Ratiometric fluorescent probe: a sensitive and reliable reporter for the CRISPR/Cas12a-based biosensing platform†
Qiang
Liu
a,
Mei
Liu
 b,
Yan
Jin
b,
Yan
Jin
 a and
Baoxin
Li
a and
Baoxin
Li
 *a
*a
aKey Laboratory of Applied Surface and Colloid Chemistry, Ministry of Education, Key Laboratory of Analytical Chemistry for Life Science of Shaanxi Province, School of Chemistry & Chemical Engineering, Shaanxi Normal University, Xi'an 710119, China. E-mail: libaoxin@snnu.edu.cn; Fax: +86-29-81530727
bCollege of Food Engineering and Nutritional Science, Shaanxi Normal University, Xi'an 710119, China
First published on 3rd May 2022
Abstract
Due to the excellent activity of trans-cleavage and target recognition, the recently discovered CRISPR/Cas12a systems provide a promising opportunity for designing fluorescence biosensing. In the reported CRISPR/Cas12a-based biosensing platform, TaqMan probe is widely used as the reporter. However, the TaqMan probe provides single-wavelength fluorescence changes, which is easily influenced by various analyte-independent confounding factors to produce false-positive signals. In this study, a ratiometric fluorescent probe was designed to act as the reporter of a CRISPR/Cas12a-based system. As a proof-of-concept, fluorescein (FAM) and tetramethylrhodamine (TAMRA) were chosen as the two fluorescence dyes to label one short ssDNA at 5′ and 3′ ends, respectively, which was designed as one ratiometric fluorescent probe. When the ratiometric probe excites at 480 nm, duo to FRET effect, the probe emitted the 580 nm-fluorescence of TAMRA. The activated Cas12a can cleave the dual-labeled ssDNA, resulting in a decrease in the TAMRA's fluorescence and an increase in the FAM's fluorescence. This dual-response fluorescent probe can act as the reporter of the CRISPR/Cas12a-based biosensing platform. Compared with the classic TaqMan, CRISPR/Cas12a-based biosensing with the ratiometric probe as the reporter not only exhibited higher sensitivity but also could distinguish and avoid false positive signals.
Introduction
The CRISPR/Cas systems provide RNA-guided adaptive immunity to degrade the nucleic acids of intruding bacteria,1 and these systems have been used as gene-editing tools.2,3 Owing to the strict complementary-dependent cleavage principle, the CRISPR/Cas systems have very high accuracy in target nucleic acid recognition. The sequence-specific recognition capability of the CRISPR/Cas systems has been proved to be versatile to develop biosensors for the detection of nucleic acid4–6 and non-nucleic acid targets.7,8 The CRISPR/Cas12a exhibits nonspecific ssDNase (single-stranded deoxyribonuclease) activity.5 Based on this principle, many biological recognizing events have been converted to DNA to activate the trans-cleavage activity of the CRISPR-Cas12a system. In most of the CRISPR/Cas12a-based fluorescence biosensors, TaqMan probe is utilized as the substrate of Cas12a.5,8–15 As one short single-stranded DNA (ssDNA) (ca. 5 nt), the classic TaqMan probes are labeled with one quencher and one fluorophore in the 3′ and 5′ terminals, respectively. In a classic TaqMan probe, the fluorophore is close to the quencher. Due to the fluorescence resonance energy transfer (FRET) between the quencher and the fluorophore, the fluorescence emission of the fluorophore is “turn-off”. After being activated, Cas12a performs a high trans-cleavage on ssDNA.5 Upon binding to the activated Cas12a, the TaqMan probe is cut, and the system emits strong fluorescence. Based on the “turn-on” fluorescence characteristic, TaqMan probe is widely used as a reporter to construct numerous homogenous CRISPR/Cas12a-based biosensors.5,8–15 The CRISPR/Cas12a-based fluorescence sensing strategy with the TaqMan probe as the reporter is simple and rapid without any washing-steps and has been used to detect various targets, including DNA,5,10,11 RNA,12,13 small molecules8,9 and enzyme activity.14,15 However, the classic TaqMan probe always provides single-wavelength fluorescence changes, and the absolute-intensity-dependent signal is easily influenced by various analyte-independent confounding factors, which seriously disturb the accuracy of target quantitation.16 Therefore, it is necessary to explore new reporters for the CRISPR/Cas12a-based biosensors.In recent years, the ratiometric fluorescence method has attracted increasing attention because of its accuracy in detecting the target concentration.17–21 Compared with the conventional fluorescence method with single-signal output, the ratiometric fluorescence method can eliminate the negative influence of complex environment through the ratio of fluorescence intensities of two wavelengths.18–21 Thus, the ratiometric fluorescence methods can effectively overcome the false-positive signal and provide more accurate detection results.18–21 There are two strategies for achieving ratiometric detection: one is the introduction of a target-insensitive fluorescence signal as the reference.22,23 Another is to use two target-insensitive signals. In the fluorescence ratiometry with two target-insensitive signals, the addition of the target often leads to specifically decrease one fluorescence signal and increase the other, which makes the ratio of the two fluorescence signals change greatly. Therefore, the ratiometry strategy with two signal changes also possesses a higher sensitivity compared with the other fluorescence method.16,24
Enlightened by the aforementioned points, we herein introduced one ratiometric fluorescence probe into the CRISPR/Cas12a-based biosensing. As a proof-of-concept, fluorescein (FAM) and tetramethylrhodamine (TAMRA) were chosen as the two fluorescence dyes to label one short ssDNA at 5′ and 3′ ends, respectively, to construct one ratiometric fluorescence probe. Due to the FRET effect, the probe emitted the 580 nm-fluorescence of TAMRA. The activated Cas12a can cleave the dual-labeled ssDNA, resulting in the decrease of the TAMRA's fluorescence and increase of the FAM's fluorescence. This dual-response fluorescence probe can act as the reporter of the CRISPR/Cas12a-based biosensing platform. Compared with the classic TaqMan probe, this probe can provide ratio signal for the CRISPR/Cas12a-based biosensing system. This ratiometric CRISPR/Cas12a-based biosensing system exhibited lower detection limit for DNA target than that with the classic TaqMan reporter. In addition, this ratiometric CRISPR/Cas12a-based biosensing system could distinguish and avoid false positive signals, improving the accuracy of the results. Furthermore, we combined this ratiometric CRISPR/Cas12a-based biosensing system with enzyme-mediated strand-displacement amplification (ESDA) to improve the detection sensitivity. Overall, this ratiometric reporter opens a new pattern for signal output of the CRISPR/Cas12a-based biosensing strategy and holds a great potential to expand to other CRISPR/Cas-assisted biosensing.
Experimental
Reagents and materials
The HPLC-purified DNA and RNA were synthesized by Sangon Biotechnology Co., Ltd (Shanghai, China). The DNA and RNA sequences are listed in Table S1.† Before use, oligonucleotide solutions were diluted to the desired concentrations with 10 mM Tris–HCl buffer (50 mM NaCl and 10 mM MgCl2, pH 7.4). Nicking endonuclease (Nt·BstNBI), Vent (exo-) DNA polymerase and Cas12a were obtained from New England Biolabs, Inc. (Ipswich, US). Millipore Milli-Q water (ca. 18 MΩ cm) was used in the whole experiments. Other chemicals were of analytical grade.Procedure of the CRISPR/Cas12a-based biosensing of target DNA
First, Cas12a protein (50 nM) was incubated with the gRNA (50 nM) in 1 × 2.1 buffer for 10 min at room temperature (ca. 20 °C). Second, 25 μL Rnase-free water, 5 μL 10 × 2.1 buffer and 5 μL ratiometric probe (1 μM) were added successively into the Cas12a–gRNA mixture. Then, 5 μL target DNA with different concentrations was added. The resulting mixture was incubated for 50 min at 37 °C. Lastly, the solution was added into 50 μL Tris–HCl buffer for fluorescence test. The fluorescence emission spectrum of the resultant reaction solution was collected on an FS5 Fluorimeter (Edinburgh, UK). The fluorescence spectra were recorded in the 508–650 nm range with an excitation wavelength (EX) of 480 nm. The ratio of the fluorescent intensity at 520 nm (F520) to the fluorescent intensity at 580 nm (F580) was used to quantify the concentration of target DNA.Procedure of ESDA with the CRISPR/Cas12a-based biosensing for DNA detection
10 μL target DNA with different concentrations and 1 μL template DNA (0.1 nM) were added into 39 μL Nt·BstNBI nicking endonuclease buffer (pH 7.9 Tris–HCl (25 mM), containing 1.25 mM dithiothreitol, 125 mM NaCl, and 12.5 mM MgCl2). Then, the above mixture was heated at 90 °C for 5 min, and was naturally cooled to room temperature (ca. 20 °C). Subsequently, 39 μL ThermoPol buffer, 1 μL dNTP (25 mM), 5 μL Nt·BstNBI (1 U μL−1) and 5 μL vent (exo-) polymerase (1 U μL−1) were added into the above mixture. After reaction for 1.5 h at 55 °C, 30 μL ESDA product was mixed with 5 μL 10× Neb buffer 2.1, 5 μL ratiometric probe (1 μM), 5 μL gRNA (50 nM) and 5 μL Cas12a protein (50 nM) were added into the mentioned solution. Then, the solution was incubated at 37 °C for 50 min. The fluorescence spectra of the reaction solution were recorded. The ratio of F520 to F580 was used to quantify the target DNA concentration.Results and discussion
Principle of ratiometric CRISPR/Cas12a-based biosensing strategy for target DNA detection
The principle of this ratiometric CRISPR/Cas12a-based biosensing is illustrated in Scheme 1. The guide RNA (gRNA) consisted of a 20 nt-long repeated sequence (black color) and a programmable target-recognizing sequence (green color), where the repeated sequence generally promotes gRNA to form the hairpin structure. The gRNA can interact with Cas12a protein to form a Cas12a/gRNA complex via sequence- and shape-specific interactions.25 The Cas12a/gRNA complex can specific bind target DNA (ssDNA) to produce gRNA-DNA heteroduplex, and then Cas12a is activated to show the ssDNase activity. The activated Cas12a can effectively cleave the ratiometric probe (reporter). The ratiometric probe is dual-labeled ssDNA with FAM and TAMRA as the fluorescence donor and acceptor, respectively. The maximum excitation wavelength (Ex) of FAM is 480 nm, and the maximum emission wavelength (Em) of FAM is 520 nm. The Em of TAMRA is 580 nm. When the ratiometric probe excites at 480 nm, the TAMRA's fluorescence at 580 (F580) is observed due to FRET. After the cleavage of the ratiometric probe, FRET is eliminated, resulting in appearance of a FAM's fluorescence at 520 nm (F520). Thus, the change of two fluorescence intensities can be monitored, and the ratio of F520 to F580 can used to quantify the amount of target DNA. When the classic TaqMan probe is acted as the reporter of the CRISPR/Cas12a system, only one signal (F520) appears in the biosensing system.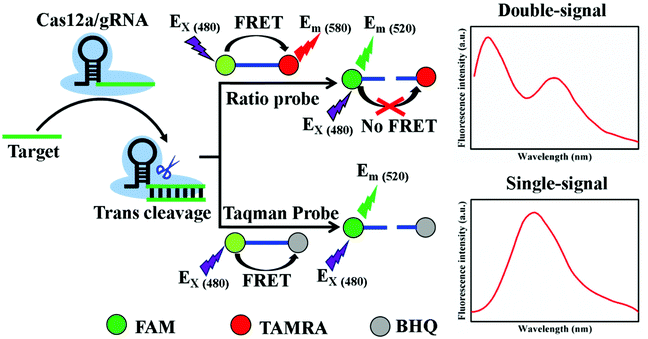 | ||
| Scheme 1 Principle of ratiometric probe or TaqMan probe as reporter in CRISPR/Cas12a-mediated biosensing platform. | ||
In order to verify the feasibility of the ratiometric reporter, we added 0.5 nM target DNA to the CRISPR/Cas12a-based biosensing system. The fluorescence emission spectra of the system were recorded in the absence and presence of the target DNA. As illustrated in Fig. 1A, without the target DNA, FAM emission (F520) was rather weak (ca. 228), whereas TAMRA fluorescence (F580) was very strong (ca. 1870), indicating that FRET is quite efficient for the FAM-TAMRA pair. With target DNA, F520 intensity increased from 228 to 2330, whereas the F580 intensity decreased from 1870 to 1470. Therefore, the signal-to-noise ratio of the ratiometric reporter was estimated to be ca. 13. In addition, we measured the fluorescence response of the biosensing system with the classic TaqMan as the reporter under the same experimental conditions. As shown in Fig. 1B, the presence of target DNA resulted in the increase of F520 from 127 to 1290. The signal-to-noise ratio of the TaqMan reporter was ca. 10. So, the sensitivity of the ratiometric probe was higher than that of the TaqMan probe.
Optimization of experimental conditions
In order to improve the performance of the CRISPR/Cas12a-based biosensing, the experimental conditions were optimized. First, we explored the effects of the length of the ratiometric probe on the response. The three DNA probes with different bases numbers (5, 8 and 11) were designed to act as the reporter of the CRISPR/Cas12a-based biosensor. The results (Fig. S1†) showed that the 8-nt probe had the highest signal-to-noise, and was used as the ratiometric probe. We also optimized the other conditions of the CRISPR/Cas12a system, including incubation time, ratiometric probe concentration, Cas12a concentration and gRNA concentration (Fig. S2†). The optimized experimental conditions were 50 min of incubation time, 1 μM of ratiometric probe, 50 nM of gRNA, and 50 nM of Cas12a.Performance of the CRISPR/Cas12a-based biosensing for target DNA
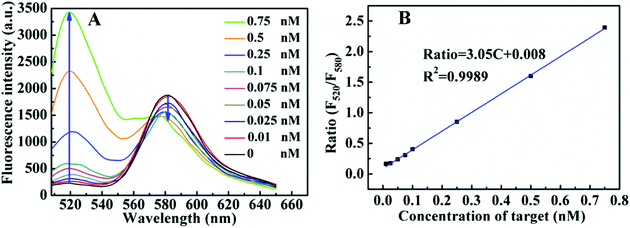
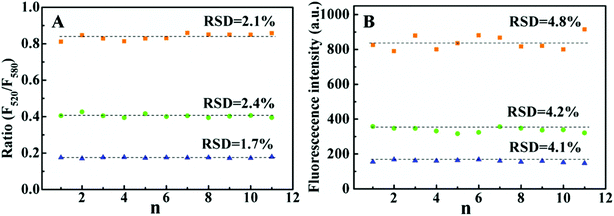
Under the optimal experiment condition, the fluorescence emission spectra of the CRISPR/Cas12a system with the ratiometric probe were obtained in the presence of target DNA with different concentrations. As shown in (Fig. 2A), with increasing target DNA concentration, F520 gradually increased, whereas F580 steadily decreased. The ratio (F520/580) increases linearly with target concentration in 0.01–0.75 nM range (Fig. 2B). The limit of detection (LOD) was calculated to be 3 pM (3σ). For a comparison, we investigated the fluorescence response of the CRISPR/Cas12a-based biosensing with the TaqMan probe as the reporter under the same conditions (Fig. S3A†). The fluorescence intensity was linearly dependent on the target DNA concentration in the range of 0.025 nM–0.25 nM (Fig. S3B†). The LOD of the TaqMan probe was estimated to be 10 pM (3σ), which was above threefold higher than that of the ratiometric probe. We also checked the precision of the two reporters. Fig. 3 shows the results of 11 parallel measurements of the CRISPR/Cas12a-based biosensing at three different concentration levels (0.025 nM, 0.1 nM, and 0.25 nM). The relative standard deviation (RSD) of the ratiometric probe was estimated to be 2.1%, 2.4% and 1.7%, respectively (Fig. 3A). The RSD of the TaqMan probe was 4.8%, 4.2%, and 4.1%, respectively (Fig. 3B). It is obvious that the precision of the ratiometric probe is better than that of the classic TaqMan probe. The ratiometric probe is more resistant to fluctuations caused by the experimental environment.Compared to the conventional single-signal probe, the ratiometric probe is expected to possess the capability of distinguish false positive signals.18–20 As illustrated in Fig. 2A, the addition of the target DNA could lead to the increase of F520, accompanying the decrease of F580. If the increase of F520 and decrease of F580 in this biosensing system do not occur simultaneously, the signal will be false-positive. To prove this, we took bovine serum albumin (BSA) as an example to study the effects of some proteins on the response of the reporter. As shown in Fig. 4A, by increasing the BSA amount from 0 to 25%, the F520 intensity of the ratiometric probe increased evidently, whereas the F580 intensity did not almost change. From the response of this ratiometric probe, it is easy to judge that the BSA-induced change is a false positive signal. However, for classic TaqMan probe, a gradually increasing false-positive signal was observed (Fig. 4B), which is undistinguishable from a true target response. In addition, the similar result was observed when Ca2+ was added into the reporter probe. Ca2+ could decrease the intensities of F520 and F580 in this ratiometric probe response (Fig. S4†). This may be attributed to that Ca2+ can bind to the phosphate backbone of the ratiometric DNA fluorescent probe,26 reducing the solubility of the DNA probe. However, as shown in Fig. 5A, the ratio of F520 to F580 remained unchanged when increasing Ca2+ from 0 to 10 mM. This is to say, the ratiometric probe can effectively avoid the false positive signal from Ca2+. Ca2+ also decreased the fluorescence of the classic TaqMan probe (Fig. 5B), but the classic TaqMan probe cannot distinguish whether the decrease of the fluorescence signal is due to the change of the Ca2+ concentration or the change of target concentration. Therefore, the ratiometric probe as a reporter of the CRISPR/Cas12a-based biosensing can distinguish and even avoid false positive signals. The ratiometric probe is suitable for an in vivo assay. Currently, there is a lack of an appropriate carrier to deliver the CRISPR/Cas12a system into living cells.27 Therefore, we did not apply the CRISPR/Cas12a ratiometric fluorescent probe in practical applications.
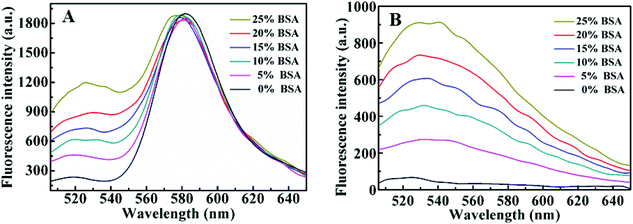 | ||
| Fig. 4 Fluorescence spectra of the ratiometric probe (A) and TaqMan probe (B) in the presence of different concentrations of BSA. | ||
 | ||
| Fig. 5 Effect of Ca2+ concentration on the fluorescence response of the ratiometric probe (A) and TaqMan probe (B) in 20 mM Tris–HCl (pH 7.4) buffer. | ||
Combining enzyme mediated strand-displacement amplification with CRISPR/Cas12a-based biosensing
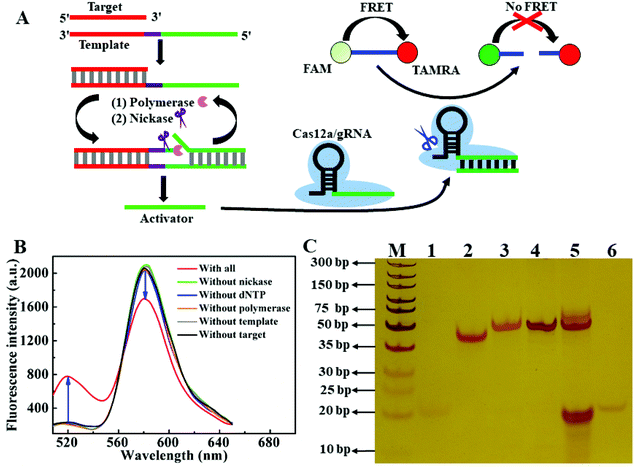
Enzyme mediated strand-displacement amplification (ESDA) is an efficient tool for amplified assay.28–30 To further enhance the sensitivity of the ratiometric CRISPR/Cas12a-based biosensing, we combined ESDA with this CRISPR/Cas12a-based biosensing. As shown in Fig. 6A, the ESDA template contained three domains: binding region of the target DNA (red), nicking site (purple) and region for generating activator (green). The hybridization of the target DNA and the template triggers the ESDA process. In the presence of dNTPs and DNA polymerase, the partial duplex (template/target) is extended along the template to form a complete dsDNA with one nicking site. Nicking endonuclease cleaves the extended DNA strand to release the activator, and simultaneously the cleavage reaction can trigger a new extension—cleavage—release cycle. As a result, the ESDA reaction continuously occurs, and many activators can be produced. The binding of the activator onto gRNA activates the trans-cleavage activity of Cas12a to cleave the ratiometric probe, generating an obvious fluorescence response. As shown in Fig. 6B, when template, polymerase, nicking endonuclease and dNTP co-existed in the reaction system, the addition of 50 pM target DNA resulted in the signal change (F520 increase and F580 decrease). However, when any of four components (dNTPs, template, nicking endonuclease and polymerase) was absent, target DNA did not induce a significant change of fluorescence. To certify whether the signal amplification would work as designed, we performed the gel electrophoresis experiments. As shown in Fig. 6C, the clear band on lane 1, 2 and 6 corresponded to target (22 nt), template (51 nt) and activator (20 nt). When the target was mixed with the template, a new upper band appeared in lane 3, which indicated the formation of a partial duplex between the target and the template. Once polymerase and dNTP were added into the system, a dark band in lane 4 emerged (51 bp), suggesting that the partial duplex was extended along the template DNA to produce the complete duplex. When nickase was unceasingly added into the system, the band at 51 bp became weak (lane 5), and in the meantime a dark band appeared at the same position as the activator (lanes 5 and 6). The gel electrophoresis results showed that a small number of targets were transformed into a large number of activators through ESDA, thereby enhancing the sensitivity of the CRISPR/Cas12a-based method.Under the optimized experiment condition (Fig. S5†), we investigated the fluorescent response of the ESDA-CRISPR/Cas12a-based biosensing system to different concentrations of the target DNA in the 1–100 pM range. As shown in Fig. 7A, by increasing the target DNA concentration, F520 gradually increased, whereas F580 steadily decreased. The ratio (F520/F580) increases linearly with target DNA concentration in the range from 1 pM to 75 pM (Fig. 7B). According to the 3σ, the LOD was estimated to be 0.3 pM, which was about 10-fold lower than that of the ratiometric CRISPR/Cas12a sensing system without ESDA. For comparison, we also investigated the fluorescence response of this biosensing system using the TaqMan probe as the reporter under the same conditions (Fig. S6†), and the LOD was calculated to be 2 pM, which was much higher than that of the ratiometric probe. These results further suggested that the ratiometric probe had higher sensitivity than the classic TaqMan probe.
Conclusions
In summary, we used two fluorophores as the donor and acceptor to design a ratiometric probe for expanding the output pattern of a single signal readout. The ratiometric probe was used as the reporter of the CRISPR/Cas12a-based biosensing system. Compared with the classic TaqMan as the reporter, the CRISPR/Cas12a-based biosensing with the ratiometric probe as the reporter not only exhibited higher sensitivity but also could distinguish and avoid false-positive signal. This study shows that the ratiometric fluorescence probe is an effective reporter for the CRISPR/Cas12a-based biosensor system, which will promote the development of the ratio fluorescence method. Furthermore, the ratiometric fluorescence probe may provide great potential for other CRISPR/Cas enzyme-assisted biosensors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by the National Natural Science Foundation of China (21974082).References
- M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna and E. Charpentier, A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity, Science, 2012, 337, 816–821 CrossRef CAS PubMed
.
- A. V. Anzalone, L. W. Koblan and D. R. Liu, Genome editing with CRISPR–Cas nucleases, base editors, transposases and prime editors, Nat. Biotechnol., 2020, 38, 824–844 CrossRef CAS PubMed
.
- G. J. Knott and J. A. Doudna, CRISPR-Cas guides the future of genetic engineering, Science, 2018, 361, 866–869 CrossRef CAS PubMed
.
- X.-W. Wang, L.-F. Hu, J. Hao, L.-Q. Liao, Y.-T. Chiu, M. Shi and Y. Wang, A microRNA-inducible CRISPR–Cas9 platform serves as a microRNA sensor and cell-type-specific genome regulation tool, Nat. Cell Biol., 2019, 21, 522–530 CrossRef PubMed
.
- J. S. Chen, E. Ma, L. B. Harrington, M. D. Costa, X. Tian, J. M. Palefsky and J. A. Doudna, CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity, Science, 2018, 360, 436–439 CrossRef CAS PubMed
.
- Y. Shan, X. Zhou, R. Huang and D. Xing, High-fidelity and rapid quantification of miRNA combining crRNA programmability and CRISPR/Cas13a trans-cleavage activity, Anal. Chem., 2019, 91, 5278–5285 CrossRef CAS PubMed
.
- Y. Zhang, Y. Wu, Y. Wu, Y. Chang and M. Liu, CRISPR-Cas systems: from gene scissors to programmable biosensors, Trends Anal. Chem., 2021, 137, 116210 CrossRef CAS
.
- M. Liang, Z. Li, W. Wang, J. Liu, L. Liu, G. Zhu, L. Karthik, M. Wang, K.-F. Wang, Z. Wang, J. Yu, Y. Shuai, J. Yu, L. Zhang, Z. Yang, C. Li, Q. Zhang, T. Shi, L. Zhou, F. Xie, H. Dai, X. Liu, J. Zhang, G. Liu, Y. Zhuo, B. Zhang, C. Liu, S. Li, X. Xia, Y. Tong, Y. Liu, G. Alterovitz, G.-Y. Tan and L.-X. Zhang, A CRISPR-Cas12a-derived biosensing platform for the highly sensitive detection of diverse small molecules, Nat. Commun., 2019, 10, 3672 CrossRef PubMed
.
- Y. Xiong, J. Zhang, Z. Yang, Q. Mou, Y. Ma, Y. Xiong and Y. Lu, Functional DNA regulated CRISPR-Cas12a sensors for point-of-care diagnostics of non-nucleic-acid targets, J. Am. Chem. Soc., 2020, 142, 207–213 CrossRef CAS PubMed
.
- Y. Li, H. Mansour, C. J. F. Watson, Y. Tang, A. J. MacNeil and F. Li, Amplified detection of nucleic acids and proteins using an isothermal proximity CRISPR Cas12a assay, Chem. Sci., 2021, 12, 2133–2137 RSC
.
- L. T. Nguyen, B. M. Smit and P. K. Jain, Enhancement of trans-cleavage activity of Cas12a with engineered crRNA enables amplified nucleic acid detection, Nat. Commun., 2020, 11, 4906 CrossRef CAS PubMed
.
- J. P. Broughton, X. D. Deng, G. X. Yu, C. L. Fasching, V. Servellita, J. Singh, X. Miao, J. A. Streithorst, A. Granados, A. Sotomayor-Gonzalez, K. Zorn, A. Gopez, E. Hsu, W. Gu, S. Miller, C. Y. Pan, H. Guevara, D. A. Wadford, J. S. Chen and C. Y. Chiu, CRISPR-Cas12-based detection of SARS-CoV-2, Nat. Biotechnol., 2020, 38, 870–874 CrossRef CAS PubMed
.
- S. Peng, Z. Tan, S. Chen, C. Lei and Z. Nie, Integrating CRISPR-Cas12a with a DNA circuit as a generic sensing platform for amplified detection of microRNA, Chem. Sci., 2020, 11, 7362–7368 RSC
.
- D.-X. Wang, J. Wang, Y.-C. Du, J.-Y. Ma, S.-Y. Wang, A.-N. Tang and D.-M. Kong, CRISPR/Cas12a-based dual amplified biosensing system for sensitive and rapid detection of polynucleotide kinase/phosphatase, Biosens. Bioelectron., 2020, 168, 112556 CrossRef CAS PubMed
.
- Y.-C. Du, S.-Y. Wang, Y.-X. Wang, J.-Y. Ma, D.-X. Wang, A.-N. Tang and D.-M. Kong, Terminal deoxynucleotidyl transferase combined CRISPR-Cas12a amplification strategy for ultrasensitive detection of uracil-DNA glycosylase with zero background, Biosens. Bioelectron., 2021, 171, 112734 CrossRef CAS PubMed
.
- P. Zhang, T. Beck and W. Tan, Design of a molecular beacon DNA probe with two fluorophores, Angew. Chem., Int. Ed., 2001, 40, 402–405 CrossRef CAS PubMed
.
- X. Huang, J. Song, B. C. Yung, X. Huang, Y. Xiong and X. Chen, Ratiometric optical nanoprobes enable accurate molecular detection and imaging, Chem. Soc. Rev., 2018, 47, 2873–2920 RSC
.
- Y. Yang, J. Huang, X. Yang, K. Quan, H. Wang, L. Ying, N. Xie, M. Ou and K. Wang, FRET nanoflares for intracellular mRNA detection: avoiding false positive signals and minimizing effects of system fluctuations, J. Am. Chem. Soc., 2015, 137, 8340–8343 CrossRef CAS PubMed
.
- L. He, D.-Q. Lu, H. Liang, S. Xie, C. Luo, M. Hu, L. Xu, X. Zhang and W. Tan, Fluorescence resonance energy transfer based DNA tetrahedron nanotweezer for highly reliable detection of tumor-related mRNA in living cells, ACS Nano, 2017, 11, 4060–4066 CrossRef CAS PubMed
.
- C. He, Z. Liu, Q. Wu, J. Zhao, R. Liu, B. Liu and T. Zhao, Ratiometric fluorescent biosensor for visual discrimination of cancer cells with different telomerase expression levels, ACS Sens., 2018, 3, 757–762 CrossRef CAS PubMed
.
- Y. Liu, T. Shen, J. Li, H. Gong, C. Chen, X. Chen and C. Cai, Ratiometric fluorescence sensor for the microRNA determination by catalyzed hairpin assembly, ACS Sens., 2017, 2, 1430–1434 CrossRef CAS PubMed
.
- G. Mao, Q. Cai, F. Wang, C. Luo, X. Ji and Z. He, One-step synthesis of Rox-DNA functionalized CdZnTeS quantum dots for the visual detection of hydrogen peroxide and blood glucose, Anal. Chem., 2017, 89, 11628–11635 CrossRef CAS PubMed
.
- H.-Q. Yin, J.-C. Yang and X.-B. Yin, Ratiometric fluorescence sensing and real-time detection of water in organic solvents with one-pot synthesis of Ru@MIL-101(Al)-NH2, Anal. Chem., 2017, 89, 13434–13440 CrossRef CAS PubMed
.
- A. Bigdeli, F. Ghasemi, S. Abbasi-Moayed, M. Shahrajabian, N. Fahimi-Kashani, S. Jafarinejad, M. A. F. Nejad and M. R. Hormozi-Nezhad, Ratiometric fluorescent nanoprobes for visual detection: design principles and recent advances-a review, Anal. Chim. Acta, 2019, 1079, 30–58 CrossRef CAS PubMed
.
- X. Wang, X. Chen, C. Chu, Y. Deng, M. Yang, Z. Ji, F. Xu, D. Huo, Y. Luo and C. Hou, Four-stage signal amplification for trace ATP detection using allosteric probe-conjugated strand displacement and CRISPR/Cpf1 trans-cleavage (ASD-Cpf1), Sens. Actuators, B, 2020, 323, 128653 CrossRef CAS
.
- M. M. Kiy, Z. E. Jacobi and J. Liu, Metal-induced specific and nonspecific oligonucleotide folding studied by FRET and related biophysical and bioanalytical implications, Chem. – Eur. J., 2012, 18, 1202–1208 CrossRef CAS PubMed
.
- D.-X. Wang, Y.-X. Wang, J. Wang, J.-Y. Ma, B. Liu, A.-N. Tang and D.-M. Kong, MnO2 nanosheets as a carrier and accelerator for improved live-cell biosensing application of CRISPR/Cas12a, Chem. Sci., 2022, 13, 4364–4371 RSC
.
- X.-L. Zhang, Y.-H. Liu, S.-M. Du, Y. Yin, L.-Q. Kong, Y.-Y. Chang, Y.-Q. Chai, Z.-H. Li and R. Yuan, Engineering a rolling-circle strand displacement amplification mediated label-free ultrasensitive electrochemical biosensing platform, Anal. Chem., 2021, 93, 9568–9574 CrossRef PubMed
.
- C. Shi, Q. Liu, C. Ma and W. Zhong, Exponential strand-displacement amplification for detection of microRNAs, Anal. Chem., 2014, 86, 336–339 CrossRef CAS PubMed
.
- J.-L. He, Z.-S. Wu, H. Zhou, H.-Q. Wang, J.-H. Jiang, G.-L. Shen and R.-Q. Yu, Fluorescence aptameric sensor for strand displacement amplification detection of cocaine, Anal. Chem., 2010, 82, 1358–1364 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2an00613h |
| This journal is © The Royal Society of Chemistry 2022 |