδC-5 (ΔδC −4.7) and ΔδC-28 (ΔδC 4.5) of the compound pairs 5/6, 7/8, 9/10, and 11/12. Thus, the inversion of C-2 and C-2′ configurations resulting in the conformational changes for the A ring and the newly formed peroxide ring (Fig. 5, Mα and Mβ) is considered as the main factor causing the obvious changes for the chemical shifts of C-5 and C-28 in each isomeric pair. Moreover, the upfield shifted C-5 signals in compounds 5, 7, 9, and 11 with α-oriented H-2 and H-2′ were likely caused by the γ-gauche effect from C-2. Similarly, the upfield shifted C-28 signals in compounds 6, 8, 10, and 12 with β-oriented H-2 and H-2′ were caused by the γ-gauche effect from C-1′ (Fig. 6).
Comparison of the NMR data (Table S4†) suggested that compound 13 is structurally related to 5/6, with the major difference being the appendage around the A ring. A new phenyl-furan moiety that fused with the A ring through C-2 and C-3 was then established by analysis of the 1D and 2D NMR data, especially the HMBC spectrum (Fig. S2B†). The remaining structural part of 13 was assigned the same as that of 5/6 based on their similar NMR data, which was corroborated by the 2D NMR spectra (Fig. S126–S129†). In a similar way, the structures of compounds 14 and 15 were assigned by 1D and 2D NMR data (see related figures in the ESI†). The absolute configuration of 14 (5R, 7R, 8R, 9R, 10S, 13S, 14S, 17R, 20R, 23R) was determined by an X-ray crystallography study using Cu Kα radiation [Flack parameter of 0.03 (5)] (Fig. 7, CCDC 2010766, see demonstration in the ESI†). The absolute configurations of 13 (5R, 7R, 8R, 9R, 10S, 13S, 14S, 17S, 20R, 22R, 23S) and 15 (5R, 8R, 9R, 10S, 13S, 14S, 17S, 20R, 22R, 23S) were then established as depicted by their similar ECD curves to that of 14 (see data under Section 2.4 in the ESI†).
Comparison of the 1H and 13C NMR data (Table S5†) revealed that the structure of 17 is closely associated with those of 1, 5, 6, and 13, with the main difference being the modification of the A ring and the C-3 appendage. A novel phenyl-enedione motif that was furnished between the A ring and the C-3 appendage was then established by the 1H–1H COSY and HMBC spectra (Fig. S2C†). The Δ3(1′) double bond was assigned to be the Z-isomer by the NOESY correlations of H-1′ with H3-28 and H3-29 (Fig. S165†). Similarly, the structures of compounds 18 and 20 with a phenyl-enedione motif were elucidated as depicted by the NMR and MS data (see related figures in the ESI†). The absolute configurations of 17 (5R, 7R, 8R, 9R, 10S, 13S, 14S, 17S, 20R, 22R, 23S), 18 (5R, 7R, 8R, 9R, 10S, 13S, 14S, 17R, 20R, 23R), and 20 (5R, 8R, 9R, 10S, 13S, 14S, 17R, 20R, 23R) were respectively assigned according to their distinct negative Cotton effects observed around 230 nm in ECD spectra (see data under Section 2.4 in the ESI†), which were supported by the biosynthetic respect.
Biosynthetic pathway
A biosynthetic pathway for the four compound classes was rationalized involving diverse chemical events (Scheme 1). Compounds 1–4 representing modified skeletal new triterpenoids might be derived by the condensation of a β-ketophenylpropionyl-CoA37 with a 3-keto-13,30-cyclodammarane triterpenoid, followed by several steps of modifications. The phenyl-peroxides 5–12 were considered as the photooxygenation products of 1–4.38–40 The phenyl-furans 13–16 were probably converted from 5–12 by a base induced cleavage of the peroxide bond followed by a dehydration-driven rearrangement.38–40 The conversion of phenyl-enediones 17–20 from 13–16 was speculated by oxidative procedures.41Bioinspired synthesis
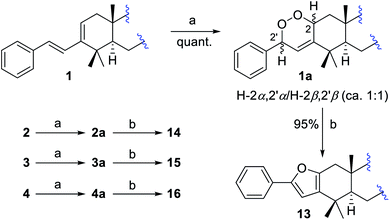
A bio-inspired three-step cascade reaction protocol was subsequently designed, which succeeded in the chemical transformation of the aforementioned four compound types, which also afforded two analogues 16 and 19 that were missed during the isolation procedures.First, a biomimetic [4 + 2] Diels–Alder cycloaddition of compounds 1–4 with photochemically generated singlet oxygen was performed to give corresponding four pairs of phenyl-endoperoxide mixtures (5–12) (Scheme 2).38–40,42 Taking compound 1 as an example, it was reacted with oxygen under 23 W fluorescent lamp irradiation in the presence of methylene blue as the photo-sensitizer, which afforded a corresponding pair of phenyl-endoperoxide mixtures 1a (5 + 6), quantitatively. Similarly, the other three pairs of phenyl-endoperoxide mixtures 2a (7 + 8), 3a (9 + 10), and 4a (11 + 12) were obtained by treating compounds 2–4 under the same conditions, respectively (Scheme 2, see demonstration in the ESI†). The 1H NMR data analyses showed that the percentages of α- and β-configured isomers were about equal in each pair of the phenyl-endoperoxide mixtures (Fig. S3–S6†).
Next, we focused on the syntheses of compounds 13–16 from the biosynthetic precursors 1a–4a. Four isomeric pairs 1a–4a were individually treated with a catalytic amount of Et3N in methanol at RT for 4 hours (Scheme 2).39,40,43 After workup, compounds 13–16 were obtained correspondingly from the diastereoisomeric mixtures 1a–4a in high yields (ca. 95%, see demonstration in the ESI†). The structure of compound 16 was established by analysis of its 1D and 2D NMR data (Fig. S10†), which was not obtained in our isolation.
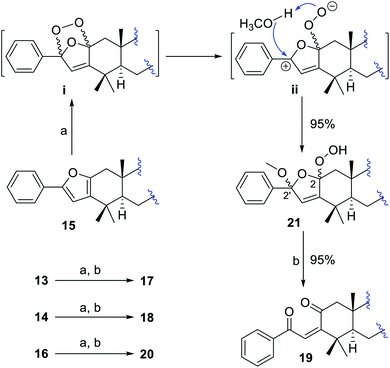
To convert phenyl-furans 13–16 into phenyl-enediones 17–20, compound 15 as a template was first treated with m-chloroperoxybenzoic acid42 and magnesium monoperphthalate41,44 respectively, and no desired product was detected in both cases. Subsequently, compound 15 was reacted with oxygen under 23 W fluorescent lamp irradiation in the presence of methylene blue as the photo-sensitizer in methanol to afford a mixture of two isomers 21a and 21b (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) in 95% total yield.45–49 The structures and percentages of 21a (2α-OOH, 2′α-OCH3) and 21b (2β-OOH, 2′β-OCH3) were determined by 1D and 2D NMR data analysis (Fig. S13†). In this reaction, only two isomers 21a and 21b were obtained, indicating that the methanol involved transformation of intermediate ii to 21 proceeded in a concerted way (Scheme 3).45 The predominant α-adduct 21a initially resulted from the formation of intermediate ivia a [4 + 2] Diels–Alder cycloaddition, which was probably caused by the relatively small steric hindrance of the α-face in the furan ring (see demonstration in the ESI†). Compound 21 was finally converted into corresponding phenyl-enedione 19 in high yield (ca. 95%) by treating with 0.02% aq. CF3COOH in MeOH at RT for 12 hours. The structure of 19 was unambiguously assigned by NMR and MS data (Fig. S14, and S178–S185†), which was not obtained during the isolation. In the same way, the phenyl-enediones 17, 18, and 20 were converted from corresponding phenyl-furans 13, 14, and 16, respectively, in high yields (Scheme 3, ca. 90%, see demonstration in the ESI†).
1) in 95% total yield.45–49 The structures and percentages of 21a (2α-OOH, 2′α-OCH3) and 21b (2β-OOH, 2′β-OCH3) were determined by 1D and 2D NMR data analysis (Fig. S13†). In this reaction, only two isomers 21a and 21b were obtained, indicating that the methanol involved transformation of intermediate ii to 21 proceeded in a concerted way (Scheme 3).45 The predominant α-adduct 21a initially resulted from the formation of intermediate ivia a [4 + 2] Diels–Alder cycloaddition, which was probably caused by the relatively small steric hindrance of the α-face in the furan ring (see demonstration in the ESI†). Compound 21 was finally converted into corresponding phenyl-enedione 19 in high yield (ca. 95%) by treating with 0.02% aq. CF3COOH in MeOH at RT for 12 hours. The structure of 19 was unambiguously assigned by NMR and MS data (Fig. S14, and S178–S185†), which was not obtained during the isolation. In the same way, the phenyl-enediones 17, 18, and 20 were converted from corresponding phenyl-furans 13, 14, and 16, respectively, in high yields (Scheme 3, ca. 90%, see demonstration in the ESI†).
This successful biomimetic transformation of four compound classes not only validated the proposed biosynthetic pathway, but also provided a strong chemical correlation network to support the structural elucidation of 1–20.
A direct UPLC-MS (ESI) analysis of a fresh ethanolic plant extract was performed to check the presence of the missing compounds 16 and 19. At the same UPLC retention times of semisynthesized 16 and 19, the diagnostic ion peaks at m/z 579.3 [M + H]+ and 613.3 [M + H]+ were detected, respectively, which verified the presence of the two compounds in the plant (Fig. S232 and S233†). Similarly, we checked the presence of some representative isolates in the four compound classes, and all of them were observed to exist in the plant extract, based on the UPLC-MS analyses (Fig. S228–S233†).
Anticancer evaluation and SAR study
Our isolation and synthesis efforts provided enough samples, which facilitated a systematic biological evaluation of compounds 1–20. The cytotoxic activities of compounds 1–20 against three human tumor cell lines (NAMALWA, A549, and Hep G2) were thus evaluated (Table 1, see demonstration in the ESI†).50,51 The most active components, 2 and 4, showed potent inhibition against the NAMALWA tumor cell line with IC50 values of 78 ± 25 and 67 ± 15 nM (the positive control doxorubicin, IC50 = 350 ± 33 nM), respectively.| Compd | Cytotoxic activity against cancer cell linesa,b | ||
|---|---|---|---|
| NAMALWA | A549 | Hep G2 | |
| a IC50 > 10 μM is considered inactive (IA). b NAMALWA = human Burkitt's lymphoma cell line, A549 = human alveolar basal epithelial cell line, and Hep G2 = human liver hepatocellular carcinoma cell line. | |||
| 1 | 0.18 ± 0.051 | 3.0 ± 0.61 | 3.7 ± 0.69 |
| 2 | 0.078 ± 0.025 | 2.4 ± 0.30 | 7.0 ± 3.2 |
| 3 | 0.15 ± 0.089 | 2.8 ± 0.26 | 2.6 ± 0.41 |
| 4 | 0.067 ± 0.015 | 2.0 ± 0.17 | 2.1 ± 0.50 |
| 1a (5 + 6) | 4.6 ± 0.25 | IA | IA |
| 2a (7 + 8) | 5.6 ± 1.8 | 4.0 ± 0.35 | 6.6 ± 2.5 |
| 3a (9 + 10) | 3.7 ± 0.45 | 5.9 ± 1.9 | 6.6 ± 3.2 |
| 4a (11 + 12) | 7.8 ± 3.5 | IA | IA |
| 13 | IA | IA | IA |
| 14 | 5.0 ± 0.098 | 6.9 ± 1.2 | 6.0 ± 1.5 |
| 15 | 3.9 ± 0.72 | 5.1 ± 2.5 | 4.2 ± 3.1 |
| 16 | IA | IA | 8.8 ± 1.0 |
| 17 | 9.8 ± 0.16 | IA | IA |
| 18 | 8.9 ± 0.37 | IA | IA |
| 19 | IA | IA | IA |
| 20 | IA | IA | IA |
| Doxorubicin | 0.35 ± 0.033 | 0.22 ± 0.039 | 0.069 ± 0.0090 |
An overview of the IC50 values of these dichapetalins in combination with their structures provided a gross idea about their structure–activity relationship (SAR), which suggested that (a) type-I dichapetalins with a phenyl-butadiene appendage (1–4) showed potent cytotoxic activities against the tested three cancer cell lines, and are more powerful against the NAMALWA cells (IC50 values 0.067–0.15 μM); (b) type-II dichapetalins (5–12) with a phenyl-endoperoxide extension, and type-III dichapetalins (13–16) with a phenyl-furan attachment, showed moderate activities or were inactive against three cancer cell lines; (c) type-IV dichapetalins (17–20) with a phenyl-enedione appendage were nearly inactive; (d) in contrast to the C-3 appendages, the substitution patterns (R1–R4) in the eastern hemispheres of the molecules seemed to have no obvious effects toward the cytotoxic activities.
Experimental
For experimental details see the ESI† material.Conclusions
In conclusion, 18 highly modified triterpenoids, representing four main compound classes, were isolated and characterized from D. gelonioides. A new strategy by using the ΔδC was proposed to assign the configuration of an isomeric pair of phenyl-endoperoxides. A biosynthetic pathway for the formation of four compound classes via cascade modifications was proposed involving diverse chemical events, which was validated by the biomimetic chemical transformations of the four compound classes efficiently in a consecutive way. The biomimetic syntheses not only afforded the desired isolates, but also yielded compounds 16 and 19 that were missed in our purification. The discovery of compounds 1–20 largely expands the structural diversity of the important dichapetalin-type triterpenoids. Particularly, this study provides new insights into the inherent biogenetic connections and the efficient synthetic methods for these four important compound classes; the biological evaluation of 1–20 revealed that the newly added appendages and/or functional groups associated with the A ring are important for cytotoxic activities.Author contributions
B. Z., X. H. G., and J. M. Y. conceived the project and designed the experiments. B. Z. and X. H. G. performed compound isolation and structure identification. M. M. Z. and H. C. L. contributed to the cytotoxic assays. B. Z. and C. Y. Z. performed the biomimetic syntheses. J. M. Y. supervised the project and provided comments on the manuscript. B. Z. and J. M. Y. wrote the manuscript. All authors discussed the results and commented on the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The financial support from the National Natural Science Foundation (No. 21532007), the Drug Innovation Major Project, MOST (2018ZX09711001-001-005), and the Biological Resources Program, CAS (KFJ-BRP-008) of People's Republic of China is gratefully acknowledged. We thank Prof. S.-M Huang, Department of Biology, Hainan University, People's Republic of China for the identification of the plant material.Notes and references
- M. Rueping, K. L. Haack, W. Ieawsuwan, H. Sundén, M. Blanco and F. R. Schoepke, Chem. Commun., 2011, 47, 3828–3830 RSC.
- J. Xu, E. J. E. Caro-Diaz, L. Trzoss and E. A. Theodorakis, J. Am. Chem. Soc., 2012, 134, 5072–5075 CrossRef CAS PubMed.
- A. Grossmann and D. Enders, Angew. Chem., Int. Ed., 2012, 51, 314–325 CrossRef CAS PubMed.
- Y. Ueda, H. Ito, D. Fujita and M. Fujita, J. Am. Chem. Soc., 2017, 139, 6090–6093 CrossRef CAS PubMed.
- B. A. Granger, I. T. Jewett, J. D. Butler, B. Hua, C. E. Knezevic, E. I. Parkinson, P. J. Hergenrother and S. F. Martin, J. Am. Chem. Soc., 2013, 135, 12984–12986 CrossRef CAS PubMed.
- H. M. Huang, P. Bellotti, P. M. Pflüger, J. L. Schwarz, B. Heidrich and F. Glorius, J. Am. Chem. Soc., 2020, 142, 10173–10183 CrossRef CAS PubMed.
- K. C. Nicolaou and J. S. Chen, Chem. Soc. Rev., 2009, 38, 2993–3009 RSC.
- B. Zhou, Y. Wu, S. Dalal, E. F. Merino, Q. F. Liu, C. H. Xu, T. Yuan, J. Ding, D. G. I. Kingston, M. B. Cassera and J. M. Yue, J. Nat. Prod., 2017, 80, 96–107 CrossRef CAS PubMed.
- X. H. Gao, Y. Y. Fan, Q. F. Liu, S. H. Cho, G. F. Pauli, S. N. Chen and J. M. Yue, Org. Lett., 2019, 21, 7065–7068 CrossRef CAS PubMed.
- K. L. Ji, Y. Y. Fan, Z. P. Ge, L. Sheng, Y. K. Xu, L. S. Gan, J. Y. Li and J. M. Yue, J. Org. Chem., 2019, 84, 282–288 CrossRef CAS PubMed.
- S. Li, J. H. Yu, Y. Y. Fan, Q. F. Liu, Z. C. Li, Z. X. Xie, Y. Li and J. M. Yue, J. Org. Chem., 2019, 84, 5195–5202 CrossRef CAS PubMed.
- Y. H. Ren, Q. F. Liu, L. Chen, S. J. He, J. P. Zuo, Y. Y. Fan and J. M. Yue, Org. Lett., 2019, 21, 1904–1907 CrossRef CAS PubMed.
- X. X. Zhu, Y. Y. Fan, L. Xu, Q. F. Liu, J. P. Wu, J. Y. Li, J. Li, K. Gao and J. M. Yue, Org. Lett., 2019, 21, 1471–1474 CrossRef CAS PubMed.
- Y. Y. Fan, S. Q. Shi, G. Z. Deng, H. C. Liu, C. H. Xu, J. Ding, G. W. Wang and J. M. Yue, Org. Lett., 2020, 22, 929–933 CrossRef CAS PubMed.
- D. D. Zhang, J. B. Xu, Y. Y. Fan, L. S. Gan, H. Zhang and J. M. Yue, J. Org. Chem., 2020, 85, 3742–3747 CrossRef CAS PubMed.
- B. Zhang, Y. Wang, S. P. Yang, Y. Zhou, W. B. Wu, W. Tang, J. P. Zuo, Y. Li and J. M. Yue, J. Am. Chem. Soc., 2012, 134, 20605–20608 CrossRef CAS PubMed.
- J. X. Zhao, Y. Y. Yu, S. S. Wang, S. L. Huang, Y. Shen, X. H. Gao, L. Sheng, J. Y. Li, Y. Leng, J. Li and J. M. Yue, J. Am. Chem. Soc., 2018, 140, 2485–2492 CrossRef CAS PubMed.
- B. Zhou, D. X. Liu, X. J. Yuan, J. Y. Li, Y. C. Xu, J. Li, Y. Li and J. M. Yue, Research, 2018, 2018, 2674182 CrossRef CAS PubMed.
- Z. P. Ge, Y. Y. Fan, W. D. Deng, C. Y. Zheng, T. Li and J. M. Yue, Angew. Chem., Int. Ed. Engl., 2021, 60, 9374–9378 CrossRef CAS PubMed.
- H. Achenbach, S. A. Asunka, R. Waibel, I. Addae-Mensah and I. V. Oppong, Nat. Prod. Lett., 1995, 7, 93–100 CrossRef CAS.
- I. Addae-Mensah, R. Waibel, S. A. Asunka, I. V. Oppong and H. Achenbach, Phytochemistry, 1996, 43, 649–656 CrossRef CAS.
- E. Weckert, G. Mattern, I. Addae-Mensah, R. Waibel and H. Achenbach, Phytochemistry, 1996, 43, 657–660 CrossRef CAS.
- D. Osei-Safo, M. A. Chama, I. Addae-Mensah, R. Waibel, W. A. Asomaning and I. V. Oppong, Phytochem. Lett., 2008, 1, 147–150 CrossRef CAS.
- P. Tuchinda, J. Kornsakulkarn, M. Pohmakotr, P. Kongsaeree, S. Prabpai, C. Yoosook, J. Kasisit, C. Napaswad, S. Sophasan and V. Reutrakul, J. Nat. Prod., 2008, 71, 655–663 CrossRef CAS PubMed.
- C. Long, Y. Aussagues, N. Molinier, L. Marcourt, L. Vendier, A. Samson, V. Poughon, P. B. Chalo Mutiso, F. Ausseil, F. Sautel, P. B. Arimondo and G. Massiot, Phytochemistry, 2013, 94, 184–191 CrossRef CAS PubMed.
- S. X. Jing, S. H. Luo, C. H. Li, J. Hua, Y. L. Wang, X. M. Niu, X. N. Li, Y. Liu, C. S. Huang, Y. Wang and S. H. Li, J. Nat. Prod., 2014, 77, 882–893 CrossRef CAS PubMed.
- Y. Ren, C. Yuan, Y. Deng, R. Kanagasabai, T. N. Ninh, V. T. Tu, H.-B. Chai, D. D. Soejarto, J. R. Fuchs, J. C. Yalowich, J. Yu and A. Douglas Kinghorn, Phytochemistry, 2015, 111, 132–140 CrossRef CAS.
- M. A. Chama, G. A. Dziwornu, R. Waibel, D. Osei-Safo, I. Addae-Mensah, J. Otchere and M. Wilson, Pharm. Biol., 2016, 54, 1179–1188 CAS.
- D. Osei-Safo, G. A. Dziwornu, R. Appiah-Opong, M. A. Chama, I. Tuffour, R. Waibel, R. Amewu and I. Addae-Mensah, Molecules, 2017, 22, 1–11 CrossRef PubMed.
- L. Fang, A. Ito, H. B. Chai, Q. Mi, W. P. Jones, D. R. Madulid, M. B. Oliveros, Q. Gao, J. Orjala, N. R. Farnsworth, D. D. Soejarto, G. A. Cordell, S. M. Swanson, J. M. Pezzuto and A. D. Kinghorn, J. Nat. Prod., 2006, 69, 332–337 CrossRef CAS PubMed.
- Y. Y. Fan, H. Zhang, Y. Zhou, H. B. Liu, W. Tang, B. Zhou, J. P. Zuo and J. M. Yue, J. Am. Chem. Soc., 2015, 137, 138–141 CrossRef CAS PubMed.
- Y. Y. Fan, L. S. Gan, H. C. Liu, H. Li, C. H. Xu, J. P. Zuo, J. Ding and J. M. Yue, Org. Lett., 2017, 19, 4580–4583 CrossRef CAS PubMed.
- M. L. Gan, M. T. Liu, L. S. Gan, S. Lin, B. Liu, Y. L. Zhang, J. C. Zi, W. X. Song and J. G. Shi, J. Nat. Prod., 2012, 75, 1373–1382 CrossRef CAS PubMed.
- Y. J. Huang, H. Lu, X. L. Yu, S. W. Zhang, W. Q. Wang, L. Y. Fen and L. J. Xuan, J. Nat. Prod., 2014, 77, 1201–1209 CrossRef CAS PubMed.
- J. H. Yu, H. Zhang, B. Zhou, F. M. Zimbres, S. Dalal, Q. F. Liu, M. B. Cassera and J. M. Yue, J. Nat. Prod., 2020, 83, 1751–1765 CrossRef CAS PubMed.
- J. Li, Y. Y. Hu, X. M. Hao, J. L. Tan, F. Li, X. R. Qiao, S. Z. Chen, C. L. Xiao, M. H. Chen, Z. G. Peng and M. L. Gan, J. Nat. Prod., 2019, 82, 1391–1395 CrossRef CAS PubMed.
- S. Noda, E. Kitazono, T. Tanaka, C. Ogino and A. Kondo, Microb. Cell Fact., 2012, 11, 1–10 CrossRef PubMed.
- K. Ono, M. Nakagawa and A. Nishida, Angew. Chem., Int. Ed., 2004, 43, 2020–2023 CrossRef CAS PubMed.
- R. J. Lee, M. R. Lindley, G. J. Pritchard and M. C. Kimber, Chem. Commun., 2017, 53, 6327–6330 RSC.
- J. M. de Souza, T. J. Brocksom, D. T. McQuade and K. T. de Oliveira, J. Org. Chem., 2018, 83, 7574–7585 CrossRef CAS PubMed.
- S. J. Hayes, D. W. Knight, A. W. T. Smith and M. J. O'Halloran, Tetrahedron Lett., 2010, 51, 720–723 CrossRef CAS.
- Y. Hirata, A. Nakazaki, H. Kawagishi and T. Nishikawa, Org. Lett., 2017, 19, 560–563 CrossRef CAS PubMed.
- T. D. Avery, G. Fallon, B. W. Greatrex, S. M. Pyke, D. K. Taylor and E. R. T. Tiekink, J. Org. Chem., 2001, 66, 7955–7966 CrossRef CAS PubMed.
- C. Dominguez, A. G. Csaky and J. Plumet, Tetrahedron Lett., 1990, 31, 7669–7670 CrossRef CAS.
- K. Gollnick and A. Griesbeck, Tetrahedron Lett., 1984, 25, 4921–4924 CrossRef CAS.
- A. Astarita, F. Cermola, M. DellaGreca, M. R. Iesce, L. Previtera and M. Rubino, Green Chem., 2009, 11, 2030–2033 RSC.
- M. R. Iesce, F. Cermola, A. Piazza, R. Scarpati and M. L. Graziano, Synthesis, 1995, 439–443 CrossRef CAS.
- C. S. Foote, M. Wuesthof, S. Wexler, I. G. Burstain, R. Denny, G. O. Schenck and K. Schultee, Tetrahedron, 1967, 23, 2583–2599 CrossRef CAS.
- K. Gollnick and A. Griesbeck, Tetrahedron, 1985, 41, 2057–2068 CrossRef CAS.
- S. F. Jin, H. L. Ma, Z. L. Liu, S. T. Fu, C. P. Zhang and Y. He, Exp. Cell Res., 2015, 339, 289–299 CrossRef CAS PubMed.
- P. Skehan, R. Storeng, D. Scudiero, A. Monks, J. Mcmahon, D. Vistica, J. T. Warren, H. Bokesch, S. Kenney and M. R. Boyd, J. Natl. Cancer Inst., 1990, 82, 1107–1112 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. CCDC 2010767 for 1, CCDC 2082769 for 3 and CCDC 2010766 for 14. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/d1sc02710g |
| ‡ The authors contribute equally to this work. |
| This journal is © The Royal Society of Chemistry 2021 |