Open Access Article
Open Access ArticleA highly sensitive and selective fluoride sensor based on a riboswitch-regulated transcription coupled with CRISPR-Cas13a tandem reaction†
Yuan
Ma
 ab,
Quanbing
Mou
a,
Peng
Yan
ab,
Quanbing
Mou
a,
Peng
Yan
 ac,
Zhenglin
Yang
d,
Ying
Xiong
a,
Deyue
Yan
ac,
Zhenglin
Yang
d,
Ying
Xiong
a,
Deyue
Yan
 b,
Chuan
Zhang
b,
Chuan
Zhang
 *b,
Xinyuan
Zhu
*b,
Xinyuan
Zhu
 *b and
Yi
Lu
*b and
Yi
Lu
 *ad
*ad
aDepartment of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801, USA. E-mail: yi-lu@illinois.edu
bSchool of Chemistry and Chemical Engineering, Frontiers Science Center for Transformative Molecules, Shanghai Jiao Tong University, 800 Dongchuan Road, Shanghai 200240, China. E-mail: xyzhu@sjtu.edu.cn; chuanzhang@sjtu.edu.cn
cKey Laboratory of Biomedical Information Engineering of Ministry of Education, School of Life Science and Technology, Xi'an Jiaotong University, 710049, Xi'an, PR China
dDepartment of Biochemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801, USA
First published on 9th August 2021
Abstract
Nucleic acid sensors have realized much success in detecting positively charged and neutral molecules, but have rarely been applied for measuring negatively charged molecules, such as fluoride, even though an effective sensor is needed to promote dental health while preventing osteofluorosis and other diseases. To address this issue, we herein report a quantitative fluoride sensor with a portable fluorometer readout based on fluoride riboswitch-regulated transcription coupled with CRISPR-Cas13-based signal amplification. This tandem sensor utilizes the fluoride riboswitch to regulate in vitro transcription and generate full-length transcribed RNA that can be recognized by CRISPR-Cas13a, triggering the collateral cleavage of the fluorophore-quencher labeled RNA probe and generating a fluorescence signal output. This tandem sensor can quantitatively detect fluoride at ambient temperature in aqueous solution with high sensitivity (limit of detection (LOD) ≈ 1.7 μM), high selectivity against other common anions, a wide dynamic range (0–800 μM) and a short sample-to-answer time (30 min). This work expands the application of nucleic acid sensors to negatively charged targets and demonstrates their potential for the on-site and real-time detection of fluoride in environmental monitoring and point-of-care diagnostics.
Introduction
Functional nucleic acids,1,2 such as aptamers,3–7 riboswitches,8,9 ribozymes,10–13 and DNAzymes,14–18 have emerged as a major class of sensors for on-site and real-time detections in applications such as environmental monitoring19,20 and point-of-care diagnostics21–24 of a wide range of targets with high selectivity.25–30 Despite the progress, these functional nucleic acid sensors work well mainly for positively charged or neutral molecules. Although anions are important analytes,31 only one nucleic acid sensor has been reported for chloride imaging, which utilizes a nucleic acid strand conjugated with an anion-specific organic molecule for sensing.32 Among these anions, fluoride is one of the most attractive targets due to its significant association with biological, medical, industrial, and environmental processes.33–35 On one hand, as an essential element for living organisms, fluoride plays an important role in dental health36 and has the potential for the treatment of osteoporosis.37 On the other hand, fluoride is easily accumulated in organisms with slow excretion,38 which may cause excess fluoride ingestion to induce pathological conditions, including fluorosis,39 urolithiasis, osteoporosis, neurological and metabolic dysfunction, gastric and kidney problems, and certain types of cancer.34,40,41 Since fluoride is widely used, the fluoride contamination in the environment and drinking water may raise major concerns. Therefore, the United States Environmental Protection Agency (EPA) sets an enforceable drinking water standard for fluoride of 4 mg mL−1 (211 μM) to prevent osteofluorosis and other diseases, and a secondary non-enforceable fluoride standard of 2 mg mL−1 (105 μM) to promote dental health.42 As a result, the optimal range of fluoride concentration between deficiency (2 mg mL−1) and toxicity (4 mg mL−1) is quite narrow. Therefore, the accurate determination of fluoride concentration in drinking water that can cover this narrow concentration range is required.To meet the above goal, several analytical methods have been developed for fluoride detection, including 19F NMR analysis, mass spectrometry, and atomic absorption spectrophotometry, standard Willard and Winter methods, fluoride selective electrodes, and ion chromatography.43,44 However, these instrumental analyses require expensive equipment, complicated procedures, and skilled operators, making these methods not affordable for on-site or point-of-care (POC) application.43 Besides, fluoride selective electrodes are easily affected by temperature, interfering ions, color of the sample, shift potential, and ion activity, and require skilled operators to ensure accuracy.43 In addition, other analytical methods are not portable, which greatly limits their applications for on-site and real-time detection.34 To address this issue, optical chemosensors have attracted great attention for developing POC fluoride sensors, due to their simple procedure and high sensitivity and selectivity, along with real-time detection.34,45 Therefore, many different optical fluoride sensors with fluorescent and colorimetric readouts have been developed based on various mechanisms, such as interactions between fluoride ions and Lewis acids,33,46 fluoride–hydrogen bond interactions,33,47 reaction based chemodosimeters,35,42,48 and interaction with other types of materials.49–51 Despite the progress made, portable fluoride detection is still not available to the general public, because very few sensors can meet all these requirements: (1) good performance in aqueous solution; (2) high selectivity for fluoride without interference from other anions; (3) detection range covering the deficiency and toxicity lines in the EPA standards; and (4) short response time.
To address this issue, we take advantage of the fluoride-binding riboswitch that has recently been discovered in bacteria and archaea.52–54 The high selectivity of the fluoride riboswitch toward fluoride over other halides or anions has been attributed to the smaller ionic radius of fluoride (0.133 nm) than other chemical species, including chloride (0.181 nm), as well as the unique hydrogen bonding abilities of fluoride.52,53 A crystal structure of the riboswitch has shown that the riboswitch folds into a 3D structure, and coordinates with three Mg2+ ions, which allows binding one F−, but not enough space to bind other anions.52,53 In the absence of fluoride, the ligand-free fluoride riboswitch transiently accesses a low-populated (∼1%) and short-lived (∼3 ms) excited conformational state, which unravels a conserved ‘linchpin’ base pair to signal transcription termination.55 In the presence of fluoride, this highly localized, fleeting process is allosterically suppressed, which activates transcription.55 Therefore, the fluoride riboswitch can activate transcription in the presence of fluoride, which can regulate gene expression encoding fluoride transporters to remove fluoride as a native defense mechanism against fluoride toxicity.55,56 Moreover, the fluoride riboswitch can regulate in vitro transcription to generate full-length transcribed RNA depending on the fluoride concentration,55 where a higher concentration of fluoride results in a larger number of full-length RNA transcripts. To convert such a highly selective fluoride-regulated transcription into a signal that can be readily detected by a portable fluorometer, we employ CRISPR-Cas13 (Clustered Regularly Interspaced Short Palindromic Repeats-associated system), which can recognize a target RNA through a sequence complementary to CRISPR RNA (crRNA), and then collaterally cleave a nearby reporter fluorophore/quencher pair with an RNA spacer, resulting in the fluorophore moving away from the quencher and thus an increase of the fluorescent signal.57–62 Due to this unique property, the CRISPR-Cas13-based sensors have been applied to detect diverse targets, such as nucleic acids,63–69 epigenetic base modification,70 viruses,71–74 bacteria,75,76 and small molecules.77 For instance, a CRISPR-Cas13-based sensor called SPRINT was able to detect 8 small molecules including fluoride.77 Despite the progress, the current nucleic acid based fluoride detection system has a narrow detection dynamic range of 1 to 100 μM, with a drastically decreased signal at fluoride concentrations above 100 μM. Since the EPA standard for fluoride is 211 μM (4 mg L−1), the detection range of the SPRINT system is above the threshold level defined by the EPA and therefore cannot be used as an on-site sensor for detecting fluoride in drinking water and differentiating whether the water is safe or not.
To address the current limitations of fluoride sensors, we report herein a Fluoride RiboswItch-regulated Transcription with Cas13a (FRITCas13a) tandem sensor, by coupling the fluoride riboswitch-regulated transcription with CRISPR-Cas13a-based cleavage of reporter RNA activated by the transcribed RNA. The system can quantitatively detect fluoride at ambient temperature in aqueous solution with a wide dynamic range (0–800 μM), a low limit of detection (LOD ≈ 1.7 μM), and a short sample-to-answer time (30 min) using a portable fluorometer, demonstrating high potential for on-site and real-time detection and quantification of fluoride in drinking water and other types of samples.
Results and discussion
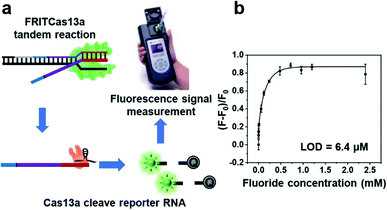
The FRITCas13a tandem sensor involves fluoride-riboswitch regulated in vitro transcription along with Cas13a-mediated collateral cleavage of the reporter RNA (Scheme 1). Specifically, the binding of fluoride by its riboswitch will activate in vitro transcription and generate full-length RNA transcripts, which carry a Cas13a targeting RNA sequence at the 3′-end from the in vitro transcription. The transcribed Cas13a targeting RNA then hybridizes with crRNA to activate the Cas13a-mediated collateral cleavage of a reporter RNA with a fluorophore (FAM) at the 5′-end and a quencher (BHQ1) at the 3′-end. Without the Cas13a-mediated collateral cleavage, the reporter RNA has a low fluorescent signal because the quencher is close to the fluorophore and thus quenched the fluorescence. In the presence of fluoride, the full-length RNA transcripts activate Cas13a to cleave the reporter RNA and release the quencher from the fluorophore and thus recover a significant fluorescence signal. In this design, the fluoride concentration in the sample is positively correlated with the copies of full-length RNA transcripts, the reporter RNA cleavage, and the fluorescence signal generation. Moreover, this FRITCas13a tandem sensor can both detect and quantify fluoride in aqueous solution. In contrast, in the absence of fluoride, the regulation effect of the fluoride riboswitch will result in partially transcribed RNA lacking the Cas13a targeting sequence at the 3′-end.55 As a result, the collateral cleavage of the reporter RNA by Cas13a will not occur and a minimal fluorescence signal will be observed.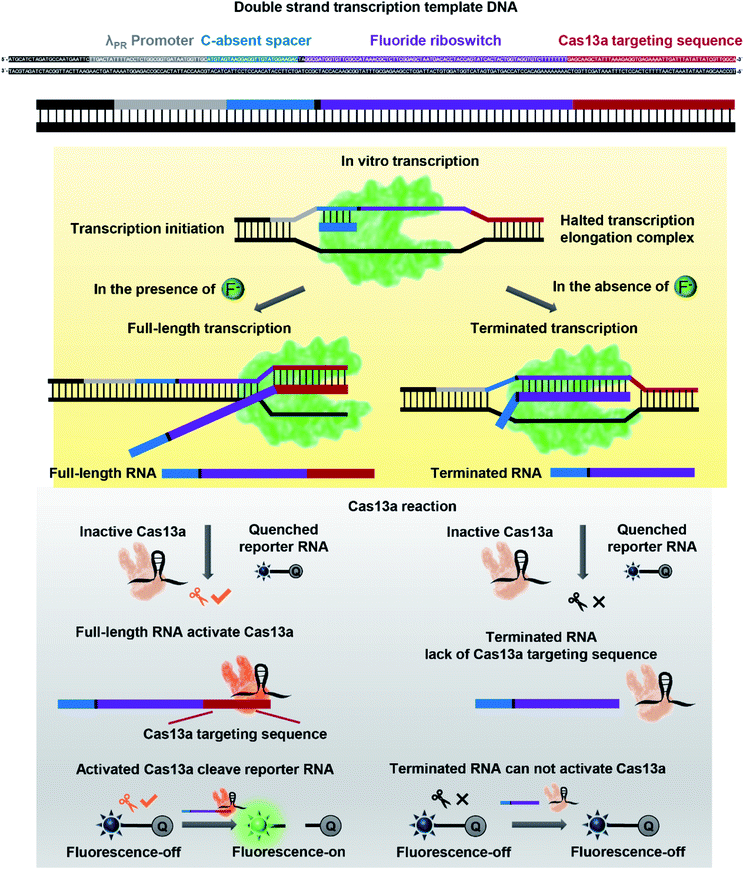 | ||
| Scheme 1 Detection strategy and workflow of a fluoride riboswitch-regulated transcription with a Cas13a (FRITCas13a) tandem sensor for detecting fluoride in aqueous solution. | ||
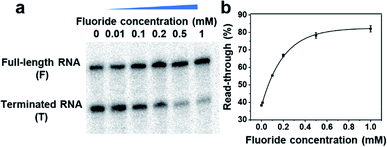
To demonstrate such a FRITCas13a tandem sensor, we take advantage of the in vitro transcription system reported by Zhang's group with a wide and tunable dynamic range between 0.01 and 100 mM.55 The DNA templates were prepared by ligation from shorter DNA units consisting of a λPR promoter, C-absent spacer, fluoride riboswitch and Cas13a targeting sequence at the 3′-end (Scheme 1, Fig. S1, Table S1†). The formation of the DNA templates was verified by 10% denaturing polyacrylamide gel electrophoresis (PAGE) gel analysis (see Fig. S2†). Moreover, the obtained DNA templates were then purified and extracted from the PAGE gel and used for in vitro transcription. To verify that the transcription is regulated by fluoride in a wide range of concentrations, fluoride-dependent transcription with different concentrations of NTP was performed. As shown in Fig. 1, S3 and S4,† the transcription readthrough, defined by full-length RNA/(full-length RNA + terminated RNA), is regulated by the fluoride concentration from 0–1 mM at different NTP concentrations of 10, 20, and 30 μM. Moreover, to develop a fluoride sensor with high sensitivity, the transcription reaction was optimized with various reaction temperatures of 25, 30, and 37 °C to increase the signal-to-noise ratio. As shown in Fig. S5 and S6,† the transcription reaction at 37 °C generated a higher readthrough ratio than those at 25 and 30 °C. Thereafter, 37 °C was chosen as the optimal transcription reaction temperature.
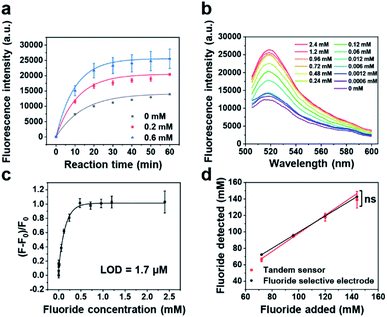
Having demonstrated that the fluoride riboswitch-regulated transcription can transform its binding of the fluoride into full-length transcribed RNA quantitively, we applied Cas13a to detect the transcribed RNA and then generate a fluorescence readout to act as a tandem sensor for fluoride detection. To achieve good analytical performance of the fluoride sensor, several parameters of the Cas13a reaction were optimized, including reaction buffer, reaction time and quenching method. First, we evaluated three commercialized or literature reported buffers,57,65 and found that buffer 1 (ref. 57) showed the best performance, due to its highest Cas13a cleavage activity and signal to noise ratio (Fig. S7†). Moreover, the kinetics of the tandem reaction were investigated by monitoring real-time fluorescence using a plate reader, which include the transcription of the full-length RNA and then the cleavage of the reporter RNA by Cas13a. As shown in Fig. 2a, 15 and 30 min were chosen as good time frames to perform detection, which not only ensure sufficient signal intensity but also reduce the sample-to-answer time. To control the reaction time more precisely, the quenching method for the tandem reaction was further studied by quenching the reaction at 65 °C or 85 °C for 10 min and then tracking real-time fluorescence using a plate reader. As shown in Fig. S8,† a steady fluorescence signal after reaction ends was obtained after quenching the reaction at 65 °C or 85 °C for 10 min. Considering that lower temperature will be beneficial for the stability of reporter RNA, the reaction quenching at 65 °C for 10 min was chosen for the subsequent experiments.
To apply the FRITCas13a tandem sensor for fluoride detection, the above optimized tandem reaction workflow was performed for 30 min with the addition of different concentrations of fluoride and then measurement of fluorescence spectra of the sensors by using a fluorometer. As shown in Fig. 2b, the fluorescence intensity increases corresponding to the elevation of fluoride concentrations. By plotting the relative fluorescence ratio ((F − F0)/F0) against the fluoride concentration, where F and F0 are derived in the presence and absence of fluoride respectively, we obtained a binding curve and the LOD was calculated to be 1.7 μM (Fig. 2c), based on 3σb/slope, where σb is the standard deviation of three blank samples. The LOD of this sensor is ∼16 fold lower than that of the EPA standard method using a fluoride selective electrode (26.3 μM).78 Moreover, to further shorten the detection time, the same test was performed for 15 min with the addition of different concentrations of fluoride. As shown in Fig. S9,† the relative fluorescence ratios ((F − F0)/F0) increase corresponding to the elevation of fluoride concentrations. Based on the 3σb/slope method, within a detection time of 15 min, the LOD of sensor was calculated to be 2.5 μM.
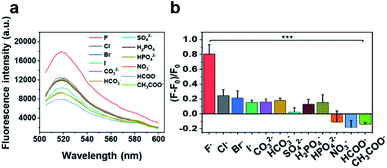
The selectivity of our sensor was also examined by performing the test in the presence of fluoride or many potentially interfering analytes at a concentration of 200 μM. As shown in Fig. 3, the sensor displays a weak or negligible response to most anions, including heavier halides (Cl−, Br−, and I−), other inorganic anions (CO32−, HCO3−, SO42−, H2PO4−, HPO42−, and NO3−), and organic anions (HCOO− and CH3COO−), while the fluorescence signal in the presence of fluoride is significantly higher than that of the other anions (p < 0.001). Moreover, considering that real environmental samples may contain anion mixtures instead of individual ones, we tested the fluoride sensor in the presence of various anion mixtures, such as a mixture of four different common anions in the environment (Cl−, NO3−, CO32−, and SO42−, 50 μM for each anion), and 11 different anion mixtures (Cl−, Br−, I−, CO32−, HCO3−, SO42−, H2PO4−, HPO42−, NO3−, HCOO−, and CH3COO−, 18 μM for each anion). As shown in Fig. S10,† the sensor displays a weak or negligible response to anion mixtures compared with the signal in the presence of fluoride (p < 0001), demonstrating that the sensor maintains high selectivity toward fluoride even in the presence of anion mixtures.
The detection of fluoride with the fluoride sensor in real samples may potentially be affected by common interferents, such as cations, anions, DNAse, RNAse, or protease. Thus, tests in the presence of those common interferents were conducted. To investigate the interference of common cations, such as Na+, K+, Mg2+, and Ca2+, we performed the test at a fluoride concentration of 200 μM in the absence or presence of each cation (100 μM). The selectivity coefficient (SC) for each of the potential interfering agents was calculated using the formula, SC = Ac+i/Ac, where Ac+i and Ac are the response for fluoride in the presence and absence of each interfering agent, respectively. As shown in Fig. S11,† the signal was not affected by the presence of Na+, K+, Mg2+, and Ca2+ as their SC approximates to 1. In addition, to understand the interference by common cation and anion mixtures in real samples, we performed the test in samples spiked with cation mixtures (Na+, K+, Mg2+, and Ca2+), or various cation and anion mixtures (Na+, K+, Mg2+, Ca2+, Cl−, NO3−, CO32−, and SO42−). As shown in Fig. S11,† both groups cause minimal interference for the fluoride detection. In addition, the fluoride sensor utilizes in vitro transcription and CRISPR-Cas13a mediated amplification, which could be potentially interfered with by DNAses, RNAses, and proteases present in the environmental sample. To address this issue, the environmental samples were heat-pretreated at 95 °C for 10 min to inactivate potential DNAses and proteases, following protocols reported previously.72,74 Moreover, since RNAses cannot be heat-inactivated, we added an RNAse inhibitor during the tandem reaction. To test the effectiveness of the pretreatment, we spiked the samples with DNAse I, RNAse A, and protease K, heated the samples at 95 °C for 10 min, and then performed the tandem reaction with a RNaseOUT™ Recombinant Ribonuclease Inhibitor, using an unspiked sample as the control. As shown in Fig. S12,† after the pretreatments, DNAse I, RNAse A, and protease K showed minor interference (6%) towards the tandem reaction signal. These results demonstrated that the method we have adopted is effective in minimizing nucleases/proteases from degrading the sensor components.
To examine the accuracy and reliability of our FRITCas13a tandem sensor in real samples, our sensor was compared with an EPA-approved method, a fluoride selective electrode, by analyzing drinking water samples spiked with different concentrations of fluoride using the standard calibration method. As shown in Fig. 2d, the fluoride concentrations detected by the FRITCas13a tandem sensor show a strong positive correlation with the added fluoride concentrations and the concentrations detected by the fluoride selective electrode. Moreover, the slope of fluoride concentration detected by the tandem sensor is 1.01 ± 0.08 (R2 = 0.9895), which is similar to the slope measured by the fluoride selective electrode 0.98 ± 0.01 (R2 = 0.9999). Moreover, the ability of the fluoride sensor to quantify fluoride in real environmental samples was tested with various types of samples, including bottled water from a supermarket, drinking water samples from tap water in Urbana (IL, US), and lake water in Urbana (IL, US). As shown in Fig. S13,† no detectable fluoride was found in the bottled water while fluoride was detected and quantified in both the tap water and the lake water by our fluoride sensor and the results were very similar to those from the standard test using the fluoride selective electrode (Fig. S13†), indicating the accuracy of our sensor. Moreover, unknown samples may contain multiple interferents, which may potentially lead to significant matrix effects and affect the accurate detection. To address this issue, the standard addition method was applied to avoid matrix effects and determine the fluoride concentration by our sensor as in a previous report.79 As a proof of concept, a tap water sample spiked with fluoride at a final concentration of 450 μM was analyzed using the standard addition method. Briefly, the samples were spiked with fluoride at different additional concentrations for the tandem reaction, and the concentration of real samples can be calculated based on the extrapolation method. As shown in Fig. S14,† according to the calibration curve obtained, the fluoride concentration of the spiked tap water sample was calculated to be 430.5 ± 11.7 μM (recovery yield of 95.7%), which demonstrates similar accuracy to the result obtained by the fluoride selective electrode (463.9 ± 5.3 μM, recovery yield of 103.1%). Therefore, the accuracy and reliability of our sensor are comparable to those of the EPA-approved fluoride selective electrode.
To demonstrate the ability of our fluoride sensor for on-site and real-time detection, we conduct the tandem reaction with the addition of different concentrations of fluoride and then use a commercially available portable fluorimeter as the fluorescence detection device (Fig. 4a). As shown in Fig. 4b, the fluorescence signal increases with the fluoride concentration from 0 to 810 μM, demonstrating a wide dynamic range for fluoride detection. Moreover, the LOD of this POC sensor is calculated to be 6.4 μM, which is 4-fold lower than 26.3 μM obtained when the EPA-certified fluoride selective electrode is used.78
Conclusions
In summary, we have developed a fluoride riboswitch-regulated transcription with Cas13a (FRITCas13a) tandem sensor for the quantitative detection of fluoride in aqueous solution. The assay uses a simple and fast workflow, with a sample-to-answer time of 30 min, which includes fluoride-riboswitch regulated transcription and Cas13a-mediated signal amplification and fluorescence output. This system can detect fluoride in aqueous solution quantitatively, with high sensitivity (LOD ≈ 1.7 μM), high selectivity to differentiate fluoride from other common anions, and a wide dynamic range of 0–800 μM. This work has expanded the application of nucleic acid sensors for anion detection, and after integration with a portable fluorometer, allows on-site and real-time detection and quantification of fluoride for both environmental monitoring and POC diagnostics.Author contributions
Y. M. designed the detection workflow, performed the experiments and wrote the manuscript. Q. M. assisted with project design and fluorescence experiments. P. Y. assisted with the portable fluorometer experiments. Z. Y. and Y. X. participated in designing Cas13a system. D. Y., C. Z., X. Z. mentored the project, revised the manuscript. Y. L. initiated idea, mentored the project, designed the workflow and revised the paper.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work was supported by Illinois-JITRI Institute (JITRI 23965) and US National Institute of Health (GM141931). Y. M. acknowledges the National Natural Science Foundation of China (21905170) and China Postdoctoral Science Foundation (2019M651479). We would like to thank Jennifer Cui for her help with the portable fluorimeter experiments.Notes and references
- J. Zhang, T. Lan and Y. Lu, Adv. Healthcare Mater., 2019, 8, e1801158 CrossRef PubMed.
- R. Micura and C. Höbartner, Chem. Soc. Rev., 2020, 49, 7331–7353 RSC.
- J. Zhang, L. P. Smaga, N. S. R. Satyavolu, J. Chan and Y. Lu, J. Am. Chem. Soc., 2017, 139, 17225–17228 CrossRef CAS PubMed.
- M. Liu, Q. Yin, Y. Chang, Q. Zhang, J. D. Brennan and Y. Li, Angew. Chem., Int. Ed. Engl., 2019, 58, 8013–8017 CrossRef CAS.
- S. Dey and J. T. Sczepanski, Nucleic Acids Res., 2020, 48, 1669–1680 CrossRef CAS PubMed.
- H. Yu, O. Alkhamis, J. Canoura, Y. Liu and Y. Xiao, Angew. Chem., Int. Ed. Engl., 2021, 60, 2–26 CrossRef.
- L. Li, S. Xu, H. Yan, X. Li, H. S. Yazd, X. Li, T. Huang, C. Cui, J. Jiang and W. Tan, Angew. Chem., Int. Ed. Engl., 2021, 60, 2221–2231 CrossRef CAS.
- P. J. McCown, K. A. Corbino, S. Stav, M. E. Sherlock and R. R. Breaker, RNA, 2017, 23, 995–1011 CrossRef CAS.
- E. Braselmann, A. J. Wierzba, J. T. Polaski, M. Chromiński, Z. E. Holmes, S. T. Hung, D. Batan, J. R. Wheeler, R. Parker, R. Jimenez, D. Gryko, R. T. Batey and A. E. Palmer, Nat. Chem. Biol., 2018, 14, 964–971 CrossRef CAS.
- J. T. Sczepanski and G. F. Joyce, Nature, 2014, 515, 440–442 CrossRef CAS PubMed.
- T. Walton, S. DasGupta, D. Duzdevich, S. S. Oh and J. W. Szostak, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 5741–5748 CrossRef CAS.
- A. Korman, H. Sun, B. Hua, H. Yang, J. N. Capilato, R. Paul, S. Panja, T. Ha, M. M. Greenberg and S. A. Woodson, Proc. Natl. Acad. Sci. U. S. A., 2020, 117 Search PubMed.
- S. S. S. Panchapakesan and R. R. Breaker, Nat. Chem. Biol., 2021, 17, 375–382 CrossRef CAS PubMed.
- H. Peng, X. F. Li, H. Zhang and X. C. Le, Nat. Commun., 2017, 8, 14378 CrossRef PubMed.
- H. Peng, A. M. Newbigging, Z. Wang, J. Tao, W. Deng, X. C. Le and H. Zhang, Anal. Chem., 2018, 90, 190–207 CrossRef CAS PubMed.
- Z. Yang, K. Y. Loh, Y. T. Chu, R. Feng, N. S. R. Satyavolu, M. Xiong, S. M. Nakamata Huynh, K. Hwang, L. Li, H. Xing, X. Zhang, Y. R. Chemla, M. Gruebele and Y. Lu, J. Am. Chem. Soc., 2018, 140, 17656–17665 CrossRef CAS PubMed.
- Y. Lin, Z. Yang, R. J. Lake, C. Zheng and Y. Lu, Angew. Chem., Int. Ed. Engl., 2019, 58, 17061–17067 CrossRef CAS.
- K. Hwang, Q. Mou, R. J. Lake, M. Xiong, B. Holland and Y. Lu, Inorg. Chem., 2019, 58, 13696–13708 CrossRef CAS PubMed.
- C. E. McGhee, K. Y. Loh and Y. Lu, Curr. Opin. Biotechnol., 2017, 45, 191–201 CrossRef CAS.
- R. J. Lake, Z. Yang, J. Zhang and Y. Lu, Acc. Chem. Res., 2019, 52, 3275–3286 CrossRef CAS.
- Y. Xiang and Y. Lu, Nat. Chem., 2011, 3, 697–703 CrossRef CAS.
- H. Li, P. Dauphin-Ducharme, G. Ortega and K. W. Plaxco, J. Am. Chem. Soc., 2017, 139, 11207–11213 CrossRef CAS.
- J. Zhang, H. Xing and Y. Lu, Chem. Sci., 2018, 9, 3906–3910 RSC.
- Y. Xiong, J. Zhang, Z. Yang, Q. Mou, Y. Ma, Y. Xiong and Y. Lu, J. Am. Chem. Soc., 2020, 142, 207–213 CrossRef CAS PubMed.
- L. Zhang, S. Guo, J. Zhu, Z. Zhou, T. Li, J. Li, S. Dong and E. Wang, Anal. Chem., 2015, 87, 11295–11300 CrossRef CAS PubMed.
- H. Li, P. Dauphin-Ducharme, N. Arroyo-Currás, C. H. Tran, P. A. Vieira, S. Li, C. Shin, J. Somerson, T. E. Kippin and K. W. Plaxco, Angew. Chem., Int. Ed. Engl., 2017, 56, 7492–7495 CrossRef CAS.
- J. Zhang and Y. Lu, Angew. Chem., Int. Ed. Engl., 2018, 57, 9702–9706 CrossRef CAS.
- J. Zhang, T. Lan and Y. Lu, TrAC, Trends Anal. Chem., 2020, 124, 115782 CrossRef CAS.
- M. Rothenbroker, E. M. McConnell, J. Gu, M. L. Urbanus, S. E. Samani, A. W. Ensminger, C. D. M. Filipe and Y. Li, Angew. Chem., Int. Ed. Engl., 2021, 60, 4782–4788 CrossRef CAS PubMed.
- J. Canoura, Z. Wang, H. Yu, O. Alkhamis, F. Fu and Y. Xiao, J. Am. Chem. Soc., 2018, 140, 9961–9971 CrossRef CAS.
- M. J. Langton, C. J. Serpell and P. D. Beer, Angew. Chem., Int. Ed. Engl., 2016, 55, 1974–1987 CrossRef CAS.
- S. Saha, V. Prakash, S. Halder, K. Chakraborty and Y. Krishnan, Nat. Nanotechnol., 2015, 10, 645–651 CrossRef CAS PubMed.
- M. Cametti and K. Rissanen, Chem. Soc. Rev., 2013, 42, 2016–2038 RSC.
- Y. Zhou, J. F. Zhang and J. Yoon, Chem. Rev., 2014, 114, 5511–5571 CrossRef CAS PubMed.
- P. P. Chen, W. Bai and Y. Y. Bao, J. Mater. Chem. C, 2019, 7, 11731–11746 RSC.
- E. A. Martínez-Mier, J. Evidence-Based Complementary Altern. Med., 2011, 17, 28–32 CrossRef.
- D. Briancon, Rev. Rhum. Engl. Ed., 1997, 64, 78–81 CAS.
- O. Barbier, L. Arreola-Mendoza and L. M. Del Razo, Chem.-Biol. Interact., 2010, 188, 319–333 CrossRef CAS.
- M. H. Akuno, G. Nocella, E. P. Milia and L. Gutierrez, J. Water Health, 2019, 17, 845–862 CrossRef CAS.
- B. K. Datta, C. Kar and G. Das, J. Chem. Sci., 2015, 127, 337–342 CrossRef CAS.
- B. Spittle, Fluoride, 2019, 52, 7–8 Search PubMed.
- R. Hu, J. Feng, D. Hu, S. Wang, S. Li, Y. Li and G. Yang, Angew. Chem., Int. Ed. Engl., 2010, 49, 4915–4918 CrossRef CAS PubMed.
- A. Dhillon, M. Nair and D. Kumar, Anal. Methods, 2016, 8, 5338–5352 RSC.
- H. Yahyavi, M. Kaykhaii and M. Mirmoghaddam, Crit. Rev. Anal. Chem., 2016, 46, 106–121 CrossRef CAS.
- Y. Jiao, B. Zhu, J. Chen and X. Duan, Theranostics, 2015, 5, 173–187 CrossRef CAS.
- F. M. Ebrahim, T. N. Nguyen, S. Shyshkanov, A. Gladysiak, P. Favre, A. Zacharia, G. Itskos, P. J. Dyson and K. C. Stylianou, J. Am. Chem. Soc., 2019, 141, 3052–3058 CrossRef CAS.
- C. H. Chen and F. P. Gabbaï, Angew. Chem., Int. Ed. Engl., 2018, 57, 521–525 CrossRef CAS.
- X. Sun, S. D. Dahlhauser and E. V. Anslyn, J. Am. Chem. Soc., 2017, 139, 4635–4638 CrossRef CAS PubMed.
- S. Guha and S. Saha, J. Am. Chem. Soc., 2010, 132, 17674–17677 CrossRef CAS PubMed.
- C. Fang, R. Dharmarajan, M. Megharaj and R. Naidu, TrAC, Trends Anal. Chem., 2017, 86, 143–154 CrossRef CAS.
- Y. Guo, J. Li, S. Chai and J. Yao, Nanoscale, 2017, 9, 17667–17680 RSC.
- J. L. Baker, N. Sudarsan, Z. Weinberg, A. Roth, R. B. Stockbridge and R. R. Breaker, Science, 2012, 335, 233–235 CrossRef CAS PubMed.
- A. Ren, K. R. Rajashankar and D. J. Patel, Nature, 2012, 486, 85–89 CrossRef CAS PubMed.
- A. Serganov and E. Nudler, Cell, 2013, 152, 17–24 CrossRef CAS.
- B. Zhao, S. L. Guffy, B. Williams and Q. Zhang, Nat. Chem. Biol., 2017, 13, 968–974 CrossRef CAS PubMed.
- M. Chawla, R. Credendino, A. Poater, R. Oliva and L. Cavallo, J. Am. Chem. Soc., 2015, 137, 299–306 CrossRef CAS.
- A. East-Seletsky, M. R. O'Connell, S. C. Knight, D. Burstein, J. H. Cate, R. Tjian and J. A. Doudna, Nature, 2016, 538, 270–273 CrossRef CAS.
- O. O. Abudayyeh, J. S. Gootenberg, P. Essletzbichler, S. Han, J. Joung, J. J. Belanto, V. Verdine, D. B. T. Cox, M. J. Kellner, A. Regev, E. S. Lander, D. F. Voytas, A. Y. Ting and F. Zhang, Nature, 2017, 550, 280–284 CrossRef PubMed.
- A. Pickar-Oliver and C. A. Gersbach, Nat. Rev. Mol. Cell Biol., 2019, 20, 490–507 CrossRef CAS PubMed.
- J. E. van Dongen, J. T. W. Berendsen, R. D. M. Steenbergen, R. M. F. Wolthuis, J. C. T. Eijkel and L. I. Segerink, Biosens. Bioelectron., 2020, 166, 112445 CrossRef CAS.
- Y. Dai, Y. Wu, G. Liu and J. J. Gooding, Angew. Chem., Int. Ed. Engl., 2020, 59, 20754–20766 CrossRef CAS PubMed.
- J. P. Broughton, X. Deng, G. Yu, C. L. Fasching, V. Servellita, J. Singh, X. Miao, J. A. Streithorst, A. Granados, A. Sotomayor-Gonzalez, K. Zorn, A. Gopez, E. Hsu, W. Gu, S. Miller, C.-Y. Pan, H. Guevara, D. A. Wadford, J. S. Chen and C. Y. Chiu, Nat. Biotechnol., 2020, 38, 870–874 CrossRef CAS.
- J. S. Gootenberg, O. O. Abudayyeh, J. W. Lee, P. Essletzbichler, A. J. Dy, J. Joung, V. Verdine, N. Donghia, N. M. Daringer, C. A. Freije, C. Myhrvold, R. P. Bhattacharyya, J. Livny, A. Regev, E. V. Koonin, D. T. Hung, P. C. Sabeti, J. J. Collins and F. Zhang, Science, 2017, 356, 438–442 CrossRef CAS PubMed.
- J. S. Gootenberg, O. O. Abudayyeh, M. J. Kellner, J. Joung, J. J. Collins and F. Zhang, Science, 2018, 360, 439–444 CrossRef CAS.
- Y. Shan, X. Zhou, R. Huang and D. Xing, Anal. Chem., 2019, 91, 5278–5285 CrossRef CAS PubMed.
- R. Bruch, J. Baaske, C. Chatelle, M. Meirich, S. Madlener, W. Weber, C. Dincer and G. A. Urban, Adv. Mater., 2019, 31, e1905311 CrossRef.
- T. Zhou, R. Huang, M. Huang, J. Shen, Y. Shan and D. Xing, Adv. Sci., 2020, 7, 1903661 CrossRef CAS PubMed.
- C. M. Ackerman, C. Myhrvold, S. G. Thakku, C. A. Freije, H. C. Metsky, D. K. Yang, S. H. Ye, C. K. Boehm, T. F. Kosoko-Thoroddsen, J. Kehe, T. G. Nguyen, A. Carter, A. Kulesa, J. R. Barnes, V. G. Dugan, D. T. Hung, P. C. Blainey and P. C. Sabeti, Nature, 2020, 582, 277–282 CrossRef CAS.
- T. Tian, B. Shu, Y. Jiang, M. Ye, L. Liu, Z. Guo, Z. Han, Z. Wang and X. Zhou, ACS Nano, 2021, 15, 1167–1178 CrossRef CAS.
- Y. Chen, S. Yang, S. Peng, W. Li, F. Wu, Q. Yao, F. Wang, X. Weng and X. Zhou, Chem. Sci., 2019, 10, 2975–2979 RSC.
- P. Qin, M. Park, K. J. Alfson, M. Tamhankar, R. Carrion, J. L. Patterson, A. Griffiths, Q. He, A. Yildiz, R. Mathies and K. Du, ACS Sens., 2019, 4, 1048–1054 CrossRef CAS PubMed.
- J. Arizti-Sanz, C. A. Freije, A. C. Stanton, B. A. Petros, C. K. Boehm, S. Siddiqui, B. M. Shaw, G. Adams, T. F. Kosoko-Thoroddsen, M. E. Kemball, J. N. Uwanibe, F. V. Ajogbasile, P. E. Eromon, R. Gross, L. Wronka, K. Caviness, L. E. Hensley, N. H. Bergman, B. L. MacInnis, C. T. Happi, J. E. Lemieux, P. C. Sabeti and C. Myhrvold, Nat. Commun., 2020, 11, 5921 CrossRef CAS PubMed.
- P. Fozouni, S. Son, M. Diaz de Leon Derby, G. J. Knott, C. N. Gray, M. V. D'Ambrosio, C. Zhao, N. A. Switz, G. R. Kumar, S. I. Stephens, D. Boehm, C. L. Tsou, J. Shu, A. Bhuiya, M. Armstrong, A. R. Harris, P. Y. Chen, J. M. Osterloh, A. Meyer-Franke, B. Joehnk, K. Walcott, A. Sil, C. Langelier, K. S. Pollard, E. D. Crawford, A. S. Puschnik, M. Phelps, A. Kistler, J. L. DeRisi, J. A. Doudna, D. A. Fletcher and M. Ott, Cell, 2021, 184, 323–333 CrossRef CAS PubMed.
- C. Myhrvold, C. A. Freije, J. S. Gootenberg, O. O. Abudayyeh, H. C. Metsky, A. F. Durbin, M. J. Kellner, A. L. Tan, L. M. Paul, L. A. Parham, K. F. Garcia, K. G. Barnes, B. Chak, A. Mondini, M. L. Nogueira, S. Isern, S. F. Michael, I. Lorenzana, N. L. Yozwiak, B. L. MacInnis, I. Bosch, L. Gehrke, F. Zhang and P. C. Sabeti, Science, 2018, 360, 444–448 CrossRef CAS PubMed.
- J. Shen, X. Zhou, Y. Shan, H. Yue, R. Huang, J. Hu and D. Xing, Nat. Commun., 2020, 11, 267 CrossRef CAS PubMed.
- J. Zhou, L. Yin, Y. Dong, L. Peng, G. Liu, S. Man and L. Ma, Anal. Chim. Acta, 2020, 1127, 225–233 CrossRef CAS.
- R. S. Iwasaki and R. T. Batey, Nucleic Acids Res., 2020, 48, e101 CrossRef CAS PubMed.
- SW-846 Test Method 9214: Potentiometric Determination of Fluoride in Aqueous Samples with Ion-Selective Electrode, https://www.epa.gov/hw-sw846/sw-846-test-method-9214-potentiometric-determination-fluoride-aqueous-samples-ion-selective, 1996.
- M. Bader, J. Chem. Educ., 1980, 57, 703–706 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc03508h |
| This journal is © The Royal Society of Chemistry 2021 |