Reusable and universal impedimetric sensing platform for the rapid and sensitive detection of pathogenic bacteria based on bacteria-imprinted polythiophene film†
Lingling
Wang‡
a,
Xiaohui
Lin‡
a,
Ting
Liu
a,
Zhaohuan
Zhang
a,
Jie
Kong
a,
Hai
Yu
a,
Juan
Yan
 a,
Donglei
Luan
a,
Yong
Zhao
*a and
Xiaojun
Bian
a,
Donglei
Luan
a,
Yong
Zhao
*a and
Xiaojun
Bian
 *abc
*abc
aCollege of Food Science and Technology, Shanghai Ocean University, Shanghai 201306, China. E-mail: yzhao@shou.edu.cn; xjbian@shou.edu.cn; Fax: +86-21-61900753; Tel: +86-21-61900753
bLaboratory of Quality and Safety Risk Assessment for Aquatic Product on Storage and Preservation (Shanghai), Ministry of Agriculture, Shanghai 201306, China
cShanghai Engineering Research Center of Aquatic-Product Processing & Preservation, Shanghai 201306, China
First published on 12th August 2022
Abstract
The rapid and sensitive detection of pathogenic bacteria is highly demanded for early warning of infectious disease epidemics and protection of human health. Herein, a reusable and universal impedimetric sensing platform based on a bacteria-imprinted polythiophene film (BIF) is proposed for the rapid and sensitive detection of pathogenic bacteria using Staphylococcus aureus (S. aureus) as a model analyte. Monomer screening among four 3-substituted thiophenes was first performed based on the imprinting factor, and 3-thiopheneethanol (TE) was eventually selected. The BIF as a recognition layer was quickly deposited in an environmentally friendly process on a glassy carbon electrode via electro-copolymerization of the S. aureus template and TE monomer followed by in situ template removal. Upon rebinding of S. aureus on the BIF, the impedance increased. Under optimal conditions, the BIF-based sensor can quantitatively detect S. aureus in a wide linear range of 10 to 107 CFU mL−1 with a low detection limit of 4 CFU mL−1. Additionally, the sensor exhibits excellent selectivity, capable of identifying S. aureus from multi-bacterial strain mixtures. It also demonstrates applicability in the analysis of real lettuce and shrimp samples with good recoveries. Most significantly, the BIF sensing interface can be reused up to five times with good signal retention. Compared with most reported methods, this sensor is more rapid with a much shorter total assay time of 30 min, including the BIF preparation, bacterial rebinding, and impedance detection. This assay may hold great potential to help in the rapid, sensitive, and label-free detection of pathogenic bacteria in fields of food safety and public health.
Introduction
Pathogenic bacterial infections represent leading causes of hospitalizations and deaths in both developed and developing countries, taking more than 6.7 million lives each year.1 Among the various bacterial strains, Staphylococcus aureus (S. aureus) is one of the most dangerous human pathogens responsible for many diseases ranging from mild skin and soft tissue infections, food poisoning to serious or even life-threatening diseases including bacteremia, sepsis, pneumonia, endocarditis, osteomyelitis, and toxic shock syndrome.2,3 Moreover, S. aureus can rapidly adapt to antibiotics and evolve into antibiotic-resistant bacteria, such as methicillin-resistant S. aureus (MRSA), that have imposed huge burdens on medical care, and posed serious threats to human life.4,5 Existing methods for tracking pathogenic bacteria such as cell culturing,6 enzyme-linked immunosorbent assay (ELISA),7,8 polymerase chain reaction (PCR),9,10 mass spectrometry,11,12 flow cytometry,13 and surface-enhanced Raman scattering (SERS)14,15 are accurate and reliable. Still, they are restricted by lengthy analysis times, insufficient sensitivity, complicated operation, or expensive instruments and reagents. Therefore, rapid, sensitive, simple, and low-cost methods for detecting pathogenic bacteria are still highly demanded.Electrochemical sensors have received considerable attention for pathogen detection due to their fast response, easy operation, low cost, and portability.16 Moreover, electrochemical sensors are robust due to their direct and label-free detection of whole bacterial cells without tedious processes, including cell lysis, nucleic acid extraction, and signal amplification. The most crucial aspect in the construction of label-free electrochemical sensors for the detection of pathogenic bacteria is the choice of suitable recognition elements and their efficient immobilization onto the electrode surface. Among various recognition elements (e.g., antibodies,4,17 aptamers,18–20 antimicrobial peptides,21etc.), antibodies are the most commonly used due to their high selectivity and binding affinity. However, antibodies are limited by their drawbacks of requiring animals for production, high cost, and low tolerance to harsh conditions such as high temperatures and salt concentrations, strong acids or bases, and organic solvents. Besides, antibodies may suffer from conformational changes or denaturation when being immobilized on the transducer surface through adsorption or covalent coupling.22
Molecularly imprinted polymers (MIPs) are synthetic antibody mimics with tailor-made binding sites complementary in physical (shape and size) and chemical (structure and functionality) features to that of a template.23 MIPs have many distinct advantages, such as easy preparation, being cost-efficient, and having long-term physical and chemical stability, which make them promising alternatives to natural antibodies.24 Owing to these unique characteristics, MIPs have been integrated with various sensing platforms for detecting simple organic molecules,25–27 and biomacromolecules like proteins,28,29 viruses,30 and bacteria.31 However, we have to acknowledge that the imprinting of whole bacterial cells is still very challenging due to their large size and complex chemical composition, leading to increased difficulty in the complete removal of the template, and the generation of imprinted sites with good accessibility and recognition capability.32
To overcome the difficulties mentioned above, various surface imprinting strategies such as stamping/microcontact,33 Pickering emulsion polymerization,34 colloidal imprinting,35,36 and electro-polymerization37 have been proposed. However, most of these are cumbersome, time-consuming, and environmentally unfriendly, using large amounts of toxic reagents such as cross-linkers, initiators, and organic solvents. In comparison, electro-polymerization offers a simple, fast, and green method for the direct synthesis of MIP films in situ on the electrode surface, avoiding the use of cross-linkers and initiators.38 By varying the electro-polymerization parameters, the thickness of polymer films can be easily controlled, and thus it is conducive to better template removal and site accessibility. Moreover, electropolymerized MIPs (e-MIPs) can be facilely integrated with an electrochemical sensing platform, omitting the step of electrode modification.25
Though synthesizing e-MIPs is a simple process, the construction of e-MIPs-based electrochemical bacterial sensors with a desired sensing performance (rapidity, sensitivity, selectivity, and simplicity) remains a challenge. One critical solution to tackling this challenge is the selection of suitable functional monomers that provide good interactions with bacterial cells (template).39,40 Since the chemical composition of the bacterial surface is complex, it is difficult to build specific covalent bindings for template-monomers, and thus noncovalent interactions such as hydrogen bonding are preferred.41
Herein, a universal and regenerable impedimetric sensor based on an electropolymerized bacteria-imprinted polythiophene film (BIF) was proposed for the rapid, sensitive, and label-free detection of pathogenic bacteria using S. aureus as a model analyte. Considering the existence of various functional groups (hydroxyl, carboxyl, phosphoryl, and amide) on the bacterial surface,42 four 3-substituted thiophenes, including 3-thiopheneacetic acid (TAA), 3-thiopheneboronic acid (TBA), 3-thiophenemethylamine (TMA), and 3-thiopheneethanol (TE), were selected as potential functional monomers for the electro-synthesis of four different BIFs. As shown in Scheme 1, the BIF was in situ formed on a glassy carbon electrode (GCE) via the cyclic voltammetric electro-copolymerization of each monomer and the S. aureus template in phosphate buffer solution (PBS) followed by template removal. The BIF preparation is speedy, green, and low cost, and can be finished within 15 min without using any harmful organic solvents. After the recognition of the target S. aureus within 10 min, the electron transfer between the [Fe(CN)6]3−/4− redox couple and GCE were blocked, leading to an increase in the BIF impedance, detected by electrochemical impedance spectroscopy (EIS). By comparing the imprinting factor of the four monomers, TE was eventually used to construct the BIF-based impedimetric sensor. Compared with most existing methods, this sensor is more sensitive and rapid with a lower limit of detection (LOD), and a much shorter total assay time. Moreover, the sensing interface can be regenerated and reused at least five times. Additionally, the applicability of the sensor was also evaluated using real lettuce and shrimp samples. The proposed assay could provide a universal and straightforward method for the rapid, sensitive, and highly selective detection of pathogenic bacteria.
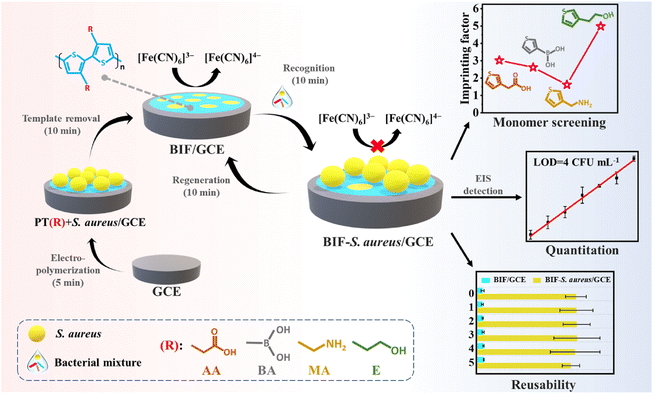 | ||
| Scheme 1 Schematic illustration of constructing a regenerable bacteria imprinted polythiophene film (BIF)-based impedimetric sensor for the rapid, sensitive, and label-free detection of S. aureus. | ||
Experimental section
Preparation of bacteria
The pure isolates of S. aureus (ATCC 25923), Listeria monocytogenes (L. monocytogenes, ATCC 19115), Escherichia coli O157:H7 (E. coli O157, ATCC 43889), and Salmonella Paratyphi B (S. Paratyphi B, CMCC 50094) were involved in the experiment. All the bacterial strains were separately grown overnight in a liquid medium at 37 °C with continuous shaking at 200 rpm, and then were quantified by the plate count method. Afterward, the bacterial culture was fixed with 100-fold diluted formaldehyde (37–40% in water). Such formaldehyde inactivated bacteria were used during the whole experiment.Electrochemical measurements
Electrochemical measurements were carried out using a CHI 660E workstation with a standard three-electrode system. A GCE (3 mm in diameter), platinum sheet electrode, and saturated calomel electrode (SCE) served as working, auxiliary, and reference electrodes, respectively. EIS of differently modified GCEs were recorded in 0.1 M KCl containing 1 mM [Fe(CN)6]3−/4− redox couple, using an open circuit voltage over a frequency range of 0.1–100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Hz with an amplitude of 5 mV. Cyclic voltammetry (CV) measurements were performed in 0.1 M KCl containing 5 mM [Fe(CN)6]3−.
000 Hz with an amplitude of 5 mV. Cyclic voltammetry (CV) measurements were performed in 0.1 M KCl containing 5 mM [Fe(CN)6]3−.
BIF preparation
A polished GCE was immersed in 5 mL PBS (0.1 M, pH 6.5) containing one of the four monomers (8 mM) and the S. aureus template (109 CFU mL−1), and CV was performed under mild stirring for 10 cycles at a scan rate of 0.1 V s−1. CV electro-polymerizations in the presence of different monomers were carried out under the same conditions except for the potential sweep range. For TE and TMA, the potential scanning was started from −0.6 to 1.0 V, while for TAA and TBA, the potential scanning range was −0.5–1.0 V, and −0.2–1.2 V, respectively. The modified electrode formed through electro-copolymerization was denoted as PT(R) + S. aureus/GCE, as shown in Scheme 1, where (R) represents the shortened form of the side chain group of each monomer. To elute the template bacteria, the PT(R) + S. aureus/GCE was soaked in CTAB (1 mM) dispersed in a diluted acetic acid solution (36%, v/v), referred to as CTAB/HAc, at 37 °C for 10 min under constant shaking (400 rpm). Such a prepared modified electrode was named BIF/GCE. As a control, a non-imprinted polythiophene film (NIF) modified electrode (NIF/GCE) was prepared via the same procedure as that of the corresponding BIF/GCE, but excluding the template bacteria from the polymerization solution.Fluorescence imaging
The target S. aureus cells (105 CFU mL−1) were stained with SYTO 9 green fluorescent dye (Invitrogen) according to the manufacturer's instructions. Detachable GCEs (Gaoss Union, China; 3 mm in diameter) were used for the formation of BIF/GCE and NIF/GCE that were separately incubated with stained S. aureus under dark conditions for 10 min. The fluorescence imaging was in situ observed on the electrode surface using a fluorescence microscope (DP80, Olympus) through an optical filter with an excitation of 460–480 nm and an emission of 495–540 nm.Electrochemical detection of S. aureus
The freshly prepared BIF/GCE was incubated with different concentrations of S. aureus in PBS (0.01 M, pH 7.4) at 37 °C for 10 min under constant shaking (250 rpm). The resulting electrode (marked as BIF-S. aureus/GCE) was rinsed with water and dried in a N2 atmosphere for immediate EIS measurements. To minimize the variability between electrodes in parallel, the EIS response of BIF/GCE towards target bacteria was described by the relative variation of the charge transfer resistance (Rct), [ΔR/R(Ω)].43 The value was calculated using the following formula:| ΔR/R = (Rcta – Rctb)/Rctb | (1) |
Optimization of experimental conditions
To obtain the optimal sensing performance, six experimental conditions, including polymerization cycles (5–15), template concentration (107–109 CFU mL−1), template elution (types of eluents and elution time from 5–15 min), recognition time (5–15 min), and oscillation speed during recognition (0–300 rpm) were optimized independently. The choice of the optimum experimental conditions, except template elution dependent on the degree of reduction in the Rct value, was based on the ΔR/R towards S. aureus at 105 CFU mL−1.Preparation of real samples
Lettuce and shrimp samples were purchased from a local supermarket. Three pieces of fresh lettuce leaves or twenty grams of shrimp meat were firstly chopped and dispersed in 200 mL 1 × PBS (pH 7.4) for homogenization. The supernatant was then filtered through a 0.22-μm-pore-size membrane filter (Millipore), and the filtrate was spiked with a known amount of S. aureus to a desired concentration. The pretreated samples were analyzed using the proposed assay.Results and discussion
Monomer selection and comparison
The choice of functional monomer determines the success of molecular imprinting and the recognition ability of MIPs to a large extent.44 Here, TAA, TBA, TMA, and TE were selected as functional monomers for BIF preparation by considering the presence of special functional groups including carboxyl, boric acid, amino, and hydroxyl groups, which may interact with the bacterial surface through hydrogen bonding. To verify if all four monomers were suitable for bacteria imprinting, EIS was carried out to monitor the impedance change during the steps of electro-copolymerization of the template (S. aureus) and the monomer, and template elution. As shown in Fig. S1A–D,† the impedance values of each electropolymerized film formed in the presence of S. aureus were much larger than those of films fabricated in the absence of the template. After elution, the impedance of each BIF decreased significantly. These results suggested that these four 3-substituted thiophene monomers can achieve the success of bacteria imprinting. To investigate their recognition capability, the EIS spectra of the four different BIFs and NIFs towards the target S. aureus (105 CFU mL−1) were recorded. The results are shown in Fig. S1E–H.† The corresponding EIS responses (bar charts) are shown in Fig. 1 (left coordinate axis). Furthermore, the imprinting effect was evaluated in terms of the imprinting factor (IF), which was calculated according to the ratio of ΔR/R for S. aureus detected on the BIF compared to that on the NIF.24 The BIF with amino groups exhibited a strong non-specific adsorption, giving the lowest IF value (1.62), while the BIF with hydroxyl groups produced the highest IF value (4.98), which would help improve the selectivity and sensitivity of the sensor. Therefore, TE was eventually employed as the monomer for the BIF preparation in the following experiment. The above results confirmed the importance of functional groups on the recognition performance of MIPs.39,45 In this case, we may speculate that the specific recognition of bacteria-imprinted polymer for pathogenic bacteria could be improved by regulating the functional groups of the polymer.Electrochemical and fluorescent characterization of bacterial imprinting and recognition
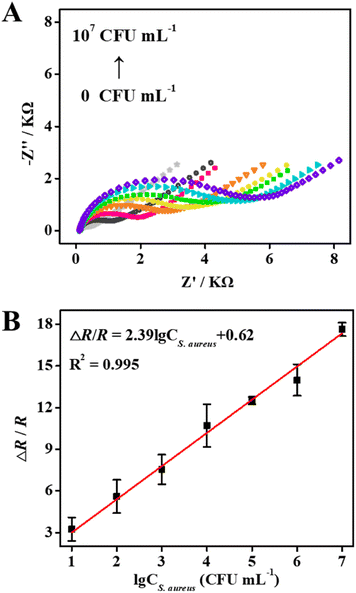
The process of bacterial imprinting and recognition using TE as the monomer was characterized by EIS and CV techniques. Fig. 2A and B show the EIS Nyquist plots of differently modified GCEs. It is known that the impedance spectrum includes a linear part and a semicircle portion corresponding to diffusion and electron-limited processes, respectively.46 The semicircle diameter equals the charge-transfer resistance (Rct) through the fitting of the Nyquist plots using the Randles equivalent circuit (inset of Fig. 2A).47 The bare GCE exhibited almost a straight line (Fig. 2A, solid square), indicating an excellent electron transfer rate on the electrode surface. After the electro-copolymerization of the TE monomer and S. aureus template on the bare GCE (PTE + S. aureus/GCE), a large semicircle domain was observed (Fig. 2A, solid circle). In comparison, a very tiny semicircle domain was found when the electro-polymerization was performed on the TE monomer alone (PTE/GCE, Fig. 2B, solid square). The result indicated that the template bacteria were successfully doped in the PTE matrices. On immersing the modified electrode (PTE + S. aureus/GCE) in CTAB/Hac for 10 min, the semicircle domain of the formed BIF/GCE (Fig. 2A, hollow circle) decreased significantly, suggesting that the template bacteria were successfully removed from the PTE matrices. Upon the capture of S. aureus (105 CFU mL−1) using the as-prepared BIF/GCE for 10 min, the semicircle domain of the modified electrode (BIF-S. aureus/GCE, Fig. 2A, solid triangle) increased obviously. The calculated Rct value of the BIF-S. aureus/GCE (ca. 3.5 kΩ) was 16 times more than that of the BIF/GCE (ca. 210 Ω). In contrast, the impedance of NIF-S. aureus/GCE (Fig. 2B, hollow circle) was just a little bit higher than that of NIF/GCE (Fig. 2B, solid triangle), signifying a negligible non-specific adsorption of the imprinted film. The result initially revealed that the formed BIF had good recognition capability toward the target S. aureus.The findings mentioned above were confirmed by CV analysis. As shown in Fig. 2C, the CV curve of the PTE + S. aureus/GCE exhibited a significant increase in peak potential difference (ΔEp) and a noticeable decrease in the peak current (Ip) compared with that of PTE/GCE. This result confirmed the successful embedding of S. aureus in the PTE matrices. After elution, the CV curve of the BIF/GCE showed a similar shape as that of the PTE/GCE, further revealing the complete removal of S. aureus from the PTE matrices. After the recognition of S. aureus, the CV curve of the BIF-S. aureus/GCE had a remarkable change with a rise in ΔEp and a reduction in Ip, compared with the BIF/GCE and NIF-S. aureus/GCE. The result again indicated that the BIF had good recognition capability toward the template bacteria with almost no non-specific adsorption.
To visualize the recognition ability of the imprinted film, the target S. aureus was stained with a green fluorescent SYTO 9 dye that is a cell-permeating nucleic acid stain. After incubation, many green spherical spots were found on the surface of the BIF/GCE (Fig. 2D), while almost no fluorescent signals could be found on the NIF/GCE (Fig. 2E). The above results further demonstrated that the BIF had good recognition capability toward the template S. aureus.
Morphological characterization
The surface morphology of the electrode during bacterial imprinting and recognition were characterized by scanning electron microscopy (SEM, JSM-7800F, JEOL, Japan) and atomic force microscopy (AFM, Bruker Corp., USA). A lot of spherical bacteria were seen on the PTE + S. aureus/GCE (Fig. 3A). Most of the bacteria adhered to each other, looking like the shape of grapes with a diameter of ca. 0.7 μm, which is consistent with the published literature.48,49 The result again implied that S. aureus was successfully doped on the PTE matrices. After elution, no more bacteria were left on the BIF/GCE (Fig. 3B), further suggesting that S. aureus was wholly removed. Similar phenomena were observed from AFM images (Fig. S2†). Using the BIF/GCE as the capture electrode, many spherical bacteria were observed on BIF-S. aureus/GCE within just 10 min (Fig. 3C), which could be more clearly seen from the magnified image as shown in Fig. 3D.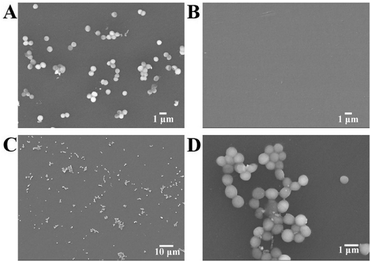 | ||
Fig. 3 SEM images of differently modified GCEs: (A) PTE + S. aureus/GCE, (B) BIF/GCE, (C) and (D) BIF-S. aureus/GCE with 1000-, and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold magnification, respectively. 000-fold magnification, respectively. | ||
Optimization of the BIF-based sensing performance
To optimize the performance of the BIF-based impedimetric sensor, a series of experimental parameters employed in the BIF preparation and the bacteria recognition were systematically optimized. With the increase of the polymerization cycle from 5 to 15 (Fig. 4A), the EIS response first changed little and then decreased obviously due to the increased difficulty in template removal when a thicker polymer film was formed. Thus 10 cycles of polymerization were employed for the BIF fabrication. With the increase of the S. aureus template amount from 107 to109 CFU mL−1 (Fig. 4B), the EIS response increased gradually due to the formation of more recognition sites. Therefore, 109 CFU mL−1 of S. aureus was applied for the BIF preparation.For the complete removal of a large bacterial template from PTE matrices, we first adopted two existing elution methods, including the successive use of lysozyme (10 mg mL−1) for 2 h and Triton X-100 (10%) for 80 min (denoted as lysozyme + Triton), and the use of SDS/HAc (5%, w/v) for 4 h. However, the template removal was not complete with a slight decrease of the impedance value. For a more efficient elution, a cationic detergent, CTAB, often used in bacterial lysis,50 was attempted for the bacterial template removal and compared with the two eluents above. As shown in Fig. 4C, the best elution effect was achieved with CTAB/HAc. In addition, the elution time with CTAB/HAc was also optimized (Fig. 4D). With the increase in the elution time from 5 to 15 min, the Rct value of the modified electrode first decreased and then changed little. Therefore, 10 min of elution with CTAB/HAc was finally used for the bacterial template removal.
To maximize the EIS response toward the target S. aureus, the recognition time and oscillation speed during recognition were also optimized. With the increase in the recognition time from 5 to 15 min, the EIS response was enhanced and then changed little (Fig. 4E). Hence, 10 min of recognition was used in the subsequent experiment. Oscillation using a metal bath was employed for S. aureus capture to prevent bacterial deposition. With an increase of the oscillation speed from 0 to 300 rpm, the EIS response first increased and then decreased (Fig. 4F). Therefore, 250 rpm was selected for S. aureus recognition.
Quantitative detection of S. aureus
With an increase of concentration from 0 to 107 CFU mL−1, the impedance of BIF/GCE gradually increased (Fig. 5A). This is because the more bacteria are rebound to the BIF, the more imprinted sites are occupied, which reduces the electron transfer between the [Fe(CN)6]3−/4− redox probe and GCE. Meanwhile, in a broad concentration ranging from 101 to 107 CFU mL−1, the EIS response had an excellent linear relationship with the logarithm of the S. aureus concentration (Fig. 5B). The linear regression equation for this sensor was expressed as ΔR/R(Ω) = 2.39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) lg
lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CS. aureus + 0.62 with a correlation coefficient (R2) of 0.995. Based on the 3σ/S rule (σ is the standard deviation of the EIS response for the blank solution, and S represents the slope of the calibration curve),49 the detection limit of the sensor was calculated to be 4 CFU mL−1, which was comparable to or even better than other reported label-free electrochemical sensors for S. aureus detection as shown in Table S1.†
CS. aureus + 0.62 with a correlation coefficient (R2) of 0.995. Based on the 3σ/S rule (σ is the standard deviation of the EIS response for the blank solution, and S represents the slope of the calibration curve),49 the detection limit of the sensor was calculated to be 4 CFU mL−1, which was comparable to or even better than other reported label-free electrochemical sensors for S. aureus detection as shown in Table S1.†
Selectivity and universality of the BIF-based sensor
The selectivity was firstly investigated by comparing the EIS response of S. aureus-templated BIF to single bacteria (the target or interfering bacteria), and bacterial mixture with or without S. aureus. As shown in Fig. 6A, the EIS response of S. aureus-templated BIF towards the target S. aureus was 5 times higher than that to the interfering bacteria (E. coli O157, S. paratyphi B, or L. monocytogenes). Moreover, the EIS response of S. aureus-templated BIF to the bacterial mixtures with S. aureus was approximately 4 times larger than that without S. aureus. This result revealed that the selectivity of this sensor is excellent. The high selectivity of the BIF against the template bacteria could be ascribed to its sensitivity to the chemical conformation of bacterial outer cell structures.37To test whether this BIF-based sensor can be applied for detecting other bacteria, three other bacteria, including E. coli O157, S. paratyphi B, and L. monocytogenes, were individually used as the template for BIF preparation. It was found that for each E. coli O157-, S. paratyphi B-, or L. monocytogenes-templated BIF, the EIS response to the corresponding template bacteria was at least 3 times more than that to the interfering bacteria (Fig. 6B). The above results showed that the BIF-based sensor could provide a universal method for the highly selective detection of bacterial pathogens.
Reusability and repeatability of the sensing interface
The imprinted sites after the bacterial rebinding could be efficiently regenerated within 10 min using the same method as the template removal. It can be seen that even after being recycled 5 times, the impedance response to the target S. aureus still retained more than 90% of the initial detection signal (Fig. 7). The result indicated that the BIF-based sensor is reusable, which is meaningful for reducing the assay cost and time. | ||
| Fig. 7 Reusability of the BIF-based sensing interface. The impedance of BIF/GCE before and after the recognition of S. aureus (105 CFU mL−1) with different reuse numbers. | ||
The repeatability was tested by monitoring the EIS responses of eight fabricated BIF/GCE electrodes toward S. aureus (105 CFU mL−1) in 0.01 M PBS (pH 7.4). It was found that the relative standard deviation % (RSD %) did not exceed 4.2%, indicating the good repeatability of the BIF-based sensor.
Detection of S. aureus in real samples
To demonstrate the practical application of the proposed BIF-based sensor for S. aureus detection in real complex samples, we have used it for the analysis of lettuce and shrimp samples artificially contaminated with different concentrations of S. aureus. As can be seen from Table 1, the average recoveries of S. aureus vary from 92.9 to 110.6%. The results implied that the BIF-based sensor had a promising potential for the determination of bacteria in real samples. The sensing strategy could also be extended to on-site, remote, or home monitoring of bacterial pathogens through combining disposable electrodes (e.g., screen-printed electrodes, carbon paper, etc.) and commercial portable electrochemical workstations.| Sample | Amounts of S. aureus (CFU mL−1) | Recovery (%) | |
|---|---|---|---|
| Added | Measured | ||
| All data are represented as the average value ± standard deviation of at least three independent experiments. | |||
| Lettuce | 101 | 10.40 ± 1.1 | 104.0 |
| 102 | 108.0 ± 24.3 | 108.0 | |
| 103 | 929.1 ± 292.4 | 92.9 | |
| Shrimp | 102 | 102.5 ± 27.7 | 102.5 |
| 103 | 1106.0 ± 261.1 | 110.6 | |
| 104 | 9922.0 ± 557.2 | 99.2 | |
Conclusion
In summary, an efficient label-free impedimetric sensing strategy based on an electropolymerized BIF has been developed for the rapid and sensitive detection of S. aureus. The results suggest that the specific recognition of MIPs can be modulated by designing suitable functional groups. In contrast to most reported label-free electrochemical sensors for S. aureus detection (Table S1†), a significant improvement on both the duration for sensing interface fabrication and the analysis time was achieved with a comparable detection limit. Moreover, the BIF sensing interface can be regenerated and reused up to five times, which is effective in reducing the assay cost and time. This assay is anticipated to provide a universal, simple, and robust method for the rapid, sensitive, and low-cost detection of pathogenic bacteria. Yet, there are still challenges in identifying live/dead bacteria, detecting complex food or whole blood samples without complicated sample pretreatment, and achieving long-term stability.Author contributions
Lingling Wang: conceptualization, investigation. Xiaohui Lin: conceptualization, writing – original draft. Ting Liu: investigation. Zhaohuan Zhang: methodology. Jie Kong: data curation. Hai Yu: investigation. Juan Yan: visualization and investigation. Donglei Luan: investigation. Yong Zhao: supervision and funding acquisition. Xiaojun Bian: conceptualization, supervision, methodology, writing – review and editing, and project administration.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge financial support from the Program of Shanghai Academic Research Leader (21XD1401200), the Agricultural Project of Shanghai Science and Technology Innovation Action Plan (19391901600), the Natural Science Foundation of Shanghai Municipal (20ZR1424100, 20ZR1423800), and Chenguang Program supported by the Shanghai Education Development Foundation and the Shanghai Municipal Education Commission (15CG54).References
- C. S. Ho, N. Jean, C. A. Hogan, L. Blackmon, S. S. Jeffrey, M. Holodniy, N. Banaei, A. A. E. Saleh, S. Ermon and J. Dionne, Nat. Commun., 2019, 10, 4927 CrossRef PubMed.
- P. N. Reddy, K. Srirama and V. R. Dirisala, Infect. Dis., 2017, 10, 1179916117703999 Search PubMed.
- W. Kong, J. Xiong, H. Yue and Z. Fu, Anal. Chem., 2015, 87, 9864–9868 CrossRef CAS PubMed.
- W. C. Hu, J. Pang, S. Biswas, K. Wang, C. Wang and X. H. Xia, Anal. Chem., 2021, 93, 8544–8552 CrossRef CAS PubMed.
- M. M. Ali, R. Silva, D. White, S. Mohammadi, Y. Li, A. Capretta and J. D. Brennan, Angew. Chem., 2022, 61, e202112346 CAS.
- V. Fabre, K. C. Carroll and S. E. Cosgrove, J. Clin. Microbiol., 2022, 60, e01005–e01021 CrossRef PubMed.
- Y. Cui, H. Wang, F. Guo, X. Cao, X. Wang, X. Zeng, G. Cui, J. Lin and F. Xu, Food Chem., 2022, 391, 133241 CrossRef CAS PubMed.
- W. Yin, L. Zhu, H. Xa, Q. Tang, Y. Ma, S.-H. Chou and J. He, Sens. Actuators, B, 2022, 366, 132005 CrossRef CAS.
- P. Athamanolap, K. Hsieh, C. M. O'Keefe, Y. Zhang, S. Yang and T.-H. Wang, Anal. Chem., 2019, 91, 12784–12792 CrossRef CAS.
- B. Wu, J. S. Hu and Y. Li, Food Microbiol., 2022, 106, 104052 CrossRef CAS PubMed.
- J. Yi, X. Wang, Y. Dai, L. Qiao and B. Liu, Anal. Chem., 2019, 91, 14220–14225 CrossRef CAS.
- Z. Chai and H. Bi, Food Chem.: X, 2022, 13, 100225 CAS.
- H. Shengbin, H. Xinyi, Z. Miaomiao, W. Lina and Y. Xiaomei, Anal. Chem., 2020, 92, 2393–2400 CrossRef PubMed.
- T. Tian, J. Yi, Y. Liu, B. Li, Y. Liu, L. Qiao, K. Zhang and B. Liu, Biosens. Bioelectron., 2022, 197, 113778 CrossRef CAS PubMed.
- E. Rho, M. Kim, S. H. Cho, B. Choi, H. Park, H. Jang, Y. S. Jung and S. Jo, Biosens. Bioelectron., 2022, 202, 113991 CrossRef CAS.
- O. Simoska and K. J. Stevenson, Analyst, 2019, 144, 6461–6478 RSC.
- N. Amin, A. S. Torralba, R. Alvarez-Diduk, A. Afkhami and A. Merkoci, Anal. Chem., 2020, 92, 4209–4216 CrossRef CAS PubMed.
- F. Zhu, X. Bian, H. Zhang, Y. Wen, Q. Chen, Y. Yan, L. Li, G. Liu and J. Yan, Biosens. Bioelectron., 2021, 176, 112943 CrossRef CAS PubMed.
- S. B. Somvanshi, A. M. Ulloa, M. Zhao, Q. Liang, A. K. Barui, A. Lucas, K. M. Jadhav, J. P. Allebach and L. A. Stanciu, Biosens. Bioelectron., 2022, 207, 114214 CrossRef CAS PubMed.
- Y. Wei, Z. Tao, L. Wan, C. Zong, J. Wu, X. Tan, B. Wang, Z. Guo, L. Zhang, H. Yuan, P. Wang, Z. Yang and Y. Wan, Biosens. Bioelectron., 2022, 211, 114282 CrossRef CAS PubMed.
- E. Fan, J. Peng, Y. He, Y. Wu, H. Ouyang, Z. Xu and Z. Fu, Sens. Actuators, B, 2019, 285, 271–276 CrossRef CAS.
- M. Amiri, A. Bezaatpour, H. Jafari, R. Boukherroub and S. Szunerits, ACS Sens., 2018, 3, 1069–1086 CrossRef CAS.
- L. Chen, X. Wang, W. Lu, X. Wu and J. Li, Chem. Soc. Rev., 2016, 45, 2137–2211 RSC.
- R. Xing, Y. Wen, Y. Dong, Y. Wang, Q. Zhang and Z. Liu, Anal. Chem., 2019, 91, 9993–10000 CrossRef CAS PubMed.
- G. Dykstra, B. Reynolds, R. Smith, K. Zhou and Y. Liu, ACS Appl. Mater. Interfaces, 2022, 14, 25972–25983 CrossRef CAS PubMed.
- X. Wang, Y. Liu, J. Liu, J. Qu, J. Huang, R. Tan, Y. Yu, J. Wu, J. Yang, Y. Li, H. Qu and J. Liu, Biosens. Bioelectron., 2022, 208, 114233 CrossRef CAS PubMed.
- P.-W. Gao, Y.-Z. Shen, C. Ma, Q. Xu and X.-Y. Hu, Analyst, 2021, 146, 6178–6186 RSC.
- P. Li, J. Pang, S. Xu, H. He, Y. Ma and Z. Liu, Angew. Chem., 2022, 61, e202113528 CAS.
- V. Ratautaite, R. Boguzaite, E. Brazys, A. Ramanaviciene, E. Ciplys, M. Juozapaitis, R. Slibinskas, M. Bechelany and A. Ramanavicius, Electrochim. Acta, 2022, 403, 139581 CrossRef CAS PubMed.
- L. Luo, F. Zhang, C. Chen and C. Cai, Anal. Chem., 2019, 91, 15748–15756 CrossRef CAS.
- Z. Zhang, Y. Guan, M. Li, A. Zhao, J. Ren and X. Qu, Chem. Sci., 2015, 6, 2822–2826 RSC.
- B. Tse Sum Bui, T. Auroy and K. Haupt, Angew. Chem., 2022, 61, e202106493 CAS.
- N. Idil, M. Hedstrom, A. Denizli and B. Mattiasson, Biosens. Bioelectron., 2017, 87, 807–815 CrossRef CAS PubMed.
- X. Shen, J. Svensson Bonde, T. Kamra, L. Bulow, J. C. Leo, D. Linke and L. Ye, Angew. Chem., Int. Ed., 2014, 53, 10687–10690 CrossRef CAS PubMed.
- J. Borovicka, W. J. Metheringham, L. A. Madden, C. D. Walton, S. D. Stoyanov and V. N. Paunov, J. Am. Chem. Soc., 2013, 135, 5282–5285 CrossRef CAS PubMed.
- J. Niu, L. Wang, T. Cui, Z. Wang, C. Zhao, J. Ren and X. Qu, ACS Nano, 2021, 15, 15841–15849 CrossRef CAS PubMed.
- S. Tokonami, Y. Nakadoi, M. Takahashi, M. Ikemizu, T. Kadoma, K. Saimatsu, Q. Dung le, H. Shiigi and T. Nagaoka, Anal. Chem., 2013, 85, 4925–4929 CrossRef CAS PubMed.
- L. M. Gonçalves, Curr. Opin. Electrochem., 2021, 25, 100640 CrossRef.
- J. Pan, W. Chen, Y. Ma and G. Pan, Chem. Soc. Rev., 2018, 47, 5574–5587 RSC.
- S. Ramanavicius and A. Ramanavicius, Adv. Colloid Interface Sci., 2022, 305, 102693 CrossRef CAS.
- J. Zhang, Y. Wang and X. Lu, Anal. Bioanal. Chem., 2021, 413, 4581–4598 CrossRef CAS PubMed.
- W. Jiang, A. Saxena, B. Song, B. B. Ward, T. J. Beveridge and S. C. B. Myneni, Langmuir, 2004, 20, 11433–11442 CrossRef CAS PubMed.
- M. Golabi, F. Kuralay, E. W. Jager, V. Beni and A. P. Turner, Biosens. Bioelectron., 2017, 93, 87–93 CrossRef CAS PubMed.
- A. G. Ayankojo, A. Tretjakov, J. Reut, R. Boroznjak, A. Opik, J. Rappich, A. Furchner, K. Hinrichs and V. Syritski, Anal. Chem., 2016, 88, 1476–1484 CrossRef CAS PubMed.
- K. R. a. R. N. Zare, ACS Nano, 2012, 6, 4314–4318 CrossRef PubMed.
- S. Dong, R. Zhao, J. Zhu, X. Lu, Y. Li, S. Qiu, L. Jia, X. Jiao, S. Song, C. Fan, R. Hao and H. Song, ACS Appl. Mater. Interfaces, 2015, 7, 8834–8842 CrossRef CAS PubMed.
- A. Ahmed, J. V. Rushworth, N. A. Hirst and P. A. Millner, Clin. Microbiol. Rev., 2014, 27, 631–646 CrossRef CAS PubMed.
- F. Yang, T. L. Chang, T. Liu, D. Wu, H. Du, J. Liang and F. Tian, Biosens. Bioelectron., 2019, 133, 147–153 CrossRef CAS PubMed.
- R. Wang, L. Wang, J. Yan, D. Luan, S. Tao, J. Wu and X. Bian, Talanta, 2021, 226, 122135 CrossRef CAS PubMed.
- G. C. Allen, M. A. Flores-Vergara, S. Krasynanski, S. Kumar and W. F. Thompson, Nat. Protoc., 2006, 1, 2320–2325 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2an01122k |
| ‡ These authors have equally contributed to this article. |
| This journal is © The Royal Society of Chemistry 2022 |