Open Access Article
Open Access ArticleA headspace collection chamber for whole body volatilomics†
Stephanie
Rankin-Turner
 a and
Conor J.
McMeniman
a and
Conor J.
McMeniman
 *ab
*ab
aW. Harry Feinstone Department of Molecular Microbiology and Immunology, Johns Hopkins Malaria Research Institute, Johns Hopkins Bloomberg School of Public Health, Johns Hopkins University, Baltimore, MD 21205, USA. E-mail: cmcmeni1@jhu.edu
bThe Solomon H. Snyder Department of Neuroscience, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA
First published on 14th October 2022
Abstract
The human body secretes a complex blend of volatile organic compounds (VOCs) via the skin, breath and bodily fluids, the study of which can provide valuable insight into the physiological and metabolic state of an individual. Methods to profile human-derived volatiles typically source VOCs from bodily fluids, exhaled breath or skin of isolated body parts. To facilitate profiling the whole body volatilome, we have engineered a sampling chamber that enables the collection and analysis of headspace from the entire human body. Whole body VOCs were collected from a cohort of 20 humans and analyzed by thermal desorption-gas chromatography/mass spectrometry (TD-GC/MS) to characterize the compounds present in whole body headspace and evaluate chemical differences between individuals. A range of compounds were detected and identified in whole body headspace including ketones, carboxylic acids, aldehydes, alcohols, and aliphatic and aromatic hydrocarbons. Considerable heterogeneity in the chemical composition of whole body odor and the concentration of its constituent compounds was observed across individuals. Amongst the most common and abundant compounds detected in human whole body odor were sulcatone, acetoin, acetic acid and C6–C10 aldehydes. This method facilitates standardized and quantitative analytical profiling of the human whole body volatilome.
Introduction
Human scent is a complex blend composed of volatile organic compounds (VOCs) emitted via the skin, breath, and bodily fluids. Across studies profiling volatile compounds emitted by the healthy human body, 1488 and 623 volatiles have been identified in breath and skin emissions respectively.1 Many additional human-derived compounds may also remain unidentified or undetected due to limitations in analytical methods. Profiling VOCs can offer a wealth of information on the physiological and metabolic state of an individual, providing insight into disease,2,3 diet and lifestyle,4,5 environmental exposure,6,7 and even the chemical attraction of arthropod disease vectors such as mosquitoes to humans.8 The volatile compounds emitted by the human body are influenced by a variety of factors, such as diet, hygiene habits, disease state and the constitution of the human microbiome. As such, human body odor can be considered specific to the individual. As a result of the growing interest in human-derived VOCs and the information that can be gleaned by their study, numerous analytical techniques have been developed for the collection and evaluation of volatile compounds from different biological matrices.The analysis of exhaled breath is amongst the most prominent areas of study in human volatilomics, driven by the desire to develop rapid, non-invasive diagnostics for disease. Metabolic changes in breath have been studied for a variety of purposes, particularly the detection of biomarkers for cancer,9,10 chronic obstructive pulmonary disease (COPD),11 asthma,12 malaria,13,14 and COVID-19.15–18 Exhaled breath is typically collected into an inert polymer bag (Tedlar), followed by collection onto tubes containing a sorbent material such as Tenax or a carbon-based material.19 Alternatively, breath samples can also be collected using specialized breathing masks which enable the capture of breath volatiles directed onto sorbent tubes.20 Collected VOCs are subsequently analyzed by thermal desorption-gas chromatography/mass spectrometry (TD-GC/MS). Though less common, direct mass spectrometry methods have been used for real-time sampling of exhaled breath, particularly selected ion flow tube mass spectrometry (SIFT-MS), secondary electrospray ionization mass spectrometry (SESI), and proton transfer reaction mass spectrometry (PTR-MS).21 Gas chromatography-ion mobility spectrometry (GC-IMS) has furthermore been applied to breath analysis, particularly in the field of rapid disease diagnostics due to its portability and low cost in comparison to benchtop mass spectrometers.22
A major contributor to human scent is the multitude of volatile compounds released from the skin. Skin emanations are derived from secretions from the eccrine, sebaceous and apocrine glands, in addition to metabolites produced by the human skin microbiome.23 As glands and microbes are distributed differently across the body, discrete areas of the body can produce distinct scent profiles. Although less commonly the subject of metabolomics investigations, skin VOCs can provide insight into the human metabolome and be utilized across numerous fields of research. The study of skin emanations has been conducted to develop analytical techniques for the detection of human scent during search and rescue operations,24–28 explore individual differences in human odor,29–31 evaluate the attraction of mosquitoes to human odor,32–34 and to fundamentally understand the biological basis of human scent.35–38 Skin VOCs are typically collected using sorbent materials placed in direct contact with the skin. Glass beads have been rubbed on the hands or feet of participants to collect skin secretions, after which compounds transferred to the beads are desorbed for analysis.33,39–41 This form of sample collection has similarly been achieved using SPME fibers,42 and polydimethylsiloxane (PDMS), including coated stir bars,43 patches,44,45 and wearable wrist bands.34,46 Aside from contact-based sampling methods, skin emanations have been explored using sampling devices and bags to collect headspace from isolated body parts, namely the hands and feet, onto SPME fibers or thermal desorption tubes.37,38,42
Most human volatilomics studies to date focus on the detection of volatiles in a particular medium, such as exhaled breath, a bodily fluid, or from an isolated part of the body. A small number of studies have made efforts to characterize human whole body emissions, however these have excluded head and breath VOC emissions,24,47,48 used techniques unsuitable for compound identification (such as low resolution ion mobility spectrometry),49 or have only been applied to single participants or small cohorts.50,51 In order to comprehensively characterize the chemical composition of whole body odor headspace, improved methods for the collection of human-derived VOCs must be developed and applied to larger cohorts in order to understand heterogeneity in the human scent signature and emission rates of constituent VOCs.
To facilitate chemical analysis of the human whole body volatilome, here we describe the development of a chamber for the controlled sampling of whole body headspace from individual humans. We engineered an acrylic sampling chamber that is flushable with purified air to provide a standardized atmosphere for the collection of human-derived VOCs from a seated human. The chamber is sealed during sampling mode to enable sufficient concentration of VOCs, has multiple ports for VOC collection and a sealable door for participant entry and exit. We applied this booth-style sampling chamber and TD-GC/MS to profile individual whole body VOC headspace of a diverse cohort of 20 human participants. We quantified emission rates of 43 select VOCs that we annotated in human whole body odor, yielding high content human scent signatures. This study lays the foundation for application of this sampling method in combination with a variety of gas chromatography and mass spectrometry techniques to comprehensively profile the human volatilome for fundamental and applied purposes.
Materials and methods
Whole body headspace collection chamber
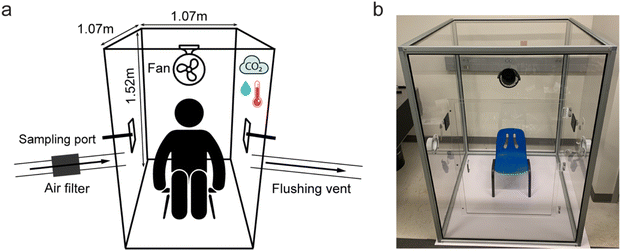
A sampling chamber was constructed to enable the collection of VOCs from the human whole body. The 1.07 m L × 1.07 m W × 1.52 m H sampling chamber with a total volume of 1734.41 L was constructed within an aluminium frame (#9030, 80/20 Inc., Columbia City, USA) with four walls and a ceiling consisting of 0.6 mm thick polymethyl methacrylate sheets (ASTM D4802 CAT. B-1, Finish 1 Type UVA, Trident Plastics, USA) (Fig. 1). The base of the chamber was a 6.4 mm thick acrylonitrile butadiene styrene white plastic sheet (Interstate Plastics, Sacramento, USA). The corners of the frame were fixed with nylon plastic 3-way corner connectors (#9150, 80/20 Inc., Columbia City, USA), and the chamber edges were sealed externally with black thermoplastic elastomer (#2117, 80/20 Inc., Columbia City, USA). A 0.61 m × 0.61 m door centered on the front wall of the chamber was fitted with magnetic discs (McMaster-Carr, Elmhurst, USA) to seal the door for participant entry and exit. On each side of the chamber were 4′′ duct flanges (#409004, DL Wholesale, Romulus USA), one of which was coupled to an inline fan and carbon filter (Model GLFANXINLINEEXPC4, iPower) using 4′′ aluminium ducting to enable rapid flushing of the chamber with filtered air prior to sampling. The alternate flange on the other side of the chamber was also connected to 4′′ aluminium ducting and served as the flushing vent. Sampling ports on each side of the chamber consisted of black plastic bulkheads and 1/4′′ OD push-to-connect fittings (#MTC 1/4-N01, MettleAir, USA), which were coupled with low-power pumps (Pocket Pump, SKC Inc., USA) to draw chamber air through 1/4′′ PTFE tubing onto Tenax-TA thermal desorption tubes (Gerstel, USA), connected using 1/4′′ Swagelok connectors. The sampling ports can be split with push-to-connect Y-unions to facilitate sampling onto additional TD tubes. During flushing mode, the sampling ports were plugged with push-to-connect plugs (#PP1/4, TechniFit). During sampling mode, the flushing port and flushing vent were plugged with steel duct end caps (#B08SBNNFJW, Europlast), and push-to-connect plugs removed from sampling ports to facilitate air sampling. A small fan (MiniFan, Comlife) was positioned internally above the chamber door to circulate air around the inside of the chamber to facilitate VOC uniformity. A monitor was positioned inside the chamber to record temperature, relative humidity and CO2 levels (IAQ Mini, CO2Meter, USA). CO2 was monitored throughout the sampling procedure both to ensure the safety of participants and to observe variation in carbon dioxide emissions between participants.Sample collection
The chamber was situated in a temperature-controlled laboratory with a total volume of 147.65 m3 that was maintained at 21 °C. This laboratory was ventilated by a HVAC system at a rate of 6 ACH (air changes per hour). Prior to use, the interior surfaces of the chamber and accessories were wiped clean with 10% ethanol and the chamber flushed with filtered air at 20 L s−1 for 1 h. Flushing of the chamber occurred immediately prior to background air sample collection and entry of participants into the chamber. Prior to sampling, Tenax-TA thermal desorption (TD) tubes (#020810-005-00, 6 × 60 mm, Gerstel, USA) were conditioned at 300 °C for 60 min in a stream of nitrogen at 50 mL min−1 using the Gerstel tube conditioner (TC2, Gerstel, USA) and then spiked with 15 ng of 2-pentadecanone (Alfa Aesar, USA) as an internal standard. Background air samples from the chamber were collected simultaneously via sampling ports in triplicate onto TD tubes at 375 mL min−1 for 30 min immediately prior to participant sampling. Participants were instructed to change into clean scrubs (65/35 polyester/cotton, SmartScrubs, Phoenix, USA) washed only in water to reduce the introduction of exogenous VOCs from non-standardized clothing. Participants were then instructed to remove their socks and enter the chamber via the chamber door.To maximise skin exposure for VOC collection, once inside the chamber participants were instructed to uncover the lower half of their legs by rolling the pant legs of the scrubs to be level with their knees. The provided scrub shirts were short sleeved, and therefore skin surfaces on the lower arms below the elbows were readily exposed. Participants were seated throughout the sampling procedure on a small high-density polyethylene and steel chair (#1173094, Lifetime, Riverdale USA). Human whole body odor samples were collected as per background sample collection for 30 minutes. Three replicate human odor samples were collected from the chamber for each participant. The full experimental workflow is detailed in Fig. S1.†
During chamber development, a pilot human subject cohort of 10 participants was used to monitor chamber oxygen levels and participant blood oxygen levels during the 30 min sampling period. Chamber oxygen levels were monitored with a Coreel 4 portable gas monitor (CoreEL Technologies) and blood oxygen levels were monitored with a CMS-50D1 fingertip oximeter (AccuMed).
Human participants
A cohort of 20 healthy adults was recruited from the Baltimore metropolitan area (MD, USA) consisting of 10 males and 10 females with an age range of 19–39 years and a median age of 26.5 years. Participants were 55% white, 10% black or African American, 25% Asian, and 10% more than one race. Participants were provided with fragrance-free shampoo and body wash (Vanicream, USA) to wash with prior to providing a whole body odor sample. All participants were requested to shower within 24 h prior to sampling. After washing, participants did not use any other cleaning products, deodorants, cosmetics, skin creams or fragrances. For 12 h prior to sampling participants were asked to refrain from the consumption of alcohol and odorous foods such as garlic, onions, and spicy foods. On the day of sampling, participants were required to provide details of their recent hygiene practices, diet, and occurrence of smoking or vaping. All individuals recruited complied with the study requirements and thus all were able to participate. It should be noted that whilst steps were taken to reduce the presence of VOCs (such as from hygiene products), traces of exogenous materials may still remain on the body without requiring participants to refrain from their use for substantially longer periods of time. Sampling for all participants occurred between the hours of 9:00–13:00. At all times during sampling, participants were visually observed by a single female study coordinator positioned adjacent to the chamber. The study was approved by the Johns Hopkins Bloomberg School of Public Health (JHSPH) Institutional Review Board (IRB no. 00014626) and all participants gave written informed consent prior to participation. No adverse events to the study protocol were reported post-participation.TD-GC/MS analysis
Samples were analyzed by thermal desorption gas chromatography/mass spectrometry (7890B GC, 5977N MSD, Agilent, USA). Tenax-TA tubes were placed in a Gerstel Thermal Desorption Unit mounted onto a Gerstel Cooled Injector System (CIS4) PTV inlet (Gerstel, USA). Analytes were desorbed in splitless mode starting at 30 °C followed by an increase of 720 °C min−1 to 280 °C and held for 3 min. Analytes were swept into the inlet which was held at −70 °C and then heated at 720 °C min−1 to desorb analytes onto a HP-INNOWAX capillary column (30 m length × 0.25 mm diameter × 0.25 μm film thickness). The GC oven was programmed with an initial temperature 40 °C with a 2 min hold followed by an increase of 10 °C min−1 to 250 °C with a 5 min hold. A helium carrier gas with a flow rate of 1.2 mL min−1 was used. The MS analyser was set to acquire over a range of m/z 30–300 and was operated in EI mode. The ion source and transfer line were set to 230 °C and 250 °C respectively.Data were deconvoluted in Agilent Unknowns Analysis software (RT window size factor of 50 and 100, absolute area 1000 counts, peak sharpness threshold 25%, min match factor 50) and exported as .csv files. Identifiable analyte peak areas occurring above the limits of detection were normalized to the internal standard, background-subtracted (manual background subtraction performed using mean analyte abundances from 3 replicate background measurements collected inside the chamber immediately prior to participant sampling), and mean values were taken from the three technical replicates. Compound identification was achieved by comparison of mass spectra with the NIST Mass Spectral Library version 2.2 and retention time matching with analytical reference standards. MetaboAnalyst 5.0 was used for the production of heat maps and chemometric analysis. The heat map was produced using data normalized to the internal standard and on a logarithmic scale.
Results and discussion
Sampling chamber development
To enable the sufficient accumulation of human body VOCs in the sampling chamber and prevent contamination with laboratory air, a sealed chamber is required. Air exchange within the chamber in sampling mode was evaluated using the CO2 concentration decay test, a method used to determine air change in a room or container based on the decreasing concentration of a tracer gas.52 A short pulse (∼5 seconds) of compressed carbon dioxide (CD USP50, Airgas, Rosedale, MD) was introduced into the sealed chamber via one of the sampling ports and the CO2 concentration monitored over time. Air exchange rate was calculated using eqn (1):53AD = 1/Δt![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ln{(C1 – CR)/(C0 – CR)} ln{(C1 – CR)/(C0 – CR)} | (1) |
During chamber development, we monitored chamber oxygen levels and participant blood oxygen concentrations to confirm safety during sampling mode within the chamber. Over the 30 minutes duration of the sampling period, chamber oxygen levels decreased an average of 0.06% (mean starting O2 concentration = 20.9%, mean ending O2 concentration = 20.84%), with no measurable decrease detected for 90% of participants (n = 10 total). At no point did chamber oxygen levels fall below the 19.5% concentration deemed to be considered an oxygen deficient environment by the Occupational Safety and Health Administration (OSHA).54 Blood oxygen concentrations decreased an average of 0.6%, always remaining within the normal range of peripheral oxygen saturation (SpO2) of 95–100% as measured by pulse oximetry,55 indicating a minimal change in participants’ blood oxygen levels throughout the sampling process (Table S1†).
To ensure reproducible sampling from the collection ports, the chamber was fitted with a fan to circulate air and aid VOC uniformity throughout the chamber. In an initial test with a single participant, we measured 6 VOCs from varied structural classes and of known human origin in whole body headspace to evaluate sampling reproducibility across three collection ports. The average relative standard deviation (RSD) of pinene, hexanal, undecane, acetoin, sulcatone, and acetic acid across three sampling ports were 11.3, 10.6, 4.9, 7.1, 8.5, and 11.0% RSD respectively, demonstrating low sampling variability across the sampling ports.
To ensure a clean chamber background prior to sample collection from humans, the interior surfaces of the chamber were wiped with 10% ethanol and the chamber flushed with filtered air. Background chamber air samples were collected in triplicate to confirm the removal of background contaminants and identify pre-existing components to disregard from human odor samples during the background subtraction stage. Some artefacts from the plexiglass sampling chamber itself were present in all background samples, such as siloxanes ubiquitous in the laboratory, however no sampling chamber contaminants interfered with peaks from human-derived analytes of interest. Headspace samples of the “scent-free” shampoo and body wash provided to participants were also collected and analyzed to determine the presence of any potential contaminants. The chemical profiles of these cleansing products were dominated by a large 1,2-hexanediol peak, the presence of which was disregarded from human odor samples.
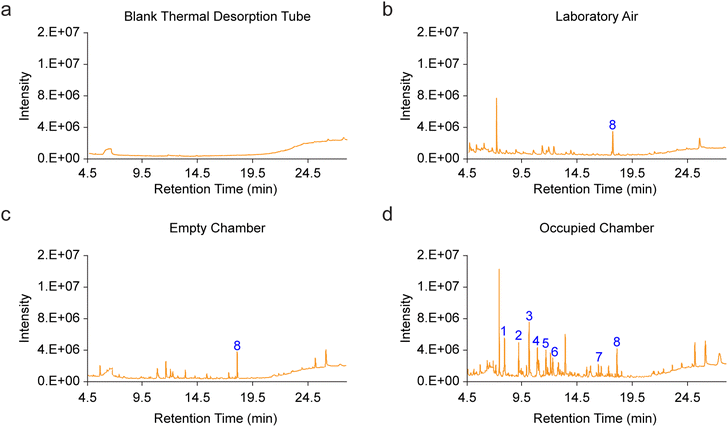
To evaluate the influence of external laboratory air on the chamber background during participant entry, background samples were collected immediately after chamber cleaning/flushing, and again after opening the chamber door for 5 seconds. This constituted the approximate time taken for a participant to enter the chamber. In these tests, there were no significant increases in detected levels of any of the human VOCs measured in this study (Fig. S2†), confirming the negligible impact of the invasion of room air into the chamber on background VOC profiles. Most of the human VOCs we profiled throughout this study were undetectable in the chamber atmosphere during this test, and we hypothesize that the low levels of background human-derived contaminants typically detected in the blank chamber such as acetic acid and 2-ethyl-1-hexanol (Fig. S2†), may possibly result from participant or investigator-related volatiles absorbed onto the chamber walls that remain after the chamber cleaning and flushing procedure. This is a particularly challenging issue to solve because using our current cleaning protocol, a human investigator must enter the chamber transiently to clean it between trials. We therefore determined that background subtraction was critical for controlled analysis of VOC emission rates from each participant. Indeed, representative chromatograms from a pre-conditioned thermal desorption tube, room air, a pre-flushed empty sampling chamber and occupied chamber (Fig. 2) highlighted the presence of multiple features from each these sources and the need for trial-by-trial background subtraction.
To evaluate the effect of sampling time on VOC detection, human whole body headspace was sampled for 10, 20 and 30 minutes in triplicate from one male participant. Across the top ten most abundant human VOCs detected, a sampling time of 30 minutes resulted in an average 61-fold increase in analyte abundance compared to a 10 minutes sampling time (Fig. S3†). As such, 30 minutes was selected as the sampling time to ensure the participant was present in the chamber for a sufficient length of time to enable the concentration and subsequent detection of as many VOCs as possible. A longer sampling time was not considered, in order to minimize the time spent in the chamber for participant comfort and safety.
As an initial evaluation of the temporal stability of whole body VOC signatures from individual humans, replicate samples were also collected from a single male participant across three timepoints (10 am, 12 pm and 2 pm). These pilot results demonstrated similar whole body VOC profiles (Fig. S4†), with the ratios of primary components remaining consistent across samples. However, a larger scale study incorporating multiple participants over longer time periods would be required to fully investigate the temporal stability of the whole body volatilome.
We engineered our whole body headspace collection chamber to be transportable using materials that can be flat-packed and assembled within a tubular aluminium frame with corner connectors. This frame conveniently has flanges that allow the walls and roof of the chamber which are made from transparent plexiglass panels to slot into position, as well as a floor made of ABS plastic. These materials were chosen due to their inert and impact resistant nature. The use of transparent plexiglass panels also allows clear observation of each participant during sampling. Previously, whole body sampling chambers have been engineered using welded stainless steel and glass.24,50,51 While these materials also provide excellent inert qualities and air tightness once sealed, we anticipate that the durability and relative light-weight nature of the materials used for construction of our whole body headspace collection chamber, will facilitate ease of its assembly and portability in a wide-variety of laboratory, clinical and field-based contexts.
Chemical constitution of human whole body headspace
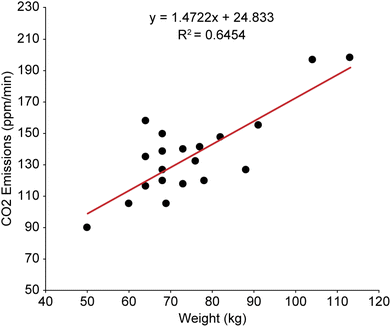
The human headspace collection chamber enabled the profiling of human whole body volatile organic compounds, in addition to facilitating quantification of exhaled CO2 and measurement of water vapor from 20 healthy human participants. Carbon dioxide is one of the primary VOCs in exhaled breath, present at approximately 4–5% by volume and produced as a by-product of cellular metabolism. The measurement of respiratory CO2 can provide clinical insight into the severity of respiratory conditions such as asthma and COPD,56 how effectively CO2 is being eliminated from the body,57 and response to medical interventions such as tracheal intubation and anaesthesia.58 Carbon dioxide is also a crucial VOC in the attraction of arthropod disease vectors, such as mosquitoes, black flies, triatomine bugs and bed bugs to humans,59–62 and variation in exhaled CO2 may play a role in the differential attractiveness of individuals to mosquitoes. For instance, Brady et al. assessed the attraction of mosquitoes to different human hosts over a 10 weeks period, demonstrating that variation of CO2 production between different human participants may be a major chemical factor driving differential attractiveness.63 Other areas of interest for the study of exhaled CO2 include exercise monitoring,64–66 the development of chemical sensors to detect trapped humans in search and rescue operations,25,67 and the monitoring of human contributions to indoor air contaminants.68In this study, the concentration of CO2 in the sampling chamber was recorded at 5 minutes intervals throughout the 30 minutes sampling period. Chamber CO2 was monitored both to ensure CO2 levels did not exceed permissible exposure limits as established by OSHA (5000 ppm as an 8 hours time-weighted average)69 and to evaluate individual differences in CO2 emissions. Across the 20 participants, CO2 emissions over the 30 minutes sampling period ranged from 89 to 197 ppm min−1, with a mean emission of 135 ppm min−1. The total CO2 concentration in the sampling chamber at the end of the 30 min sampling period ranged from 3130 ppm to 6407 ppm with a mean total of 4482 ppm. There was a moderate correlation between body weight and CO2 emission rates (Fig. 3), although variation in CO2 emissions were observed between participants of similar weights.
Temperature and humidity within the sampling chamber were also recorded at the beginning of the experiment and at 5 minutes intervals throughout the sampling process. The laboratory in which the human headspace collection chamber was situated was maintained at a constant temperature and the mean starting temperature inside the chamber was 21.1 °C. By the end of the 30 minutes sampling period, the interior temperature of the chamber increased by an average of 2.5 °C (range of 1.7 to 3.2 °C increase). Average starting relative humidity in the sampling chamber was 56.9% with an average increase of 19.3% (range of 12 to 30%) by the end of the 30 minutes sampling period. In this study, the sorbent tubes used for VOC sampling contained only hydrophobic Tenax-TA which does not retain water and thus the increase in relative humidity was not deemed problematic for downstream VOC analysis.
A large number of VOCs were detected in human whole body headspace belonging to a range of compounds classes, including aldehydes, ketones, carboxylic acids, alcohols, and aliphatic and aromatic hydrocarbons. Across the 20 participants, we detected a minimum of 797 and maximum of 1140 features, with a mean of 983 features detected in human headspace. On average, 326 of these features had a possible match in NIST library. In this study, the identity of approximately 13% of features with a NIST match have been confirmed. See Table S2† for a breakdown of the number of features detected in the headspace of each participant.
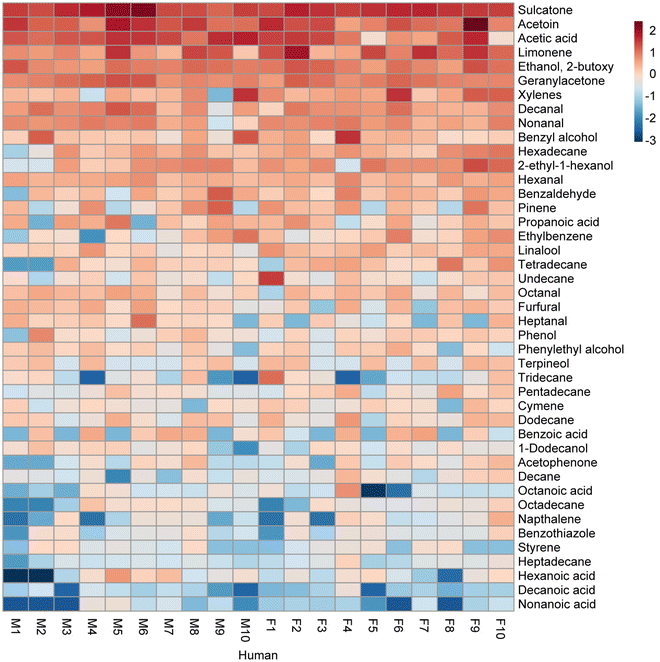
Whole body headspace exhibited heterogeneous chemistry across the 20 participants, as detailed in Fig. S5,† which shows the relative abundance of 43 confirmed VOCs annotated in headspace samples. These compounds were selected based on their frequency of detection and availability of analytical standards to validate their identity. The majority of these 43 detected compounds were conserved in human whole body headspace, present in all or many of the participants (Table 1), though distinct differences were observed in the relative abundance of the compounds detected. Furthermore, the quantity of individual compounds detected varied considerably across participants (Fig. 4).
| Compound | CAS no. | Chemical class | Detection frequency | Mean emission rate (μg h−1) | Possible origin |
|---|---|---|---|---|---|
| 6-Methyl-5-hepten-2-one (sulcatone) | 110-93-0 | Ketone | 100% | 14.56 | Skin (squalene oxidation, breath)73 |
| 3-Hydroxy-2-butanone (acetoin) | 513-86-0 | Ketone | 100% | 10.48 | Breath, skin (microbial metabolite)85 |
| Limonene | 138-86-3 | Terpene | 100% | 5.65 | Breath, skin (diet, cleaning products)57 |
| Ethanol, 2-butoxy- | 111-76-2 | Alcohol | 100% | 3.19 | Skin (microbial metabolite)106 |
| 6,10-Dimethyl-5,9-undecadien-2-one (geranylacetone) | 3796-70-1 | Ketone | 100% | 3.07 | Skin (squalene oxidation)73 |
| Decanal | 112-31-2 | Aldehyde | 100% | 1.98 | Breath, skin (fatty acid degradation, microbial metabolite)73,85 |
| Nonanal | 124-19-6 | Aldehyde | 100% | 1.72 | Breath, skin (fatty acid degradation)31,73 |
| Benzyl alcohol | 100-51-6 | Alcohol | 100% | 1.63 | Breath, skin (toluene metabolism, microbial metabolite)5,85 |
| Hexanal | 66-25-1 | Aldehyde | 100% | 1.04 | Breath, skin (fatty acid degradation)31,73 |
| Undecane | 1120-21-4 | Hydrocarbon | 100% | 0.74 | Breath, skin (lipid peroxidation, microbial metabolite)104,106 |
| Linalool | 78-70-6 | Terpenoid | 100% | 0.67 | Breath, skin (unknown) |
| Dodecane | 112-40-3 | Hydrocarbon | 100% | 0.44 | Breath, skin (microbial metabolite)85,106 |
| Pentadecane | 629-62-9 | Hydrocarbon | 100% | 0.32 | Breath, skin (lipid peroxidation, microbial metabolite)85,104 |
| Xylenes | 1330-20-7 | Aromatic | 95% | 2.45 | Breath (exogenous)45 |
| Hexadecane | 544-76-3 | Hydrocarbon | 95% | 1.11 | Breath, skin (lipid peroxidation)104 |
| Benzaldehyde | 100-52-7 | Aldehyde | 95% | 1.01 | Breath, skin (benzyl alcohol oxidation, microbial metabolite)85,106,107 |
| Ethylbenzene | 100-41-4 | Aromatic | 95% | 0.72 | Breath, skin (exogenous) |
| Octanal | 124-13-0 | Aldehyde | 95% | 0.61 | Breath, skin (fatty acid oxidation)73 |
| Phenol | 108-95-2 | Aromatic | 95% | 0.39 | Breath, skin (microbial metabolite)85 |
| 1-Dodecanol | 112-53-8 | Alcohol | 95% | 0.21 | Breath, skin (microbial metabolite)85 |
| Octanoic acid | 124-07-2 | Acid | 95% | 0.18 | Breath, skin (sebaceous gland secretions)108 |
| Decane | 124-18-5 | Hydrocarbon | 95% | 0.18 | Breath, skin (lipid peroxidation, microbial metabolite)85,104,106 |
| Heptadecane | 629-78-7 | Hydrocarbon | 95% | 0.09 | Breath, skin (lipid peroxidation)104 |
| Acetic acid | 64-19-7 | Acid | 90% | 6.95 | Breath, skin (human metabolism, microbial metabolite)49,57,106 |
| Propanoic acid | 79-09-4 | Acid | 90% | 0.81 | Breath, skin (microbial metabolite)85 |
| Tetradecane | 629-59-4 | Hydrocarbon | 90% | 0.65 | Breath, skin (lipid peroxidation, microbial metabolite)85,104 |
| Furfural | 98-01-1 | Aldehyde | 90% | 0.51 | Breath, skin (unknown) |
| Phenylethyl alcohol | 60-12-8 | Alcohol | 90% | 0.35 | Skin (microbial metabolite)85 |
| Cymene | 527-84-4 | Aromatic | 90% | 0.26 | Breath (diet)109 |
| Hexanoic acid | 142-62-1 | Acid | 90% | 0.23 | Breath, skin (microbial metabolite)110 |
| Benzothiazole | 95-16-9 | Thiazole | 90% | 0.13 | Breath, skin (unknown) |
| 2-Ethyl-1-hexanol | 104-76-7 | Alcohol | 85% | 1.82 | Breath, skin (exogenous)111 |
| Tridecane | 629-50-5 | Hydrocarbon | 85% | 0.34 | Breath, skin (lipid peroxidation, microbial metabolite)85,104 |
| Acetophenone | 98-86-2 | Ketone | 85% | 0.22 | Breath, skin (unknown) |
| Octadecane | 593-45-3 | Hydrocarbon | 85% | 0.17 | Breath, skin (lipid peroxidation)104 |
| Decanoic acid | 334-48-5 | Acid | 85% | 0.04 | Breath and skin (sebaceous gland secretions)108 |
| Heptanal | 111-71-7 | Aldehyde | 80% | 0.49 | Breath, skin (fatty acid degradation)31 |
| Naphthalene | 91-20-3 | Aromatic | 80% | 0.16 | Skin (microbial metabolite, exogenous)112 |
| Pinene | 80-56-8 | Terpene | 75% | 0.98 | Breath, skin (exogenous)37 |
| Nonanoic acid | 112-05-0 | Acid | 75% | 0.04 | Breath, skin (sebaceous gland secretions)108 |
| Benzoic acid | 65-85-0 | Acid | 65% | 0.41 | Breath, skin (benzyl alcohol oxidation)107 |
| Styrene | 100-42-5 | Aromatic | 65% | 0.12 | Breath, skin (microbial metabolite)85 |
| Terpineol | 8000-41-7 | Terpenoid | 55% | 0.35 | Breath, skin (exogenous)57 |
The compounds we detected in whole body headspace represent a broad range of VOCs of endogenous and microbial origin from skin emissions and exhaled breath. Many of these are of known interest in various areas of metabolomics. Squalene is an abundant component of human sebum, a lipid-rich substance secreted by the sebaceous glands.70 Although not a volatile compound, squalene is a major contributor to human odor due to the number of volatiles that have been demonstrated to originate from the reaction of squalene with ozone present in the atmosphere. Numerous studies have demonstrated that ozone oxidation of squalene results in the production of various VOCs including 6-methyl-5-hepten-2-one (sulcatone), 6,10-dimethyl-5,9-undecadien-2-one (geranylacetone), acetone, hydroxyacetone and 4-oxopentanal.71–73 Sulcatone and geranylacetone were amongst the most abundant compounds detected in whole body headspace, present in 100% of participants. Both compounds are of particular interest in the study of mosquito host-seeking and are potentially important human odor cues in the differential attraction of mosquitoes to different individuals.74,75
Similarly, long-chain aldehydes are known products of the reaction between numerous unsaturated fatty acids found on the skin and ozone.73 In this study, several aldehydes were detected, in particular hexanal, heptanal, octanal, nonanal and decanal, which were found in the whole body headspace of almost all participants. Aldehydes are of particular interest in clinical metabolomics, with numerous studies detecting changes in the aforementioned aldehydes in breath and skin emissions with various diseases including lung cancer,76,77 breast cancer,78 colorectal cancer,79 COPD,80 and malaria.81–83 The whole body headspace of all participants contained substantial amounts of 3-hydroxy-2-butanone (acetoin), a known bacterial metabolite produced during the catabolism of pyruvate. Acetoin has previously been detected in the headspace of several types of bacteria, including Staphylococcus epidermidis84,85 and Staphylococcus aureus85,86 both of which are present on human skin. The high abundance of acetoin in human headspace may therefore primarily be a bacterial metabolite, though acetoin has also been detected in exhaled breath and has been highlighted as a potential breath biomarker for lung cancer.87–89
Short chain fatty acids (SCFAs) have been previously reported to be abundant components of human odor, produced via the catabolism of skin lipids into long-chain fatty acids and subsequently highly volatile SCFAs by Staphylococcus, Propionibacteria and Corynebacteria.90 Furthermore, VFAs can also be produced from the conversion of branched aliphatic amino acids by Staphylococcus bacteria.91 In this study, acetic acid was amongst the most abundant compounds present and was detected across 90% of the participants. Other volatile fatty acids were also detected, including propanoic and hexanoic acid, though these were present in considerably lower concentrations than acetic acid.
In addition to SCFAs, longer chain saturated fatty acids were detected at relatively low abundance, including octanoic, nonanoic and decanoic acid. Although readily produced by skin bacteria, volatile fatty acids are also present in breath and are linked to oral malodor.92 Finally, the presence of fatty acids in human odor may also be a product of oxidation. Pleik et al. conducted a study on degradation products in fingerprint residues, demonstrating decanoic acid was produced from the oxidation of decanal, which was amongst the most abundant aldehydes detected in whole body headspace using the sampling chamber.93 Organic acids in exhaled breath have gained some clinical interest in recent years, with changes in the levels of certain carboxylic acids being associated with lung cancer94 and gastrointestinal cancer.95,96 Furthermore, carboxylic acids are known attractants to anthropophilic mosquitoes, and the further study of these in human odor could provide important insight into mosquito host-seeking.97,98 Benzoic acid, an aromatic carboxylic acid detected in the whole body headspace of 55% of participants in this study, is a breath- and skin-derived compound that has been linked to both stress response and respiratory disease. Martin et al. demonstrated the upregulation of benzoic acid in skin VOCs following psychological stress,45 whereas Dallinga et al. identified benzoic acid in exhaled breath is a biomarker for distinguishing asthma patients from healthy controls.99
Although a large proportion of the compounds profiled using this method can be attributed to endogenous and microbial volatiles released from the skin, many VOCs detected in whole body headspace are also likely derived from exhaled breath (Table 1). Numerous aromatic hydrocarbons and terpenes were identified, including xylene, ethylbenzene, styrene, limonene, pinene, and cymene, all of which have been frequently detected in exhaled breath in previous studies.1 Aliphatic hydrocarbons were also detected in this study, which have been previously detected in both exhaled breath and skin emissions.1 Although volatile hydrocarbons are found in the exhaled breath of healthy individuals, several of the compounds, including styrene, xylene and ethylbenzene, are known biomarkers of environmental exposure. The elevated presence of some of these aromatic compounds has been found in the breath and biological fluids of chemical plant workers,100 petrochemical industry workers,101 and smokers.102,103 In this study, none of the participants were smokers, however two participants did report the occasional use of electronic cigarettes (participants M4 and F7). No notable differences were observed in the whole body VOCs of these participants. Similarly, elevated levels of certain alkanes in exhaled breath have been described as biomarkers of oxidative stress.104 Many other compounds known to be released from human skin are also present in exhaled breath, including aldehydes, ketones and carboxylic acids.9 As such, the human whole body volatilome is a complex mix of both exhaled breath and skin VOCs. An interesting area for future research will therefore be to modify the chamber design in such a way to enable breathing outside of the chamber (e.g. via a tight mask over the mouth and nose that is directly vented outside the chamber) to discern the relative contribution of skin and breath VOCs to the whole body volatilome.
In this study only thermal desorption tubes with Tenax-TA sorbent were utilized, which is not suitable for the trapping of highly volatile species.105 For instance, acetone and isoprene, which are well known to be two of the most abundant VOCs in human breath, were not well captured using this sorbent. Although they are detected using Tenax-TA alone, they were not included in this study to avoid use of an unsuitable sorbent that could affect reproducibility. In future studies, the use of additional sorbents such as Carboxen and Carbopack may enable the collection and detection of a broader range of volatile compounds. Furthermore, the use of alternative techniques could be employed to improve analyte identification and quantification. The use of two-dimensional GC (GC × GC) would provide an additional degree of separation, enabling the improved separation and detection of co-eluting analytes and reducing the need for deconvolution for analyte separation and detection. Furthermore, direct mass spectrometry, such as proton-transfer reaction MS (PTR-MS), could be utilized for the real-time analysis of compounds difficult to detect by traditional GC/MS, such as highly volatile compounds. The use of such instrumentation can furthermore enable VOC analysis without the need to collect samples onto sorbent tubes, reducing the potential for loss or reaction of analytes. In addition to PTR-MS, gas-specific monitors could be used to measured highly volatile compounds such as ammonia.
Such integrative approaches may further be used to comprehensively characterize the whole human body volatilome and understand the temporal stability of individual scent signatures over time, enabling the identification of both core and variable components of the human volatilome. A comprehensive understanding of the qualitative and quantitative composition of whole body VOC signatures may also assist to demonstrate what body parts and associated gut, oral and skin microbiome communities contribute most to human body headspace. Additional participant monitoring could also be implemented, including skin conductivity, skin hydration, and heart rate. Future studies could also benefit from increased control of participant conditions prior to sampling including defined dietary regimes and standardized hygiene practices. Restricted use of hygiene products and intake of certain foods for longer time periods prior to sampling may help to reduce exogenous volatiles. Furthermore, slight differences in the time between washing and sampling could introduce variability in the type and abundance of VOCs detected, thus stricter control of these activities could be implemented. It should also be noted that the cotton scrubs and the seated position of participants that we implemented for scent collection, may have excluded or altered the release rates of VOCs from certain areas of the body covered by these substrates, thus influencing compounds profiled in this study protocol. We employed standardized cotton scrubs to minimize potential introduction of exogenous volatiles during sampling and provided participants with seating to maximize their comfort and minimize potential for venous pooling, commonly associated with standing, throughout the 30 minutes sampling period. However, if of interest, future studies may choose to evaluate the effects of clothing and body position on the chemical composition of the whole body volatilome.
Our whole body headspace collection chamber was housed within a temperature-controlled laboratory and operated as a sealed system during sampling mode to maximize VOC concentration for analysis by TD-GC/MS. However, one caveat of our simple chamber design was that temperature and humidity were not regulated inside the chamber atmosphere upon participant entry. Further experiments are therefore needed to evaluate the effect of changes in temperature and humidity on the chemical constitution of the whole body volatilome. Considerations for further iterative design features of this chamber may include implementation of internal control mechanisms to dynamically regulate temperature, humidity, and air exchange inside the chamber atmosphere, with these parameters and sampling times optimized according to the downstream analytical technique being employed. Using our current chamber design, basal temperature and humidity conditions inside the chamber will likely mirror environmental conditions wherever sampling occurs. This will be particularly pertinent in field-based contexts where environmental conditions fluctuate. Future testing of the headspace collection chamber under ambient conditions (i.e. outside of a temperature controlled laboratory) will help to discern whether our current chamber design suffices for whole body volatilomic profiling in the field, or if installation of portable mechanisms to dynamically regulate the temperature and humidity in the headspace collection chamber atmosphere are necessary for sampling in these settings. In addition, installation of a chamber anteroom that is flushed with purified air prior to participant entry into the main sampling chamber, as well as evaluation of supplemental chamber cleaning protocols may help to further reduce exogenous and human-derived background contaminants in this method.
This study demonstrates the utility of our headspace collection chamber design for standardized sampling and quantitative analysis of human whole body volatilome in a laboratory setting. The high-content scent signatures derived using this method highlight the most frequent and abundant components of whole body VOC emissions, as well as the complexity and heterogeneity of human body odor. This method could be a powerful addition to several fields of research in human volatilomics, including clinical metabolomics to detect changes in the chemistry of human scent in response to disease and stress, exposomics to study evaluated human exposure to environmental contaminants, and chemical ecology to identify VOCs that attract arthropod disease vectors to human hosts.
Conclusion
Human odor is a complex blend of volatile compounds released via the skin, breath and bodily fluids, but characterizing whole body odor in its entirety has been challenging due to limitations in available sampling configurations. This study aimed to develop a controlled and standardized method for headspace collection from seated humans to facilitate chemical analysis of the human whole body volatilome. A booth-style sampling chamber was engineered and used in a laboratory setting to profile whole body volatiles from a pilot cohort of 20 human participants. Human headspace samples were collected onto Tenax-TA thermal desorption tubes and analyzed by TD-GC/MS to identify and quantify human odor components. This approach enabled the characterization of a broad range of endogenous and microbial skin and breath-derived volatiles, including ketones, aldehydes, hydrocarbons, carboxylic acids and alcohols. Many compounds were identified across all participants, whereas some were only present in the headspace of select participants at varying levels. Such inter-individual variation in VOC frequency and abundance highlights both common and heterogeneous features of human scent chemistry. This new analytical approach to profile the human whole body volatilome could be readily used to characterize the contribution of the human microbiome to VOCs detected in whole body headspace, and for varied applications in clinical metabolomics, exposomics, chemical ecology, security and forensics to yield high-content human scent signatures.Author contributions
S. R. T. and C. J. M. together conceptualized the research, developed the methodology, wrote and edited the paper, and visualized the data. S. R. T. performed the experiments and analyzed the data. All authors have approved the final version of the manuscript.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We thank Diego Giraldo and Margot Wohl for comments on the manuscript and Terry Shelley at the JHU Center for Neuroscience Research Machine Shop for expert fabrication services supported by NINDS Center grant (NS050274). This research was supported by funding from the USAID Combating Zika and Future Threats Grand Challenge initiative (AID-OAA-F-16-00061) and Innovative Vector Control Consortium (P105) to C. J. M. We further acknowledge generous support to C. J. M. and S. R. T. from Johns Hopkins Malaria Research Institute (JHMRI) and Bloomberg Philanthropies.References
- N. Drabińska, C. Flynn, N. Ratcliffe, I. Belluomo, A. Myridakis, O. Gould, M. Fois, A. Smart, T. Devine and B. D. L. Costello, J. Breath Res., 2021, 15(3), 034001 CrossRef PubMed.
- M. Shirasu and K. Touhara, J. Biochem., 2011, 150, 257–266 CrossRef CAS PubMed.
- T. Chen, T. Liu, T. Li, H. Zhao and Q. Chen, Clin. Chim. Acta, 2021, 515, 61–72 CrossRef CAS PubMed.
- A. Krilaviciute, M. Leja, A. Kopp-Schneider, O. Barash, S. Khatib, H. Amal, Y. Y. Broza, I. Polaka, S. Parshutin, A. Rudule, H. Haick and H. Brenner, J. Breath Res., 2019, 13, 026006 CrossRef CAS PubMed.
- A. Peralbo-Molina, M. Calderón-Santiago, B. Jurado-Gámez, M. D. Luque de Castro and F. Priego-Capote, Sci. Rep., 2017, 7, 1421 CrossRef CAS PubMed.
- H. Vereb, A. M. Dietrich, B. Alfeeli and M. Agah, Environ. Sci. Technol., 2011, 45, 8167–8175 CrossRef CAS PubMed.
- L. C. A. Amorim and Z. de L. Cardeal, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2007, 853, 1–9 CrossRef CAS PubMed.
- J. Martinez, A. Showering, C. Oke, R. T. Jones and J. G. Logan, Philos. Trans. R. Soc., B, 2021, 376, 20190811 CrossRef CAS PubMed.
- X. Sun, K. Shao and T. Wang, Anal. Bioanal. Chem., 2016, 408, 2759–2780 CrossRef CAS PubMed.
- J. Zhou, Z.-A. Huang, U. Kumar and D. D. Y. Chen, Anal. Chim. Acta, 2017, 996, 1–9 CrossRef CAS PubMed.
- A. Christiansen, J. R. Davidsen, I. Titlestad, J. Vestbo and J. Baumbach, J. Breath Res., 2016, 10, 034002 CrossRef PubMed.
- A. Azim, C. Barber, P. Dennison, J. Riley and P. Howarth, Eur. Respir. J., 2019, 54, 1900056 CrossRef CAS PubMed.
- A. Z. Berna, J. S. McCarthy, R. X. Wang, K. J. Saliba, F. G. Bravo, J. Cassells, B. Padovan and S. C. Trowell, J. Infect. Dis., 2015, 212, 1120–1128 CrossRef CAS PubMed.
- C. L. Schaber, N. Katta, L. B. Bollinger, M. Mwale, R. Mlotha-Mitole, I. Trehan, B. Raman and A. R. Odom John, J. Infect. Dis., 2018, 217, 1553–1560 CrossRef CAS PubMed.
- A. Z. Berna, E. H. Akaho, R. M. Harris, M. Congdon, E. Korn, S. Neher, M. M'Farrej, J. Burns and A. R. Odom John, ACS Infect. Dis., 2021, 7, 2596–2603 CrossRef CAS PubMed.
- H. Chen, X. Qi, L. Zhang, X. Li, J. Ma, C. Zhang, H. Feng and M. Yao, J. Breath Res., 2021, 15(4), 047104 CAS.
- W. Ibrahim, R. L. Cordell, M. J. Wilde, M. Richardson, L. Carr, A. Sundari Devi Dasi, B. Hargadon, R. C. Free, P. S. Monks, C. E. Brightling, N. J. Greening and S. Siddiqui, ERJ Open Res., 2021, 7, 00139–02021 Search PubMed.
- B. N. Zamora-Mendoza, L. Díaz de León-Martínez, M. Rodríguez-Aguilar, B. Mizaikoff and R. Flores-Ramírez, Talanta, 2022, 236, 122832 CrossRef CAS PubMed.
- O. Lawal, W. M. Ahmed, T. M. E. Nijsen, R. Goodacre and S. J. Fowler, Metabolomics, 2017, 13, 110 CrossRef PubMed.
- M. Basanta, T. Koimtzis, D. Singh, I. Wilson and C. L. P. Thomas, Analyst, 2007, 132, 153–163 RSC.
- T. Bruderer, T. Gaisl, M. T. Gaugg, N. Nowak, B. Streckenbach, S. Müller, A. Moeller, M. Kohler and R. Zenobi, Chem. Rev., 2019, 119, 10803–10828 CrossRef CAS PubMed.
- W. Vautz, J. Franzke, S. Zampolli, I. Elmi and S. Liedtke, Anal. Chim. Acta, 2018, 1024, 52–64 CrossRef CAS PubMed.
- D. P. Elpa, H.-Y. Chiu, S.-P. Wu and P. L. Urban, Trends Endocrinol. Metab., 2021, 32, 66–75 CrossRef CAS PubMed.
- P. Mochalski, K. Unterkofler, H. Hinterhuber and A. Amann, Anal. Chem., 2014, 86, 3915–3923 CrossRef CAS PubMed.
- R. Huo, A. Agapiou, V. Bocos-Bintintan, L. J. Brown, C. Burns, C. S. Creaser, N. A. Devenport, B. Gao-Lau, C. Guallar-Hoyas, L. Hildebrand, A. Malkar, H. J. Martin, V. H. Moll, P. Patel, A. Ratiu, J. C. Reynolds, S. Sielemann, R. Slodzynski, M. Statheropoulos, M. A. Turner, W. Vautz, V. E. Wright and C. L. P. Thomas, J. Breath Res., 2011, 5, 046006 CrossRef CAS PubMed.
- W. Vautz, R. Slodzynski, C. Hariharan, L. Seifert, J. Nolte, R. Fobbe, S. Sielemann, B. C. Lao, R. Huo, C. L. P. Thomas and L. Hildebrand, Anal. Chem., 2013, 85, 2135–2142 CrossRef CAS PubMed.
- M. Ueland, S. Harris and S. L. Forbes, Forensic Sci. Int., 2021, 323, 110781 CrossRef CAS PubMed.
- S. Stadler, P.-H. Stefanuto, M. Brokl, S. L. Forbes and J.-F. Focant, Anal. Chem., 2013, 85, 998–1005 CrossRef CAS PubMed.
- A. M. Curran, S. I. Rabin, P. A. Prada and K. G. Furton, J. Chem. Ecol., 2005, 31, 1607–1619 CrossRef CAS PubMed.
- A. M. Curran, P. A. Prada and K. G. Furton, J. Forensic Sci., 2010, 55, 50–57 CrossRef CAS PubMed.
- S. Haze, Y. Gozu, S. Nakamura, Y. Kohno, K. Sawano, H. Ohta and K. Yamazaki, J. Invest. Dermatol., 2001, 116, 520–524 CrossRef CAS PubMed.
- U. R. Bernier, D. L. Kline, C. E. Schreck, R. A. Yost and D. R. Barnard, J. Am. Mosq. Control Assoc., 2002, 18, 186–195 CAS.
- U. R. Bernier, M. M. Booth and R. A. Yost, Anal. Chem., 1999, 71, 1–7 CrossRef CAS PubMed.
- M. Wooding, E. R. Rohwer and Y. Naudé, J. Sep. Sci., 2020, 43, 4202–4215 CrossRef CAS PubMed.
- Y. Xu, S. J. Dixon, R. G. Brereton, H. A. Soini, M. V. Novotny, K. Trebesius, I. Bergmaier, E. Oberzaucher, K. Grammer and D. J. Penn, Metabolomics, 2007, 3, 427–437 CrossRef CAS.
- M. Gallagher, C. J. Wysocki, J. J. Leyden, A. I. Spielman, X. Sun and G. Preti, Br. J. Dermatol., 2008, 159, 780–791 CrossRef CAS PubMed.
- P. Mochalski, J. King, K. Unterkofler, H. Hinterhuber and A. Amann, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2014, 959, 62–70 CrossRef CAS PubMed.
- Z.-M. Zhang, J.-J. Cai, G.-H. Ruan and G.-K. Li, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2005, 822, 244–252 CrossRef CAS PubMed.
- N. O. Verhulst, H. Beijleveld, Y. T. Qiu, C. Maliepaard, W. Verduyn, G. W. Haasnoot, F. H. J. Claas, R. Mumm, H. J. Bouwmeester, W. Takken, J. J. A. van Loon and R. C. Smallegange, Infect., Genet. Evol., 2013, 18, 87–93 CrossRef CAS PubMed.
- Y. T. Qiu, R. C. Smallegange, S. Hoppe, J. J. A. van Loon, E.-J. Bakker and W. Takken, Med. Vet. Entomol., 2004, 18, 429–438 CrossRef CAS PubMed.
- P. Doležal, P. Kyjaková, I. Valterová and Š. Urban, J. Chromatogr. A, 2017, 1505, 77–86 CrossRef PubMed.
- L. Dormont, J.-M. Bessière, D. McKey and A. Cohuet, J. Exp. Biol., 2013, 216, 2783–2788 CAS.
- D. J. Penn, E. Oberzaucher, K. Grammer, G. Fischer, H. A. Soini, D. Wiesler, M. V. Novotny, S. J. Dixon, Y. Xu and R. G. Brereton, J. R. Soc., Interface, 2007, 4, 331–340 CrossRef PubMed.
- S. Riazanskaia, G. Blackburn, M. Harker, D. Taylor and C. L. P. Thomas, Analyst, 2008, 133, 1020 RSC.
- H. J. Martin, M. A. Turner, S. Bandelow, L. Edwards, S. Riazanskaia and C. L. P. Thomas, J. Breath Res., 2016, 10, 046012 CrossRef CAS PubMed.
- A. P. Roodt, Y. Naudé, A. Stoltz and E. Rohwer, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2018, 1097–1098, 83–93 CrossRef CAS PubMed.
- J. G. Logan, M. A. Birkett, S. J. Clark, S. Powers, N. J. Seal, L. J. Wadhams, A. J. Mordue and J. A. Pickett, J. Chem. Ecol., 2008, 34, 308–322 CrossRef CAS PubMed.
- Z. Zhao, J. L. Zung, A. L. Kriete, A. Iqbal, M. A. Younger, J. Benjamin, D. Merhof, S. Thiberge, M. Strauch, C. S. Mcbride, C. Dynamics, N. York and C. Vision, bioRxiv, DOI:10.1101/2020.11.01.363861.
- P. Mochalski, H. Wiesenhofer, M. Allers, S. Zimmermann, A. T. Güntner, N. J. Pineau, W. Lederer, A. Agapiou, C. A. Mayhew and V. Ruzsanyi, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2018, 1076, 29–34 CrossRef CAS PubMed.
- Z. Zou, J. He and X. Yang, Build. Environ., 2020, 181, 107137 CrossRef.
- R. I. Ellin, R. L. Farrand, F. W. Oberst, C. L. Crouse, N. B. Billups, W. S. Koon, N. P. Musselman and F. R. Sidell, J. Chromatogr. A, 1974, 100, 137–152 CrossRef CAS PubMed.
- S. Cui, M. Cohen, P. Stabat and D. Marchio, Build. Environ., 2015, 84, 162–169 CrossRef.
- S. Batterman, Int. J. Environ. Res. Public Health, 2017, 14, 1–22 Search PubMed.
- Occupational Safety and Health Administration, Permit-required confined spaces (29 CFR 1910.146), 2004 Search PubMed.
- S. DeMeulenaere, J. Nurse Pract., 2007, 3, 312–317 CrossRef.
- O. P. Singh, T. A. Howe and M. Malarvili, J. Breath Res., 2018, 12, 026003 CrossRef PubMed.
- A. Amann and D. Smith, Volatile Biomarkers, 2013 Search PubMed.
- M. S. Siobal, Respir. Care, 2016, 61, 1397–1416 CrossRef PubMed.
- V. Harraca, C. Ryne, G. Birgersson and R. Ignell, J. Exp. Biol., 2012, 215, 623–629 CrossRef CAS PubMed.
- M. T. Gillies, Bull. Entomol. Res., 1980, 70, 525–532 CrossRef.
- C. J. McMeniman, R. A. Corfas, B. J. Matthews, S. A. Ritchie and L. B. Vosshall, Cell, 2014, 156, 1060–1071 CrossRef CAS PubMed.
- S. W. Schofield and J. F. Sutcliffe, J. Med. Entomol., 1996, 33, 102–108 CrossRef CAS PubMed.
- J. Brady, C. Costantini, N. Sagnon, G. Gibson and M. Coluzzi, Ann. Trop. Med. Parasitol., 1997, 91, S121–S122 CrossRef.
- L. M. Heaney and M. R. Lindley, Metabolomics, 2017, 13, 139 CrossRef.
- D. Smith, A. Pysanenko and P. Španěl, Rapid Commun. Mass Spectrom., 2009, 23, 1419–1425 CrossRef CAS PubMed.
- L. M. Heaney, S. Kang, M. A. Turner, M. R. Lindley and C. L. P. Thomas, Molecules, 2022, 27, 370 CrossRef CAS PubMed.
- A. Agapiou, A. Amann, P. Mochalski, M. Statheropoulos and C. L. P. Thomas, TrAC, Trends Anal. Chem., 2015, 66, 158–175 CrossRef CAS.
- X. Tang, P. K. Misztal, W. W. Nazaroff and A. H. Goldstein, Environ. Sci. Technol., 2016, 50, 12686–12694 CrossRef CAS PubMed.
- Occupational Safety and Health Administration, Sampling and Analytical Methods: Carbon Dioxide in Workplace Atmospheres, 1990 Search PubMed.
- M. Picardo, M. Ottaviani, E. Camera and A. Mastrofrancesco, Dermatoendocrinol., 2009, 1, 68–71 CrossRef CAS PubMed.
- L. Petrick and Y. Dubowski, Indoor Air, 2009, 19, 381–391 CrossRef CAS PubMed.
- C. J. Weschler, A. Wisthaler, S. Cowlin, G. Tamás, P. Strøm-Tejsen, A. T. Hodgson, H. Destaillats, J. Herrington, J. Zhang and W. W. Nazaroff, Environ. Sci. Technol., 2007, 41, 6177–6184 CrossRef CAS PubMed.
- A. Wisthaler and C. J. Weschler, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 6568–6575 CrossRef CAS PubMed.
- J. G. Logan, N. M. Stanczyk, A. Hassanali, J. Kemei, A. E. G. Santana, K. A. L. Ribeiro, J. A. Pickett and A. J. Mordue, Malar. J., 2010, 9, 1–10 CrossRef PubMed.
- H. M. Leal, J. K. Hwang, K. Tan and W. S. Leal, Sci. Rep., 2017, 7, 17965 CrossRef PubMed.
- P. Fuchs, C. Loeseken, J. K. Schubert and W. Miekisch, Int. J. Cancer, 2009, 126(11), 2663–2670 Search PubMed.
- D. Poli, M. Goldoni, M. Corradi, O. Acampa, P. Carbognani, E. Internullo, A. Casalini and A. Mutti, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2010, 878, 2643–2651 CrossRef CAS PubMed.
- J. Li, Y. Peng, Y. Liu, W. Li, Y. Jin, Z. Tang and Y. Duan, Clin. Chim. Acta, 2014, 436, 59–67 CrossRef CAS PubMed.
- D. F. Altomare, M. Di Lena, F. Porcelli, L. Trizio, E. Travaglio, M. Tutino, S. Dragonieri, V. Memeo and G. de Gennaro, Br. J. Surg., 2012, 100, 144–150 CrossRef PubMed.
- M. Corradi, I. Rubinstein, R. Andreoli, P. Manini, A. Caglieri, D. Poli, R. Alinovi and A. Mutti, Am. J. Respir. Crit. Care Med., 2003, 167, 1380–1386 CrossRef PubMed.
- A. Robinson, A. O. Busula, M. A. Voets, K. B. Beshir, J. C. Caulfield, S. J. Powers, N. O. Verhulst, P. Winskill, J. Muwanguzi, M. A. Birkett, R. C. Smallegange, D. K. Masiga, W. Richard Mukabana, R. W. Sauerwein, C. J. Sutherland, T. Bousema, J. A. Pickett, W. Takken, J. G. Logan and J. G. De Boer, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E4209–E4218 CAS.
- C. M. De Moraes, C. Wanjiku, N. M. Stanczyk, H. Pulido, J. W. Sims, H. S. Betz, A. F. Read, B. Torto and M. C. Mescher, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 5780–5785 CrossRef CAS PubMed.
- H. Pulido, N. M. Stanczyk, C. M. De Moraes and M. C. Mescher, Sci. Rep., 2021, 11, 13928 CrossRef CAS PubMed.
- N. O. Verhulst, R. Andriessen, U. Groenhagen, G. Bukovinszkiné Kiss, S. Schulz, W. Takken, J. J. A. van Loon, G. Schraa and R. C. Smallegange, PLoS One, 2010, 5, e15829 CrossRef CAS PubMed.
- S. Fitzgerald, E. Duffy, L. Holland and A. Morrin, Sci. Rep., 2020, 10, 1–12 CrossRef PubMed.
- G. Preti, E. Thaler, C. W. Hanson, M. Troy, J. Eades and A. Gelperin, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2009, 877, 2011–2018 CrossRef CAS PubMed.
- A. Bajtarevic, C. Ager, M. Pienz, M. Klieber, K. Schwarz, M. Ligor, T. Ligor, W. Filipiak, H. Denz, M. Fiegl, W. Hilbe, W. Weiss, P. Lukas, H. Jamnig, M. Hackl, A. Haidenberger, B. Buszewski, W. Miekisch, J. Schubert and A. Amann, BMC Cancer, 2009, 9, 348 CrossRef PubMed.
- G. Song, T. Qin, H. Liu, G.-B. Xu, Y.-Y. Pan, F.-X. Xiong, K.-S. Gu, G.-P. Sun and Z.-D. Chen, Lung Cancer, 2010, 67, 227–231 CrossRef PubMed.
- X.-A. Fu, M. Li, R. J. Knipp, M. H. Nantz and M. Bousamra, Cancer Med., 2014, 3, 174–181 CrossRef CAS PubMed.
- A. G. James, J. Casey, D. Hyliands and G. Mycock, World J. Microbiol. Biotechnol., 2004, 20, 787–793 CrossRef CAS.
- A. G. James, D. Hyliands and H. Johnston, Int. J. Cosmet. Sci., 2004, 26, 149–156 CrossRef CAS PubMed.
- S. R. Porter and C. Scully, Br. Med. J., 2006, 333, 632–635 CrossRef CAS PubMed.
- S. Pleik, B. Spengler, T. Schäfer, D. Urbach, S. Luhn and D. Kirsch, J. Am. Soc. Mass Spectrom., 2016, 27, 1565–1574 CrossRef CAS PubMed.
- L. Callol-Sanchez, M. A. Munoz-Lucas, O. Gomez-Martin, J. A. Maldonado-Sanz, C. Civera-Tejuca, C. Gutierrez-Ortega, G. Rodriguez-Trigo and J. Jareno-Esteban, J. Breath Res., 2017, 11(2), 026004 CrossRef CAS PubMed.
- S. Kumar, J. Huang, N. Abbassi-Ghadi, P. Španěl, D. Smith and G. B. Hanna, Anal. Chem., 2013, 85, 6121–6128 CrossRef CAS PubMed.
- S. R. Markar, T. Wiggins, S. Antonowicz, S.-T. Chin, A. Romano, K. Nikolic, B. Evans, D. Cunningham, M. Mughal, J. Lagergren and G. B. Hanna, JAMA Oncol., 2018, 4, 970 CrossRef PubMed.
- B. G. J. Knols, J. J. A. van Loon, A. Cork, R. D. Robinson, W. Adam, J. Meijerink, R. De Jong and W. Takken, Bull. Entomol. Res., 1997, 87, 151–159 CrossRef CAS.
- R. C. Smallegange, Y. T. Qiu, G. Bukovinszkiné-Kiss, J. J. A. Van Loon and W. Takken, J. Chem. Ecol., 2009, 35, 933–943 CrossRef CAS PubMed.
- J. W. Dallinga, C. M. H. H. T. Robroeks, J. J. B. N. van Berkel, E. J. C. Moonen, R. W. L. Godschalk, Q. Jöbsis, E. Dompeling, E. F. M. Wouters and F. J. van Schooten, Clin. Exp. Allergy, 2009, 40, 68–76 Search PubMed.
- F. Brugnone, L. Perbellini, G. B. Faccini, F. Pasini, B. Danzi, G. Maranelli, L. Romeo, M. Gobbi and A. Zedde, Am. J. Ind. Med., 1989, 16, 385–399 CrossRef CAS PubMed.
- B. Heibati, K. J. Godri Pollitt, J. Y. Charati, A. Ducatman, M. Shokrzadeh, A. Karimi and M. Mohammadyan, Ecotoxicol. Environ. Saf., 2018, 149, 19–25 CrossRef CAS PubMed.
- E. Papaefstathiou, M. Stylianou, C. Andreou and A. Agapiou, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2020, 1160, 122349 CrossRef CAS PubMed.
- W. Filipiak, V. Ruzsanyi, P. Mochalski, A. Filipiak, A. Bajtarevic, C. Ager, H. Denz, W. Hilbe, H. Jamnig, M. Hackl, A. Dzien and A. Amann, J. Breath Res., 2012, 6, 036008 CrossRef CAS PubMed.
- A. Van Gossum and J. Decuyper, Eur. Respir. J., 1989, 2, 787–791 CAS.
- K. Dettmer and W. Engewald, Anal. Bioanal. Chem., 2002, 373, 490–500 CrossRef CAS PubMed.
- C. L. Jenkins and H. D. Bean, Metabolites, 2020, 10, 347 CrossRef CAS PubMed.
- J. Boehnlein, A. Sakr, J. L. Lichtin and R. L. Bronaugh, Pharm. Res., 1994, 11, 1155–1159 CrossRef CAS PubMed.
- A. Girod, R. Ramotowski and C. Weyermann, Forensic Sci. Int., 2012, 223, 10–24 CrossRef CAS PubMed.
- B. de Lacy Costello, A. Amann, H. Al-Kateb, C. Flynn, W. Filipiak, T. Khalid, D. Osborne and N. M. Ratcliffe, J. Breath Res., 2014, 8, 014001 CrossRef CAS PubMed.
- E. Fredrich, H. Barzantny, I. Brune and A. Tauch, Trends Microbiol., 2013, 21, 305–312 CrossRef CAS PubMed.
- L. Dormont, J.-M. Bessière and A. Cohuet, J. Chem. Ecol., 2013, 39, 569–578 CrossRef CAS PubMed.
- S. U. Morath, R. Hung and J. W. Bennett, Fungal Biol. Rev., 2012, 26, 73–83 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2an01227h |
| This journal is © The Royal Society of Chemistry 2022 |