Ultra-bright carbon quantum dots for rapid cell staining†
Tongtong
Zhu‡
ab,
Lei
Cao‡
ab,
Zhenqiao
Zhou
b,
Hanzhou
Guo
c,
Mingfeng
Ge
*b,
Wen-Fei
Dong
 ab and
Li
Li
ab and
Li
Li
 *b
*b
aSchool of Biomedical Engineering (Suzhou), Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei 230026, P. R. China
bCAS Key Laboratory of Biomedical Diagnostics, Suzhou Institute of Biomedical Engineering and Technology, Chinese Academy of Science (CAS), Suzhou 215163, P. R. China. E-mail: gemf@sibet.ac.cn; lil@sibet.ac.cn
cChangchun Guoke Medical Engineer and Technology Development Co., Ltd, Changchun 13003, China
First published on 11th April 2022
Abstract
Cellular imaging using carbon dots is an important research method in several fields. Herein, green-emissive carbon quantum dots (G-CDs) with a pretty high absolute quantum yield (QY) were fabricated via a one-step solvothermal method by using m-phenylenediamine and concentrated hydrochloric acid. G-CDs displayed strong green fluorescence with excitation/emission peaks at 460/500 nm, and their absolute quantum yield was as high as 58.65%. Further experiments suggested that the G-CDs we prepared have good solubility, excellent biocompatibility, and the capacity of rapidly imaging HeLa and 4T1 cells. Over expectations, the G-CDs could penetrate cells in only 10 s and the confocal images showed that the G-CDs could target the nucleus of cells. Moreover, by using 920 nm as the excitation wavelength, two-photon imaging has been successfully applied to 4T1 cells, overcoming the inherent limitations of single-photon imaging. The extremely high absolute quantum efficiency, ultra-fast imaging speed, and two-photon imaging capability make the G-CDs have good application potential in biomedical analysis and the clinical diagnostic field.
1. Introduction
Biological imaging is an important research method to understand the tissue structure of organisms and clarify the various physiological functions of organisms. Due to the advantages of high sensitivity, high resolution, intuitive imaging, fast imaging speed, and non-destructive detection, biological imaging has been widely used in scientific research and biomedical diagnosis.1 Biological imaging also has significant and practical applications in exploring the pathogenesis, clinical manifestations, and genetic lesions of diseases, understanding the corresponding physiological and pathological information, diagnosing diseases, and developing new medical methods. Currently, biological imaging includes fluorescence imaging,2 magnetic resonance imaging (MRI),3 computed tomography (CT),4 emission computed tomography (ECT),5 photothermal imaging (PTI),6 Raman imaging (RI),7 ultrasound imaging (USI)8 and photoacoustic imaging (PAI).9 Among them, fluorescence imaging has higher signal intensity, longer duration, and less experimental cost, along with the feasibility for imaging in vivo to ex vivo. Therefore, fluorescence biological imaging has been widely used.10 In fluorescence imaging, the intensity of the emitted fluorescence signal after excitation of the fluorescent substance has a linear relationship with the amount of fluorescein within a certain range.11 Fluorescently labeled molecules have also been attracting research interest in recent years. Such molecules include fluorescent proteins, fluorescent organic dyes, and quantum dots (QDs), which can be used to label genes, proteins, antibodies, and chemical compounds.12,13Among the reported fluorescently labeled molecules, carbon quantum dots (CDs) have attracted enormous attention owing to their inherent advantages including easy preparation, unique optical properties, excellent photostability, and good biocompatibility.14,15 Therefore, CDs are widely used as new fluorescent probes in cell imaging.16 However, current CD imaging suffers from some disadvantages, such as low absolute quantum efficiency, long fluorescence imaging time, and the rare possibility for two-photon imaging. A low absolute quantum efficiency implies that for a clear and bright image, high CD concentrations are required, which directly impacts cell viability. Moreover, most cell imaging using CDs takes 1–24 hours,17–23 while the cellular environment changes rapidly, which makes it difficult to rapidly image and detect intracellular activities. In addition, single-photon fluorescence imaging exhibits low spatial distribution, poor tissue permeability, and shallow imaging depth. To meet the spatial and temporal resolution required for tissue depth imaging and better imaging performance, using CDs for two-photon imaging (TP) seems to be a good measure to solve this issue. Two-photon absorption/excitation refers to the process in which a molecule simultaneously absorbs two photons and transitions from the ground state to an excited state with twice the photon energy under strong light excitation. In TP fluorescence imaging, the absorbed near-infrared light leads to ultraviolet light emission. Long-wave infrared light is not easily scattered by cells, penetrates deep into the sample, and can be used to detect thick samples. Moreover, the long-wave light source causes less optical damage to living organisms.24,25 The absorbed long-wavelength photons have a wavelength twice that of the single-photon excitation light of the fluorescent molecule. In this way, fluorescent molecules that require single-photon ultraviolet light excitation can be excited by two photons in the near-infrared or even infrared wavelength range. Therefore, the two-photon technique can significantly reduce phototoxicity in the detection of living samples.26
In this study, strongly green and fluorescent carbon quantum dots (G-CDs) were synthesized via a one-step method by using m-phenylenediamine (m-PD) and concentrated hydrochloric acid (Scheme 1). The absolute QY of the synthesized G-CDs reaches up to 58.65%, which was higher than those reported in most published studies (Table 1). The G-CDs showed good biocompatibility and displayed excellent imaging effects even at low concentrations. Most importantly, G-CDs can enter cells within 10 s, which provides a strong foundation for fast fluorescence imaging. Moreover, the G-CDs can also be used for two-photon imaging. With a near-infrared (920 nm) excited light source, the G-CDs exhibit strong green fluorescence and showed excellent imaging performance in 4T1 cells. All the above results provide a potentially excellent biological imaging tool for early diagnosis and clinical treatment.
 | ||
| Scheme 1 A schematic illustration of the synthesis of G-CDs and cell imaging under one-photon and two-photon conditions. | ||
| Precursors | Imaging probe | Quantum yield | Reaction time | Ref. |
|---|---|---|---|---|
| Cysteine | Cell | 38.7% | 12 h | 17 |
| Citric acid, ammonia | Cell | 44.3% | 4 h | 18 |
| Thiomalic acid, diethylenetriamine | Hg2+ | 17.59% | 24 h | 19 |
| EDTA, urea | Apoferritin | 42.3% | 1 h | 20 |
| Citric acid, cystamine dihydrochloride | Cr6+ | 39.7% | 6 h | 21 |
| Casein | Hg2+ | 31.8% | 4 h | 22 |
| L-Cystine, o-phenylenediamine | Ag+ | 35.7% | 4 h | 23 |
| m-Phenylenediamine | Cell | 87.6% | 10 s | This work |
2. Experimental section
2.1 Reagents and materials
m-Phenylenediamine (m-PD), and metal ions were all purchased from Aladdin Ltd. All of the other chemicals were of analytical grade and used without further purification and ultrapure water was used throughout.2.2 Instruments
The morphologies and the size of the G-CDs were recorded using a TECNAI G20 high-resolution transmission electron microscope (TEM) (FEI, USA) at an acceleration voltage of 100 kV. The X-ray diffraction (XRD) pattern was recorded in the 2θ interval from 5° to 45° with an X-ray diffractometer (Bruker, Germany). The X-ray photoelectron spectroscopy (XPS) spectra were recorded using an ESCALAB 250 Xi spectrometer (Thermo Fisher, USA). Moreover, the Fourier transform infrared (FTIR) spectra were recorded with a VERTEX 70 FT-IR spectrometer (Bruker, Germany). A UV-vis absorption spectrophotometer (Agilent Cary 300 Scan) was used for obtaining the UV-vis absorption spectra of the G-CDs. All fluorescence spectra were recorded with a F97Pro FL spectrophotometer (Lengguang Technology, Shanghai) equipped with a 1.0 cm quartz cell. Fluorescence imaging was performed by using an Olympus FV1000 confocal laser scanning microscope (Olympus, Japan). Furthermore, the absolute QY was determined using integrating spheres fitted within an Edinburgh Analytical FLS1000 Instrument. The Nikon A1 + Confocal laser fluorescence microscope was used for cell imaging. TP images were acquired with the Olympus BX61 with a 20× water objective (Olympus, Japan), which was observed under femtosecond pulse excitation at 920 nm.2.3 Synthesis of the G-CDs
G-CDs were synthesized via a one-step solvothermal method. In brief, m-PD (0.2 g) was dissolved in C2H5OH (20 mL) followed by adding 672 μL of concentrated HCl under stirring. Then the resulting mixture was treated hydrothermally in a Teflon-lined stainless steel autoclave at 180 °C for 2 h. After cooling down to room temperature, the products were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 min to remove the non-fluorescent precipitates then purified via silica column chromatography using a mixture of methylene dichloride and methanol as the eluent (CH2Cl2/CH3OH, 10
000 rpm for 20 min to remove the non-fluorescent precipitates then purified via silica column chromatography using a mixture of methylene dichloride and methanol as the eluent (CH2Cl2/CH3OH, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v). The G-CDs were selected followed by evaporating the solvent and further drying under vacuum to collect solid powder for further characterization.
1 v/v). The G-CDs were selected followed by evaporating the solvent and further drying under vacuum to collect solid powder for further characterization.
2.4 Quantum yield calculations of the G-CDs
To obtain accurate quantum efficiency results, both relative quantum efficiency and absolute quantum efficiency data of G-CDs were measured. Quinine sulfate in 0.1 M H2SO4 solution was chosen as the reference. The relative quantum yield of quinine sulfate was 54% at 365 nm excitation, with the relative quantum yield of G-CDs calculated using the following equation:where QY is the relative quantum yield, F represents the integral of the fluorescence intensity over the range from 430 nm to 530 nm, A represents the UV-vis absorption, η represents the refractive index, and the subscript “CD” and “S” refer to the G-CDs and quinine sulfate, respectively. In order to minimize self-absorption effects, the absorptions of G-CDs and quinine sulfate were limited to be below 0.10.
2.5 Cytotoxicity assay
The cytotoxicity of G-CDs was measured using the standardized WST-1 method. Firstly, 4T1 cells (or HeLa cells) were placed in a 96-well plate and incubated for 24 hours, then the G-CD solutions of different concentrations (0–100 μg mL−1) were added to incubate for 24 hours. Secondly, 20 μL of WST-1 was added to each well and then incubated again for 40 min. Finally, the absorbance of the mixture was read using a microplate at 450 nm. The incubation conditions were both 37 °C and 5% CO2.2.6 Cell imaging
For cell imaging, the 4T1 cells (or HeLa cells) were incubated in a glass bottom dish for 24 h. Then, the medium was abandoned and 2 mL of a brand new culture medium containing 40 μg cm−1 G-CD solution was added into the dish. The co-incubation time was divided into four different time gradients: 10 seconds, 1 minute, 10 minutes, and 180 minutes, then all liquid was removed after incubation and washed three times with PBS. DAPI counterstaining was performed with an incubation time of 1 hour, and then fixed with paraformaldehyde solution for 15 minutes. Finally, the fluorescence of 4T1 cells (or HeLa cells) was observed by confocal microscopy using a 40× objective.3. Results and discussion
3.1 Characterization of G-CDs
The as-prepared G-CDs were characterized by using tunneling electron microscopy (TEM), X-ray diffraction (XRD), X-ray photoelectron spectroscopy (XPS), and Fourier transform infrared (FTIR) spectroscopy. The high-resolution TEM image shown in Fig. 1A demonstrates that the G-CDs exhibited a near spherical morphology and had a distinct crystallinity with a lattice spacing of 0.21 nm. The size of the G-CDs was measured by DLS and found to be around 5 nm (Fig. S1†), which is consistent with the TEM image. The zeta potential of G-CDs was evaluated and found to be +12.3 mV (Fig. S2†). As shown in Fig. 1B, the G-CDs showed a distinct XRD pattern with a peak at 2θ = 22° consistent with the spacing on the TEM image, indicating the main graphene structure of G-CDs.27 Moreover, the surface chemical bands and elemental composition of the G-CDs were characterized by XPS as shown in Fig. 1C. The three major peaks at 284.75, 399.05, and 531.85 eV corresponded to C 1s, N 1s, and O 1s, respectively. The composition of C, N, and O elements is 71.92%, 14.10%, and 13.97%. The high-resolution C 1s spectrum (Fig. S3†) can be deconvoluted into three peaks at 284.74, 286.03, and 288.93 eV, corresponding to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/C–C, C
C/C–C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, and O–C
O, and O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds, respectively.28,29 Meanwhile, the high-resolution N 1 s spectrum shows two peaks at 399.16 and 401.17 eV, which represented the presence of pyridinic N and amino N, respectively.30,31 Finally, the two main bands at 531.82 eV and 532.31 eV in the high-resolution O 1s spectrum could be identified as C
O bonds, respectively.28,29 Meanwhile, the high-resolution N 1 s spectrum shows two peaks at 399.16 and 401.17 eV, which represented the presence of pyridinic N and amino N, respectively.30,31 Finally, the two main bands at 531.82 eV and 532.31 eV in the high-resolution O 1s spectrum could be identified as C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–OH.32 The chemical bonds on the surface of the G-CDs were measured by using FTIR spectroscopy. The FTIR spectrum of the G-CDs is shown in Fig. 1D, and the result indicates that the CD presence of –NH2 (3324 cm−1), C–H (2932 cm−1), C
O and C–OH.32 The chemical bonds on the surface of the G-CDs were measured by using FTIR spectroscopy. The FTIR spectrum of the G-CDs is shown in Fig. 1D, and the result indicates that the CD presence of –NH2 (3324 cm−1), C–H (2932 cm−1), C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C (1619 cm−1), –NO2 (1508 cm−1), and C–N–C (1331 cm−1), which is consistent with the result of XPS. Thus, the results of XPS and FTIR show that G-CDs contain functional groups such as amino and carboxyl groups, which enhance the hydrophilicity and stability of G-CDs.
C (1619 cm−1), –NO2 (1508 cm−1), and C–N–C (1331 cm−1), which is consistent with the result of XPS. Thus, the results of XPS and FTIR show that G-CDs contain functional groups such as amino and carboxyl groups, which enhance the hydrophilicity and stability of G-CDs.
 | ||
| Fig. 1 (A) Transmission electron microscopy (TEM) images of the G-CDs. (B) XRD spectra of the G-CDs. (C) XPS full scan spectrum of the G-CDs. (D) FT-IR spectrum of the G-CDs. | ||
3.2 Optical properties of the G-CDs
The UV–vis and fluorescence spectra of the G-CDs were also recorded. As shown in Fig. 2A, the G-CDs exhibited the maximum absorption bands at 248 nm, which represents the π–π* transition of the aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds.33 The weak absorption peak at approximately 300 nm was attributed to the n–π* electron transition, which indicates the presence of C
C bonds.33 The weak absorption peak at approximately 300 nm was attributed to the n–π* electron transition, which indicates the presence of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N and C
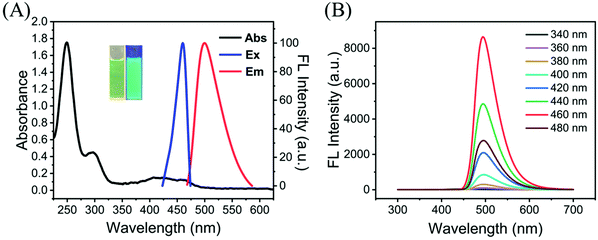
N and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O on the surface of G-CDs.34 In addition, the maximum excitation and emission wavelengths of the G-CDs were 460 and 500 nm, respectively. To determine the excitation-independent emission characteristics of the G-CDs, the excitation wavelengths were varied from 340 to 480 nm as shown in Fig. 2B and from 720 to 900 nm in Fig. S4.† No shift in the emission wavelengths of the G-CDs was observed, which indicated the excitation-independent fluorescence of the G-CDs. As shown in Fig. 3, full-scan spectra and the Commission International de L'Eclairage (CIE) coordinates (0.169, 0.502) provide more detailed information, also demonstrating the excitation-independent properties of G-CDs. Excitation-independent features guarantee the stability of the fluorescence emission and accurate results in a wide range of excitation bands. In addition, it can also be found from the fluorescence spectra that G-CDs have the characteristic of narrow full width at half maximum (FWHM), which can increase high color-purity displays and serve as cell imaging materials. The absolute quantum yield of G-CDs was also notably up to 58.65% (Fig. S5†). Moreover, the quantum yield of the G-CDs was as high as 87.6% by using quinine sulfate (ΦR = 0.54) solution dissolved in 0.1 M sulfuric acid as the reference. This quantum yield is higher than those reported in most published work and can effectively overcome the autofluorescence of cells (Table 1). Therefore, based on its excellent fluorescence characteristics, we believe that the excellent fluorescence characteristics of G-CDs guarantee its performance in cell imaging applications.
O on the surface of G-CDs.34 In addition, the maximum excitation and emission wavelengths of the G-CDs were 460 and 500 nm, respectively. To determine the excitation-independent emission characteristics of the G-CDs, the excitation wavelengths were varied from 340 to 480 nm as shown in Fig. 2B and from 720 to 900 nm in Fig. S4.† No shift in the emission wavelengths of the G-CDs was observed, which indicated the excitation-independent fluorescence of the G-CDs. As shown in Fig. 3, full-scan spectra and the Commission International de L'Eclairage (CIE) coordinates (0.169, 0.502) provide more detailed information, also demonstrating the excitation-independent properties of G-CDs. Excitation-independent features guarantee the stability of the fluorescence emission and accurate results in a wide range of excitation bands. In addition, it can also be found from the fluorescence spectra that G-CDs have the characteristic of narrow full width at half maximum (FWHM), which can increase high color-purity displays and serve as cell imaging materials. The absolute quantum yield of G-CDs was also notably up to 58.65% (Fig. S5†). Moreover, the quantum yield of the G-CDs was as high as 87.6% by using quinine sulfate (ΦR = 0.54) solution dissolved in 0.1 M sulfuric acid as the reference. This quantum yield is higher than those reported in most published work and can effectively overcome the autofluorescence of cells (Table 1). Therefore, based on its excellent fluorescence characteristics, we believe that the excellent fluorescence characteristics of G-CDs guarantee its performance in cell imaging applications.
3.3 Stability of G-CDs
As we know, the pH value, metal ions and bioactive compounds could affect the intensity of CDs. Thus, the effect of different pH values on the fluorescence intensity was also tested to ensure that G-CDs have good imaging performance in common cellular environments. Since most of the intracellular pH is between 7.35–7.45, and a few fluctuate around this value, a wide range of experimental conditions from pH 4 to 9 were chosen to test the pH dependence of the fluorescence intensity. In Fig. 4A, the results show that when the pH changes within the intracellular range (pH 4.0–9.0), the fluorescence emitted by G-CDs is not quenched and remains at a high level. This means that in most pH environments produced by cells, G-CDs exhibit excellent fluorescence performance.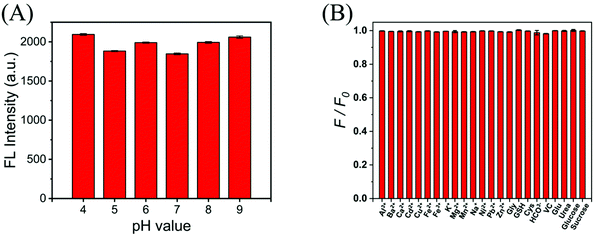 | ||
| Fig. 4 (A) The fluorescence intensity of the G-CD solution in the presence of different pH (pH 4.0–9.0). (B) FL intensity response (F/F0) of the G-CDs to potential interfering substances. | ||
In addition, the potential interference of high concentrations of metal ions (Al3+, Ba2+, Ca2+, Cd2+, Cu2+, Fe2+, Fe3+, K+, Mg2+, Mn2+, Na+, Ni2+, Pb2+, Zn2+, and HCO3−), some amino acids and biothiols in cells was also measured along with their effect on the fluorescence intensity of G-CDs. As shown in Fig. 4B, no marked changes in fluorescence intensity were observed for G-CDs with respect to these interfering substances. No significant changes were observed in the G-CD fluorescence intensity with respect to these interfering substances. Moreover, in the range of 10–60 °C, the fluorescence intensity of G-CDs changed little (Fig. S7†), showing good stability. All the results above suggest that in a typical cellular environment, G-CDs can emit bright green fluorescence, which provides support for intracellular imaging.
3.4 Cytotoxicity and intracellular imaging of G-CDs
In addition to the pH effects and ionic interference, cytotoxicity is also an important parameter for determining the biological suitability of G-CDs. A standardized WST-1 assay was used to assess the cell viability following the addition of G-CDs. As shown in Fig. 5, after co-incubation with G-CDs for 24 h, the survival rates of 4T1 and HeLa cells were still higher than 82% and 85%, even when the concentration of G-CDs has reached 100 µg ml−1. When the concentration of G-CDs was 40 µg ml−1, the cell viability was above 90% in both 4T1 cells and HeLa cells, illustrating that the G-CDs are biofriendly nanomaterials with low cytotoxicity and good biocompatibility. These results indicate that G-CDs have little effect on cell viability and are useful in cell imaging.Subsequently, G-CDs were applied for imaging in 4T1 and HeLa cells. First, these cells were incubated with 2 mL of complete culture and a humidity atmosphere of 5% CO2 at 37 °C for 24 h. After incubating the G-CDs with 4T1 and HeLa cells at 37 °C and a concentration of 40 µg ml−1 for different times, we obtained clear and bright cell images using confocal microscopy. Interestingly, as shown in Fig. 6, the G-CDs can enter the cells within 10 s, which allows for clear and rapid imaging. Considering that most carbon dots require co-incubation for 1–24 h for cell imaging, the imaging capability of G-CDs within 10 s dramatically speeds up the process. Moreover, as shown in Table 1, not only are the reaction times much faster than those of previous reports, but the quantum yield is also much higher than most CDs. We speculate that the good imaging performance can be attributed to the small particle size, extremely high absolute quantum efficiency, and low cytotoxicity of the G-CDs, which can then penetrate cells in a short time and exhibit strong green fluorescence. Thus, our results significantly facilitate the understanding of the physiological state of cells through rapid and conclusive information. In addition, the cell imaging of G-CDs had little dependence on the incubation time. In time-gradient cell imaging experiments of both types of cells, we determined the imaging stability of the G-CDs. The fluorescence intensity under the fluorescence field of cell imaging at 1 min showed no significant difference from those at 10 min and 180 min, and all samples showed clear and bright images. Furthermore, the normal morphology of 4T1 and HeLa cells under bright field indicates that the G-CDs possess biocompatibility. The cell activity can still be maintained above 90% when the concentration of G-CDs is 40 µg ml−1, which is consistent with the cytotoxicity results. In summary, imaging stability and low toxicity allow for the use of G-CDs in observing cell states for a long time. Combined with the ability to rapidly image within 10 s, G-CDs show great application potential for both short- and long-term imaging.
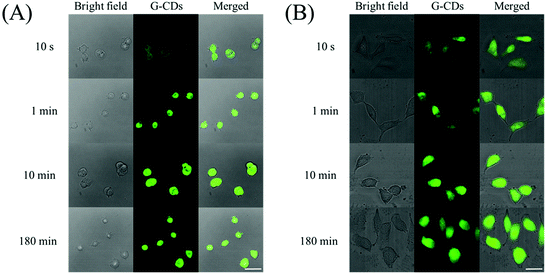 | ||
| Fig. 6 The one-photon fluorescence imaging of 4T1 cells (A) and HeLa cells (B) under incubation with G-CDs (40 µg mL−1) for 10 seconds, 1 minute, 10 minutes, and 180 minutes. The scale bar is 20 µm. | ||
3.5 Study on the properties of target nuclei in cell imaging
The images acquired by confocal microscopy can also be analyzed for colocalization. Biologically, colocalization refers to the same structure in which two or more different molecules are located in the cells, and is often used to study the positional relationship and possible interactions of two fluorescent molecules in tissues or cells. To further investigate the imaging characteristics of G-CDs, DAPI was used to restore the 4T1 and HeLa cells after 60 min of incubation with G-CDs. DAPI is a fluorescent dye capable of strongly binding to DNA, which can pass through the intact cell membrane for the staining of cells. As shown in Fig. 7, green and blue colors were used to distinguish the G-CDs and DAPI, respectively. The green and blue areas have a large overlap, which means that most of the G-CDs accumulated in the nucleus, while a small part existed in the cytoplasm.Considering that the visual interpretation of image data is qualitative, describing its statistical significance is difficult, and potential information could possibly be missed. Therefore, a software-based systematic analysis of colocalization images is required. Two software packages, Color2 and JACoP, were used to analyze the fluorescence colocalization results.35–38 The closer the image of the scatter plot is to the diagonal line, the higher the degree of colocalization. Moreover, Pearson's coefficient can quantitatively express the closeness and direction of the linear relationship between two continuous variables, and the overlap expresses the degree of coincidence of the staining results of the two fluorescent dyes. The closer the values of the Pearson's coefficient and overlap are to 1, the better is the fluorescence colocalization effect. Using Color2 software, we drew scatter plots corresponding to HeLa cells (corresponding to Fig. 7A) and 4T1 cells (corresponding to Fig. 7B) in Fig. 7, respectively. Fig. S7† shows that the scatter plots of HeLa and 4T1 cells are evenly distributed on both sides of the diagonal line, and the Pearson's coefficient of A and B is 0.80 and 0.84, respectively. Meanwhile, using the JACoP software, the Pearson's coefficient of Fig. 7A and B in Fig. 7 was 0.803 and 0.838, respectively. The overlap coefficients were 0.962 and 0.994 as shown in Fig. 7A and B. The results of confocal experiments and fluorescence colocalization analysis suggested that the G-CDs had strong target nuclear properties. This broadens the application of the G-CDs in biomedicine.
The results of confocal experiments and fluorescence colocalization analysis suggested that the G-CDs had strong target nuclear properties. According to the analysis of TEM images (Fig. 1A) and DLS results (Fig. S1†), the particle size of G-CDs is about 5 nm, which may help them enter the nucleus through nuclear pores with a size of about 10 nm. Meanwhile, some well-known commercialized nuclear-targeting reagents such as DAPI and Hoechst 33258 contain a benzene ring structure, considering that the precursor of G-CDs is m-PD, which may contribute to the targeting of G-CDs to the nucleus. In the characterization of G-CDs, it has been found that the surface has –NH2 and –OH, which are positively charged groups. In addition, the zeta potential of G-CDs was also investigated and the result was +12.3 mV (Fig. S2†). The backbone of the DNA double helix is composed of phosphate groups and deoxyribose, and the phosphate groups are negatively charged after hydrolysis. Although the actual mechanism of the nuclear targeting ability of G-CDs remains to be further investigated, it is fully assumed that G-CDs have a high affinity for electronegative regions of DNA through electrostatic interactions. For example, hydrogen bonds can be formed between the amino groups on the surface of G-CDs and the oxygen atoms in DNA thymine, which provide strong support for the target nuclear properties of G-CDs.
3.6 Two-photon imaging of G-CDs
The G-CDs have achieved good single-photon fluorescence imaging in both 4T1 and HeLa cells, especially in the ability to quickly image and target cell nuclei. However, owing to the limitations of single-photon fluorescence imaging, this method still has a low spatial distribution rate, poor permeability organization, and significant depth limitation. Considering that the excitation wavelength of the G-CDs is 460 nm, and the energy generated by two photons at 920 nm is equal to one photon at 460 nm, the two-photon imaging experiment was performed. Notably, the near-infrared excitation light source can effectively reduce the interference of background fluorescence, minimize the damage to cells, and allow for long-term observation. As shown in Fig. 8, the TP fluorescence imaging of 4T1 cells after incubation for 3 h showed the same effect as that of ordinary single-photon fluorescence imaging, and the fluorescence area was clear and bright, confirming the application of the G-CDs to two-photon imaging. | ||
| Fig. 8 The two-photon fluorescence imaging of 4T1 cells incubated with 40 µg ml−1 G-CDs at 37 °C for 3 h, which was observed under a femtosecond pulse excitation at 920 nm. | ||
Therefore, we argue that the G-CDs can exhibit clear green fluorescence under both single- and two-photon excitation conditions, which provides a powerful tool for fields such as fluorescence imaging, tissue depth imaging, and spatial high-resolution imaging.
4. Conclusion
In this study, the G-CDs were synthesized via a one-step solvothermal method, with m-phenylenediamine and concentrated HCl as reagents. The G-CDs exhibit a small particle size with good dispersion, excellent biocompatibility, and low cytotoxicity. The absolute quantum yield of G-CDs is as high as 58.65%, which provides strong support for the excellent luminescence properties and good imaging effects of G-CDs. At both 10 s and 180 min of incubation, the G-CDs showed bright and clear green fluorescence in 4T1 and HeLa cells at a concentration of 40 µg ml−1, which translated to excellent, rapid, and stable imaging. Compared with most carbon dots whose incubation time is between 1 and 24 hours, G-CDs can enter cells hundreds or thousands of times faster, and can quickly observe the state of cells, which greatly facilitates various scientific research. In addition, DAPI counterstaining and fluorescence colocalization experiments showed that the G-CDs are also useful in targeting the nucleus. Both the molecular size around 5 nm and the surface positive electricity provide strong support for G-CDs to target the nucleus. Furthermore, the G-CDs have also been successfully used in two-photon imaging, overcoming the limitation of most carbon dots in single-photon imaging. Using near infrared light as excitation can effectively improve the tissue penetration depth and reduce autofluorescence interference, which is of great significance for its clinical application. In conclusion, the high absolute quantum efficiency, rapid imaging, colocalization, and successful two-photon imaging make the G-CDs a highly promising tool in the promotion of fields such as cellular imaging and biological clinical diagnosis.Author contributions
Tongtong Zhu: conceptualization, methodology, investigation, writing of the original draft. Lei Cao: conceptualization, methodology, investigation, writing of the original draft. Zhenqiao Zhou: software, validation. Hanzhou Guo: software, validation. Mingfeng Ge: methodology, data curation, validation, writing-review and editing. Wen-Fei Dong: methodology, data curation, validation, writing-review and editing. Li Li: methodology, data curation, validation, writing-review and editing.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This research was funded by the National Key R&D Program of China (Grand No. 2020YFC2004600), the National Natural Science Foundation of China (Grand No. 21803075, 91959112, 81902166, 62027825, and 82172077), the Science Foundation of the Chinese Academy of Sciences (No. 2020SYHZ0041), the Instrument Developing Project of Chinese Academy of Science (YJKYYQ20200038), the Primary Research & Development Plan of Jiangsu Province (BE2019683), and the Science and Technology Department of Jinan City (2018GXRC016).References
- Y. Ma, Q. Chen, X. Pan and J. Zhang, Top. Curr. Chem., 2021, 379, 1–21 CrossRef PubMed.
- Y. Zhang, L. K. Schroeder, M. D. Lessard, P. Kidd, J. Chung, Y. Song, L. Benedetti, Y. Li, J. Ries and J. B. Grimm, Nat. Methods, 2020, 17, 225–231 CrossRef CAS PubMed.
- Z. Zhou, L. Yang, J. Gao and X. Chen, Adv. Mater., 2019, 31, 1804567 CrossRef PubMed.
- S.-P. Kwon, S. Jeon, S.-H. Lee, H. Y. Yoon, J. H. Ryu, D. Choi, J.-Y. Kim, J. Kim, J. H. Park and D.-E. Kim, Biomaterials, 2018, 150, 125–136 CrossRef CAS PubMed.
- Q. Fan, K. Cheng, X. Hu, X. Ma, R. Zhang, M. Yang, X. Lu, L. Xing, W. Huang and S. S. Gambhir, J. Am. Chem. Soc., 2014, 136, 15185–15194 CrossRef CAS PubMed.
- Q. Tang, W. Si, C. Huang, K. Ding, W. Huang, P. Chen, Q. Zhang and X. Dong, J. Mater. Chem. B, 2017, 5, 1566–1573 RSC.
- Z. Liu, X. Li, S. M. Tabakman, K. Jiang, S. Fan and H. Dai, J. Am. Chem. Soc., 2008, 130, 13540–13541 CrossRef CAS PubMed.
- H. S. Min, S. Son, D. G. You, T. W. Lee, J. Lee, S. Lee, J. Y. Yhee, J. Lee, M. H. Han and J. H. Park, Biomaterials, 2016, 108, 57–70 CrossRef CAS PubMed.
- J. Kang, D. Kim, J. Wang, Y. Han, J. M. Zuidema, A. Hariri, J. H. Park, J. V. Jokerst and M. J. Sailor, Adv. Mater., 2018, 30, 1800512 CrossRef PubMed.
- J. A. Thomas, Chem. Soc. Rev., 2015, 44, 4494–4500 RSC.
- S. Dunst and P. Tomancak, Genetics, 2019, 211, 15–34 CrossRef CAS PubMed.
- S. Qu, X. Wang, Q. Lu, X. Liu and L. Wang, Angew. Chem., Int. Ed., 2012, 51, 12215–12218 CrossRef CAS PubMed.
- K. Ghosal and A. Ghosh, Mater. Sci. Eng., C, 2019, 96, 887–903 CrossRef CAS PubMed.
- R. Atchudan, T. N. J. I. Edison, M. G. Sethuraman and Y. R. Lee, Appl. Surf. Sci., 2016, 384, 432–441 CrossRef CAS.
- F. P. Mutuyimana, J. Liu, M. Na, S. Nsanzamahoro, Z. Rao, H. Chen and X. Chen, Microchim. Acta, 2018, 185, 1–10 CrossRef CAS PubMed.
- Y. Choi, S. Kim, M. H. Choi, S. R. Ryoo, J. Park, D. H. Min and B. S. Kim, Adv. Funct. Mater., 2014, 24, 5781–5789 CrossRef CAS.
- P. Ni, Q. Li, C. Xu, H. Lai, Y. Bai and T. Chen, Appl. Surf. Sci., 2019, 494, 377–383 CrossRef CAS.
- C. Wang, T. Hu, Z. Wen, J. Zhou, X. Wang, Q. Wu and C. Wang, J. Colloid. Interf. Sci, 2018, 521, 33–41 CrossRef CAS PubMed.
- S. Chandra, A. R. Chowdhuri, T. K. Mahto, D. Laha and S. K. Sahu, Nano-Struct. Nano-Objects, 2017, 12, 10–18 CrossRef CAS.
- C. Han, R. Wang, K. Wang, H. Xu, M. Sui, J. Li and K. Xu, Biosens. Bioelectron., 2016, 83, 229–236 CrossRef CAS PubMed.
- J. Chen, J. Liu, J. Li, L. Xu and Y. Qiao, J. Colloid Interface Sci., 2017, 485, 167–174 CrossRef CAS PubMed.
- S. Xu, Y. Liu, H. Yang, K. Zhao, J. Li and A. Deng, Anal. Chim. Acta, 2017, 964, 150–160 CrossRef CAS PubMed.
- M. Zhang, R. Su, J. Zhong, L. Fei, W. Cai, Q. Guan, W. Li, N. Li, Y. Chen and L. Cai, Adv. Nano Res., 2019, 12, 815–821 CrossRef CAS.
- L. Guo and M. S. Wong, Adv. Mater., 2014, 26, 5400–5428 CrossRef CAS PubMed.
- L. Guo, M. Chan, D. Xu, D. Tam, F. Bolze, P. Lo and M. Wong, ACS Chem. Biol., 2015, 10, 1171–1175 CrossRef CAS PubMed.
- L. Wu, J. Liu, P. Li, B. Tang and T. D. James, Chem. Soc. Rev., 2021, 50, 702–734 RSC.
- Y. Dong, H. Pang, H. B. Yang, C. Guo, J. Shao, Y. Chi, C. M. Li and T. Yu, Angew. Chem., Int. Ed., 2013, 52, 7800–7804 CrossRef CAS PubMed.
- H. Ding, S.-B. Yu, J.-S. Wei and H.-M. Xiong, ACS Nano, 2016, 10, 484–491 CrossRef CAS PubMed.
- N. Wang, A.-Q. Zheng, X. Liu, J.-J. Chen, T. Yang, M.-L. Chen and J.-H. Wang, ACS Appl. Mater. Interfaces, 2018, 10, 7901–7909 CrossRef CAS PubMed.
- Y. Liu, W. Duan, W. Song, J. Liu, C. Ren, J. Wu, D. Liu and H. Chen, ACS Appl. Mater. Interfaces, 2017, 9, 12663–12672 CrossRef CAS PubMed.
- H. Zhang, Y. Chen, M. Liang, L. Xu, S. Qi, H. Chen and X. Chen, Anal. Chem., 2014, 86, 9846–9852 CrossRef CAS PubMed.
- K. Jiang, S. Sun, L. Zhang, Y. Lu, A. Wu, C. Cai and H. Lin, Angew. Chem., Int. Ed., 2015, 54, 5360–5363 CrossRef CAS PubMed.
- H. Li, Y. Xu, J. Ding, L. Zhao, T. Zhou, H. Ding, Y. Chen and L. Ding, Microchim. Acta, 2018, 185, 1–7 CrossRef PubMed.
- X. Luo, W. Zhang, Y. Han, X. Chen, L. Zhu, W. Tang, J. Wang, T. Yue and Z. Li, Food Chem., 2018, 258, 214–221 CrossRef CAS PubMed.
- S. Bolte and F. P. J. Cordelières, J. Microsc., 2006, 224, 213–232 CrossRef CAS PubMed.
- K. W. Dunn, M. M. Kamocka and J. H. McDonald, Am. J. Physiol.: Cell Physiol., 2011, 300, C723–C742 CrossRef CAS PubMed.
- E. Manders, F. Verbeek and J. Aten, J. Microsc., 1993, 169, 375–382 CrossRef CAS PubMed.
- S. V. Costes, D. Daelemans, E. H. Cho, Z. Dobbin, G. Pavlakis and S. Lockett, Biophys. J., 2004, 86, 3993–4003 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2an00325b |
| ‡ These authors contributed equally |
| This journal is © The Royal Society of Chemistry 2022 |