Open Access Article
Open Access ArticleA comprehensive analysis in one run – in-depth conformation studies of protein–polymer chimeras by asymmetrical flow field-flow fractionation †
Bibifatima
Kaupbayeva
ab,
Hironobu
Murata
b,
Krzysztof
Matyjaszewski
bc,
Alan J.
Russell
abcd,
Susanne
Boye
 *e and
Albena
Lederer
*e and
Albena
Lederer
 *ef
*ef
aDepartment of Biological Sciences, Carnegie Mellon University, 4400 Fifth Avenue, Pittsburgh, PA 15213, USA
bCenter for Polymer-Based Protein Engineering, Carnegie Mellon University, 5000 Forbes Avenue, Pittsburgh, PA 15213, USA
cDepartment of Chemistry, Carnegie Mellon University, 4400 Fifth Avenue, Pittsburgh, PA 15213, USA
dDepartment of Chemical Engineering, Carnegie Mellon University, 5000 Forbes Avenue, Pittsburgh, PA 15213, USA
eCenter Macromolecular Structure Analysis, Leibniz-Institut für Polymerforschung Dresden e.V., Hohe Straße 6, Dresden 01069, Germany. E-mail: lederer@ipfdd.de; alederer@sun.ac.za
fStellenbosch University, Department of Chemistry and Polymer Science, Private Bag X1, Matieland 7602, South Africa
First published on 27th September 2021
Abstract
Polymer-based protein engineering has enabled the synthesis of a variety of protein–polymer conjugates that are widely applicable in therapeutic, diagnostic and biotechnological industries. Accurate characterizations of physical–chemical properties, in particular, molar masses, sizes, composition and their dispersities are critical parameters that determine the functionality and conformation of protein–polymer conjugates and are important for creating reproducible manufacturing processes. Most of the current characterization techniques suffer from fundamental limitations and do not provide an accurate understanding of a sample's true nature. In this paper, we demonstrate the advantage of asymmetrical flow field-flow fractionation (AF4) coupled with multiple detectors for the characterization of a library of complex, zwitterionic and neutral protein–polymer conjugates. This method allows for determination of intrinsic physical properties of protein–polymer chimeras from a single, rapid measurement.
1 Introduction
Protein–polymer conjugates are exceptional macromolecules that possess characteristics of both biotic and abiotic worlds. Synthetic polymers are conjugated to proteins to enhance the native properties of the protein, improve activity and increase stability in non-native environments. This has been a significant area of interest since the synthesis of the first protein–polymer chimera reported in 1977. This conjugate, and most since, was generated by covalently attaching poly(ethylene)glycol (PEG) to bovine serum albumin (BSA).1 PEGylation is used to shield the antibody binding epitopes within a protein, while also providing protection from proteolytic enzymes.2,3 Preventing antibody binding is a crucial step in evading the immune system thereby increasing the plasma circulation time. The ability of a polymer to shield the surface of a protein from immune complex formation, led to medicinal application of protein–polymer conjugates.4In recent years, considerable attention has been paid to the creation of “smart conjugates” by incorporating stimuli responsive polymers to proteins.5–7 Examples include temperature-5,7,8 and pH-responsive9 polymers that can change conformation in response to trigger. “Smart conjugates” have been employed for sensing and detection.10–12 Polymer-based protein engineering has also been used to increase protein stability at high temperatures, pH extremes, in organic solvents and in ionic liquids. These properties are highly attractive for industrial applications of protein–polymer chimeras.
Protein–polymer conjugates can be synthesized using two alternative strategies: “grafting to” and “grafting from”. In the “grafting to” approach, end-functionalized polymers are synthesized first to a desired molar mass and then grafted covalently to a protein surface.13 This approach, while widely used, can be a somewhat random process where grafting density and attachment sites are difficult to control. Additionally, purification of these conjugates can be laborious due to the difficulties associated with the removal of unreacted polymers.13–15 Polymer synthesis has also evolved over the past few decades, with the development of controlled radical polymerization (CRP).16,17 Using CRP considerably widens the synthetic horizon, allowing the incorporation of wide range of monomers, introduce plethora of functional groups and monomers with complex architecture. Atom-transfer radical polymerization (ATRP) and reversible addition–fragmentation chain transfer (RAFT) have been shown to be ideal methods to prepare protein–polymer chimeras. “Grafting from” approach consists of modifying proteins with small initiator groups9 or chain transfer agents,18 followed by polymer synthesis using the protein as macroinitiator. Comparatively, “grafting from” allows the conjugates to be prepared with higher grafting densities,19 higher yield9 and finer modification site control.20,21 Additionally, CRP methods provide precise control of polymer composition, functionality and topology.22 Therefore, not only linear homopolymers, but also random copolymers, gradient copolymers and block copolymers as well as branched structures can be grown from proteins.8 Due to the process of growing polymers from a protein surface, simpler purification processes, such as dialysis, are also possible.
While the synthetic strategies to make protein–polymer conjugates have evolved over the past decade, the development of precise and accurate characterization techniques has lagged considerably behind. Synthetic advances cannot achieve the desired impact without reliable characterization of the products.23 Due to their hybrid nature, the characterization of intact protein–polymer conjugates is very difficult, and we have had to rely on controlled decomposition and characterization of the resulting synthetic and biologic fragments. Thus, the synthetic polymer chains are typically cleaved from the protein surface, followed by molar mass and dispersity (Đ) characterization using size exclusion chromatography (SEC).9 Once the single chain's molar mass is determined and assuming that the number of polymers per protein is known, the whole conjugates molar mass can be estimated. This method has several drawbacks which may affect the molar mass calculation: (i) SEC molar masses are mostly estimated in reference to calibration standards, which may have different solution properties, conformation, architecture, (ii) different chemical structures may lead to different interaction with separation column's material than the cleaved polymer chains.24 Absolute molar masses can be obtained by the application of multi-angle static light scattering detector (MALS). However, limited size range, shear degradation and sample interaction of column-based SEC are still existent (see ESI, Fig. S14†). Various complementary assay techniques, as well as batch dynamic light scattering (DLS) studies, are also used to provide information about the quality of conjugation. With these techniques, however, only average values are obtained, and the co-existence of complex protein–polymer structures remains undetected. Thus, novel and versatile techniques are needed to gain a deeper insight into the true nature of protein–polymer chimeras.
An attractive alternative to column-based SEC is the asymmetrical flow field-flow fractionation (AF4) as a channel-based separation technique that separates molecules depending on differences in their Brownian motion. This gentle technique does not use a densely packed column for separation and is non-destructive to delicate samples such as bioconjugates due to significantly reduced shear forces.25–28 Channel-based separation can more accurately reflect physical properties of the sample in solution. When coupling AF4 to MALS, the absolute, intrinsic molar mass can be obtained based on the Rayleigh ratio.29,30 Since MALS does not require the use of calibration standards, protein–polymer conjugate molar mass, radius and Đ can be determined as intact entities, without cleaving the polymers prior to analysis (Fig. 1). MALS can provide information on radius of gyration (Rg) and combining that with hydrodynamic radius (Rh) that is obtained from online DLS detector, can further provide an insight into conformation properties such as scaling behavior and ρ parameter (Rg/Rh), as well as information about structural complexity.31 In previous studies, the great potential of AF4 in combination with up to five detectors for characterization of complex macromolecular architectures and particles has been demonstrated.28,32,33 Light scattering detectors in combination with concentration and chemical structure sensitive detectors (UV-Vis) also enable the determination of the chemical composition distribution. Previous studies show the potential of AF4-MALS for the separation and characterization of conjugated structures and enabled a detailed study of the complexity of avidin–polymer conjugation. These studies began to unravel a variety of co-existing architectures that were driven by changes of parameters such as type of polymer, degree of biotinylation, molar ratios, etc.25,34–36
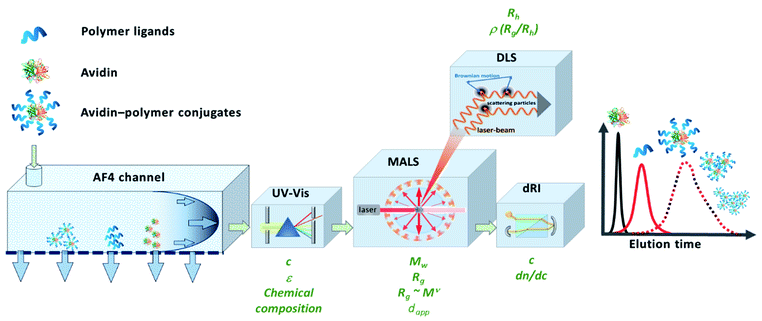 | ||
| Fig. 1 Schematic representation of the instrumental setup of asymmetrical flow field-flow fractionation with fourfold detection system (AF4-D4) for the in-depth characterization of the molecular properties of avidin–polymer conjugates. Adapted with permission from J. Engelke, et al.33 Copyright 2021 American Chemical Society. | ||
Herein, we demonstrate the potential of using AF4 with four-fold detection (AF4-D4, Fig. 1) for the characterization of avidin–polymer intrinsic physical properties. The library of polymers was prepared by using surface-initiated ATRP and variation of the number of polymer chains (grafting density), polymer length (degree of polymerization, DP) and polymer type. The comprehensive study of molecular properties like molar mass, radius and conformations by AF4-D4 enabled a correlation between the synthetic strategies and the physical properties of the resulting polymer–protein chimeras.
2 Experimental
2.1 Materials
Avidin from egg white was purchased from Lee Biosolutions (Maryland Heights, MO). Bicinchoninic acid solution, copper(II) chloride, sodium ascorbate and poly(ethylene glycol)methyl ether methacrylate (OEGMA, average Mn 500) were purchased from Sigma Aldrich (St. Louis, MO), CBMA was purchased from TCI (Portland, OR). Protein surface active ATRP initiators were prepared as described previously.9,19 4-(Bis(N,N-diethylaminoethyl)aminoethyl)morpholine (MMA) synthesized in a previous report was used as ligand.2.2 Synthetic procedures
Double-headed ATRP initiator was synthesized as previously described.19 Avidin–Br2 conjugates were synthesized by mixing avidin (350 mg, 0.022 mmol protein, 0.22 mmol primary amine groups) with double-headed bromine-functionalized ATRP initiator (1.53 mmol, 915.7 mg (7 eq.) dissolved in 2 mL DMSO) in 170 mL of 0.1 M sodium phosphate buffer, pH 8.0. The reaction was stirred at 4 °C for 2 h and then dialyzed against 25 mM sodium phosphate, pH 8 using dialysis tubing with a molecular weight cut off of 15 kDa, for 24 h at 4 °C.
2.3 Analytical methods
3 Results and discussion
3.1 Polymer-based protein engineering of avidin with zwitterionic and neutral polymers
Avidin–polymer conjugates were synthesized with varying polymer grafting density, polymer chain length and polymer type using grafting-from ATRP (Fig. 2). The avidin surface was first modified with either single or double-headed initiators as described previously.19 The number of modified amino groups was determined using MALDI-ToF mass spectrometry. Single-grafted avidin–polymer conjugates were synthesized from 35 single-headed initiator modified avidin (Fig. S2†) while double-grafted conjugates were produced from 29 double-headed initiator attached avidin (Fig. S3†). Two different chain lengths (short and long) of two different polymer types (zwitterionic poly(carboxybetaine methacrylate) and neutral poly(oligo(ethylene glycol) methacrylate)) were selected to be polymerized to avidin and analyzed by AF4-D4. To obtain a suitable comparison of the properties and solution behavior of polymers and protein–polymer conjugates, the non-conjugated polymers were also investigated using AF4-D4 (Table S1†). SEC analysis and bicinchoninic acid (BCA) protein assay of single-grafted polymers and avidin–polymer conjugates results were reported previously.37 Avidin is a tetrameric protein with four identical subunits. For SEC analysis, when estimating overall avidin–polymer conjugate molar mass, the calculations used monomeric avidin with polymers. Due to nondestructive nature of AF4, in a channel-based separation, avidin conjugates elute in their tetrameric form. Therefore, intrinsic conjugate molar masses determined from AF4-D4 are those of tetrameric avidin with polymers. For an easier comparison of AF4 results to conventional SEC and BCA assays, we recalculated the molar masses from SEC and BCA for tetrameric avidin–polymer conjugates (Table S2†). The polymer chain length was controlled by variation of the monomer to initiator ratio in ATRP solution (targeted DPs of 50 and 200). Conjugates were purified via dialysis and then lyophilized.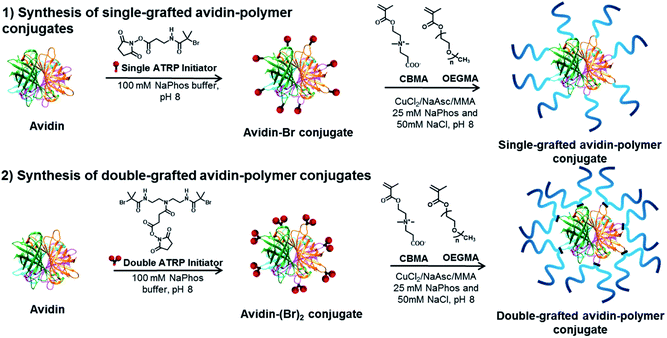 | ||
| Fig. 2 Surface-initiated synthesis of single-grafted and double-grafted avidin conjugates using zwitterionic pCBMA and neutral pOEGMA polymer ligands. | ||
3.2 Avidin–polymer conjugates molar mass estimation using SEC and BCA protein assay
After purification, conjugates were characterized using BCA to determine protein content in the conjugates and then used to estimate conjugate molar mass and polymer DP. Targeted DPs were 50 and 200 and measured DPs from BCA assay were 32 and 62 for single-grafted avidin-pCBMA and 17 and 26 for double-grafted avidin-pCBMA, while for single-grafted avidin-pOEGMA DPs were 14 and 26 and for double-grafted avidin–pOEGMA conjugates they were 10 and 19 (Table S2†).Acid induced polymer cleavage followed by SEC was performed to determine polymer's molar mass and Đ. For pCBMA DPs of 41 and 172 in single-grafted and 21 and 206 in double-grafted conjugates were found, while pOEGMA show DPs of 19 and 104 in single-grafted and 21 and 197 in double-grafted conjugates (Table S2†).
Our goal herein was to develop a characterization approach that could provide accurate and precise physical properties for tightly and weakly controlled ATRP reactions. The very large and disperse complexes that were generated in the more complex conjugates were particularly useful to test the limits of BCA, SEC, and later AF4.
It was interesting that BCA assay and SEC showed significantly different DPs for longer polymer chains. The difference can be ascribed to different characterization principles and assumptions in result interpretation. Indeed, this is one of the central problems of current ways to characterize protein–polymer conjugates. In SEC, polymer–column enthalpic interactions (Fig. S14†) and calculation of molar mass in reference to a calibration standard can lead to a significant bias of the resulting data. It is critical to note that SEC separates according to size and not to molar mass. To accurately convert size into molar mass, calibration standards must be of identical conformation and density as the analyte.24 Due to the lack of calibration standards for protein–polymer chimeras the polymers have to be cleaved from protein by acid treatment. Conjugates with high-grafting densities and long polymer chains can prevent the full cleavage and result in some polymers that are still linked to peptide fragments. This will increase heterogeneity of the analyte and thereby affect both Đ and the molar mass calculations.23
In the BCA assay, characterization relies on accurate determination of protein concentration in the sample. Using protein concentration one can calculate the molar ratio of protein to polymer and conjugate's molar mass. Here the challenge is to accurately determine the protein content of conjugates that have high grafting polymer densities and long polymer chains, which then can result in misinterpretations of results.
3.3 AF4-D4 studies of avidin–protein conjugates
The complete library of single-grafted polymers with varied physical and chemical properties and the corresponding conjugates have been characterized by AF4-D4, the results are summarized in Table 1. Additionally, more detailed interpretations and all fractograms can be found in the ESI.†
| M n (kg mol−1) | M w (kg mol−1) | Đ (Mw/Mn) | Polymer chains per avidinb | R g (nm) | R h (nm) | ρ(Rg/Rh)c | Apparent density (kg m−3) | Recoverye (%) | ||
|---|---|---|---|---|---|---|---|---|---|---|
| a Calculated with determined dn/dc = 0.154 mL g−1 for pCBMA conjugates and 0.139 mL g−1 for pOEGMA conjugates. b Average number of functionalities per avidin molecule (calculated by Mn of conjugate C, subtracted by Mn of macroinitiator (68 kg mol−1) and divided by Mn of specific polymer F). c Determined at the peak maximum of DRI signal. d Single polymer segments per double-headed initiator. e Calculated by dn/dc and exact sample load. | ||||||||||
| Zwitterionic polymers | ||||||||||
| C1 | Single-grafted avi-pCBMAshort | 383 | 517 | 1.35 | 7.9 | 16.3 | 15.0 | 0.97 | 22.0 | 93 |
| C2 | Single-grafted avi-pCBMAlong | 741 | 1250 | 1.69 | 6.3 | 23.0 | 24.6 | 0.97 | 18.9 | 92 |
| C3 | Double-grafted avi-pCBMAshort | 159 | 386 | 2.43 | n.d. | 17.2 | 16.5 | 0.94 | 14.0 | 95 |
| C4 | Double-grafted avi-pCBMAlong | 395 | 867 | 2.19 | n.d. | 24.7 | 23.6 | 1.05 | 10.6 | 88 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||||||
| Neutral polymers | ||||||||||
| C5 | Single-grafted avi-pOEGMAshort | 488 | 797 | 1.63 | 6.5 | 19.1 | 20.0 | 0.92 | 21.1 | 90 |
| C6 | Single-grafted avi-pOEGMAlong | 641 | 1540 | 2.40 | 3.5 | 31.2 | 32.5 | 1.07 | 9.3 | 92 |
| C7 | Double-grafted avi-pOEGMAshort | 298 | 904 | 3.03 | 2.3 (4.6d) | 28.9 | 28.0 | 0.91 | 6.6 | 91 |
| C8 | Double-grafted avi-pOEGMAlong | 2320 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800 |
5.09 | 7.8 (15.6d) | 109 | 66.6 | 1.38 | 1.7 | 97 |
In Fig. 3, estimated conjugate molar masses of SEC and BCA assay are compared with the absolute molar masses determined by AF4-D4.
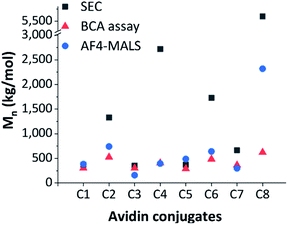 | ||
| Fig. 3 Comparison of differently determined conjugate molar masses, estimated Mn by SEC (grey square), estimated Mn by BCA assay (red triangle) and absolute Mn by AF4-D4 (blue circle). | ||
The molar masses obtained by SEC were significantly different from those determined by the BCA assay and the AF4-D4 (which were similar to each other). This strong discrepancy in SEC data may have been due to the incomplete hydrolytic cleavage of the polymer ligands and the resulting overestimation of the molar masses. Furthermore, molar mass determination is affected by the relative calibration with pullulan standards. The conformational properties of this polysaccharide are not comparable to those of zwitterionic pCBMA or neutral pOEGMA. The quantification of avidin by BCA and the estimation of related conjugated polymer amount was in better agreement with the AF4-D4 results. Only conjugate C8 showed deviation in the molar mass determined by these two methods. A deeper look into conformational properties helped clarify these inconsistencies, as discussed below.
It should be noted that the average number of effectively coupled polymer chains per avidin was less than the number of available sites for all conjugates, as determined by MALDI (accessibility of primary amines). ATRP of free single-grafted polymers and single-grafted avidin–polymer conjugates should yield molecules with similar polymer properties.
In addition, the synthesis and conjugation processes are statistical in nature, leading to different chain lengths of the polymers as indicated from the broad molar mass distribution determined via AF4-D4. Physical properties such as chain stiffness or flexibility, which are determined by the chemical nature, can have an influence on the dispersity and the resulting conjugate architecture. Therefore, the determination of the conformational properties is essential when seeking to understand the resulting conjugate dispersity. In former studies,25,35 it was demonstrated that differences in chemical composition and structural parameters caused a broad heterogeneity and dispersity instead of well-defined architectures.
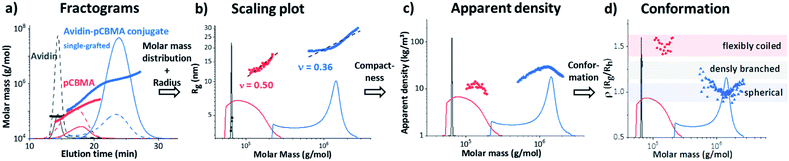
The fractograms in Fig. 4a show the separation of the individual components (native avidin, pCBMAlong (F2) and avidin-pCBMAlong conjugate (C2)) detected at different elution times. The later the elution, the lower the diffusion coefficient and, accordingly, the larger the hydrodynamic volume or molar mass. In addition to the absolute molar masses and radii (Rg and Rh) obtained from MALS and DLS detection, information about conformations at the molecular level was also obtained. For example, changes in the radii of gyration as a function of molar mass provided information about the scaling properties of the conjugate in the respective solvent. The slope ν (0.50) of the scaling plot for pCBMA in Fig. 4b was typical for statistical coils in theta-solvent (see ESI†).31 After the formation of conjugates, the scaling parameter of C2 (ν = 0.36) revealed a more compact conformation that approximated a spherical shape. Due to the small size of native avidin and the narrow dispersity, no scaling plot could be generated.
Further data processing allowed the calculation of the apparent densities. These densities provided information about molecular compactness. Compared to the pure polymer chain, the conjugate exhibited a significantly higher dapp (Fig. 4c). For both, the polymer and the conjugate, it was noticeable that the dapp increased with increasing molar masses, passed through a maximum and then dropped again. Closer inspection of the ratio of Rg and Rh, known as the ρ parameter, revealed the conformation of the conjugates even more precisely and confirmed our previous observations (Fig. 4d). While ρ(Rg/Rh) for the F2 was in the range for flexibly coiled chains, the conjugate possessed values of around 1. According to the literature, these ρ(Rg/Rh) values are typical for spheres with rather heterogeneous surfaces. This behavior has also been observed for polymeric vesicles such as polymersomes.32,38,39
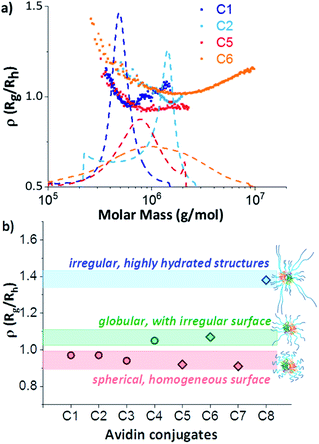
The ρ(Rg/Rh) parameters (ρ = 0.97) of the zwitterionic conjugates (C1 and C2) and the neutral C5 in this area were characteristic for globular, sphere-like conformations with a well-defined surface (Fig. 5). An increased heterogeneous surface was observed for the neutral avidin–pOEGMA conjugate with long polymer ligands (C6). Here, the ρ parameter was slightly higher (ρ = 1.07), which was characteristic for a compact, globular shape possessing a rough surface.
This behaviour was an indication that the conformational properties of the polymer chains had an influence on the conjugate shape. Although this may seem obvious, this is the first time that a single run technique could tease apart the intricacies of the protein–polymer conjugate formation. The scaling plots (Fig. S9†) of the individual polymers F2 and F4 confirmed this assumption. While the polymer chains of the zwitterionic pCBMA (F2) were clearly more flexible and more coiled (ν = 0.5), the neutral POEGMA (F4) showed a more elongated conformation (ν = 0.75). In turn, the double grafted F6 of similar chemical origin as F4 shows again the typical reduction of the scaling parameter due to the compact, branching conformation. Finally, the more flexible polymer chains of C2 compensated for different degrees of polymerisation and generated a uniform conjugate surface. In contrast, the differences in chain length of the rigid polymer molecules after conjugation formed a rather rough surface for C6.
This becomes even clearer when considering the double-grafted conjugate structures, which contained higher numbers of polymer chains grown-from double-headed initiators. The formation of double-grafted avidin-conjugates with short polymers (C3 and C7) led to comparable spherical conformations as observed for single-grafted C1 and C5 independent on the polymer chemistry. However, an increase of radii and a decrease in apparent density was observed for the double-grafted C3 and C7 (Fig. S11 and S13†).
A completely different behaviour was observed in the case of double-grafted, neutral avidin conjugate with long pOEGMA (C8). Here, molar masses, dispersity and radii were significantly higher compared to the other conjugates, but their apparent density was extremely low (Table 1). The reason for this can be found in their specific structure and conformation. The ρ(Rg/Rh) parameter of C8 was significantly higher (1.38). This value is typical for irregular, clustered or branched chain conformations. In addition, an extremely low apparent density was observed. These two observations lead to the conclusion, that structures with a large hydrated shell around the avidin–polymer conjugate had been generated. A comparable behaviour was demonstrated for a similar polymer-conjugate system by molecular dynamic simulations.40 A few highly hydrated, very long and stiff polymer chains, in addition to shorter chains resulted in high radii with low apparent densities. These structures can be classified as micelles, but with a heterogeneous polymer chain distribution (Fig. 5b). Additionally, the very broad distribution and high molar masses indicated the formation of a heterogeneous network, constructed of several avidin molecules. One reason for that could be aggregation. We systematically investigated the interaction of non-conjugated polymers with each other and with avidin by mixing variation of free polymer chains with native avidin. No shifts to higher molar masses or radii were observed and native avidin and free polymers eluted separately (Fig. S6d†). Thus, the interaction between single polymer chains and conjugates was excluded.
However, as mentioned earlier, the average number of available grafting sites for the polymer ligands per avidin molecule was 35 single-headed initiators and 29 double-headed initiators, respectively. Steric reasons could also have led to structures with longer polymer chains. At the same time, unreacted initiators were possibly still present.
Unfortunately, a statistical calculation of the polymer or avidin distribution within the conjugates was not possible due to absorption of both components at the applied wavelength (λ = 280 nm) of the UV detector. Here, a labelling of the protein could help. However, the introduction of a marker group could also change the solution properties and AF4 membrane interactions significantly. Furthermore, fluorescence effects in the wavelength range of the laser must be avoided when using light scattering detection.
4 Conclusions
AF4-D4 shows that the formation of polymer–protein conjugates depends on the physicochemical properties of the polymer chains. As an example, two polymer types (zwitterionic pCBMA as well as neutral pOEGMA) with different chain lengths, single-grafted and double-grafted conjugate architectures were generated and characterized. To correlate the conformation properties with the chemical nature, asymmetrical flow field flow fractionation in combination with a multidetection system was used. The AF4-D4 hyphenation allows to generate the data needed for determination of a series of structural parameters, which can be correlated only if they are collected simultaneously. The data evaluation enabled a comprehensive elucidation of the structures with the help of the conformational properties and contributed to a remarkable understanding of the conjugation processes. It has been shown that polymer flexibility and hydration have a major influence on the properties in solution such as shape and apparent density.Data availability
All relevant data is included in the paper and ESI.†Author contributions
BK and HM prepared the polymers and conjugate samples and performed the SEC, BCA and MALDI-ToF experiments. SB performed the AF4 experiments and the conformation studies. BK and SB analysed the data and wrote the manuscript. KM and AR supervised the synthetic work, MALDI-ToF and BCA analyses. AL supervised the AF-D4 analyses and interpretation. All authors contributed to discussing the results and reviewing the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank Mrs P. Treppe (IPF Dresden) for performing dn/dc determinations and supporting the AF4 experiments. Additionally, we acknowledge financial support from DTRA grant HDTRA1-20-1-0014.Notes and references
- A. Abuchowski, J. R. McCoy, N. C. Palczuk, T. van Es and F. F. Davis, J. Biol. Chem., 1977, 252, 3582–3586 CrossRef CAS.
- J. Milton Harris and R. B. Chess, Nat. Rev. Drug Discovery, 2003, 2, 214–221 CrossRef PubMed.
- P. L. Turecek, M. J. Bossard, F. Schoetens and I. A. Ivens, J. Pharm. Sci., 2016, 105, 460–475 CrossRef CAS PubMed.
- Y. Wu, D. Y. W. Ng, S. L. Kuan and T. Weil, Biomater. Sci., 2015, 3, 214–230 RSC.
- I. Cobo, M. Li, B. S. Sumerlin and S. Perrier, Nat. Mater., 2015, 14, 143–149 CrossRef CAS PubMed.
- K. L. Heredia, D. Bontempo, T. Ly, J. T. Byers, S. Halstenberg and H. D. Maynard, J. Am. Chem. Soc., 2005, 127, 16955–16960 CrossRef CAS PubMed.
- C. Cummings, H. Murata, R. Koepsel and A. J. Russell, Biomacromolecules, 2014, 15, 763–771 CrossRef CAS PubMed.
- C. Cummings, H. Murata, R. Koepsel and A. J. Russell, Biomaterials, 2013, 34, 7437–7443 CrossRef CAS PubMed.
- H. Murata, C. S. Cummings, R. R. Koepsel and A. J. Russell, Biomacromolecules, 2013, 14, 1919–1926 CrossRef CAS PubMed.
- J. M. Paloni, E. A. Miller, H. D. Sikes and B. D. Olsen, Biomacromolecules, 2018, 19, 3814–3824 CrossRef CAS PubMed.
- Y. Liu, T. K. Nevanen, A. Paananen, K. Kempe, P. Wilson, L. S. Johansson, J. J. Joensuu, M. B. Linder, D. M. Haddleton and R. Milani, ACS Appl. Mater. Interfaces, 2019, 11, 3599–3608 CrossRef CAS PubMed.
- M. Jain, R. G. Vaze, S. C. Ugrani and K. P. Sharma, RSC Adv., 2018, 8, 39029–39038 RSC.
- G. N. Grover and H. D. Maynard, Curr. Opin. Chem. Biol., 2010, 14, 818–827 CrossRef CAS PubMed.
- S. Jevsevar, M. Kunstelj and V. G. Porekar, Biotechnol. J., 2010, 5, 113–128 CrossRef CAS PubMed.
- B. Treetharnmathurot, C. Ovartlarnporn, J. Wungsintaweekul, R. Duncan and R. Wiwattanapatapee, Int. J. Pharm., 2008, 357, 252–259 CrossRef CAS PubMed.
- J. S. Wang and K. Matyjaszewski, Macromolecules, 1995, 28, 7901–7910 CrossRef CAS.
- M. Kato, M. Kamigaito, M. Sawamoto and T. Higashimura, Macromolecules, 1995, 28, 1721–1723 CrossRef CAS.
- M. Li, H. Li, P. De and B. S. Sumerlin, Macromol. Rapid Commun., 2011, 32, 354–359 CrossRef CAS PubMed.
- B. Kaupbayeva, H. Murata, A. Lucas, K. Matyjaszewski, J. S. Minden and A. J. Russell, Biomacromolecules, 2019, 20, 1235–1245 CrossRef CAS PubMed.
- S. Carmali, H. Murata, E. Amemiya, K. Matyjaszewski and A. J. Russell, ACS Biomater. Sci. Eng., 2017, 3, 2086–2097 CrossRef CAS PubMed.
- S. Carmali, H. Murata, K. Matyjaszewski and A. J. Russell, Biomacromolecules, 2018, 19, 4044–4051 CrossRef CAS PubMed.
- K. Matyjaszewski and N. V. Tsarevsky, J. Am. Chem. Soc., 2014, 136, 6513–6533 CrossRef CAS PubMed.
- S. L. Baker, B. Kaupbayeva, S. Lathwal, S. R. Das, A. J. Russell and K. Matyjaszewski, Biomacromolecules, 2019, 20, 4272–4298 CrossRef CAS PubMed.
- J. Engelke, J. Brandt, C. Barner-Kowollik and A. Lederer, Polym. Chem., 2019, 10, 3410–3425 RSC.
- S. Boye, F. Ennen, L. Scharfenberg, D. Appelhans, L. Nilsson and A. Lederer, Macromolecules, 2015, 48, 4607–4619 CrossRef CAS.
- B. Kaupbayeva, S. Boye, A. Munasinghe, H. Murata, K. Matyjaszewski, A. Lederer, C. M. Colina and A. J. Russell, Bioconjug. Chem., 2021, 32, 821–832 CrossRef CAS PubMed.
- U. L. Muza, S. Boye and A. Lederer, Anal. Sci. Adv., 2021, 2, 5–108 Search PubMed.
- S. Lathwal, S. S. Yerneni, S. Boye, U. L. Muza, S. Takahashi, N. Sugimoto, A. Lederer, S. R. Das, P. G. Campbell and K. Matyjaszewski, Proc. Natl. Acad. Sci. U.S.A., 2021, 118, e2020241118 CrossRef CAS PubMed.
- J. C. Giddings, Science, 1993, 260, 1456–1465 CrossRef CAS PubMed.
- F. A. Messaud, R. D. Sanderson, J. R. Runyon, T. Otte, H. Pasch and S. K. R. Williams, Prog. Polym. Sci., 2009, 34, 351–368 CrossRef CAS.
- W. Burchard, in Light Scattering from Polymers, Springer Berlin Heidelberg, Berlin, Heidelberg, 1983, pp. 1–124 Search PubMed.
- H. Gumz, S. Boye, B. Iyisan, V. Krönert, P. Formanek, B. Voit, A. Lederer and D. Appelhans, Adv. Sci., 2019, 6(7), 1801299 CrossRef PubMed.
- J. Engelke, S. Boye, B. T. Tuten, L. Barner and C. Barner-Kowollik, ACS Macro Lett., 2020, 9, 1569–1575 CrossRef CAS.
- F. Ennen, S. Boye, A. Lederer, M. Cernescu, H. Komber, B. Brutschy, B. I. Voit and D. Appelhans, Polym. Chem., 2014, 5, 1323–1339 RSC.
- J. Fingernagel, S. Boye, A. Kietz, S. Höbel, K. Wozniak, S. Moreno, A. Janke, A. Lederer, A. Aigner, A. Temme, B. Voit and D. Appelhans, Biomacromolecules, 2019, 20, 3408–3424 CrossRef CAS PubMed.
- J. Daeg, X. Xu, L. Zhao, S. Boye, A. Janke, A. Temme, J. Zhao, A. Lederer, B. Voit, X. Shi and D. Appelhans, Biomacromolecules, 2020, 21, 199–213 CrossRef CAS PubMed.
- H. Murata, S. L. Baker, B. Kaupbayeva, D. J. Lewis, L. Zhang, S. Boye, A. Lederer and A. J. Russell, J. Polym. Sci. Part A Polym. Chem., 2020, 58, 42–47 CAS.
- S. Moreno, S. Boye, A. Lederer, A. Falanga, S. Galdiero, B. Voit and D. Appelhans, Biomacromolecules, 2020, 21, 5162–5172 CrossRef CAS PubMed.
- X. Wang, S. Moreno, S. Boye, P. Wang, X. Liu, A. Lederer, B. Voit and D. Appelhans, Adv. Sci., 2021, 2004263 CrossRef PubMed.
- A. Munasinghe, S. L. Baker, P. Lin, A. J. Russell and C. M. Colina, Soft Matter, 2020, 16, 456–465 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, synthetic procedures, analytical data. See DOI: 10.1039/d1sc03033g |
| This journal is © The Royal Society of Chemistry 2021 |