Open Access Article
Open Access ArticleControlling enzyme reactions by supramolecular protection and deprotection of oligosaccharide substrates†
Milad
Zangiabadi
and
Yan
Zhao
 *
*
Department of Chemistry, Iowa State University, Ames, Iowa 50011-3111, USA. E-mail: zhaoy@iastate.edu; Tel: +1-515-294-5845
First published on 9th August 2022
Abstract
Protection/deprotection is a powerful strategy in the total synthesis of complex organic molecules but similar tools are nearly absent in enzymatic reactions. We here report supramolecular protective receptors that outcompete an enzyme in the binding of oligosaccharides. The strong binding inhibits the enzymatic reaction and addition of an even stronger ligand for the receptor releases the substrate. These receptors could be used to control products from the same substrate/enzyme mixture and regulate enzymatic reactions reversibly.
Protecting groups are indispensable tools in modern organic synthesis.1 Although the need for selective protection seems low in enzymatic reactions due to the extraordinary substrate selectivity of enzymes, enzymatic selectivity has its own limitations. Exoglycosidases, for example, hydrolyze glycans typically one residue at a time from the nonreducing end.2 If one wants to cleave two or three residues at a time, different enzymes would be needed if they exist at all. One way to control enzyme reactions is through compartmentation, which can improve efficiency of cascade reactions of a multienzyme system and limit cross reactivities.3,4 To control a single-enzyme reaction, we postulated that a supramolecular protective receptor would be sufficient.
Our model substrate for the proposed protection/deprotection is a malto-oligosaccharide and maltase cleaves one glucose residue at a time from its nonreducing end.5 Supramolecular protectors for peptides have been reported6–13 but tools for glycans are difficult to obtain due to their much weaker binding.14,15
Our glycan-protecting receptors were prepared via molecular imprinting,16–18 which has been used to produce different types of glycan-binding materials.19–27 To shield part of a glycan while having the rest accessible to the enzyme, however, the receptor must be nanosized and water-soluble, making traditional imprinting methods unsuitable. Our general method (Scheme 1) starts with solubilization of a suitable template in the mixed micelle of cross-linkable surfactants 1 and 2, together with divinyl cross-linkers and a hydrophobic radical initiator (2,2-dimethoxy-2-phenylacetophenone or DMPA).28 With a high local concentration of terminal alkyne and azide, the micellar surface is readily cross-linked by the highly efficient click reaction. UV-irradiation then cross-links the micellar core by free radical polymerization, around the template to form a template-complementary imprinted site. The doubly cross-linked micelle is further functionalized on the surface by click reaction with monoazide 3, to enhance its hydrophilicity and facilitate purification of the resulting molecularly imprinted nanoparticles (MINPs) by simple solvent washing.
Glycans are strongly solvated in water and thus will not enter a micelle. To help the micelle “catch” the glycan, we converted maltose into neoglycoconjugates 4a–d as the templates. Amphiphilicity of the templates helps their incorporation into the micelles. Removal of the template after imprinting would “release” the template and allow the imprinted receptor to protect/shield the glycan. We reasoned that the template should work well as a “deprotecting” ligand later on, as it is expected to bind to the MINP protector more strongly than the glycan to be protected.
Natural glycan-binding proteins use an extensive array of hydrogen bonds to bind their guests.14 To create such a feature in our receptor, we employed a mixture of divinyl benzene (DVB) and N,N′-methylenebisacrylamide (MBAm) for the free radical cross-linking. The water-insoluble radical initiator (DMPA) resides inside the micelle. During core-cross-linking, the propagating radical is confined in the micelle and thus could polymerize only those MBAm molecules diffused to surface of the micelle.29 Cross-linking would fix those MBAm molecules hydrogen-bonded to the template, creating a complementary binding site (Scheme 1, upper right panel). Although hydrogen bonds are compromised by solvent competition for molecular recognition in water, they are stabilized inside the hydrophobic core of a micelle30 or at the surfactant/water interface.31
Preparation of the MINPs and their characterization are reported in ESI.† The cross-linking was monitored by 1H NMR spectroscopy (Fig. S1, S3 and S5, ESI†). The particle size was determined by dynamic light scattering (DLS, Fig. S2, S4 and S6, ESI†) and confirmed by transmission microscopy (TEM, Fig. S7, ESI†).
Success of the “catch-and-release” strategy is supported by the strong binding of not only template 4a but also free maltose by MINP(4a)—i.e., MINP prepared with 4a as the template (Table 1). The binding constant for maltose (Ka = 8.4 × 104 M−1) compares favorably with the reported value (660 M−1) for hydrogen-bond-based small-molecule receptors32 and is 10–30% of that by maltose-binding periplasmic protein of Escherichia coli.33,34 The binding should be strong enough to outcompete maltase that has a Km value of 13.4 mM for maltose and 6.2 mM for maltotriose.5
| Entry | Template | Guest | K a (×104 M−1) | −ΔG (kcal mol−1) | BRb |
|---|---|---|---|---|---|
| a The titrations were performed in duplicates with the indicated errors in HEPES buffer (10 mM, pH 7.4) at 298 K (Fig. S8 and S10, ESI). b BR is the binding ratio between the template (or surrogate) to free maltose. c The binding constant was estimated from ITC due to the weak binding. | |||||
| 1 | 4a | 4a | 16.6 ± 1.3 | 7.12 | — |
| 2 | 4a | Maltose | 8.4 ± 0.4 | 6.71 | 2.0 |
| 3 | 4b | 4b | 53 ± 3 | 7.80 | — |
| 4 | 4b | Maltose | 7.0 ± 0.3 | 6.61 | 7.5 |
| 5 | 4c (n = 7) | 4c (n = 7) | 32 ± 2 | 7.50 | — |
| 6 | 4c (n = 7) | Maltose | 9.1 ± 0.4 | 6.76 | 3.5 |
| 7 | 4d | 4d | 862 ± 11 | 9.45 | — |
| 8 | 4d | Maltose | 10.3 ± 0.6 | 6.83 | 84 |
| 9 | 4d | 15 | 513 ± 17 | 9.15 | 50 |
| 10 | Maltose | Maltose | ∼0.003c | — | — |
| 11 | None | Maltose | ∼0.002c | — | — |
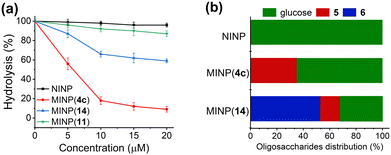
As the acyl group of the template was varied, the MINP receptor bound its own template in the order of 4b > 4c (n = 7) > 4a, thus correlating with the overall hydrophobicity of the template. Meanwhile, MINP(4a–c) exhibited very similar binding for free maltose (Table 1, entries 2, 4, 6). For template 4c containing a linear acyl chain, binding of the resulting MINP peaked at C8 (Fig. 1a). When the acyl chain is too short, the hydrophobic driving force most likely is insufficient for the micelle to “catch” the template. When the chain is too long, it is possible that the glycan gets pushed by the long hydrophobic tail into water while the tail is anchored in the hydrophobic core of the micelle. Since molecular imprinting of the glycan is most effective at the surfactant/water interface (where the majority of MBAm molecules would polymerize), moving the glycan away from this region is expected to be detrimental to the imprinting.
 | ||
| Fig. 1 (a) Dependence of the MINP–maltose binding on the chain length of the acyl group of the template 4c (Fig. S9, ESI†). (b) Relative binding constants (Krel) of different sugars by MINP(4a–c) normalized to that of maltose by the same receptor (see Table S1, ESI† for details). The titrations were performed in duplicates in HEPES buffer (10 mM, pH 7.4) at 298 K (Fig. S11–S13, ESI†). | ||
Poor binding was observed when maltose was used directly as the template (entry 10), indicating the importance of the hydrophobic hydrazide in the “catching” of the glycan for imprinting. When the template was eliminated altogether in the preparation, the resulting nonimprinted nanoparticles (NINPs) showed negligible binding (entry 11). The imprinting factor for maltose, measured by the imprint/nonimprint ratio in Ka, was >3500–4500 for MINP(4a–c).
Among the three receptors, MINP(4c) outperformed the other two in selectivity, evident from its weaker binding of other oligosaccharide guests relative to maltose (Fig. 1b). Among the guests tested (5–10 in Scheme 1), cellobiose (7) showed the highest cross-reactivity (Fig. 1b)—a reasonable result given its structural similarity to maltose. Although the α/β selectivity was modest, the selectivity for chain length (5vs.6), glycosylation site (1,4 in 5vs. 1,6 in 8), and sugar building blocks (5vs.9 or 10) was much stronger. One benefit of using hydrogen bonds for sugar binding is their pH-insensitivity. The binding constant for maltose by MINP(4c) stayed largely unchanged over pH 6.5–9 (Fig. S14 and S15, ESI†). In contrast, binding between phenylboronic acid and its sugar guests is known to change over an order of magnitude from pH 6.5 to 8.5.35
The “catch-and-release” strategy also worked well for other disaccharides, as shown by the molecular imprinting of the lactose-derived template 11 (Table S2, ESI†). Poor results were obtained, however, when maltotriose-derived 12 was the template (Table 2). Normally, as long as good host–guest complementary is produced during imprinting, a larger template should afford a stronger binding (to its own imprinted receptor), due to a larger binding interface. However, the binding of the trisaccharide by MINP(12) was less than half of that of the disaccharide by MINP(4c) under comparable conditions (Ka = 4.47 × 104vs. 9.1 × 104 M−1). This result suggests that the C8 hydrophobic tail was unable to help the micelle “catch” the glycan effectively. A trisaccharide, having a longer sugar chain, can easily extend itself into the aqueous phase even while the C8 acyl chain is anchored in the micellar core. Poor imprinting would result as hypothesized earlier when the glycan moves away from the surfactant/water interface.
| Entry | Guest | MINP(12) | MINP(12) with FM 13 | ||
|---|---|---|---|---|---|
| K a (×104 M−1) | K rel | K a (×104 M−1) | K rel | ||
| a The titrations were performed in duplicates with the indicated errors in HEPES buffer (10 mM, pH 7.4) at 298 K (Fig. S17 and S18, ESI). | |||||
| 1 | Maltotriose | 4.47 ± 0.13 | 1.0 | 33.1 ± 2.4 | 1.0 |
| 2 | Maltose | 0.93 ± 0.04 | 0.21 | 7.25 ± 0.21 | 0.22 |
| 3 | Cellobiose | 0.82 ± 0.02 | 0.18 | 6.41 ± 0.24 | 0.19 |
| 4 | Gentiobiose | 0.95 ± 0.06 | 0.21 | 7.53 ± 0.13 | 0.23 |
| 5 | Lactose | 0.81± 0.01 | 0.18 | 3.44 ± 0.32 | 0.10 |
| 6 | Maltulose | 0.74 ± 0.05 | 0.16 | 5.61 ± 0.25 | 0.17 |
To better “catch” the trisaccharide, we included boroxole 13 as the functional monomer (FM), which forms anionic boronate 14in situ,36–38 stabilized by the cationic micelle.28 With hydrophobic groups at both ends, the amphiphilic template–FM complex should easily migrate into a micelle to be imprinted. MINP(12) prepared with the boroxole FM, indeed, displayed much stronger binding for maltotriose (Table 2), with a binding constant (Ka = 33.1 × 104 M−1) ∼1/3 of that by maltotriose-binding protein of Thermus thermophilus.39 Importantly, the binding selectivity (Krel) was maintained while the binding affinity for maltotriose increased.
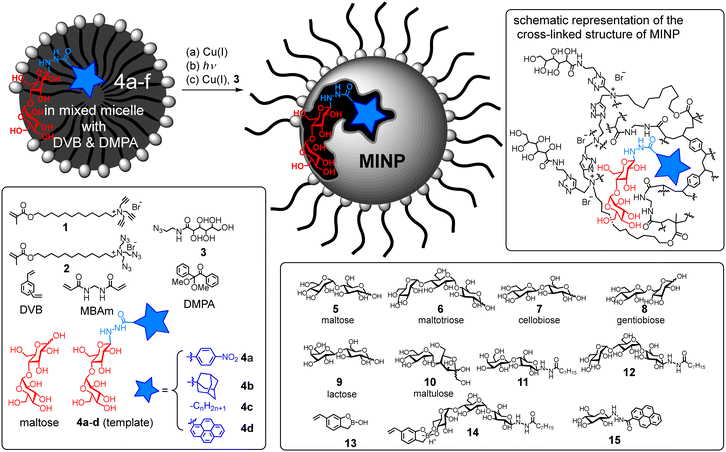
The strong binding of our MINP receptors allowed them to be used as protective agents for their targeted glycans. Fig. 2a shows that maltase hydrolyzed maltose completely in 20 min in the presence of NINPs. The hydrolysis was measured enzymatically by a commercial glucose assay kit (ESI†). In contrast, MINP(14) and, in particular, MINP(4c) inhibited the hydrolysis in a concentration-dependent manner. At a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, the latter was able to suppress the enzymatic hydrolysis to <20%. Not surprisingly, MINP(11) designed for binding lactose gave little protection to maltose.
1 ratio, the latter was able to suppress the enzymatic hydrolysis to <20%. Not surprisingly, MINP(11) designed for binding lactose gave little protection to maltose.
Not only so, these MINP protectors could be used to alter product distribution in an enzymatic reaction. Fig. 2b shows that maltase hydrolyzed maltotetraose completely into glucose after 2 h with NINPs in the solution. In the presence of 2 equivalents of MINP(4c) (n = 7), nearly 40% of the product was maltose. In the presence of MINP(14) designed to shield maltotriose, over 50% of the product was maltotriose. Hence, the MINP protectors were able to shield their targeted glycans and protect them from enzymatic degradation. In this way, different products can be produced with the same substrate and the same enzyme.
The above studies gave us a good understanding of the “catch-and-release” imprinting and yielded strong protectors for both maltose and maltotriose. If we want to deprotect the MINP-bound glycan, however, these MINPs are still lacking, because the template as the deprotecting ligand needs to outcompete the protected glycan in the MINP binding. Yet, the template/glycan binding ratio (BR) was only 2.0–7.5 for MINP(4a–c) (Table 1).
Fortunately, to increase the BR value, all we had to do was to use a more hydrophobic hydrazide, since the free sugar does not have the hydrophobic group. Template 4d, containing a pyrenyl group, afforded a large BR of 84 (Table 1, entry 8). To avoid having the same sugar structure in the deprotecting ligand as in the protected glycan, we synthesized 15, which has the same pyrene-containing hydrazide conjugated to glucose instead of maltose. With the large hydrophobic pyrenyl group, this template surrogate bound to MINP(4d) still much more strongly than maltose, affording a BR of 50 (entry 9).
With a large BR ratio, MINP(4d) could be used to protect/deprotect a glycan reversibly. Fig. 3 shows that 1 equivalent of the protective receptor shielded maltose from maltase very well. Anytime when the stronger-binding deprotecting ligand 15 was added, equivalent amounts of maltose were deprotected/released from the MINP protector and underwent enzymatic hydrolysis. The protection/deprotection could be repeated until all the maltose was consumed in the reaction mixture.
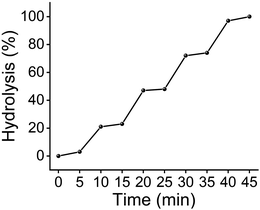 | ||
| Fig. 3 Hydrolytic yields of maltose over time, with 25 μM of deprotecting ligand 15 added at 5, 15, 25, and 35 min. [maltose] = [MINP(4d)] = 100 μM. [maltase] = 10 units per mL. | ||
Enzymatic reactions in cells are regulated in multiple ways including allostery, oligomerization, and compartmentation.40 In this work, we report selective nanoparticle receptors for glycans and demonstrated their abilities to shield the targeted glycans from hydrolytic enzymes. These materials can be used to control product distribution in enzymatic reactions and, in combination with a stronger ligand for the nanoparticles, to modulate enzymatic reactions reversibly.
We thank NSF (DMR2002659) for financial support.
Conflicts of interest
There are no conflicts to declare.Notes and references
- P. G. M. Wuts, T. W. Greene and T. W. Greene, Greene's protective groups in organic synthesis, Wiley, Hoboken, New Jersey, 5th edn, 2014 Search PubMed.
- M. Sinnott, Carbohydrate chemistry and biochemistry: structure and mechanism, RSC Publishing, Cambridge, 2nd edn, 2013, pp. 299–477 Search PubMed.
- A. H. Chen and P. A. Silver, Trends Cell Biol., 2012, 22, 662–670 CrossRef CAS PubMed.
- S. Schmidt, K. Castiglione and R. Kourist, Chem. – Eur. J., 2018, 24, 1755–1768 CrossRef CAS PubMed.
- K. Matsusaka, S. Chiba and T. Shimomura, Agric. Biol. Chem., 1977, 41, 1917–1923 CAS.
- T. Kodadek, Biopolym. Peptide Sci., 2002, 66, 134–140 CrossRef CAS PubMed.
- L. A. Logsdon and A. R. Urbach, J. Am. Chem. Soc., 2013, 135, 11414–11416 CrossRef CAS PubMed.
- E. Faggi, Y. Perez, S. V. Luis and I. Alfonso, Chem. Commun., 2016, 52, 8142–8145 RSC.
- H. Peacock, C. C. Thinnes, A. Kawamura and A. D. Hamilton, Supramol. Chem., 2016, 28, 575–581 CrossRef CAS.
- J. Mosquera, B. Szyszko, S. K. Y. Ho and J. R. Nitschke, Nat. Commun., 2017, 8, 14882 CrossRef CAS PubMed.
- L. Tapia, N. Solozabal, J. Solà, Y. Pérez, W. T. Miller and I. Alfonso, Chem. – Eur. J., 2021, 27, 9542–9549 CrossRef CAS PubMed.
- X. Li, T. M. Palhano Zanela, E. S. Underbakke and Y. Zhao, J. Am. Chem. Soc., 2021, 143, 639–643 CrossRef CAS PubMed.
- X. Li, K. Chen and Y. Zhao, Angew. Chem., Int. Ed., 2021, 60, 11092–11097 CrossRef CAS PubMed.
- B. Wang and G.-J. Boons, Carbohydrate recognition: biological problems, methods, and applications, Wiley, Hoboken, NJ, 2011 Search PubMed.
- N. R. Council, Transforming glycoscience: a roadmap for the future, National Academies Press, Washington, DC, 2012 Search PubMed.
- G. Wulff, Chem. Rev., 2002, 102, 1–28 CrossRef CAS PubMed.
- K. Haupt and K. Mosbach, Chem. Rev., 2000, 100, 2495–2504 CrossRef CAS PubMed.
- L. Ye and K. Mosbach, Chem. Mater., 2008, 20, 859–868 CrossRef CAS.
- G. Wulff and S. Schauhoff, J. Org. Chem., 1991, 56, 395–400 CrossRef CAS.
- G. Wulff, Angew. Chem., Int. Ed. Engl., 1995, 34, 1812–1832 CrossRef CAS.
- S. Wang, D. Yin, W. Wang, X. Shen, J.-J. Zhu, H.-Y. Chen and Z. Liu, Sci. Rep., 2016, 6, 22757 CrossRef CAS PubMed.
- S. Shinde, Z. El-Schich, A. Malakpour, W. Wan, N. Dizeyi, R. Mohammadi, K. Rurack, A. Gjörloff Wingren and B. Sellergren, J. Am. Chem. Soc., 2015, 137, 13908–13912 CrossRef CAS PubMed.
- A. Stephenson-Brown, A. L. Acton, J. A. Preece, J. S. Fossey and P. M. Mendes, Chem. Sci., 2015, 6, 5114–5119 RSC.
- B. Demir, M. M. Lemberger, M. Panagiotopoulou, P. X. Medina Rangel, S. Timur, T. Hirsch, B. Tse Sum Bui, J. Wegener and K. Haupt, ACS Appl. Mater. Interfaces, 2018, 10, 3305–3313 CrossRef CAS PubMed.
- J. Pan, W. Chen, Y. Ma and G. Pan, Chem. Soc. Rev., 2018, 47, 5574–5587 RSC.
- Y. Dong, W. Li, Z. Gu, R. Xing, Y. Ma, Q. Zhang and Z. Liu, Angew. Chem., Int. Ed., 2019, 58, 10621–10625 CrossRef CAS PubMed.
- P. X. Medina Rangel, S. Laclef, J. Xu, M. Panagiotopoulou, J. Kovensky, B. Tse Sum Bui and K. Haupt, Sci. Rep., 2019, 9, 3923 CrossRef PubMed.
- R. W. Gunasekara and Y. Zhao, J. Am. Chem. Soc., 2017, 139, 829–835 CrossRef CAS PubMed.
- M. Zangiabadi and Y. Zhao, Nano Lett., 2020, 20, 5106–5110 CrossRef CAS PubMed.
- J. S. Nowick, J. S. Chen and G. Noronha, J. Am. Chem. Soc., 1993, 115, 7636–7644 CrossRef CAS.
- K. Ariga and T. Kunitake, Acc. Chem. Res., 1998, 31, 371–378 CrossRef CAS.
- P. Stewart, C. M. Renney, T. J. Mooibroek, S. Ferheen and A. P. Davis, Chem. Commun., 2018, 54, 8649–8652 RSC.
- G. Gilardi, L. Q. Zhou, L. Hibbert and A. E. G. Cass, Anal. Chem., 1994, 66, 3840–3847 CrossRef CAS PubMed.
- M.-H. Seo, J. Park, E. Kim, S. Hohng and H.-S. Kim, Nat. Commun., 2014, 5, 3724 CrossRef CAS PubMed.
- J. Yan, G. Springsteen, S. Deeter and B. Wang, Tetrahedron, 2004, 60, 11205–11209 CrossRef CAS.
- M. Dowlut and D. G. Hall, J. Am. Chem. Soc., 2006, 128, 4226–4227 CrossRef CAS PubMed.
- M. Bérubé, M. Dowlut and D. G. Hall, J. Org. Chem., 2008, 73, 6471–6479 CrossRef PubMed.
- H. Kim, Y. J. Kang, S. Kang and K. T. Kim, J. Am. Chem. Soc., 2012, 134, 4030–4033 CrossRef CAS PubMed.
- M. J. Cuneo, A. Changela, L. S. Beese and H. W. Hellinga, J. Mol. Biol., 2009, 389, 157–166 CrossRef CAS PubMed.
- A. Sols, in Current Topics in Cellular Regulation, ed. B. L. Horecker and E. R. Stadtman, Academic Press, 1981, vol. 19, pp. 77–101 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Synthetic procedures, characterization data, ITC titration curves, additional data, and NMR spectra. See DOI: https://doi.org/10.1039/d2cc03239b |
| This journal is © The Royal Society of Chemistry 2022 |