Open Access Article
Open Access ArticleExpanding the reactivity of inorganic clusters towards proteins: the interplay between the redox and hydrolytic activity of Ce(IV)-substituted polyoxometalates as artificial proteases†
Shorok A. M.
Abdelhameed
 a,
Hong Giang T.
Ly
a,
Hong Giang T.
Ly
 ab,
Jens
Moons
a,
Francisco
de Azambuja
ab,
Jens
Moons
a,
Francisco
de Azambuja
 a,
Paul
Proost
c and
Tatjana N.
Parac-Vogt
a,
Paul
Proost
c and
Tatjana N.
Parac-Vogt
 *a
*a
aKU Leuven, Department of Chemistry, Celestijnenlaan 200F, 3001 Leuven, Belgium
bDepartment of Chemistry, College of Natural Sciences, Can Tho University, Can Tho, Vietnam
cKU Leuven Department of Microbiology, Immunology, and Transplantation, Herestraat 49, 3000 Leuven, Belgium. E-mail: tatjana.vogt@kuleuven.be
First published on 16th July 2021
Abstract
The ability of soluble metal-oxo clusters to specifically interact with protein surfaces makes them attractive as potential inorganic drugs and as artificial enzymes. In particular, metal-substituted polyoxometalates (MS-POMs) are remarkably selective in hydrolyzing a range of different proteins. However, the influence of MS-POMs' redox chemistry on their proteolytic activity remains virtually unexplored. Herein we report a highly site-selective hydrolysis of hemoglobin (Hb), a large tetrameric globular protein, by a Ce(IV)-substituted Keggin polyoxometalate (CeIVK), and evaluate the effect of CeIVK's redox chemistry on its reactivity and selectivity as an artificial protease. At pH 5.0, incubation of Hb with CeIVK resulted in strictly selective protein hydrolysis at six Asp-X bonds, two of which were located in the α-chain (α(Asp75-Leu76) and α(Asp94-Pro95)) and five at the β-chain (β(Asp51-Ala52), β(Asp68-Ser69), β(Asp78-Asp79), β(Asp98-Pro99) and β(Asp128-Phe129)). However, increasing the pH of the reaction mixture to 7.4 decreased the CeIVK hydrolytic reactivity towards Hb, resulting in the cleavage of only one peptide bond (β(Asp128-Phe129)). Combination of UV-Vis, circular dichroism and Trp fluorescence spectroscopy indicated similar interactions between Hb and CeIVK at both pH conditions; however, 31P NMR spectroscopy showed faster reduction of CeIVK into the hydrolytically inactive CeIIIK form in the presence of protein at pH 7.4. In agreement with these results, careful mapping of all hydrolyzed Asp-X bonds on the protein structure revealed that the lower reactivity toward the α-chain was consistent with the presence of more redox-active amino acids (Tyr and His) in this subunit in comparison with the β-chain. This points towards a link between the presence of the redox-active sites on the protein surface and efficiency and selectivity of redox-active MS-POMs as artificial proteases. More importantly, the study provides a way to tune the redox and hydrolytic reactivity of MS-POMs towards proteins through adjustment of reaction parameters like temperature and pH.
Introduction
Over the last decade different approaches have been introduced in the field of proteomics in order to increase the speed and accuracy of high throughput sequencing of proteins.1,2 The well-established bottom-up approach depends on protein digestion using natural proteases like trypsin. Although reliable and selective, trypsin produces many short peptide fragments (1–3 kDa) which largely complicates data analysis. On the other hand, the top-down approach, which aims to analyze the protein without a prior digestion step, faces several technical challenges caused mainly by the large size and complexity of protein samples, which limit its widespread application.3 In this context, the middle-down approach, which relies on the analysis of larger protein fragments (3–10 kDa), has emerged as an interesting alternative.4 Analysis of longer peptide fragments results in a better sequence coverage; however, there are still a limited number of natural proteases suitable for the middle-down approach,1,5 which often operate under a limited range of pH and temperature. Therefore, chemical agents which selectively cleave proteins producing middle size fragments are still highly desired.Over the last few years, we have extensively developed metal-substituted polyoxometalates (MS-POMs) as artificial proteases.6–12 Polyoxometalates are a large and diverse group of water-soluble metal–oxygen anionic clusters with distinct chemical and physical properties.13–15 Lacunary POMs, having a defect in their structure, can bind a wide range of metals, generating metal-substituted POMs.16,17 Screening of different MS-POMs revealed that HfIV-, ZrIV- and CeIV-POMs show the highest activity toward peptide bond hydrolysis, presumably due to their pronounced Lewis acidity, flexible geometries and high coordination numbers (up to 12 for Ce vs. 8 for ZrIV and HfIV). Moreover, combining the hydrolytic activity of the embedded metal with the capability of POMs to electrostatically interact with the protein surface endows HfIV, ZrIV and CeIV POMs with high selectivity as artificial proteases. Unlike natural proteases, they are inexpensive, readily available, and stable under a wide range of pH and temperature. In addition, they generally produce 5–15 kDa peptide fragments suitable for middle-down proteomics, thus becoming promising artificial alternatives to current protein digestion methods.18
One intriguing aspect of the reactivity between CeIV-POMs and proteins is that, unlike for redox-stable ZrIV- and HfIV-POMs, the hydrolysis reaction is usually accompanied by a reduction of the embedded CeIV to CeIII.19–21 The resulting CeIII-POM, generated in situ, is essentially hydrolytically inactive due to lower Lewis acidity of CeIII ions as compared to CeIV.22 Recently, we have shown that isolated amino acids – specifically, cysteine, tryptophan, tyrosine, histidine and phenylalanine – can reduce a CeIV-POM to a CeIII-POM in solution.23 Interestingly, the rate of CeIV-POM reduction was significantly decreased when the redox-active amino acid was incorporated in a di/tripeptide moiety, indicating that the accessibility of the MS-POM to the redox-active amino acid residue plays an important role in redox reaction kinetics. In general, CeIV-POM hydrolysis of peptide bonds is much slower than the concurrent redox reactions.19,23 Thus, redox-active amino acids in those peptides reduce a CeIV-POM into an inactive CeIII-POM before the activation of the peptide bond occurs, posing a sound challenge to the development of Ce-POM's peptidase potential.
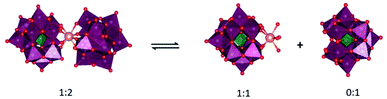
Interestingly, in sharp contrast to this poor activity toward peptides, CeIV-POM exhibited a high hydrolytic activity towards different proteins.19–21 Hen egg white lysozyme (HEWL), transferrin and cytochrome c proteins have redox-active amino acids on their surface that can reduce CeIV to CeIII; nevertheless, their hydrolysis by CeIVK was still observed. One of the hypotheses for this observation was that proteins are able to facilitate the dissociation of CeIVK 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 species to the more hydrolytically active species 1
2 species to the more hydrolytically active species 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 1) under physiological pH, which was not the case in the presence of peptides.24 Furthermore, CeIV-POM accessibility to redox-active amino acids in protein substrates likely has a great impact on the redox reaction rate, which is in agreement with the slower redox reaction observed previously with di/tripeptides in comparison with isolated redox-active amino acids.23
1 (Fig. 1) under physiological pH, which was not the case in the presence of peptides.24 Furthermore, CeIV-POM accessibility to redox-active amino acids in protein substrates likely has a great impact on the redox reaction rate, which is in agreement with the slower redox reaction observed previously with di/tripeptides in comparison with isolated redox-active amino acids.23
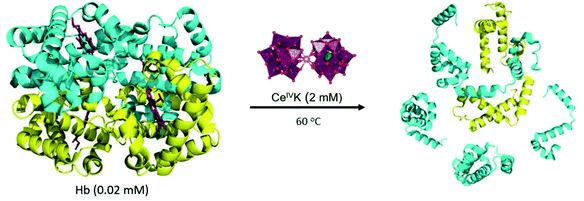
Despite such indications, the redox activity of CeIV-POMs towards proteins, and the interplay between redox and hydrolytic reactions have not been addressed in detail. Although interactions between POMs and proteins have been well studied on a molecular level,18,25–27 the redox chemistry between inorganic clusters and proteins is a virtually unexplored field. Therefore, we set out to investigate the redox reactivity of a CeIV-POM towards a protein substrate and evaluate how such reactivity impacts the hydrolytic reaction that leads to protein fragmentation. For such study we have selected hemoglobin (Hb), a large tetrameric protein consisting of two α- and two β-chain subunits with a total molecular weight of 64.5 kDa.28 Hb is an excellent model for this study due to its large size which can allow multiple CeIV-POM binding sites on the protein surface, and its many surface-exposed tryptophan, tyrosine, and histidine residues, which can engage in electron-transfer reactions,29 allowing us to probe and compare both reaction manifolds with a single protein substrate.
Results and discussion
Redox reactivity of CeIVK towards hemoglobin
We started our study by monitoring the reduction of CeIVK (2 mM) (Fig. 1) in the presence of hemoglobin (Hb, 20 μM) under different pH and temperature values (Fig. 2). The reduction of CeIVK over time was followed by 31P NMR spectroscopy, which conveniently allows for observation of CeIVK (−13.4 ppm) and CeIIIK (−18.7 ppm) through their characteristic 31P NMR peaks.17 The reaction was probed at pH 5.0 and 7.4, at 25 °C, 37 °C and 60 °C, since these are the most common conditions used in MS-POM mediated protein hydrolysis studies.21,24,30,31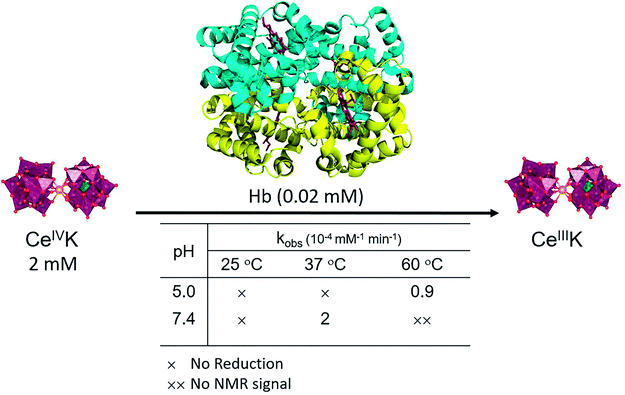 | ||
| Fig. 2 Reduction of CeIVK in the presence of Hb and the observed rate constants (kobs) at different temperature and pH values. | ||
The rate of CeIVK reduction in the presence of Hb is highly dependent on the pH and temperature of the reaction (Fig. 2). At 25 °C, no reduction could be noticed over 3 days, neither at pH 5.0 nor at 7.4, in contrast to what was observed in the presence of free amino acids and di/tripeptides.23 At 37 °C no reduction of CeIVK took place at pH 5.0; however, it was detected to occur at pH 7.4. Based on the disappearance of the CeIVK signal in the 31P NMR (Fig. S1 and S2†), an observed rate constant (kobs) of 2 × 10−4 mM−1 min−1 with a half-life (t½) of 42 h was calculated for the CeIVK (2 mM) reduction in the presence of Hb (0.02 mM) at pH 7.4. At 60 °C, CeIVK was reduced with kobs = 0.9 × 10−4 mM−1 min−1 (t½ = 93 h) at pH 5.0 (Fig. 3). However, in the sample containing CeIVK/Hb at pH 7.4, loss of the CeIVK signal in the 31P NMR spectrum was observed at 60 °C, precluding the detection of any other changes in the reaction mixture. The loss of the 31P NMR signal could be due to the fast exchange at 60 °C, occurring between the free CeIVK and the large CeIVK/Hb complex which has a long correlation time and fast relaxation, resulting in broadening of NMR signals.32 Nevertheless, these results clearly show that both pH and temperature influence the rate of CeIVK reduction in the presence of Hb, as the kobs at pH 7.4 and 37 °C is approximately two times higher than the rate measured at pH 5.0 and 60 °C. These trends are in agreement with our previous findings with isolated amino acids, which have shown that the reduction of CeIVK is faster at higher temperatures, but slower at more acidic pH ranges.23
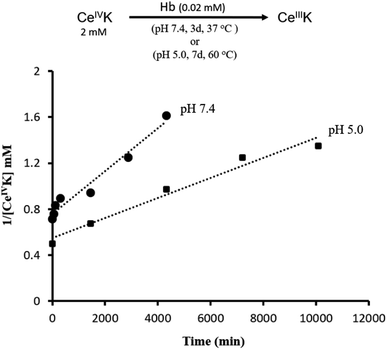 | ||
| Fig. 3 Plot of 1/[CeIVK] (mM) versus time (min) in the presence of Hb, at pH 7.4 and 37 °C, and pH 5.0 and 60 °C, showing second order kinetic in CeIVK. | ||
The absence of CeIVK reduction in the presence of Hb at 25 °C allowed us to investigate its interaction with Hb, without interference from the redox reaction. Due to the presence of two accessible Trp residues (Trp14 residues of α- and β-chains), Trp fluorescence spectroscopy was used to probe the binding strength between the POM and protein (Fig. S3A and S3B†). Similar to previous studies,21,33 addition of CeIVK resulted in the quenching of Trp fluorescence, implying an ongoing CeIVK/Hb interaction. The values of association constant (ka) and the number of CeIVK bound molecules (n) were determined using the derived Stern–Volmer equation giving values of ka = 2.95 × 106 M−1 and n = 1.22 at pH 5.0 (Fig. S3C†). At pH 7.4 (Fig. S3D†), the values of ka and n were 4.8 × 105 M−1 and 1.07, respectively. These results evidence a slightly stronger interaction between CeIVK and Hb at pH 5.0 than at pH 7.4, probably due to the stronger positive charge on the protein surface at more acidic pH. The isoelectric point (pI) of Hb is 6.8, which means that at pH 5.0 the surface of the protein is predominantly positively charged, thus resulting in stronger interaction with the negatively charged CeIVK compared to that at pH 7.4.34,35 Furthermore, 1H NMR, UV-Vis and circular dichroism (CD) spectroscopy confirmed that CeIVK causes conformational changes in the Hb secondary structure causing partial unfolding of the protein (Fig. S4–S6†).36,37 Therefore, the absence of CeIVK reduction at 25 °C cannot be attributed to the lack of interaction with the protein surface. Moreover, such results indicate that the absence of redox reaction at pH 5.0 and 37 °C is also due to the slower redox reactivity at lower pH values, and not due to the absence of CeIVK/Hb interactions.
Since Hb is a heme protein, and the oxidation state of iron can be detected by using UV-Vis,38,39 we have probed whether the iron of the heme group might be responsible for the reduction of CeIVK using UV-Vis spectra of Hb solutions recorded in the absence and presence of CeIVK under different conditions. Four oxidation states are known for the iron-heme group in Hb: deoxyHb (FeII-Hb), oxyHb (Fe(II)O2-Hb), metHb (FeIII-Hb) and FerriHb (FeIV-Hb) with corresponding Soret absorption bands at λmax = 430, 415, 405 and 418 nm, respectively.40Fig. 4 show that, in the absence of CeIVK, the Soret band is at 405 nm, which can be attributed to an FeIII-Hb form. Incubation of Hb alone for 3 h at 60 °C and pH 7.4 did not change the position of the Soret band, though its intensity decreased as a consequence of partial unfolding of the protein caused by high temperature and small protein precipitation which was observed in solution. Upon CeIVK addition, directly after mixing, a slight red shift to 411 nm was detected. A similar small red shift was also observed in the presence of the redox-inactive Zr(IV)-Keggin (ZrIVK), which indicates that it is most likely caused by POM/Hb interactions, while the heme iron (FeIII) oxidation state remained unchanged.41 Monitoring this mixture over 5 h at 60 °C and pH 7.4 did not affect the position of the band, and only led to a partial decrease in its intensity, presumably due to changes in the protein's secondary structure and its partial unfolding arising from CeIVK/Hb interactions, which were also observed by CD spectroscopy. Additionally, the pH of solution was stable throughout the reaction, which supports the view that the observed changes mainly originate from CeIVK/Hb interactions and not from pH changes. Similar results were obtained when Hb was titrated with different concentrations of CeIVK at pH 5.0 and 25 °C (Fig. S5B†). These observations are in agreement with the literature, which previously reported that the shift in the Soret band from 405 to 411 is due to the changes in the coordination environment around Fe(III) of the heme group.38 Together, these results strongly suggest that the CeIVK reduction is not related to the traditional redox chemistry of the heme group, which in turn indicates that redox-active amino acid residues are likely involved.
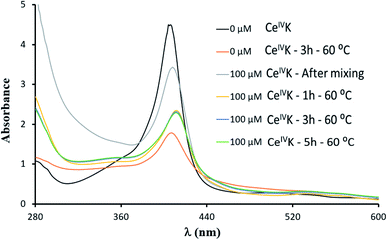 | ||
| Fig. 4 UV-Vis absorbance spectra of Hb (10 μM) in the absence and presence of CeIVK (100 μM) at pH 7.4 and 60 °C. | ||
Hydrolysis of hemoglobin in the presence of CeIVK
In the following step the ability of CeIVK to hydrolyze Hb was studied under different reaction conditions (Fig. 5). Reaction aliquots were taken at different time increments and analyzed using a denaturing polyacrylamide gel in the presence of Tricine (SDS-Tricine-PAGE). The SDS-PAGE chromatogram showed the presence of only monomer bands due to the Hb denaturation via sodium dodecyl sulphate (SDS) which results in breaking of non-covalent interactions.42Fig. 6 shows the SDS-PAGE image of Hb and its fragmentation in the presence of CeIVK at pH 5.0 and 60 °C. In addition to the intact α and β chains at ca. 16 kDa, the appearance of new bands with molecular weight lower than 16 kDa indicated that Hb was hydrolyzed in the presence of CeIVK in the course of the reaction. The selective hydrolysis in the presence of CeIVK is also clear in Fig. 6, which shows that all fragments appeared after 48 hours, and their intensity increased over longer reaction times. In contrast, in control experiments without CeIVK or in the presence of a lacunary Keggin-type POM (PW9) only the intact protein was observed. The addition of cerium ammonium nitrate (CAN) salt to Hb resulted in the precipitation of Hb which led to a very faint band on the SDS-PAGE. Interestingly, no hydrolysis was observed even in the presence of CeIIIK, strongly suggesting that complete reduction of CeIVK in situ would lead to full loss of catalytic activity.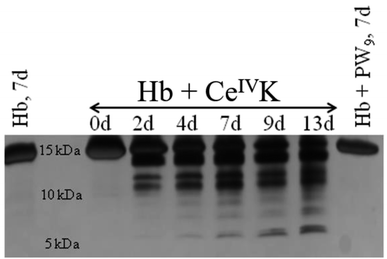 | ||
| Fig. 6 Silver stained SDS-PAGE gel for the hydrolysis of Hb (0.02 mM) at pH 5.0 and 60 °C in the presence of CeIVK (2 mM) at different time increments or PW9 (2 mM). | ||
Additionally, the hydrolytic activity of CeIVK towards Hb was followed at pH 7.4 and only one hydrolytic fragment (14.0 kDa) could be observed over 6 days of incubation at 60 °C (Fig. S7†). Thus, the hydrolytic activity is much higher at pH 5.0 than at pH 7.4, which could be due to several factors. Firstly, the isoelectric point (pI) of Hb is 6.8, which means that at pH 5.0 the surface of the protein is predominantly positively charged and interacts more strongly with the negatively charged POM surface than at pH 7.4. This is in agreement with the tryptophan fluorescence results discussed above, which point to a stronger interaction at lower pH, and should result in more cleavages.34,35 Secondly, at pH < 6, Hb tends to dissociate and form dimers (αβ) or even monomers, making cleavage sites more accessible to CeIVK.43,44 Thirdly, since at more acidic pH the reduction of CeIVK is much slower this maintains the integrity of CeIVK active species longer than at neutral pH.23
Selectivity of hemoglobin hydrolysis by CeIVK
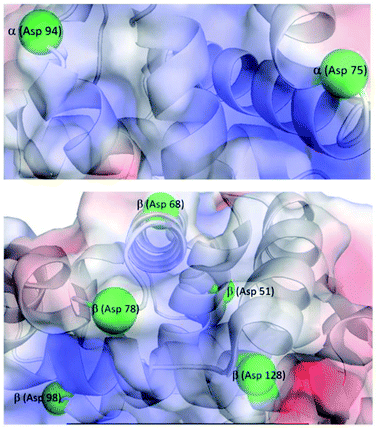
The specific cleavage sites of Hb fragments generated by CeIVK were probed by blotting the fragments on a PVDF membrane and the NH2-terminal sequence of each Coomassie stained fragment was defined by Edman degradation. The estimated molecular mass of the bands on the SDS-Tricine-PAGE gel and the results obtained from Edman degradation unambiguously confirmed that at pH = 5.0 Hb was hydrolyzed at seven peptide bonds (Table S1 and Fig. S8†). Remarkably, Hb was exclusively cleaved only at Asp-X peptide bonds, two of which were found in the α-chain (α(Asp75-Leu76) and α(Asp94-Pro95)) and the other five in the β-chain (β(Asp51-Ala52), β(Asp68-Ser69), β(Asp78-Asp79), β(Asp98-Pro99) and β(Asp128-Phe129)). Interestingly, only one peptide bond, Asp128-Phe129, found in the β-chain of Hb, was cleaved at pH 7.4. Several studies have shown that the interactions between proteins and MS-POMs are either electrostatic in nature, occurring between the negative surface of the MS-POM and the positive patches of the proteins, or may occur via interactions between the substituted metal and the side chain of amino acids.24-26,45,46Fig. 7 shows that all hydrolyzed Asp-X bonds are present near the accessible surface of the protein and in the vicinity of the positively charged regions of Hb, which is consistent with the more efficient hydrolysis observed at lower pH, as decreasing pH leads to protonation of side chains in some amino acid residues. Moreover, five sites from the seven cleavage sites are located in the random coil region and are easier to hydrolyze than the more structured protein regions.11,41,47The hydrolysis of Hb by ZrIVK reported previously resulted in the same selectivity towards Asp-X bonds; however, CeIVK is only able to hydrolyze Hb at seven Asp-X sites, while ZrIVK was found to cleave Hb at eleven Asp-X sites.41 For the α-chain subunit, ZrIVK hydrolyzed at seven Asp-X positions, but CeIVK cleaved only two Asp-X peptide bonds. On the other hand, both ZrIVK and CeIVK hydrolyzed β-chain subunits at six Asp-X sites (Table S2†). As discussed below, the fewer cleavage sites observed for CeIVK might be related to the loss of activity imparted by in situ reduction to inactive CeIIIK, which is not the case for the redox stable ZrIVK complex.
Redox vs. hydrolytic activity of CeIVK
This study reveals the correlation between the redox activity and the hydrolytic activity of CeIVK towards Hb. As shown above, the reduction of CeIVK decreases its hydrolytic activity due to the formation of the hydrolytically inactive CeIIIK. According to the results, reduction of CeIVK is always faster than the hydrolysis reaction, and this could be attributed to several points. Firstly, unlike the hydrolytic reaction, the redox reaction does not need a direct interaction between the acceptor and the donor since the transfer of electrons can be mediated by the solvent;48 secondly, MS-POMs usually bind proteins by pointing their hydrolytically active metal center towards the solvent, requiring an extra ‘reorientation step’ to turn the active metal center of MS-POMs towards the backbone of the protein.32,49,50 Thirdly, CeIVK 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 species needs to dissociate and form first the hydrolytically active 1
2 species needs to dissociate and form first the hydrolytically active 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 species (Fig. 1) to be able to hydrolyze proteins, while this is not necessary for the redox reaction.23
1 species (Fig. 1) to be able to hydrolyze proteins, while this is not necessary for the redox reaction.23
The rate of CeIVK reduction or the rate of electron transfer (ET) mainly depends on the distance between the electron donor (redox-active amino acids) and acceptor (CeIVK). A direct interaction between the electron donor and acceptor will lead to a high ET rate which explains the fast reduction of CeIVK in the presence of free amino acids or simple peptides. However, in the presence of proteins, direct interaction between CeIVK and redox-active amino acids might be hindered, as the redox-active amino acids are not always surface-exposed. Therefore, the ET process in proteins is usually a long-range ET process that happens via different pathways depending on the distance between the electron donor and acceptor, rather than via direct electron transfer between the electron donor and acceptor observed with free amino acids and simple peptides.51–53
The lower hydrolytic activity of CeIVK (7 cleavage sites) in comparison with ZrIVK (11 cleavage sites) towards Hb might be due to the presence of surface-exposed redox-active amino acids, which could cause the reduction of CeIVK and lead to loss of its hydrolytic activity due to the formation of CeIIIK (Fig. 8). Furthermore, the presence of more cleavage sites on the β-chain than the α-chain could be attributed not only to the presence of more surface-exposed redox-active amino acids but more specifically to the presence of more His residues in the random coil region on the α-chain than the β-chain (Table 1). This could make CeIVK more susceptible to reduction upon binding to the α-chain, as it has been reported before that MS-POMs favor binding near the random coil regions of proteins.41,47 Although we previously reported the low redox activity of His towards the reduction of CeIVK due to its high redox potential,23 His is known to play a crucial rule in long-range ET in proteins which could facilitate the reduction of CeIVK in the presence of Hb.54 Moreover, the His imidazole side chain (IM) is able to bind to different types of metal strongly, and among all the redox-active amino acids, His showed the strongest binding with CeIVK.55 Table S3† shows the 1H and 13C NMR chemical shift values of the free His in the absence and presence of CeIVK, revealing that the highest chemical shift values (Δδ) are related to the IM ring, which indicates a strong binding between CeIVK and the imidazole ring. Therefore, it is plausible that CeIVK bind to the surface-exposed imidazole rings which will facilitate the process of electron transfer and eventually the formation of CeIIIK. These observations support the above-mentioned results of the fast reduction of CeIVK at pH 7.4 since pKa of IM is ∼6 which facilitates the binding between CeIV ions and the IM side chain at physiological pH.
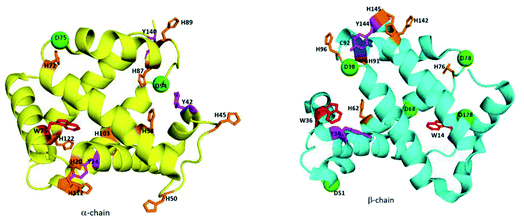 | ||
| Fig. 8 Positions of the redox-active amino acids and the hydrolysable Asp residues in Hb; Trp (red), Tyr (magenta), Cys (blue), His (orange) and hydrolysable Asp (D, green). | ||
| Redox-active amino acids | α-Chain | β-Chain |
|---|---|---|
| Cys (C) | — | C92 |
| Tyr (Y) | Y24, Y42, Y140 | Y34, Y144 |
| Trp (W) | W14 | W14, W36 |
| His (H) | H20*, H45*, H50*, H58, H72*, H87, H89*, H103, H112*, H122 | H62, H76*, H91, H96*, H145, H142 |
Finally, taking into account the possible equilibrium between CeIVK 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and CeIVK 1
2 and CeIVK 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 species, the obtained evidence makes it difficult to unambiguously assign whether both monomeric and dimeric forms, or just one of them is responsible for the observed redox behavior. While both species can in principle be reduced in solution, previously reported data for the CeIVK/CeIIIK redox pair indicate that the reduction of the 1
1 species, the obtained evidence makes it difficult to unambiguously assign whether both monomeric and dimeric forms, or just one of them is responsible for the observed redox behavior. While both species can in principle be reduced in solution, previously reported data for the CeIVK/CeIIIK redox pair indicate that the reduction of the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 species is more favored than that of the 1
1 species is more favored than that of the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 dimeric complex (0.74 vs. 0.42 V).56 In our previous study of the CeIVK 1
2 dimeric complex (0.74 vs. 0.42 V).56 In our previous study of the CeIVK 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 reduction in the presence of amino acids, no evidence was observed for the dissociation of the dimeric complex, which indicates that the 1
2 reduction in the presence of amino acids, no evidence was observed for the dissociation of the dimeric complex, which indicates that the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 complex might be redox-active even without dissociation.23 In the current study, the stronger POM–protein interaction observed at lower pH should also lead to more pronounced dissociation of CeIVK 1
2 complex might be redox-active even without dissociation.23 In the current study, the stronger POM–protein interaction observed at lower pH should also lead to more pronounced dissociation of CeIVK 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2.24 However, under such conditions the slower reduction of CeIVK was also observed. Thus, these facts alone are not sufficient to conclude that in the presence of a protein the CeIVK 1
2.24 However, under such conditions the slower reduction of CeIVK was also observed. Thus, these facts alone are not sufficient to conclude that in the presence of a protein the CeIVK 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 form is more redox-active than the CeIVK 1
2 form is more redox-active than the CeIVK 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 form. The detailed redox investigation of CeIVK 1
1 form. The detailed redox investigation of CeIVK 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 redox activity in the presence of a protein is hampered by the fast equilibrium between 1
1 redox activity in the presence of a protein is hampered by the fast equilibrium between 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and 1
2 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 forms, and by the low concentration of the CeIVK 1
1 forms, and by the low concentration of the CeIVK 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 species at near-neutral pH values. In addition, the reduction of monomeric CeIVK 1
1 species at near-neutral pH values. In addition, the reduction of monomeric CeIVK 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to CeIIIK 1
1 to CeIIIK 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 would reform the expected CeIIIK 1
1 would reform the expected CeIIIK 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, as detected by 31P NMR spectroscopy (Fig. S2†).
2, as detected by 31P NMR spectroscopy (Fig. S2†).
Conclusion
In summary, the interplay between the redox and hydrolytic activities of a Ce(IV)-substituted POM as an artificial protease was probed using a large model protein with many surface-exposed tryptophan, tyrosine, and histidine residues, which are known to engage in electron transfer reactions. The combined kinetic and spectroscopic studies suggest that the redox activity of CeIVK governs its hydrolytic activity towards proteins. Careful inspection of the redox-active sites on the surface of Hb suggests a crucial role of His residues in the reduction of CeIVK, which ultimately influences its hydrolytic reactivity, and explains the more extensive hydrolysis of the β-chain. Moreover, in agreement with the key role attributed to histidine residues, a profound effect of pH on the efficiency of both redox and hydrolytic reactivities of CeIVK as an artificial protease has been observed. At pH 7.4, fast reduction of CeIVK to hydrolytically inactive CeIIIK was observed, resulting in the decrease of hydrolytic activity and hydrolysis of only one Asp-X peptide bond in hemoglobin. However, slower CeIVK reduction at pH 5.0 allowed for seven Asp-X peptide bonds in Hb to be hydrolyzed. Together, these results indicate that efficiency and selectivity of redox-active MS-POMs may be controlled through careful tuning of reaction parameters like temperature and pH, which can be further exploited in their development as artificial proteases in middle-down proteomics applications. To the best of our knowledge, this is the first study that correlates the redox chemistry and the hydrolytic reactivity of Ce-containing inorganic clusters toward proteins, thereby enhancing the general understanding of their redox activity towards biomolecules. Considering the wide range of reported biological applications of inorganic clusters,57–61 this study also contributes to their development as inorganic drugs with antibacterial, antiviral and antitumor properties.Data availability
All data supporting the findings of this study are available within the paper and its ESI files.Author contributions
S. A. M. A., H. G. T. L. and J. M. performed the experiments reported in this work. F. d. A. , P. P. and T. N. P. V. helped with the design and interpretation of the experiments. The manuscript was written through contributions of S. A. M. A., F. d. A., H. G. T. L. and T. N. P. V. All authors have given approval to the final version of the manuscript.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
We thank KU Leuven and the Research Foundation – Flanders (FWO) for funding. S. A. M. A., F. d. A. (195931/1281921N) and H. G. T. L. thank the FWO for fellowships.References
- L. Tsiatsiani and A. J. R. Heck, Proteomics beyond Trypsin, FEBS J., 2015, 282(14), 2612–2626, DOI:10.1111/febs.13287.
- F. Lermyte, Y. O. Tsybin, P. B. O'connor and J. A. Loo, Top or Middle? Up or Down? Toward a Standard Lexicon for Protein Top-Down and Allied Mass Spectrometry Approaches, J. Am. Soc. Mass Spectrom., 2019, 30, 1149–1157, DOI:10.1007/s13361-019-02201-x.
- A. Armirotti and G. Damonte, Achievements and Perspectives of Top-down Proteomics, Proteomics, 2010, 10(20), 3566–3576, DOI:10.1002/pmic.201000245.
- P. B. Pandeswari and V. Sabareesh, Middle-down Approach: A Choice to Sequence and Characterize Proteins/Proteomes by Mass Spectrometry, RSC Adv., 2019, 9, 313, 10.1039/c8ra07200k.
- A. Cristobal, F. Marino, H. Post, H. W. P. Van Den Toorn, S. Mohammed and A. J. R. Heck, Toward an Optimized Workflow for Middle-Down Proteomics, Anal. Chem., 2017, 89(6), 3318–3325, DOI:10.1021/acs.analchem.6b03756.
- G. Absillis and T. N. Parac-Vogt, Peptide Bond Hydrolysis Catalyzed by the Wells-Dawson Zr(α2-P2W17O61)2 Polyoxometalate, Inorg. Chem., 2012, 51(18), 9902–9910, DOI:10.1021/ic301364n.
- H. G. T. Ly, G. Absillis and T. N. Parac-Vogt, Amide Bond Hydrolysis in Peptides and Cyclic Peptides Catalyzed by a Dimeric Zr(IV)-Substituted Keggin Type Polyoxometalate, Dalton Trans., 2013, 42(30), 10929–10938, 10.1039/c3dt50705j.
- K. Stroobants, G. Absillis, E. Moelants, P. Proost and T. N. Parac-Vogt, Regioselective Hydrolysis of Human Serum Albumin by Zr IV-Substituted Polyoxotungstates at the Interface of Positively Charged Protein Surface Patches and Negatively Charged Amino Acid Residues, Chem.–Eur. J., 2014, 20(14), 3894–3897, DOI:10.1002/chem.201303622.
- K. Stroobants, V. Goovaerts, G. Absillis, G. Bruylants, E. Moelants, P. Proost and T. N. Parac-Vogt, Molecular Origin of the Hydrolytic Activity and Fixed Regioselectivity of a ZrIV-Substituted Polyoxotungstate as Artificial Protease, Chem.–Eur. J., 2014, 20(31), 9567–9577, DOI:10.1002/chem.201402683.
- A. Sap, G. Absillis and T. N. Parac-Vogt, Selective Hydrolysis of Oxidized Insulin Chain B by a Zr(IV)-Substituted Wells–Dawson Polyoxometalate, Dalton Trans., 2014, 44(4), 1539–1548, 10.1039/c4dt01477d.
- H. G. T. Ly, G. Absillis, R. Janssens, P. Proost and T. N. Parac-Vogt, Highly Amino Acid Selective Hydrolysis of Myoglobin at Aspartate Residues as Promoted by Zirconium(IV)-Substituted Polyoxometalates, Angew. Chem., Int. Ed., 2015, 54(25), 7391–7394, DOI:10.1002/anie.201502006.
- T. Quanten, T. De Mayaer, P. Shestakova and T. N. Parac-Vogt, Selectivity and Reactivity of ZrIV and CeIV Substituted Keggin Type Polyoxometalates Toward Cytochrome c in Surfactant Solutions, Front. Chem., 2018, 6, 372, DOI:10.3389/fchem.2018.00372.
- M. T. Pope and A. Müller, Polyoxometalate Chemistry: An Old Field with New Dimensions in Several Disciplines, Angew. Chem., Int. Ed. Engl., 1991, 30(1), 34–48, DOI:10.1002/anie.199100341.
- M. Ammam, Polyoxometalates: Formation, Structures, Principal Properties, Main Deposition Methods and Application in Sensing, J. Mater. Chem. A, 2013, 1(21), 6291, 10.1039/c3ta01663c.
- N. I. Gumerova and A. Rompel, Synthesis, Structures and Applications of Electron-Rich Polyoxometalates, Nat. Rev. Chem., 2018, 112, DOI:10.1038/s41570-018-0112 , Nature Publishing Group..
- J. J. Carbó, C. Bo and J. M. Poblet, Structure and Reactivity of Polyoxometalates, 2013, vol. 9. DOI: DOI:10.1016/B978-0-08-097774-4.00937-2.
- M. Arefian, M. Mirzaei, H. Eshtiagh-Hosseini and A. Frontera, A Survey of the Different Roles of Polyoxometalates in Their Interaction with Amino Acids, Peptides and Proteins, Dalton Trans., 2017, 46(21), 6812–6829, 10.1039/C7DT00894E.
- F. de Azambuja, J. Moons and T. N. Parac-Vogt, The Dawn of Metal-Oxo Clusters as Artificial Proteases: From Discovery to the Present and Beyond, Acc. Chem. Res., 2021, 54, 1673–1684, DOI:10.1021/acs.accounts.0c00666.
- K. Stroobants, E. Moelants, H. G. T. Ly, P. Proost, K. Bartik and T. N. Parac-Vogt, Polyoxometalates as a Novel Class of Artificial Proteases: Selective Hydrolysis of Lysozyme under Physiological PH and Temperature Promoted by a Cerium(IV) Keggin-Type Polyoxometalate, Chem.–Eur. J., 2013, 19(8), 2848–2858, DOI:10.1002/chem.201203020.
- A. Sap, L. Van Tichelen, A. Mortier, P. Proost and T. N. Parac-Vogt, Tuning the Selectivity and Reactivity of Metal-Substituted Polyoxometalates as Artificial Proteases by Varying the Nature of the Embedded Lewis Acid Metal Ion, Eur. J. Inorg. Chem., 2016, 2016(32), 5098–5105, DOI:10.1002/ejic.201601098.
- J. Moons, L. S. Van Rompuy, A. Rodriguez, S. A. M. Abdelhameed, W. Simons and T. N. Parac-Vogt, Hydrolysis of Transferrin Promoted by a Cerium(IV)-Keggin Polyoxometalate, Polyhedron, 2019, 170, 570–575, DOI:10.1016/J.POLY.2019.06.010.
- T. Takarada, M. Yashiro and M. Komiyama, Catalytic Hydrolysis of Peptides by Cerium(IV), Chem–Eur. J., 2000, 6(21), 3906–3913, DOI:10.1002/1521-3765(20001103)6:21<3906::aid-chem3906>3.3.co;2-a.
- S. A. M. Abdelhameed, L. Vandebroek, F. de Azambuja and T. N. Parac-Vogt, Redox Activity of Ce(IV)-Substituted Polyoxometalates toward Amino Acids and Peptides, Inorg. Chem., 2020, 59, 10569–10577, DOI:10.1021/acs.inorgchem.0c00993.
- L. Vandebroek, E. De Zitter, H. G. T. Ly, D. Conić, T. Mihaylov, A. Sap, P. Proost, K. Pierloot, L. Van Meervelt and T. N. Parac-Vogt, Protein-Assisted Formation and Stabilization of Catalytically Active Polyoxometalate Species, Chem.–Eur. J., 2018, 24(40), 10099–10108, DOI:10.1002/chem.201802052.
- A. Bijelic and A. Rompel, Polyoxometalates: More than a Phasing Tool in Protein Crystallography, ChemTexts, 2018, 4(3), 10, DOI:10.1007/s40828-018-0064-1.
- A. Bijelic and A. Rompel, Ten Good Reasons for the Use of the Tellurium-Centered Anderson-Evans Polyoxotungstate in Protein Crystallography, Acc. Chem. Res., 2017, 50(6), 1441–1448, DOI:10.1021/acs.accounts.7b00109.
- M. Zhao, X. Chen, G. Chi, D. Shuai, L. Wang, B. Chen and J. Li, Research Progress on the Inhibition of Enzymes by Polyoxometalates, Inorg. Chem. Front., 2020, 7(22), 4320–4332, 10.1039/d0qi00860e.
- R. Bhomia, V. Trivedi, N. J. Coleman and J. C. Mitchell, The Thermal and Storage Stability of Bovine Haemoglobin by Ultraviolet-Visible and Circular Dichroism Spectroscopies, J. Pharm. Anal., 2016, 6, 242–248, DOI:10.1016/j.jpha.2016.02.004.
- R. Aranda IV, H. Cai, C. E. Worley, E. J. Levin, R. Li, J. S. Olson, G. N. Phillips and M. P. Richards, Structural Analysis of Fish versus Mammalian Hemoglobins: Effect of the Heme Pocket Environment on Autooxidation and Hemin Loss, Proteins: Struct., Funct., Bioinf., 2009, 75(1), 217–230, DOI:10.1002/prot.22236.
- L. S. Van Rompuy, N. D. Savić, A. Rodriguez and T. N. Parac-Vogt, Selective Hydrolysis of Transferrin Promoted by Zr-Substituted Polyoxometalates, Molecules, 2020, 25(15), 3472, DOI:10.3390/molecules25153472.
- H. G. T. Ly, G. Absillis, R. Janssens, P. Proost and T. N. Parac-Vogt, Highly Amino Acid Selective Hydrolysis of Myoglobin at Aspartate Residues as Promoted by Zirconium(IV)-Substituted Polyoxometalates, Angew. Chem., Int. Ed., 2015, 54(25), 7391–7394, DOI:10.1002/anie.201502006.
- A. Sap, E. De Zitter, L. Van Meervelt and T. N. Parac-Vogt, Structural Characterization of the Complex between Hen Egg-White Lysozyme and Zr IV-Substituted Keggin Polyoxometalate as Artificial Protease, Chem.–Eur. J., 2015, 21(33), 11692–11695, DOI:10.1002/chem.201501998.
- H. G. T. Ly and T. N. Parac-Vogt, Spectroscopic Study of the Interaction between Horse Heart Myoglobin and Zirconium(IV)-Substituted Polyoxometalates as Artificial Proteases, ChemPhysChem, 2017, 18(18), 2451–2458, DOI:10.1002/cphc.201700680.
- A. Conway-Jacobs and L. M. Lewin, Isoelectric Focusing in Acrylamide Gels: Use of Amphoteric Dyes as Internal Markers for Determination of Isoelectric Points, Anal. Biochem., 1971, 43(2), 394–400, DOI:10.1016/0003-2697(71)90269-7.
- P. Novák and V. Havlíček, Protein Extraction and Precipitation, in Proteomic Profiling and Analytical Chemistry: The Crossroads, Elsevier Inc., 2nd edn, 2016, pp. 52–62. DOI: DOI:10.1016/B978-0-444-63688-1.00004-5.
- S. M. Kelly, T. J. Jess and N. C. Price, How to Study Proteins by Circular Dichroism, Biochim. Biophys. Acta, 2005, 1751, 119–139, DOI:10.1016/j.bbapap.2005.06.005.
- H. Zhang, Y. Liu, R. Zhang, R. Liu and Y. Chen, Binding Mode Investigations on the Interaction of Lead(II) Acetate with Human Chorionic Gonadotropin, J. Phys. Chem. B, 2014, 118(32), 9644–9650, DOI:10.1021/jp505565s.
- W. Liu, X. Guo and R. Guo, The Interaction between Hemoglobin and Two Surfactants with Different Charges, Int. J. Biol. Macromol., 2007, 41(5), 548–557, DOI:10.1016/j.ijbiomac.2007.07.006.
- L. M. Moreira, A. L. Poli, A. J. Costa-Filho and H. Imasato, Ferric Species Equilibrium of the Giant Extracellular Hemoglobin of Glossoscolex paulistus in Alkaline Medium: HALS Hemichrome as a Precursor of Pentacoordinate Species, Int. J. Biol. Macromol., 2008, 42(2), 103–110, DOI:10.1016/j.ijbiomac.2007.10.001.
- L. Gebicka and E. Banasiak, Flavonoids as Reductants of Ferryl Hemoglobin, Acta Biochim. Pol., 2009, 56(3), 509–513 CrossRef CAS PubMed.
- H. G. T. Ly, T. T. Mihaylov, P. Proost, K. Pierloot, J. N. Harvey and T. N. Parac-Vogt, Chemical Mimics of Aspartate-Directed Proteases: Predictive and Strictly Specific Hydrolysis of a Globular Protein at Asp–X Sequence Promoted by Polyoxometalate Complexes Rationalized by a Combined Experimental and Theoretical Approach, Chem.–Eur. J., 2019, 25(63), 14370–14381, DOI:10.1002/chem.201902675.
- H. Sakai, Y. Masada, S. Takeoka and E. Tsuchida, Characteristics of Bovine Hemoglobin as a Potential Source of Hemoglobin-Vesicles for an Artificial Oxygen Carrier 1, 2002, vol. 131 Search PubMed.
- A. R. Fanelli, E. Antonini and A. Caputo, Hemoglobin and Myoglobin, Adv. Protein Chem., 1964, 19(C), 73–222, DOI:10.1016/S0065-3233(08)60189-8.
- Y. X. Huang, Z. J. Wu, B. T. Huang and M. Luo, Pathway and Mechanism of PH Dependent Human Hemoglobin Tetramer-Dimer- Monomer Dissociations, PLoS One, 2013, 8(11), 81708, DOI:10.1371/journal.pone.0081708.
- L. Vandebroek, L. Van Meervelt and T. N. Parac-Vogt, Direct Observation of the ZrIV Interaction with the Carboxamide Bond in a Noncovalent Complex between Hen Egg White Lysozyme and a Zr-Substituted Keggin Polyoxometalate, Acta Crystallogr., Sect. C Struct. Chem., 2018, 74(11), 1348–1354, DOI:10.1107/S2053229618010690.
- L. Wu, J. Liang, L. Wu and Á. J. Liang, Polyoxometalates and Their Complexes Toward Biological Application, 2017. DOI: DOI:10.1007/978-981-10-6059-5_13.
- A. Solé-Daura, A. Rodríguez-Fortea, J. M. Poblet, D. Robinson, J. D. Hirst and J. J. Carbó, Origin of Selectivity in Protein Hydrolysis by Zr(IV)-Containing Metal Oxides as Artificial Proteases, ACS Catal., 2020, 10, 13455–13467, DOI:10.1021/acscatal.0c02848.
- K. L. Nash and J. C. Sullivan, Chapter 102 Kinetics of Complexation and Redox Reactions of the Lanthanides in Aqueous Solutions, Handbook on the Physics and Chemistry of Rare Earths, Elsevier, 1991, pp. 347–391. DOI: DOI:10.1016/S0168-1273(05)80008-X.
- A. Solé-Daura, V. Goovaerts, K. Stroobants, G. Absillis, P. Jiménez-Lozano, J. M. Poblet, J. D. Hirst, T. N. Parac-Vogt and J. J. Carbó, Probing Polyoxometalate-Protein Interactions Using Molecular Dynamics Simulations, Chem.–Eur. J., 2016, 22(43), 15280–15289, DOI:10.1002/chem.201602263.
- L. Vandebroek, Y. Mampaey, S. Antonyuk, L. Van Meervelt and T. N. Parac-Vogt, Noncovalent Complexes Formed between Metal-Substituted Polyoxometalates and Hen Egg White Lysozyme, Eur. J. Inorg. Chem., 2019, 2019(3–4), 506–511, DOI:10.1002/ejic.201801113.
- A. Shah, B. Adhikari, S. Martic, A. Munir, S. Shahzad, K. Ahmad and H.-B. Kraatz, Electron Transfer in Peptides, Chem. Soc. Rev., 2015, 44, 1015, 10.1039/c4cs00297k.
- J. Lin, I. A. Balabin and D. N. Beratan, Chemistry: The Nature of Aqueous Tunneling Pathways between Electron-Transfer Proteins, Science, 2005, 310(5752), 1311–1313, DOI:10.1126/science.1118316.
- D. N. Beratan, J. N. Onuchic, J. R. Winkler and H. B. Gray, Electron-Tunneling Pathways in Proteins, Science, 1992, 258, 1740–1741, DOI:10.1126/science.1334572.
- M. Di Donato and A. Peluso, A Possible Role of Histidine Residues in Long-Range Electron Transfer in Proteins, Theor. Chem. Acc., 2004, 111(2–6), 303–310, DOI:10.1007/s00214-003-0536-1.
- N. Tsud, R. G. Acres, M. Iakhnenko, D. Mazur, K. C. Prince and V. Matolín, Bonding of Histidine to Cerium Oxide, J. Phys. Chem. B, 2013, 117(31), 9182–9193, DOI:10.1021/jp404385h.
- N. Haraguchi, Y. Okaue, T. Isobe and Y. Matsuda, Stabilization of Tetravalent Cerium upon Coordination of Unsaturated Heteropolytungstate Anions, Inorg. Chem., 1994, 33(6), 1015–1020, DOI:10.1021/ic00084a008.
- T. Yamase, Anti-Tumor, -Viral, and -Bacterial Activities of Polyoxometalates for Realizing an Inorganic Drug, J. Mater. Chem., 2005, 15(45), 4773, 10.1039/b504585a.
- A. Bijelic, M. Aureliano and A. Rompel, The Antibacterial Activity of Polyoxometalates: Structures, Antibiotic Effects and Future Perspectives, Chem. Commun., 2018, 54(54), 1153–1169 RSC.
- A. Bijelic, M. Aureliano and A. Rompel, Polyoxometalates as Potential Next-Generation Metallodrugs in the Combat Against Cancer, Angew. Chem., Int. Ed., 2019, 58(10), 2980–2999, DOI:10.1002/anie.201803868.
- L. S. Van Rompuy and T. N. Parac-Vogt, Interactions between Polyoxometalates and Biological Systems: From Drug Design to Artificial Enzymes, Curr. Opin. Biotechnol., 2019, 58, 92–99, DOI:10.1016/J.COPBIO.2018.11.013.
- A. G. Enderle, M. Bosso, R. Groß, M. Heiland, M. Bollini, M. J. Culzoni, F. Kirchhoff, J. Münch and C. Streb, Increased in Vitro Anti-HIV Activity of Caffeinium-Functionalized Polyoxometalates, ChemMedChem, 2021, 16, 1–5, DOI:10.1002/cmdc.202100281.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc02760c |
| This journal is © The Royal Society of Chemistry 2021 |