Open Access Article
Open Access ArticleRational design of a dual-reactive probe for imaging the biogenesis of both H2S and GSH from L-Cys rather than D-Cys in live cells†
Haishun
Ye
a,
Longhuai
Cheng
 b,
Xiaoqiang
Tu
a,
Da-Wei
Wang
*b and
Long
Yi
b,
Xiaoqiang
Tu
a,
Da-Wei
Wang
*b and
Long
Yi
 *a
*a
aState Key Laboratory of Organic-Inorganic Composites and Beijing Key Lab of Bioprocess, Beijing University of Chemical Technology (BUCT), Beijing 100029, China. E-mail: yilong@mail.buct.edu.cn
bState Key Laboratory of Elemento-Organic Chemistry and Department of Chemical Biology, College of Chemistry, National Pesticide Engineering Research Center, Nankai University, Tianjin, 300071, China. E-mail: wangdw@nankai.edu.cn
First published on 17th May 2022
Abstract
Biothiols and their interconversion are involved in cellular redox homeostasis as well as many physiological processes. Here, a dual-reactive dual-quenching fluorescent probe was rationally developed based on thiolysis reactions of 7-nitrobenzoxadiazole (NBD) tertiary amine and 7-cyanobenzoxadiazole (CBD) arylether for imaging of the biothiol interconversion. We demonstrate that the NBD-CBD probe exhibits very weak background fluorescence due to the dual-quenching effects, and can be dual-activated by H2S and GSH with an over 500-fold fluorescence increase at 525 nm. In addition, the probe shows high sensitivity, excellent selectivity, and good biocompatibility, all of which promote the simultaneous detection of both H2S and GSH in live cells. Importantly, probe 1 was successfully employed to reveal the biogenesis of both H2S and GSH from L-Cys rather than from D-Cys, and therefore, D-Cys would be solely converted into H2S, which may help understand the more H2S generation from D-Cys than from L-Cys in live cells.
Introduction
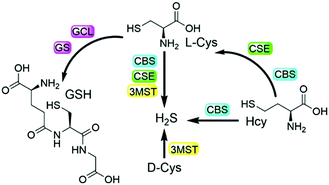
Biothiols include small molecules of glutathione (GSH), cysteine (Cys), homocysteine (Hcy), and hydrogen sulfide (H2S), all of which play important roles in many physiological and pathological processes.1–7 For example, biological H2S is considered to be a nitric oxide (NO)-like signaling molecule that is involved in the regulation of angiogenesis, vasodilation, metabolic bioenergetics, etc.2 GSH is the most abundant biothiol in cells (1–10 mM), which majorly contributes to maintaining redox homeostasis and defending against toxins.3 As shown in Scheme 1, endogenous H2S can be enzymatically produced from Cys by cystathionine β-synthase (CBS), cystathionine γ-lyase (CSE), and 3-mercaptopyruvate sulfurtransferase (3MST)/cysteine aminotransferase (CAT),4a,b while GSH can be enzymatically generated from L-Cys by the consecutive actions of glutamate cysteine ligase (GCL) and GSH synthetase (GS).4c,d On the other hand, Hcy can be enzymatically transformed to generate cystathionine and then to Cys, as well as to H2S.4e Based on the metabolism and biotransformation of these pathways, the concentrations of biothiols are in dynamic equilibrium in live systems. In addition, abnormal levels of one or more different biothiols are closely related to many diseases including Alzheimer's disease, liver cirrhosis, renal/cardiovascular diseases, and cancers.5,6 Moreover, protein post-translational modifications (PTMs) are also involved in interconversion of biothiols. For example, the S-glutathionylation of CBS under oxidative stress and GSH can promote the generation of endogenous H2S.7 Therefore, the development of facile and reliable tools to study the interconversion and inherent crosstalk of these biothiols is of great value for related fundamental research.Fluorescence-based methods are highly suitable and sensitive for in situ and real-time visualization of biomolecules. In the past decade, numerous fluorescent probes have been developed for the detection of biothiols in live systems.8 In addition, probes with multiple reaction sites have been rationally designed for distinguishing different biothiols from each other with different emission channels.9 It is noted that a strategy of using multi-reactive multi-quenching probes has been demonstrated to be useful for highly sensitive and selective detection of biothiols and other analytes.10,11 Attachment of multi-reactive quenchers to the same fluorophore results in low-background fluorescence of the probe due to the multi-quenching fashions through multiplication effects,11 which can be used to enhance the sensitivity and selectivity of the detection in complex biological environments. We as well as others further employed such a multi-quenching strategy to develop probes that require multiple activations from different analytes,12 which can investigate the cooperative relationship of the analytes and differentiate cancer cells. Herein, we report the rational design and preparation of a dual-quenching probe for imaging the biogenesis of both H2S and GSH from L-Cys rather than D-Cys in live cells.
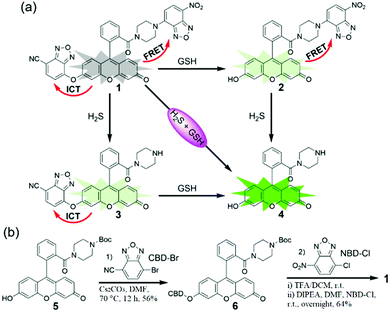
As shown in Scheme 2, we combined the NBD and CBD moieties into a fluorescein to access the dual-quenching probe 1. The thiolysis reactions of NBD amine10 and CBD arylether13 are specific sensing motifs for H2S and GSH, respectively. The CBD moiety can quench the fluorescence through the intramolecular charge transfer (ICT) effect,13 while the NBD amine moiety can switch off the fluorescence of the fluorescein through the fluorescence resonance energy transfer (FRET) effect.14 As a result, probe 1 is highly quenched from the dual-quenching effects, and dual activations of this probe should result in significant fluorescence enhancement (Scheme 2a).
Probe 1 was prepared by a facile three-step synthesis from commercially available reagents (Scheme 2b). The coupling of fluorescein and boc-piperazine provided compound 5, which was modified by CBD–Br to generate compound 6. After deprotection of the boc group and further coupling with NBD–Cl, probe 1 was obtained in good yield. Structural identification was confirmed by 1H NMR, 13C NMR, and HRMS (see ESI†).
With the probe in hand, we investigated the absorbance spectra of probe 1 in the presence of H2S and/or GSH in PBS buffer (50 mM, pH 7.4). The concentration-dependent absorbance of probe 1 in 0.5% DMSO-containing PBS suggested the good water solubility of the probe up to 80 μM (Fig. S1, ESI†). After treatment with both H2S (using Na2S as an equivalent) and GSH, a sharp absorbance peak appeared at 500 nm, which is assigned to the fluorescein product 4 (Fig. S2a, ESI†). When H2S alone was added, a new peak with low absorbance at 500 nm was also observed (Fig. S2b, ESI†), and there was an obvious overlap between the absorbance profile of NBD and the emission profile of 4 (Fig. S2c, ESI†), both of which support the existence of an intramolecular FRET effect in 2. When 1 was treated by GSH alone, the absorption peak was obviously red-shifted from 470 nm to 500 nm, implying an ICT sensing mechanism (Fig. S2d, ESI†).
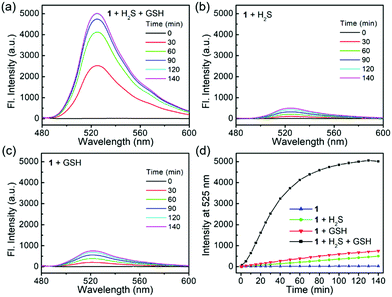
Probe 1 is essentially non-fluorescent (Φ1 = 0.18%) due to the FRET-ICT dual-quenching effects. After dual activations with H2S (100 μM) and GSH (2 mM), the fluorescence of the probe increased significantly, with over 500-fold turn-on at 525 nm (Fig. 1a) and generating high bright 4 (Φ4 = 70.6%). When 1 was treated by H2S or GSH alone, only slight fluorescence enhancements were observed (Fig. 1b and c), and the by-products NBD–SH and CBD–SG were non-fluorescent.8o,13 Time-dependent emissions at 525 nm suggested that probe 1 was stable in the absence of analytes and had obviously larger signal enhancement for H2S + GSH than that of any single analyte (Fig. 1d). In addition, the data (Table S1, ESI†) suggest that the turn-on folds of the dual-reactive probe could be approximate multiplications of each turn-on fold of both single-reactive probes,11 highlighting the advantage of the dual-reactive dual-quenching method.
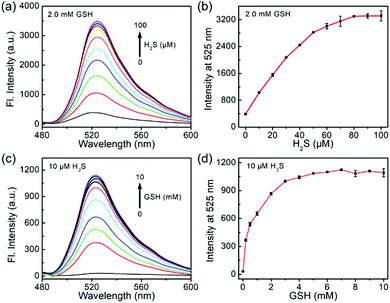
To study the sensitivity of the dual-reactive probe, we incubated 1 with different levels of H2S or GSH for 1 h in the presence of a certain concentration of another activator, after which the emission profiles were measured (Fig. 2). When probe 1 was treated with different concentrations of H2S (0–100 μM) in the presence of GSH (2 mM), the emission at 525 nm was linearly enhanced to the concentrations of H2S from 0 to 60 μM (Fig. 2a and b). Similarly, when 1 was treated with various levels of GSH (0–10 mM) in the presence of H2S (10 μM), we observed a fluorescence enhancement in a concentration-dependent fashion from 0–4 mM (Fig. 2c and d). Taken together, probe 1 can be used to detect the co-existence of H2S and GSH.
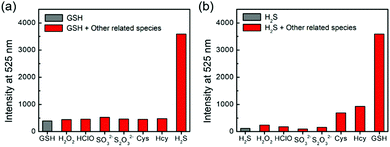
One major requirement for a fluorescent probe is that it should exhibit a selective response toward the targeted analytes but not for other competing biological species. To this end, probe 1 was incubated with different reactive sulfur species (SO32− and S2O32−), other biothiols (Cys and Hcy) and reactive oxygen species (H2O2 and HOCl) in the presence of H2S and/or GSH for 1 h. As shown in Fig. 3, the co-incubation of GSH or H2S and small-molecules triggered a slight fluorescence increase. Therefore, probe 1 could be suitable for sensing the dual activations from H2S and GSH in the presence of other biological analytes.
We also validated the sensing reactions of 1 by HRMS (Fig. S3, ESI†). Product 4 from the dual-activated reactions was observed as [M + H]+ 401.1490 (calcd for C24H21N2O4+, 401.1496). The H2S-triggered product 3 and GSH-triggered product 2 were also observed as [M + H]+ 544.1621 (calcd for C31H22N5O5+, 544.1615) and [M + H]+ 564.1513 (calcd for C30H22N5O7+, 564.1514), respectively. We further used HPLC to monitor and validate the reaction mechanism (Fig. S4, ESI†). These results confirmed probe 1 for the dual-reactive detection of H2S and GSH.
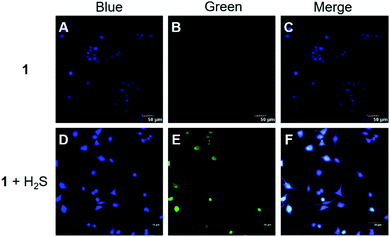
Encouraged by the above results, fluorescence imaging experiments were carried out to evaluate the potential of probe 1 for imaging in live HeLa cells (human cervical cancer cells). We first evaluated the cytotoxicity of 1 by the MTT assay. The results (Fig. S5, ESI†) suggested that probe 1 had a slight effect on cells after 24 h of incubation, and the cell viability at 50 μM of probe 1 was still above 85%. HeLa cells contain nearly no endogenous H2S15 but a moderate level of endogenous GSH (5.4 mM).16 As expected, HeLa cells treated with only probe 1 showed weak fluorescence, and when the cells were pre-incubated with 100 μM Na2S and then with probe 1, obvious green fluorescence was observed (Fig. 4). Therefore, the co-existence of intracellular H2S and GSH can trigger the fluorescent turn-on response of probe 1, which is consistent with the results in buffers.
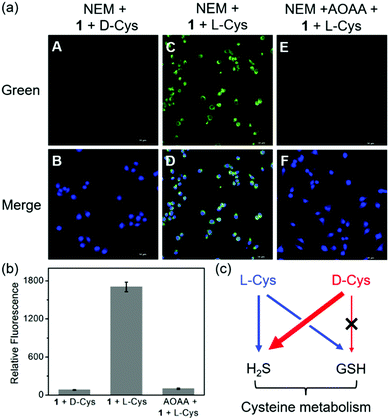
In addition to the H2S biosynthesis, L-Cys is also a substrate for GSH synthesis.4dD-Cys, which should be not a substrate for GSH due to the different chirality, can be metabolized to generate H2S.4b On the other hand, our previous work indicated the imaging of more H2S production from D-Cys than that from L-Cys.17 We envisioned that metabolic differences of D-Cys and L-Cys in live cells may be validated by our dual-reactive probe 1 from live-cell imaging. To this end, we removed the endogenous GSH by the thiol blocking reagent N-ethylmaleimide (NEM) of HeLa cells and then checked the intracellular fluorescence in the presence of chiral Cys using probe 1. As shown in Fig. 5a, the green fluorescence was hardly detected in the D-Cys-treated cells, while strong fluorescence was observed in L-Cys-treated cells. In a control, we employed aminooxyacetic acid (AOAA, 200 μM), an inhibitor of enzymatic H2S synthesis,2b to show the reduced green fluorescence from L-Cys-treated cells (Fig. 5b). These results suggest that D-Cys should be mainly metabolized into H2S, whereas L-Cys could be expressed as both H2S and GSH (Fig. 5c), resulting in dual activations of the probe.
Conclusions
In summary, we rationally designed and synthesized the first dual-reactive fluorescent probe based on NBD amine and CBD arylether for dual-activated detection from H2S + GSH, which is also the first example of the combination between CBD arylether and another sensing motif. Probe 1 shows very weak background fluorescence due to the dual-quenching effects, and can be dual-activated by the co-existence of H2S and GSH with a 500-fold fluorescence increase at 525 nm. Moreover, probe 1 exhibits high sensitivity, excellent selectivity, and good biocompatibility, all of which enable us to detect the intracellular H2S and GSH. Based on this probe tool, we demonstrate the biogenesis of both H2S and GSH from L-Cys rather than from D-Cys in live cells. These metabolic differences of chiral Cys help understand that the treatment of D-Cys has been shown to elevate the H2S level more effectively than L-Cys in live cells.17 Moreover, we believe that such multi-reactive multi-quenching probes are useful for investigating the crosstalk and relationship of other biological molecules in the future.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Prof. Zhen Xi at Nankai University for kind support. We acknowledge the support of NSFC (22177010).Notes and references
- (a) G. Yang, L. Wu, B. Jiang, W. Yang, J. Qi, K. Cao, Q. Meng, A. K. Mustafa, W. Mu, S. Zhang, S. H. Snyder and R. Wang, Science, 2008, 332, 587 CrossRef PubMed; (b) X. Cao, L. Ding, Z. Xie, Y. Yang, M. Whiteman, P. K. Moore and J.-S. Bian, Antioxid. Redox Signaling, 2019, 31, 1–38 CrossRef CAS PubMed; (c) M. R. Filipovic, J. Zivanovic, B. Alvarez and R. Banerjee, Chem. Rev., 2018, 118, 1253–1337 CrossRef CAS PubMed; (d) M. Trujillo, B. Alvarez and R. Radi, Free Radical Res., 2016, 50, 150–171 CrossRef CAS PubMed; (e) F. Rabe von Pappenheim, M. Wensien, J. Ye, J. Uranga, I. Irisarri, J. de Vries, L.-M. Funk, R. A. Mata and K. Tittmann, Nat. Chem. Biol., 2022, 18, 368–375 CrossRef CAS PubMed; (f) P. Bora, S. Manna, M. A. Nair, R. R.-M. Sathe, S. Singh, V. S. Sreyas Adury, K. Gupta, A. Mukherjee, D. K. Saini, S. S. Kamat, A. B. Hazra and H. Chakrapani, Chem. Sci., 2021, 12, 12939–12949 RSC; (g) C. M. Levinn, M. M. Cerda and M. D. Pluth, Acc. Chem. Res., 2019, 52, 2723–2731 CrossRef CAS PubMed; (h) X. Ni, S. S. Kelly, S. Xu and M. Xian, Acc. Chem. Res., 2021, 54, 3968–3978 CrossRef CAS PubMed.
- (a) R. Wang, Physiol. Rev., 2012, 92, 791–896 CrossRef CAS PubMed; (b) C. Szabo, Biochem. Pharmacol., 2018, 149, 5–19 CrossRef CAS PubMed; (c) Y. Liu, M. Lu, L. Hu, P. T. Wong, G. D. Webb and J. Bian, Antioxid. Redox Signaling, 2012, 17, 141–185 CrossRef CAS PubMed; (d) K. Modis, Y. Ju, A. Ahmad, A. A. Untereiner, Z. Altaany, L. Wu, C. Szabo and R. Wang, Pharmacol. Res., 2016, 113, 116–124 CrossRef CAS PubMed; (e) V. S. Khodade, S. C. Aggarwal, B. M. Pharoah, N. Paolocci and J. P. Toscano, Chem. Sci., 2021, 12, 8252–8259 RSC.
- (a) K. Yang, F. Cao, L. Tao, Y. Zhu and Y. Xue, Front. Physiol., 2022, 13, 840293 CrossRef PubMed; (b) T. Liu, L. Sun, Y. Zhang, Y. Wang and J. Zheng, J. Biochem. Mol. Toxicol., 2022, 36, e22942 CAS; (c) R. A. Cairns, I. S. Harris and T. W. Mak, Nat. Rev. Cancer, 2011, 11, 85–95 CrossRef CAS PubMed; (d) C. Hwang, A. J. Sinskey and H. F. Lodish, Science, 1992, 257, 1496–1502 CrossRef CAS PubMed.
- (a) H. Kimura, Amino Acids, 2011, 41, 113 CrossRef CAS PubMed; (b) N. Shibuya, S. Koike, M. Tanaka, M. Ishigami-Yuasa, Y. Kimura, Y. Ogasawara, K. Fukui, N. Nagahara and H. Kimura, Nat. Commun., 2013, 4, 1366 CrossRef PubMed; (c) B. D. Paul, J. I. Sbodio and S. H. Snyder, Trends Pharmacol. Sci., 2018, 39, 513–524 CrossRef CAS PubMed; (d) S. C. Lu, Biochim. Biophys. Acta, Gen. Subj., 2013, 1830, 3143–3153 CrossRef CAS PubMed; (e) M. H. Stipanuk, Annu. Rev. Nutr., 2004, 24, 539–577 CrossRef CAS PubMed.
- (a) C. Szabo, Nat. Rev. Drug Discovery, 2016, 15, 185–203 CrossRef CAS PubMed; (b) C. Szabo and A. Papapetropoulos, Pharmacol. Rev., 2017, 69, 497–564 CrossRef CAS PubMed; (c) J. Dorszewska, M. Prendecki, A. Oczkowska, M. Dezor and W. Kozubski, Curr. Alzheimer Res., 2016, 13, 952–963 CrossRef CAS PubMed; (d) L. Kennedy, J. K. Sandhu, M. Harper and M. Cuperlovic-Culf, Biomolecules, 2020, 10, 1429 CrossRef CAS PubMed; (e) C. Coletta, K. Modis, B. Szczesny, A. Brunyanszki, G. Olah, E. C.-S. Rios, K. Yanagi, A. Ahmad, A. Papapetropoulos and C. Szabo, Mol. Med., 2015, 21, 1–14 CrossRef CAS PubMed.
- (a) S. Seshadri, A. Beiser, J. Selhub, P. F. Jacques, I. H. Rosenberg, R. B. D'Agostino, P. W. Wilson and P. A. Wolf, N. Engl. J. Med., 2002, 346, 476–483 CrossRef CAS PubMed; (b) Y. Chen, H. Dong, D. C. Thompson, H. G. Shertzer, D. W. Nebert and V. Vasiliou, Food Chem. Toxicol., 2013, 60, 38–44 CrossRef CAS PubMed; (c) Y. Xiong, C. Xiao, Z. Li and X. Yang, Chem. Soc. Rev., 2021, 50, 6013–6041 RSC; (d) O. Karmin and Y. L. Siow, Curr. Med. Chem., 2018, 25, 367–377 CrossRef CAS PubMed.
- W.-N. Niu, P. K. Yadav, J. Adamec and R. Banerjee, Antioxid. Redox Signaling, 2015, 22, 350–361 CrossRef CAS PubMed.
- (a) X. Chen, Y. Zhou, X. Peng and J. Yoon, Chem. Soc. Rev., 2010, 39, 2120–2135 RSC; (b) L.-Y. Niu, Y.-Z. Chen, H.-R. Zheng, L.-Z. Wu, C.-H. Tung and Q.-Z. Yang, Chem. Soc. Rev., 2015, 44, 6143–6160 RSC; (c) V. S. Lin, W. Chen, M. Xian and C. J. Chang, Chem. Soc. Rev., 2015, 44, 4596–4618 RSC; (d) Y. Yue, F. Huo and C. Yin, Chem. Sci., 2021, 12, 1220–1226 RSC; (e) Z. Xu, X. Huang, X. Han, D. Wu, B. Zhang, Y. Tan, M. Cao, S.-H. Liu, J. Yin and J. Yoon, Chem, 2018, 4, 1609–1628 CrossRef CAS; (f) I. Ismail, Z. Chen, L. Sun, X. Ji, H. Ye, X. Kang, H. Huang, H. Song, S. G. Bolton, Z. Xi, M. D. Pluth and L. Yi, Chem. Sci., 2020, 11, 7823–7828 RSC; (g) H. Z. Liu, W. T. Song, S. R. Zhang, K. S. Chan, Z. J. Guo and Z. Shen, Chem. Sci., 2020, 11, 8495–8501 RSC; (h) X. Tian, L. C. Murfin, L. Wu, S. E. Lewis and T. D. James, Chem. Sci., 2021, 12, 3406–3426 RSC; (i) R. Zhang, J. Yong, J. Yuan and Z. P. Xu, Coord. Chem. Rev., 2020, 408, 213182 CrossRef CAS; (j) Z. Liu, W. Zhou, J. Li, H. Zhang, X. Dai, Y. Liu and Y. Liu, Chem. Sci., 2020, 11, 4791–4800 RSC; (k) M. Yang, J. Fan, J. Du and X. Peng, Chem. Sci., 2020, 11, 5127–5141 RSC; (l) X. Wang, J. Zha, W. Zhang, W. Zhang and B. Tang, Analyst, 2020, 145, 6119–6124 RSC; (m) M. D. Pluth, Y. Zhao and M. M. Cerda, Methods Enzymol., 2020, 641, 149–164 CAS; (n) A. P.-A. Dos Santos, J. K. da Silva, J. M. Neri, A. C.-O. Neves, D. F. de Lima and F. G. Menezes, Org. Biomol. Chem., 2020, 18, 9398–9427 RSC; (o) C. Jiang, H. Huang, X. Kang, L. Yang, Z. Xi, H. Sun, M. D. Pluth and L. Yi, Chem. Soc. Rev., 2021, 50, 7436–7495 RSC; (p) J. M. An, S. Kang, E. Huh, Y. Kim, D. Lee, H. Jo, J. F. Joung, V. J. Kim, J. Y. Lee, Y. S. Dho, Y. Jung, J. K. Hur, C. Park, J. Jung, Y. Huh, J.-L. Ku, S. Kim, T. Chowdhury, S. Park, J. S. Kang, M. S. Oh, C.-K. Park and D. Kim, Chem. Sci., 2020, 11, 5658–5668 RSC; (q) H. Zong, J. Peng, X.-R. Li, M. Liu, Y. Hu, J. Li, Y. Zang, X. Li and T. D. James, Chem. Commun., 2020, 56, 515–518 RSC; (r) Y. Zou, M. Li, Y. Xin, T. Duan, X. Zhou and F. Yu, ACS Sens., 2020, 5, 242–249 CrossRef CAS PubMed; (s) X. Zhang, W. Qu, H. Liu, Y. Ma, L. Wang, Q. Sun and F. Yu, Anal. Chim. Acta, 2020, 1109, 37–43 CrossRef CAS PubMed.
- (a) J. Liu, Y.-Q. Sun, Y. Huo, H. Zhang, L. Wang, P. Zhang, D. Song, Y. Shi and W. Guo, J. Am. Chem. Soc., 2014, 136, 574–577 CrossRef CAS PubMed; (b) H. Zhang, R. Liu, J. Liu, L. Li, P. Wang, S. Q. Yao, Z. Xu and H. Sun, Chem. Sci., 2016, 7, 256–260 RSC; (c) C.-X. Yin, K.-M. Xiong, F.-J. Huo, J. C. Salamanca and R. M. Strongin, Angew. Chem., Int. Ed., 2017, 56, 13188–13198 CrossRef CAS PubMed; (d) Z. Xu, T. Qin, X. Zhou, L. Wang and B. Liu, Trends Anal. Chem., 2019, 121, 115672 CrossRef CAS; (e) D. Qiao, T. Shen, M. Zhu, X. Liang, L. Zhang, Z. Yin, B. Wang and L. Shang, Chem. Commun., 2018, 54, 13252–13255 RSC; (f) J. Li, C. Yin, Y. Zhang, Y. Yue, J. Chao and F. Huo, Dyes Pigm., 2020, 172, 107826 CrossRef CAS; (g) Z. Chai, Q. Wu, K. Cheng, X. Liu, L. Jiang, M. Liu and C. Li, Chem. Sci., 2021, 12, 1095–1100 RSC; (h) H. Huang, X. Ji, Y. Jiang, C. Zhang, X. Kang, J. Zhu, L. Sun and L. Yi, Org. Biomol. Chem., 2020, 18, 4004–4008 RSC; (i) J. Liu, M. Liu, H. Zhang, X. Wei, J. Wang, M. Xian and W. Guo, Chem. Sci., 2019, 10, 10065–10071 RSC; (j) J. Zhang, Y. Zhang, Q. Guo, G. Wen, H. Xiao, S. Qi, Y. Wang, H. Zhang, L. Wang and H. Sun, ACS Sens., 2022, 7, 1105–1112 CrossRef CAS PubMed.
- (a) H. Zhang, C. Zhang, R. Liu, L. Yi and H. Sun, Chem. Commun., 2015, 51, 2029–2032 RSC; (b) C. Zhang, L. Wei, C. Wei, J. Zhang, R. Wang, Z. Xi and L. Yi, Chem. Commun., 2015, 51, 7505–7508 RSC; (c) G. Zheng, Z. Li, Q. Duan, K. Cheng, Y. He, S. Huang, H. Zhang, Y. Jiang, Y. Jia and H. Sun, Sens. Actuators, B, 2020, 310, 127890 CrossRef CAS; (d) H. Tong, J. Zhao, X. Li, Y. Zhang, S. Ma, K. Lou and W. Wang, Chem. Commun., 2017, 53, 3583–3586 RSC; (e) F.-F. Wang, Y.-J. Liu, B.-B. Wang, L.-X. Gao, F.-L. Jiang and Y. Liu, Dyes Pigm., 2018, 152, 29–35 CrossRef CAS; (f) B.-J. Wang, R.-J. Liu, J. Fang, Y.-W. Wang and Y. Peng, Chem. Commun., 2019, 55, 11762–11765 RSC; (g) Y. Jiang, G. Zheng, Q. Duan, L. Yang, J. Zhang, H. Zhang, J. He, H. Sun and D. Ho, Chem. Commun., 2018, 54, 7967–7970 RSC.
- C. Zhang, R. Wang, L. Cheng, B. Li, Z. Xi and L. Yi, Sci. Rep., 2016, 6, 30148 CrossRef CAS PubMed.
- (a) C. Zhang, Q.-Z. Zhang, K. Zhang, L.-Y. Li, M. D. Pluth, L. Yi and Z. Xi, Chem. Sci., 2019, 10, 1945–1952 RSC; (b) C. Liu, R. Zhang, W. Zhang, J. Liu, Y. L. Wang, Z. Du, B. Song, Z. P. Xu and J. Yuan, J. Am. Chem. Soc., 2019, 141, 8462–8472 CrossRef CAS PubMed; (c) C. Du, S. Fu, X. Wang, A. C. Sedgwick, W. Zhen, M. Li, X. Li, J. Zhou, Z. Wang, H. Wang and J. L. Sessler, Chem. Sci., 2019, 10, 5699–5704 RSC; (d) J. Xie, R. Mu, M. Fang, Y. Cheng, F. Senchyna, A. Moreno, N. Banaeibcd and J. Rao, Chem. Sci., 2021, 12, 9153–9161 RSC.
- X. Tu, L. He, H. Huang, H. Ye, L. Sun and L. Yi, Chem. Commun., 2021, 57, 8802–8805 RSC.
- (a) R. Wang, Z. Li, C. Zhang, Y. Li, G. Xu, Q.-Z. Zhang, L.-Y. Li, L. Yi and Z. Xi, ChemBioChem, 2016, 17, 962–968 CrossRef CAS PubMed; (b) R. Chen, H. Ye, T. Fang, S. Liu, L. Yi and L. Cheng, Org. Biomol. Chem., 2022, 20 10.1039/D2OB00442A.
- Y. Zhao, A. K. Steiger and M. D. Pluth, J. Am. Chem. Soc., 2019, 141, 13610–13618 CrossRef CAS PubMed.
- (a) X. Jiang, Y. Yu, J. Chen, M. Zhao, H. Chen, X. Song, A. J. Matzuk, S. L. Carroll, X. Tan, A. Sizovs, N. Cheng, M. Wang and J. Wang, ACS Chem. Biol., 2015, 10, 864–874 CrossRef CAS PubMed; (b) Z. Liu, X. Zhou, Y. Miao, Y. Hu, N. Kwon, X. Wu and J. Yoon, Angew. Chem., Int. Ed., 2017, 56, 5812–5816 CrossRef CAS PubMed.
- (a) L. Wei, L. Yi, F. B. Song, C. Wei, B. F. Wang and Z. Xi, Sci. Rep., 2014, 4, 4521 CrossRef; (b) L. Wei, Z. Zhu, Y. Li, L. Yi and Z. Xi, Chem. Commun., 2015, 51, 10463–10466 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details and additional figures. See DOI: https://doi.org/10.1039/d2cb00105e |
| This journal is © The Royal Society of Chemistry 2022 |