Emerging mRNA technologies: delivery strategies and biomedical applications
Yufen
Xiao†
b,
Zhongmin
Tang†
b,
Xiangang
Huang†
 b,
Wei
Chen
b,
Jun
Zhou
b,
Haijun
Liu
b,
Chuang
Liu
b,
Wei
Chen
b,
Jun
Zhou
b,
Haijun
Liu
b,
Chuang
Liu
 b,
Na
Kong
*ab and
Wei
Tao
b,
Na
Kong
*ab and
Wei
Tao
 *b
*b
aLiangzhu Laboratory, Zhejiang University Medical Center, Hangzhou, Zhejiang 311121, China. E-mail: kongna@zju.edu.cn
bCenter for Nanomedicine and Department of Anesthesiology, Brigham and Women's Hospital, Harvard Medical School, Boston, Massachusetts 02115, USA. E-mail: wtao@bwh.harvard.edu
First published on 19th April 2022
Abstract
The great success achieved by the two highly-effective messenger RNA (mRNA) vaccines during the COVID-19 pandemic highlights the great potential of mRNA technology. Through the evolution of mRNA technology, chemistry has played an important role from mRNA modification to the synthesis of mRNA delivery platforms, which allows various applications of mRNA to be achieved both in vitro and in vivo. In this tutorial review, we provide a summary and discussion on the significant progress of emerging mRNA technologies, as well as the underlying chemical designs and principles. Various nanoparticle (NP)-based delivery strategies including protein-mRNA complex, lipid-based carriers, polymer-based carriers, and hybrid carriers for the efficient delivery of mRNA molecules are presented. Furthermore, typical mRNA delivery platforms for various biomedical applications (e.g., functional protein expression, vaccines, cancer immunotherapy, and genome editing) are highlighted. Finally, our insights into the challenges and future development towards clinical translation of these mRNA technologies are provided.
Key learning points(1) Summary of the latest developments in mRNA technologies.(2) A systematic review of emerging mRNA delivery platforms and their design principles. (3) Discussion of various applications of mRNA therapeutics. (4) Current challenges and future developments for the clinical translation of mRNA therapeutics. |
1. Introduction
Messenger RNA (mRNA) was first discovered in the early 1960s and delivered into cells for protein expression in the 1970s.1 Unlike DNA-based technology for protein expression, mRNA does not infect the original genome, as it does not need to enter the nucleus to fulfill its functions. Therefore, the concept of using various mRNAs for expressing a variety of proteins to tackle different diseases has attracted the attention of the scientific community. During the past decades, although some studies have advanced the development of mRNA technology in terms of mRNA modification and design of delivery platforms, these efforts to enhance mRNA stability and form the basis for the real application majorly remain in academia. Until the outbreak of the global COVID-19 pandemic, the effectiveness of mRNA nanoparticle (NP)-based vaccines in preventing the spread of viruses has finally brought mRNA technology into the public eye from academia,2 and the amount of research on mRNA technology from both academia and industry has also increased greatly.The application of mRNA therapeutics in different diseases usually includes the following processes: choosing the right target,3 designing the corresponding mRNA sequence and modifying it to improve its stability,4 and then using a suitable delivery platform to deliver mRNA to the right place(s) for correct protein expression.5 Obviously, the application of mRNA requires knowledge from the fields of biology, materials science, chemistry, pharmacology, and medicine. While the design and stability of mRNA itself are prerequisites, the efficiency of mRNA delivery platforms is also critical, as that determines the efficacy of mRNA therapeutics in real applications.
At present, mRNA alone can have little effect on the body without the protection and help of delivery platforms. Consequently, various materials have previously been employed for mRNA delivery, including lipids,6 lipid-like materials,7 polymers,8 inorganic materials,9 cell membranes, and hybrid systems.10 Among them, lipids and lipid-like materials containing a polar head group, a hydrophobic chain, and a linker are the most commonly used. They interact with the negatively charged mRNA through electrostatic interactions to ensure the functional integrity of mRNA and facilitate its endosomal escape. The lipid materials used in mRNA delivery can be further categorized into cationic lipids, ionizable lipids, and zwitterionic lipids. Unlike cationic lipids which contain a permanent positively charged polar head group, ionizable lipids can undergo pH-responsive charge changes. They can be neutrally charged under physiological conditions but positively charged under low pH, laying the foundation for efficient endosomal escape.11 Lipid materials used in the two approved COVID-19 mRNA vaccines (SM-102 and ALC-0315) are both ionizable lipids. The scope of application of mRNA now covers not only tumor treatment4,12 (directly inducing the expression of therapeutic proteins or enhancing the immune response), but also virus vaccines13 and gene editing,14etc. Moreover, designing specific ionizable lipids or combining different ionizable lipids could achieve organ-specific delivery of mRNA for the treatment of disease.3
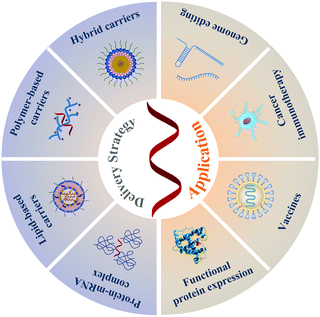
In this tutorial review, emphasizing the chemistry viewpoint, we summarize the significant progress of emerging mRNA technologies in various biomedical applications that are achieved by versatile delivery strategies (Scheme 1). Through examining the history of the development of mRNA technology, we can understand not only the chemical modification of mRNA and the exploration of delivery platforms, but also the clear importance of controlling chemical composition and structure in delivery platforms. Representative applications of mRNA in the field of biomedicine are also presented in detail. Most importantly, we also offer our understanding and thoughts on this field and its future development. We expect that this tutorial review could help highlight the fundamental role of chemistry in the rapid and successful development of mRNA therapeutics, and lessons learned may eventually inspire more mRNA innovations from the fields of materials chemistry and biomedicine.
2. Non-viral delivery strategies
As a single-stranded long polynucleotide with 2000–2500 bases by average, mRNA is subject to rapid degradation by omnipresent extracellular ribonucleases even before entering target cells to perform its transcription functions. Moreover, the cell membrane further hampers the diffusion of mRNA into the cytoplasm by electrostatic repulsion, thus preventing the mRNA transfection. Strategies including viral vectors and non-viral vehicles have therefore been developed to enhance the internalization of mRNA by cells. In this review, we focus on non-viral strategies for mRNA delivery in this section.2.1 Naked mRNA
The delivery of naked mRNA has been utilized in a number of in vivo studies, especially as vaccines encoding specific antigens. The administration of naked mRNA via intradermal, subcutaneous, intramuscular, or intranodal injection can lead to the expression of antigens and the subsequent induction of immune responses. To this end, Kreiter et al. developed naked RNA vaccines that were successfully internalized by dendritic cells (DCs) to express the related antigens, which induced a specific CD8+ T cell response.15However, naked mRNA is far from sufficient for other applications where continuous expression of proteins with high levels is required. Phua et al. studied the transfection efficiency of naked mRNA and mRNA lipid nanoparticles (LNPs) via various administration routes.16In vitro transfection in primary DCs demonstrated that LNPs could efficiently transfect DCs with >50% efficiency, whereas naked mRNA failed in transfection with DCs. For in vivo transfection studies, results varied with the administration routes. Phua et al. found that subcutaneous injection of naked mRNA performed more efficiently than LNPs. However, when intranasally administered, LNPs achieved transfection after 4 h and the signals had decayed by 24 h, while no signals could be detected in the nasal cavity of mice administered naked mRNA. Similar results were also achieved after intravenous (i.v.) injection, where short-lived luciferase expression was observed after transfection with Fluc mRNA-encapsulated LNPs; however, no signals were observed in the naked mRNA group. The failure of naked mRNA might be caused by tissue barriers, whereas LNPs efficiently protected the mRNA from degradation, underlining the vital role of vehicles in mRNA transfection.
2.2 Protein–mRNA complex
Natural positively charged proteins can be complexed with negatively charged mRNA, resulting in the self-assembly of protein–mRNA complex via electrostatic interaction. The positively charged surface further enhances cellular uptake and hence increases transfection efficiency.Protamine is a positively charged protein that is rich in arginine. Fotin-Mleczek et al. reported that protamine-complexed mRNA vaccines strongly enhanced immune stimulation by activating the TLR7 receptor.17 Complexing protamine with mRNA encoding tumor-associated antigens (TAAs) successfully achieved antigen expression and immune stimulation, establishing a strong antitumor immune response in mice. Moreover, Weide et al. performed a clinical trial of protamine-stabilized mRNAs encoding various tumor-related antigens in metastatic melanoma patients.18 After intradermal injection, no obvious toxicity except for inflammation at the injection site was observed. A decrement of regulatory T cells and increment of vaccine-directed T cells were noted in some patients, demonstrating the exciting potential of protamine-mRNA vaccines.
Recently, Segel et al. reported the use of mammalian retrovirus-like protein PEG10 to deliver mRNA.19 The PEG10 bound to and secreted its own mRNA into the virus-like particle (VLP). Using an enhanced cross-linking and immunoprecipitation (eCLIP) assay, they found that PEG10 strongly bound to and packaged a number of mRNAs, including its own mRNA. In addition, PEG10 was able to deliver both single-guide RNA (sgRNA) and Streptococcus pyogenes Cas9 (SpCas9) with 40% efficiency in human embryonic kidney 293FT (HEK293FT) cells, further illustrating the capability of this PEG10-based platform to bind, stabilize, and deliver mRNAs.
2.3 Lipid-based carriers
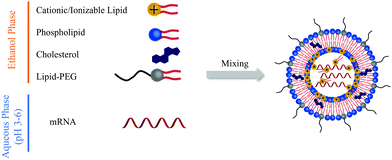
Lipid-based carriers including LNPs and lipoplexes have been widely applied for nucleic acid delivery. With proper engineering, mRNA can be effectively encapsulated into LNPs and lipoplexes, preventing its degradation by ribonucleases and facilitating its cellular internalization and endosomal escape. Cationic or ionizable lipids can be complexed with mRNA; cholesterol can improve stability and promote membrane fusion; poly(ethylene) glycol (PEG)–lipid can control size and stability to avoid aggregation; phospholipids can mediate mRNA encapsulation; and mixture of the above components at precise molar ratios can generate LNPs with all the above-mentioned functionalities for mRNA delivery. As presented in Fig. 1, typically, the mRNA is first dispersed in 10 mM citrate buffer (pH 3–6) as an aqueous phase. Other components are dissolved in ethanol. The two phases are then mixed by springe pumps at a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, resulting in the formulation of mRNA LNPs after dialysis against phosphate-buffered saline (PBS).
1 ratio, resulting in the formulation of mRNA LNPs after dialysis against phosphate-buffered saline (PBS).
The principle of mRNA delivery originates from the siRNAs/miRNAs delivery. However, since mRNA is much larger than siRNAs/miRNAs, the formulation for mRNA delivery is particularly critical and challenging. Under the circumstances, Kauffman et al. studied the effect of weight ratio of ionizable lipid C12-200 to mRNA, the molar ratio of the four components, the phospholipid identity, and different phospholipids structures with differing head groups and degrees of tail saturation on delivery efficiency.11 They found that an increased weight ratio of C12-200 to mRNA from 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 10
1 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 would benefit the transfection. Moreover, the higher C12-200 amount with higher positive charges had a stronger binding affinity with mRNA and thus increased the encapsulation efficiency. However, further increasing the weight ratio beyond 10
1 would benefit the transfection. Moreover, the higher C12-200 amount with higher positive charges had a stronger binding affinity with mRNA and thus increased the encapsulation efficiency. However, further increasing the weight ratio beyond 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 brought no improvement in transfection efficacy. The phospholipid also played a vital role in transfection efficacy; the formulation using 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) exhibited a higher transfection efficacy than 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), which might result from the different phospholipid structure. Specifically, DOPE is a conical lipid that can form a less stable hexagonal phase with the organelle membrane, thus promoting endosomal escape and mRNA release. Whereas DSPC is a cylindrical lipid that tends to form the lamellar phase, which is more structurally stable and blocks mRNA release. On the other hand, increasing the phospholipid content could reduce the LNP size and thus improve the transfection efficacy. Cheng et al. optimized the LNPs by changing the molar ratios of the four components (Fig. 2A).20 They studied how the ratios of dendrimer-based ionizable lipid 5A2-SC8 within the four components affected transfection efficacy. As displayed in Fig. 2B and C, varying the molar ratio of 5A2-SC8 within the four components had a distinct effect on in vitro transfection efficacy, and less 5A2-SC8 assisted in mRNA release and improved the transfection efficacy. However, very low 5A2-SC8 was insufficient for in vivo transfection, indicating that a balance of 5A2-SC8 and other components was essential. Cheng et al. also found that increasing the percentage of DOPE would improve transfection efficacy. The LNP formulations without lipid–PEG exhibited very low transfection efficacy, because lipid–PEG could improve the stability of LNP by preventing its aggregation.
1 brought no improvement in transfection efficacy. The phospholipid also played a vital role in transfection efficacy; the formulation using 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) exhibited a higher transfection efficacy than 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), which might result from the different phospholipid structure. Specifically, DOPE is a conical lipid that can form a less stable hexagonal phase with the organelle membrane, thus promoting endosomal escape and mRNA release. Whereas DSPC is a cylindrical lipid that tends to form the lamellar phase, which is more structurally stable and blocks mRNA release. On the other hand, increasing the phospholipid content could reduce the LNP size and thus improve the transfection efficacy. Cheng et al. optimized the LNPs by changing the molar ratios of the four components (Fig. 2A).20 They studied how the ratios of dendrimer-based ionizable lipid 5A2-SC8 within the four components affected transfection efficacy. As displayed in Fig. 2B and C, varying the molar ratio of 5A2-SC8 within the four components had a distinct effect on in vitro transfection efficacy, and less 5A2-SC8 assisted in mRNA release and improved the transfection efficacy. However, very low 5A2-SC8 was insufficient for in vivo transfection, indicating that a balance of 5A2-SC8 and other components was essential. Cheng et al. also found that increasing the percentage of DOPE would improve transfection efficacy. The LNP formulations without lipid–PEG exhibited very low transfection efficacy, because lipid–PEG could improve the stability of LNP by preventing its aggregation.
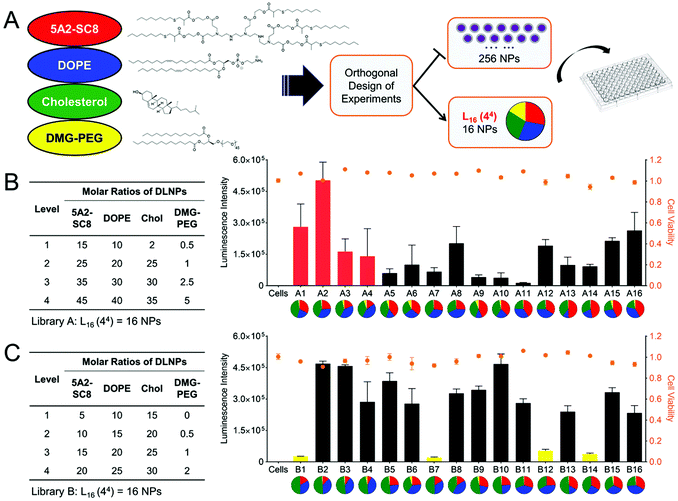 | ||
| Fig. 2 Optimization of LNP formulations by changing the molar ratio of the four components. (A) Schematic illustration of LNP formulation. (B and C) LNP libraries based on Design of Experiment calculations and their corresponding Luc mRNA transfection efficacy. Reproduced with permission from ref. 20. Copyright 2018, Wiley-VCH. | ||
In addition to optimizing the LNP formulations for higher transfection efficiency, the exploration of novel cationic or ionizable lipids has great potential to further improve transfection. As mentioned above, cationic or ionizable lipids usually contain an amino head, a hydrophobic tail, and a linker between them. Thus, the introduction of different functional groups into the lipid head or tail may improve transfection efficacy. For example, Zhang et al. constructed a library of ionizable lipids that had the dendritic amino core as the lipid head and functionalized N1,N3,N5-tris(2-aminoethyl)benzene-1,3,5-tricarboxamide (TT) as the lipid tail.21 As shown in Fig. 3A, different types of lipid tails, i.e., linear saturated and unsaturated carbon chains, as well as linear and branched ester chains, were reacted with TT via reductive amination. LNPs with linear ester chains containing lipids showed much lower luciferase expression compared with the original LNP with TT3 containing lipids (Fig. 3B). This might indicate that the introduction of ester bonds decreased transfection efficacy. Moreover, decrements were also observed in double bond-containing lipid groups. Thus, the unsaturation of lipid might cause a decline in transfection efficacy. In contrast, LNPs with branched ester chains containing lipids exhibited excellent transfection efficacy, and this difference might be the result of their different degradation rates and liver clearance rates. The LNPs with a linear ester chain (FTT9) showed a quick clearance: 80% in 6 h. In contrast, LNPs with a branched ester chain (FTT5) showed a much slower clearance with a half-life of 48 h (Fig. 3C and D). Miao et al. also studied how the alkyne and ester groups in the ionizable lipid affected mRNA delivery (Fig. 3E).22 They incorporated biodegradable alkyne rather than alkene groups into the hydrophobic tails, which enhanced endosomal escape and systemic tolerability of LNPs. Ester linkages were further incorporated to accelerate tissue clearance. When combined with cKK-E12, the addition of novel biodegradable alkyne lipids showed synergistic effects in improving transfection efficacy (Fig. 3F). Lee et al. also explored the effect of unsaturated lipids in LNPs.23 They synthesized various lipids with alkenyl thiols as unsaturated hydrophobic tails and compared their transfection abilities with saturated lipids (Fig. 4A). Parameters including the degree of saturation, double bond position, and configuration were investigated, and seven representative amino cores with different chemical structures were selected to build the lipids. However, transfection varied and no relationship between saturation and transfection efficacy was detected (Fig. 4B). Moreover, the changes in lipid saturation degree did not affect in vivo biodistribution, while the changes in double bond position and configuration affected both transfection efficacy and biodistribution (Fig. 4C and D), which might be attributed to the rigidity of the lipids and their ability to promote membrane permeability and endosomal escape. Thus, the relationship between saturation and transfection efficiency remains unclear.
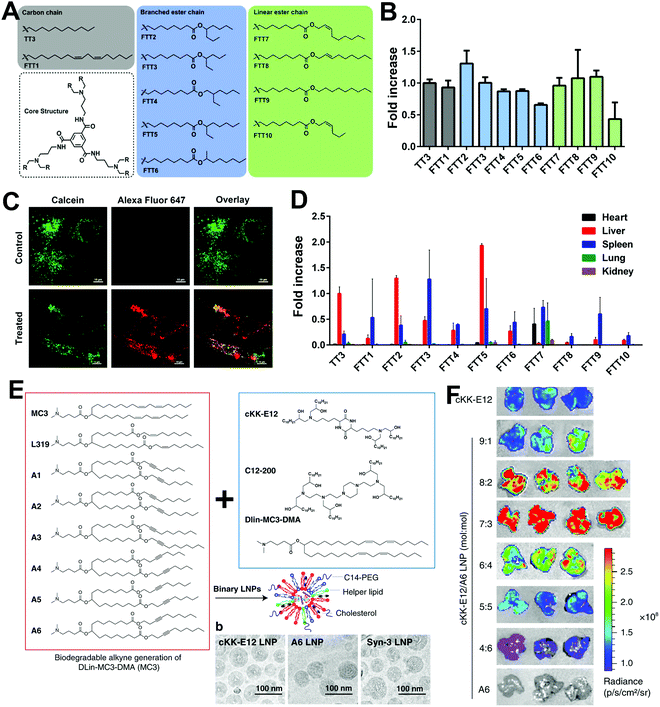 | ||
| Fig. 3 The effect of saturation degrees of novel ionizable lipids on mRNA transfection. (A) Chemical structures of TT and FTT ionizable lipids with different lipid tails. (B) In vitro mRNA transfection in Hep3B cells treated with different LNP formulations. (C) Endosomal escape performance of FTT5 LNPs. (D) Biodistribution of LNPs via i.v. injection. Reproduced with permission from ref. 21. Copyright 2020, American Association for the Advancement of Science. (E) LNPs constructed by alkyne and ester-containing ionizable lipids and representative cryo-EM images. (F) Enhanced transfection efficacy in the liver was achieved by the incorporating unsaturated lipid A6 and adjusting LNP formulations. Reproduced with permission from ref. 22. Copyright 2020, Springer Nature. | ||
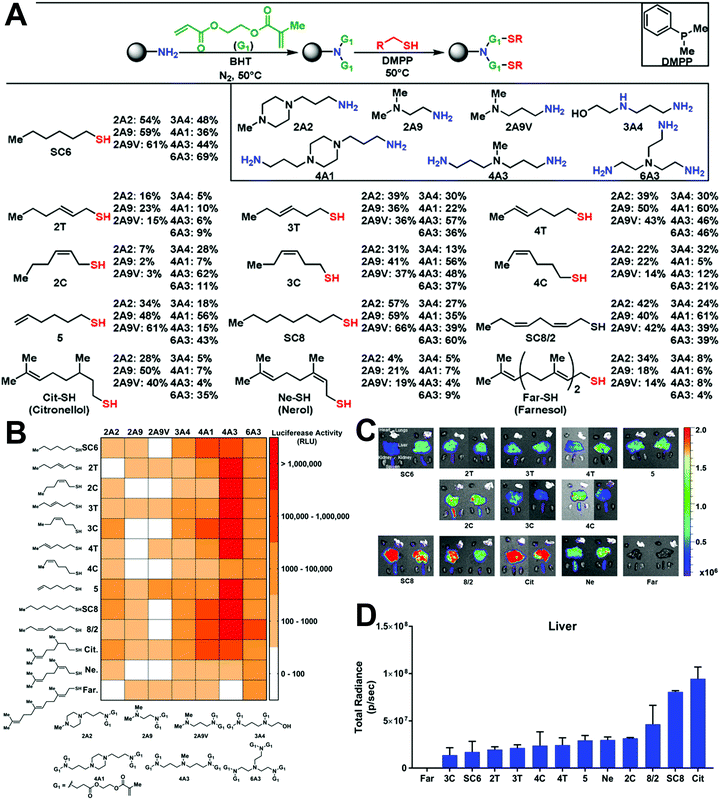 | ||
| Fig. 4 The role of lipid saturation in mRNA transfection. (A) Chemical structures of ionizable lipids with unsaturated thiols as lipid tails. (B) Heat map of Luc expression using LNPs with different lipids. (C and D) Biodistribution of LNPs with different lipids. The degree of saturation, position, and configuration affected transfection efficacy and organ distribution. Reproduced with permission from ref. 23. Copyright 2021, Wiley-VCH. | ||
Natural products are also good candidates to act as delivery vehicles due to their excellent biocompatibility. Hou et al. constructed a series of vitamin-derived LNPs for the delivery of AMP-CatB mRNA.24 As displayed in Fig. 5A, five vitamin-derived lipids, i.e., VB3-lipid, VC-lipid, VD-lipid, VE-lipid, VH-lipid, were synthesized by esterification with tertiary amines containing carboxylic acid. The obtained lipids could be ionized under acidic conditions and provided positive charges to interact with mRNAs. VC-lipid-based LNPs were 20-fold more effective than other LNPs, and 10-fold more effective than lipofectamine 3000 (Fig. 5B). In addition, Hou et al. also optimized the formulation with different weight ratios of VC-lipid/mRNA, and the molar ratio of VC-lipid/DOPE/cholesterol to achieve successful endosomal escape (Fig. 5C). In another study, Li et al. synthesized a lipid library using different amino heads and a novel lipid tail named OCholB through Michael addition (Fig. 5D).25 This new lipid tail contained a biodegradable disulfide bond to provide glutathione (GSH) responsiveness and a cholesteryl moiety to improve biocompatibility. Aminoglycosides are antibiotics used against Gram-negative pathogens. Owing to the presence of amino sugars, aminoglycosides are capable of disrupting the lipopolysaccharide moieties and penetrating cell membranes, which can greatly benefit endosomal escape. Inspired by the unique properties of aminoglycosides, Yu et al. explored alkyl lipid-modified aminoglycosides for the delivery of mRNA.26 As presented in Fig. 5E, they constructed amikacin (AM), hygromycin (HG), geneticin (GN), and gentamicin (GT) with alkyl chains containing 8–14 carbons by ring-opening or Michael addition reaction. Among them, the most effective was GT with nine carbon tails and AM with eight carbon tails, indicating that moderate carbon tails might be suitable to adjust both hydrophobicity and flexibility.
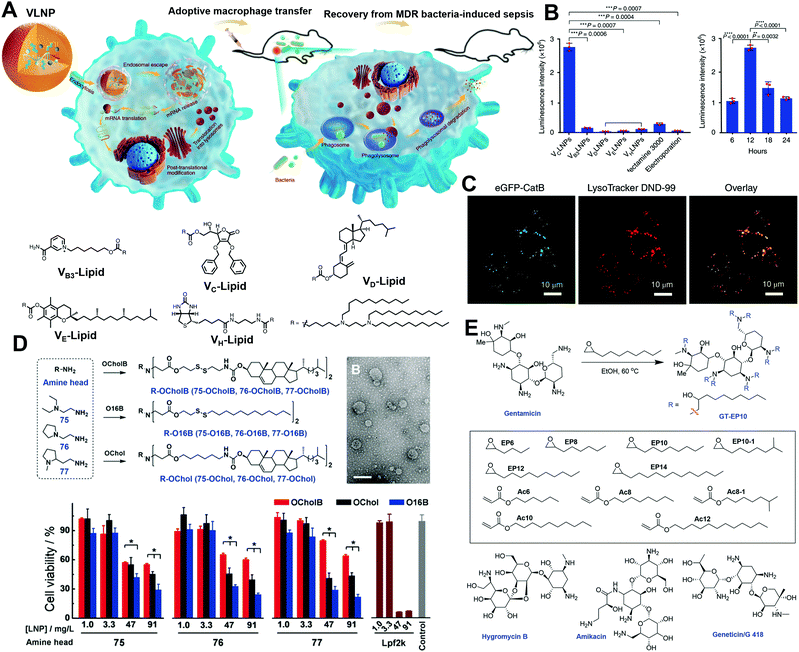 | ||
| Fig. 5 Design of natural product-derived cationic/ionizable lipids. (A) Adoptive macrophages transfected by vitamin-derived LNPs were transferred to mice for anti-bacterial application. (B) Luc expression by different vitamin-derived LNPs. VC-LNP exhibited remarkably high Luc expression in RAW264.7 cells. (C) Endosomal escape activity of VC-LNPs. Reproduced with permission from ref. 24. Copyright 2020, Springer Nature. (D) Synthesis of cholesterol-containing lipids with improved biocompatibility. Reproduced with permission from ref. 25. Copyright 2020, Wiley-VCH. (E) Synthesis of alkyl lipid-modified aminoglycosides by ring-opening or Michael addition reactions. Reproduced with permission from ref. 26. Copyright 2020, Wiley-VCH. | ||
In addition to cationic and ionizable lipids, which have already attracted much attention, zwitterionic phospholipids have also received notice for their critical role in endosomal escape by membrane fusion. As shown in Fig. 6, Liu et al. developed a series of pH-switchable ionizable phospholipids (iPhos) with multi tails to explore their mechanisms in mRNA delivery.27 The iPhos were composed of an ionizable amine head, a phosphate head, and several hydrophobic tails, synthesized by ring-opening of dioxaphospholane oxide with different amines (Fig. 6A). The amine heads and phosphate group endowed the iPhos with zwitterionic properties that could withstand the reversible ionizable process when the pH switched between neutral and acidic (Fig. 6B). Having entered the endosomes, the tertiary amine would be protonated to assist in the endosomal escape. Moreover, Liu et al. found that a single zwitterion head performed better than multiple zwitterions, which could be ascribed to steric hindrance in membrane phase transformation. In addition, iPhos with three tails tended to insert into the endosomal membranes to form a cone shape and facilitate the formation of the unstable hexagonal II phase (Fig. 6C). More interestingly, the iPhos exhibited organ selectivity in vivo. The iPhos with shorter hydrophobic tails, i.e., 9–12 carbons, accumulated in the liver (Fig. 6D), whereas iPhos with longer tails tended to be in the spleen, suggesting that iPhos could also be applied as selective organ targeting (SORT) lipids.
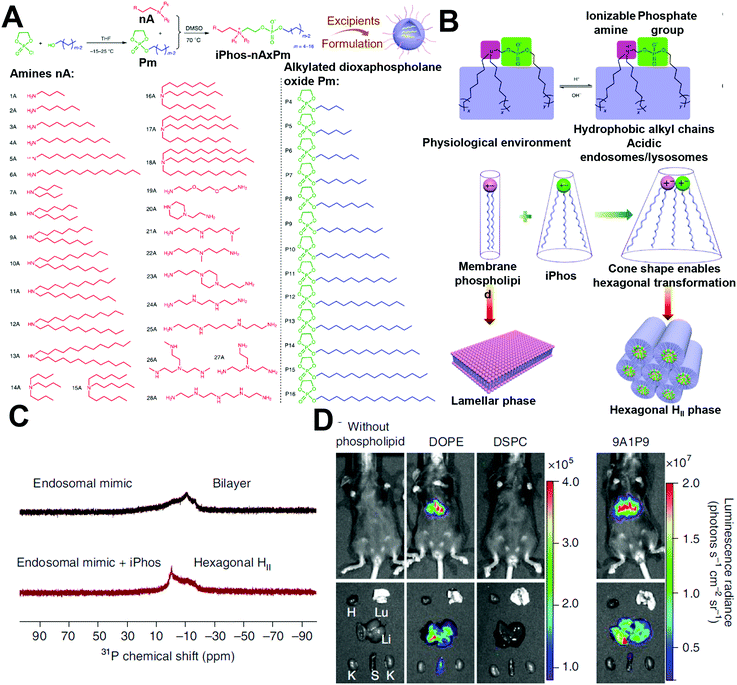 | ||
| Fig. 6 The role of phosphate lipids in mRNA delivery. (A) Synthesis of the iPhos by ring-opening reactions. (B and C) The iPhos with a switchable zwitterionic head and three tails aided in endosomal escape and exhibited higher transfection efficacy by transforming the endosomal membranes into hexagonal structures. (D) The iPhos exhibited superior in vivo transfection efficacy than commonly used phosphate lipids. Reproduced with permission from ref. 27. Copyright 2021, Springer Nature. | ||
2.4 Polymer-based carriers
Polymeric NPs represent another successful delivery system, with considerable potential in mRNA-based therapeutics. Cationic polymers have been constructed to complex with mRNA into nanoparticles, forming mRNA polyplexes. Polyethylenimine (PEI) and poly(L-lysine) (PLL) are among the earliest materials applied in RNA delivery. However, their unsatisfying in vivo transfection efficacy and toxicity limit further development. Recently, functional and biodegradable polymers have also received increasing attention, as they could be less toxic than PEI and chemically designable to afford better transfection efficacy.As discussed above, a reversible charge between neutral and acidic pH is beneficial for endosomal escape. Thus, charge-altering releasable transport (CART) (which can undergo charge-altering, charge-reversing, or charge-neutralizing in different pHs) offers opportunities for delivering mRNA by assisting in mRNA release in the cytoplasm. Waymouth and Wender et al. explored several CARTs with different chemical structures to successfully deliver and release mRNA. For example, they prepared cationic oligo(α-amino ester)s with alkyl chains.31 Under basic conditions, partial ammonium was deprotonated, resulting in intramolecular cyclization, followed by rearrangement of nitrogen (Fig. 7A). After complexing with mRNAs, CARTs were capable of controlling the intracellular release of mRNA by self-immolative degradation (i.e., rearrangement of nitrogen), with >90% transfection efficacy. In addition to these lipophilic oligocarbonate-based CARTs, Waymouth and Wender et al. also developed serine ester-based CARTs (Ser-CARTs).28 The introduction of oligo (serine ester) block not only improved biocompatibility, but also accelerated the degradation rate due to the byproduct of serine peptides. Furthermore, the same group developed a CART library with various side-chain lipids.7 By mixing the CARTs at different ratios, expression was enhanced in Jurkat cells, A20 immortalized B cells, EL4 immortalized T cells, and primary T cells. Moreover, by synthesizing statistical polymers with two side-chain lipids, lymphocyte transfection was improved. Through analysis of the performance of their CART library and their mixture, these researchers found that the improved mRNA transfection was related to the increased uptake and endosomal escape. Unsaturation increased the fluidity of lipids, which enhanced membrane fusion and triggered endocytosis. Moreover, the introduction of two unsaturated lipids assisted in forming a hexagonal phase with membranes. The membrane fusion and hexagonal phase formation finally contributed to the high transfection efficacy through enhanced uptake and endosomal escape.
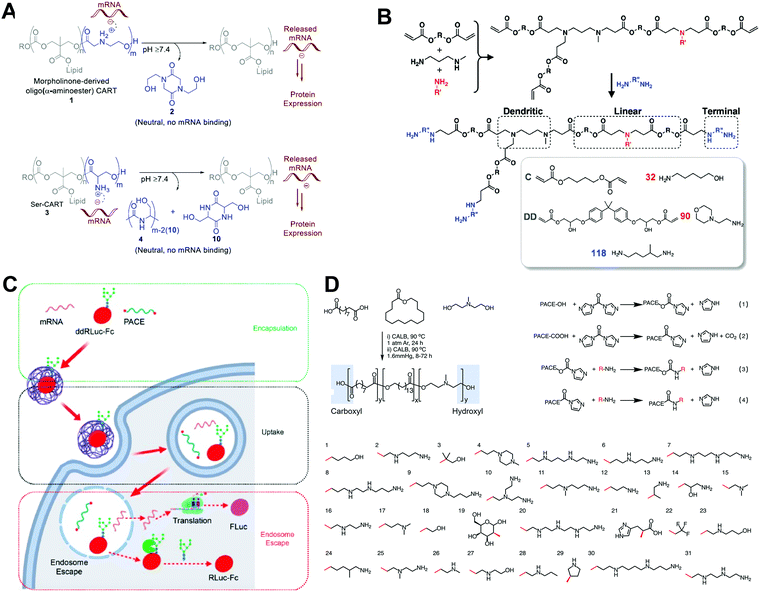 | ||
| Fig. 7 Polymer-based carriers for mRNA delivery. (A) Rearranging Mechanism of CARTs via O–N acyl shifts. Reproduced with permission from ref. 28. Copyright 2019, American Chemical Society. (B) Synthesis of hyperbranched PBAEs by trifunctional amine, amine, and diacrylate. Reproduced with permission from ref. 29. Copyright 2019, Wiley-VCH. (C and D) Screening of PACEs with different terminal groups and quantifying their transfection efficacy with encapsulation, uptake, and endosomal escape. Reproduced with permission from ref. 30. Copyright 2020, American Chemical Society. | ||
Poly(β-amino esters) (PBAEs) are positively charged biodegradable polymers composed of ester bonds. PBAEs are usually synthesized by a Michael addition polymerization between amines and a diacrylate, followed by the addition of a diamine to end the reaction. Owing to their facile synthesis, PBAEs hold great potential in gene delivery. For instance, Palmiero et al. developed a polycaprolactone (PCL)-based PBAE polymer for mRNA delivery.8 A premixing protocol, dissolving PBAE and mRNA in different solvents before mixing, was used to obtain a higher transfection efficacy than direct mixing. The degree of PCL affected the lipophilicity of the polymer, which in turn affected transection efficiency. The benefit was obtained while increasing the CL content. However, it was counterbalanced by the decrement of ionizable amines, which were essential to condense mRNA and form mRNA/PBAE complex. Fornaguera et al. introduced oligopeptide end-modified PBAEs (OM-PBAEs) to condense and complex mRNA.32 The lysine-modified PBAE enhanced interaction with cell membranes with strong positive charges, and the histidine-modified PBAE exhibited a “proton sponge” effect for enhanced endosomal escape. Thus, the combination of lysine- and histidine-modified PBAEs further improved transfection efficiency. In this way, Fornaguera et al. achieved mRNA transfection in antigen-presenting cells in vitro and spleen in vivo by optimizing the lysine and histidine composition. Patel et al. synthesized hyperbranched PBAEs to complex with mRNA and form stable and concentrated polyplexes.29 By introducing a trifunctional amine, they generated hyperbranched PBAEs (Fig. 7B). Compared with linear PBAEs, hyperbranched PBAEs had superior amino content and were thus able to generate a greater positive surface charge. These enabled better stability during inhalation to deliver mRNA to the lung epithelium.
Jiang et al. developed a library of polymers to quantitate endosomal escape.30 They prepared a library of biodegradable poly(amine-co-ester)s (PACEs) with different terminal groups. These PACEs underwent a hydrolysis reaction after delivering mRNA. They studied the relationship between transfection efficacy and encapsulation, internalization, and endosomal escape (Fig. 7C and D). Almost all PACEs showed a high transfection efficacy when their encapsulation efficiency exceeded 80%, indicating that encapsulation was the first essential step in mRNA transfection. However, the correlation between internalization and transfection was much weaker. Endosomal escape was the most important step for effective transfection. Adding ddRluc-Fc probe into the system, they directly measured endosomal escape, and a robust correlation (R2 = 0.7603) was obtained.
Zhang et al. designed an ionizable amphiphilic Janus dendrimer (IAJD) for the delivery of mRNA.33 Unlike the general four-component LNPs systems, this one-component IAJD system was prepared with simple methods. Six libraries of IAJDs were synthesized according to modular-orthogonal methodologies. Libraries 5 and 6, which were twin-twin and hybrid twin-mix IAJDs, performed poorly, whereas some of the single-single IAJDs showed high RNA transfection. In addition, Zhang et al. found that the cation–π interaction affected transfection by adjusting pKa, which might open new avenues for the design of ionizable lipids.
2.5 Hybrid carriers
Hybrid carriers containing both lipid and polymer exhibit the advantages of both.10 Decoration of mRNA-loaded polymeric NPs with lipid could further enhance their stability and pharmacokinetics by prolonging the circulation time after intravenous injection, through evading uptake by the reticuloendothelial system. In addition, mRNA delivery systems formed by organic/inorganic hybrid NPs, such as metal–organic framework (MOF), gold nanoparticles, and graphene oxide-PEI complexes are also addressed below.Choi et al. prepared polyethyleneimine (PEI)-conjugated graphene oxide (GO) for mRNA delivery to target induced pluripotent stem cells (iPSC).34 The introduction of GO not only reduced the cytotoxicity of the positively charged PEI, but also increased the loading capacity due to the high specific surface area of GO, and protected the loaded mRNA against RNase. Sun et al. constructed a cationic supramolecule-modified zirconium (Zr)-based MOF to load mRNA and protect it from enzymatic degradation, appreciably increasing in vitro mRNA transfection.35 The UiO66-Zr-MOF was coated with ethanolamine-modified poly(glycidyl methacrylate)[PGMA(EA)] to endow the MOF with a positive charge to complex with negatively charged mRNA; it exhibited enhanced mRNA transfection efficacy, approximately 20-fold higher than the linear PGMA(EA). In another study by Wang et al., mesoporous material was applied for mRNA delivery.9 As demonstrated in Fig. 8A, they constructed PEI-modified mesoporous organosilica as an mRNA delivery vector. The large-pore structure increased the PEI modification ratio (Fig. 8B). Moreover, tetrasulfide was incorporated to deplete GSH and activate the mammalian target of the rapamycin complex 1 (mTORC1) pathway. Thus, high mRNA transfection was achieved by the above two strategies (Fig. 8C).
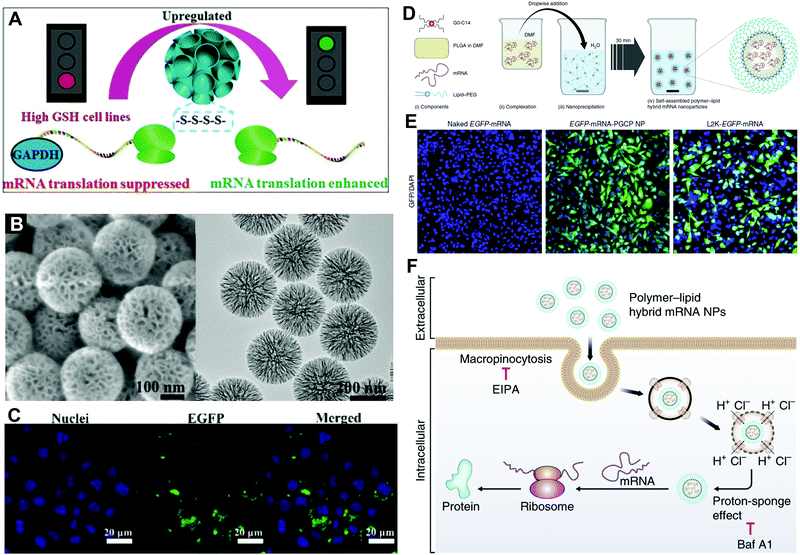 | ||
| Fig. 8 Hybrid carriers for mRNA delivery. (A) Schematic illustration of tetrasulfide-incorporated and PEI-modified mesoporous organosilica nanoparticles with enhanced mRNA translation. (B) SEM and TEM images of mesoporous organosilica nanoparticles. (C) In vitro EGFP-mRNA transfection in RAW264.7 cells. Reproduced with permission from ref. 9. Copyright 2020, Wiley-VCH. (D) Self-assembly of polymer–lipid hybrid mRNA NPs. (E) In vitro EGFP-mRNA transfection in PC3 cells treated with naked mRNA, hybrid mRNA NPs, and Lip2K mRNA NPs. (F) The hybrid mRNA NPs underwent cellular uptake and intracellular transport by proton-sponge effect before mRNA release and expression in the cytoplasm. Reproduced with permission from ref. 5. Copyright 2018, Springer Nature. | ||
The lipid/polymer hybrid NPs are also among the most promising carriers for mRNA delivery. For example, Islam et al. employed the cationic lipid G0-C14 to complex with mRNA, the FDA-approved poly(lactic-co-glycolic acid) (PLGA) to construct a robust NP core, and 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy (polyethylene glycol)] (DSPE-PEG) as a coating for stability.5 Hybrid mRNA NPs were obtained by centrifugation to remove organic solvents and encapsulated mRNA (Fig. 8D). The hybrid NPs exhibited higher EGFP in vitro transfection than Lip2K in PC3 cells. As demonstrated in Fig. 8F, the hybrid NPs entered the cell by micropinocytosis and released the loaded mRNA by proton-sponge effect, which achieved higher delivery and transfection efficacy. Furthermore, Kong et al. applied the poly(disulfide amide) (PDSA) to construct a polymer core, which responded to the redox microenvironment in tumor cells and assisted in the redox-responsive release.4 By applying different mRNAs into the system, both studies achieved efficient tumor suppression both in vitro and in vivo against different types of cancer.
3. Application of mRNA-based therapeutics
mRNA-based therapies play a significant role in the fight against various diseases. Recently, mRNA has emerged as an exciting strategy to translate proteins for the treatment of cancers,4 cardiovascular diseases,36 and infectious diseases,24 as well as genome editing and protein replacement.37 It is also worth mentioning that the applications of mRNA technologies are not limited to the areas mentioned above, but are expanding to many other biomedical fields.3.1 Functional protein expression
The expression of functional proteins in target cells is the most straightforward application of mRNA drugs. It is typically used to treat rare genetic metabolic diseases by supplementing unexpressed proteins or proteins that do not function in a regulated or tissue-specific manner. For example, decreased levels of the brain-derived neurotrophic factor (BDNF), an endogenous protein that may augment neuronal survival and function, have been reported in various neurodegenerative diseases. Therefore, upregulation of BDNF levels using mRNA could be promising for the prevention of neuronal death. For instance, Fukushima et al. developed a polyplex nanomicelle containing BDNF-encoding mRNA (BDNF-mRNA) for the treatment of ischemic neuronal death.38 In a transient global ischemia (TGI) model, the rapid rise of BDNF levels in the hippocampus was achieved by intraventricular injection of nanomicelles encapsulating BDNF-mRNA, leading to a significant increment in the survival of hippocampal neurons after TGI. Immunohistochemical analysis revealed that the nanomicelles were mainly taken up by astrocytes, which secreted BDNF to provide a supportive microenvironment for the survival of neurons under ischemic conditions.Besides mRNAs encoding neurotrophic factors, mRNAs encoding tumor-suppressor proteins could hold great promise in cancer therapy. One representative tumor suppressor protein is phosphatase and tensin homolog deleted on chromosome 10 (PTEN). To validate the therapeutic potential of PTEN-encoding mRNA (PTEN-mRNA), a polymer-lipid hybrid NP was developed for the in vivo delivery of PTEN-mRNA to tumors.5 The PTEN proteins of the PTEN-null prostate cancer cells were successfully restored by using the PTEN-mRNA-loaded NPs. More importantly, the restoration of PTEN proteins in mice with PTEN-null prostate cancer via PTEN-mRNA-loaded NPs dramatically inhibited tumor growth. The mechanistic study suggested that the upregulation of PTEN proteins inhibited the phosphatidylinositol 3-kinase (PI3K)–AKT pathway and promoted apoptosis. Similar to PTEN, p53 is another representative tumor suppressor protein. A redox-responsive polymer-lipid hybrid NP platform was constructed to deliver p53-encoding mRNA (p53-mRNA).4 As displayed in Fig. 9A, the p53-mRNA NPs not only significantly suppressed the growth of p53-null hepatocellular carcinoma (HCC) cells and non-small cell lung cancer (NSCLC) cells via promoting the cell cycle arrest and apoptosis, but also increased the sensitivity of tumor cells to the mammalian target of rapamycin (mTOR) inhibitor everolimus, which had shown negligible therapeutic effect for advanced HCC and NSCLC when applied alone. In multiple mouse models of HCC and NSCLC, the p53-mRNA NPs synergistically delayed the growth of tumors in the presence of everolimus. Moreover, intravesical delivery of mRNA encoding lysin-specific demethylase 6A (KDM6A), another typical tumor suppressor protein, by mucoadhesive NPs in mice bearing orthotopic Kdm6a-null bladder cancer (BCa) also showed effectiveness in inhibiting the BCa metastasis.10
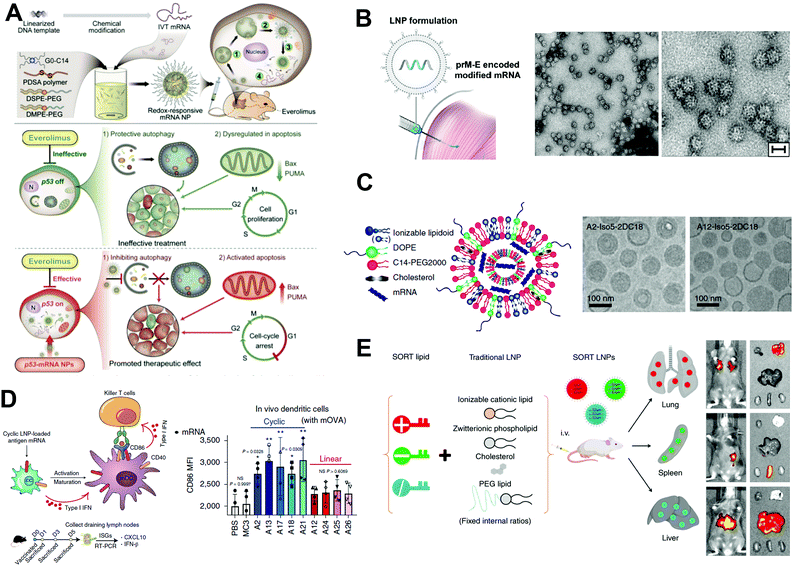 | ||
| Fig. 9 Application of mRNA-based therapeutics. (A) p53-mRNA hybrid NPs suppress tumor growth by promoting cell cycle arrest and apoptosis. Reproduced with permission from ref. 4. Copyright 2019, American Association for the Advancement of Science. (B) Zika virus (prM-E) encoded mRNA is encapsulated into NPs for intramuscular vaccination. HeLa cells were transfected with prM-E mRNA, and secreted virus-like subviral particles (SVPs) in the supernatant were isolated and subjected to electron microscopy. Low- and high-magnification images of SVPs. Scale bar, 30 nm. Reproduced with permission from ref. 13. Copyright 2017, Elsevier Inc. (C) Schematic illustration of mRNA-loaded LNPs and representative cryogenic electron microscopy images of LNPs. Reproduced with permission from ref. 12. Copyright 2019, Springer Nature. (D) Schematic illustration of mRNA LNPs in activating immune-related cells through STING pathway, and corresponding in vitro mRNA transfection. Reproduced with permission from ref. 12. Copyright 2019, Springer Nature. (E) The in vivo delivery profile of traditional lipid NPs can be easily altered via the addition of a SORT molecule. Reproduced with permission from ref. 3. Copyright 2020, Springer Nature. | ||
3.2 Vaccines
Vaccines stimulate active immune responses, protecting millions from disease. Compared with conventional vaccines, mRNA vaccines hold greater potential in preventing both infectious and non-infectious diseases owing to their efficient expression of membrane-bound proteins and polyprotein antigens, thereby mimicking the expression of antigens during natural infection. The most successful mRNA vaccines are the two FDA-approved COVID-19 vaccines developed by Moderna (Spikevax) and BioNTech/Pfizer (BNT-162), which have been deployed worldwide and greatly decreased infection rates, saving millions of lives. Both vaccines showed highly protective efficiency (∼95%) against SARS-CoV-2 via the delivery of nucleoside-modified mRNA that encoded the full-length spike glycoprotein of SARS-CoV-2 using similar LNPs. From a mechanical point of view, after two doses of the vaccinations, these two vaccines induced notably high levels of antigen-specific antibodies and promoted a strong T helper 1 cell response, providing protection against SARS-CoV-2. Despite the great success achieved, the stability of current mRNA vaccines could be further improved. At present, both BNT-162 and Spikevax must be stored in the required cold chain which has to be adequately monitored, and after being removed from the cold chain, both types of mRNA vaccines have to be used up within critical timeframes.39 For example, Pfizer's mRNA vaccine demands the storage in −70 °C ultra-cold freezers (this storage requirement was updated to the temperature ranging of anywhere between![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) −25 and −15 °C in a recently submitted data to FDA), and Moderna's mRNA vaccine has to be stored and delivered at −20 °C, respectively. These strict storage conditions limit the long-distance shipment and usage to avoid freeze–thaw. Besides, the decrement in protect efficiency with time and the decreased protection efficiency against new variants may also become problems.
−25 and −15 °C in a recently submitted data to FDA), and Moderna's mRNA vaccine has to be stored and delivered at −20 °C, respectively. These strict storage conditions limit the long-distance shipment and usage to avoid freeze–thaw. Besides, the decrement in protect efficiency with time and the decreased protection efficiency against new variants may also become problems.
The rapid development of these COVID-19 mRNA vaccines has greatly benefited from the experience and knowledge acquired from long-term investigations of other viruses. For example, Richner et al. prepared an LNP-based mRNA vaccine for the prevention of Zika virus infection (Fig. 9B).13 Modified mRNAs that encoded wild-type or variant Zika virus structural genes were formulated into LNPs. After intramuscular administration of two doses to mice, the Zika mRNA vaccine induced high neutralizing antibody titers, providing effective protection against the virus. In another study by Alberer et al., an mRNA rabies vaccine (CV7201) was investigated in a first-in-human phase I clinical trial.40 This mRNA vaccine used the cationic protamine to complex and deliver mRNAs encoding rabies virus glycoprotein. The phase I clinical trial data showed the safety and immunogenicity of the CV7201 vaccine in a reasonable dosage. In addition, the influenza virus has threatened global health for decades, and effective vaccines against this virus and its variants are strongly needed. To that end, mRNA vaccines against H10N8 and H7N9 influenza viruses were studied in phase I randomized clinical trials.41 The H10N8 and H7N9 vaccines displayed satisfactory safety profiles as well as induced robust humoral immune responses. Intramuscular injection of 100 μg of H10N8 mRNA vaccine resulted in hemagglutination inhibition (HAI) titers ≥1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 in all participants and microneutralization (MN) titers ≥1
40 in all participants and microneutralization (MN) titers ≥1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 in 87% of participants. Intramuscular injection of an optimal dose of 25 μg of H7N9 mRNA vaccine-induced HAI titers ≥1
20 in 87% of participants. Intramuscular injection of an optimal dose of 25 μg of H7N9 mRNA vaccine-induced HAI titers ≥1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 in 96.3% of the participants and MN titers ≥1
40 in 96.3% of the participants and MN titers ≥1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 in 100% of the participants. It is worth noting that a two-dose prime-boost vaccine strategy has been applied for all these mRNA vaccines to generate potent and long-term protection against various viruses, in which the first dose primes the immune response, and the second one boosts the immune response.
20 in 100% of the participants. It is worth noting that a two-dose prime-boost vaccine strategy has been applied for all these mRNA vaccines to generate potent and long-term protection against various viruses, in which the first dose primes the immune response, and the second one boosts the immune response.
Monoclonal antibody (mAb) therapy against virus infection also arose attention due to their nearimmediate action. To that end, Kose et al. constructed mRNA encoding neutralizing human mAbs CHKV-24.42 By delivering CHKV-24 mRNA with LNPs, the mean human mAb level significantly improved from 10.1 to 35.9 μg mL−1 in mice, demonstrating a protective effect against chikungunya virus (CHIKV) infection with a prolonged survive rate. Moreover, August et al. from Moderna, Inc. released a phase I clinical trial report of LNP-encapsulated mRNA-1944 encoding CHKV-24.6 After i.v. injection, mild to moderate adverse effects were observed. The levels of CHKV-24 IgG exhibited dose-dependent behavior and neutralizing activity were detected.
3.3 Cancer immunotherapy
Cancer immunotherapy aims to elicit an antitumor immune response, thereby suppressing tumor growth and leading to effective clinical outcomes. The adaptive immune system protects us from infections either through the production of cytotoxic T cells or the generation of humoral antibodies. The activation of the antitumor immune response can be achieved by the delivery of mRNA encoding TAAs. For example, Oberli et al. developed an LNP platform encapsulating TAA-mRNA for cancer immunotherapy.43 In a mouse model with aggressive B16F10 melanoma, administration of a single dose of this TAA-mRNA LNP resulted in potent cytotoxic CD8 T cell activation, leading to a significant reduction in tumor size and increment in overall survival rate. In addition, a more potent immune response could be achieved via introducing adjuvant lipopolysaccharides (LPS). In some cases, the ionizable lipids used for the construction of LNPs can also function as an immune adjuvant to further boost the immune response induced by TAA-mRNA. For example, Maio et al. constructed an LNP platform consisting of heterocyclic lipids to deliver TAA-mRNAs for cancer immunotherapy (Fig. 9C).12 The LNPs containing heterocyclic lipids, which elicited an immune response through the intracellular stimulator of interferon genes (STING) pathway (Fig. 9D), were identified via screening more than 1,000 LNP formulations. This top-performing LNP was then used to encapsulate mRNA that can encode tyrosinase-related protein 2 (Trp2)-a crucial TAA of B16F10 melanoma. In a B16F10 melanoma mouse model, the administration of three doses of this Trp2-mRNA-loaded LNPs induced a strong adaptive immune response, as evidenced by the dramatically elevated cytotoxic CD8 T cell numbers, attributed to delayed tumor growth and prolonged survival of the mice.Another strategy to activate antitumor immune response is to deliver mRNA encoding bispecific T cell-engaging antibody, a special type of antibody that can recruit cytotoxic T cells to the tumor, resulting in target-dependent polyclonal T cell activation and consequent tumor cell lysis.44 After three low doses (5 μg) of the antibody-mRNA, the growth of the ES-2 tumor xenografts was effectively suppressed. In addition, mRNAs encoding immune system modulators such as cytokines IL-23, IL-36γ, and OX40L have also been delivered to mice to stimulate potent immune responses, enabling effective tumor destruction.45
Reversing immunosuppression is another emerging strategy. By delivering mRNA encoding a single-domain antibody, for example, BisCCL2/5i, via LNPs, the antibody level significantly improved.46 Once delivered to the liver, the mRNA LNP was capable of suppressing the M2 tumor-associated macrophages (TAMs) polarization and activating M1 phenotype polarization, displaying prolonged survival in mouse models with liver cancer.
3.4 Genome editing
Genome editing offers promising therapies for genetic diseases by precisely inserting, removing, or replacing DNA sequences at a specific locus in the genomic DNA. The revolutionary discovery of the CRISPR/Cas9 system (clustered regularly interspaced short palindromic repeat-associated protein 9) has led to the explosive rise of gene editing. Co-delivery of Cas9 mRNA and sgRNA can achieve transient expression of Cas9 protein, reduce off-target effects, and overcome the limitations of the traditional CRISPR/Cas9 delivery by the plasmid.47 To this end, Miller et al. prepared a zwitterionic amino lipid (ZAL) NP platform for the co-delivery of Cas9 mRNA and sgRNA for CRISPR/Cas9 gene editing in vitro and in vivo.14 The unique structure of the ZAL-NPs resulted in high gene editing efficiency (>90%) in vitro at relatively low sgRNA doses (15 nM). The in vivo gene editing capability was measured in a genetically engineered mouse model, whose cells permanently expressed a homozygous Rosa26 promoter Lox-Stop-Lox tdTomato (tdTO) cassette. The selective disruption of LoxP led to the deletion of the Stop cassette, allowing the expression of tdTO. Indeed, i.v. injection of ZAL-NPs encapsulated Cas9 mRNA and sgLoxP resulted in significant expression of floxed tdTO in most cells of the major organs including liver, kidneys, and lungs. Notably, the strongest expression of tdTO (in the liver) could be ascribed to the preferable accumulation of NPs in the liver, as with most other NPs. Although various LNP formulations have been explored to deliver Cas9 mRNA and sgRNA, some may have toxicity issues. To address this problem, a reducible lipid was synthesized to prepare LNPs for the co-delivery of Cas9 mRNA and sgRNA.37 The leading reducible lipid BAMEAO16B contained disulfide bonds connecting the ionizable head and lipid tails. Upon internalization by cells, the reductive intracellular environment triggers the cleavage of the lipids, promoting the release of mRNA from LNPs and reducing potential toxicity. Indeed, efficient in vitro genome editing was observed only 24 h post-treatment of the Cas9 mRNA/sgRNA NPs. After intravenous administration of Cas9 mRNA/sgRNA NPs, the level of proprotein convertase subtilisin/Kexin type 9 in mouse serum decreased by 80%, suggesting the high in vivo gene editing efficiency of this platform.So far, the liver has been the main target of most gene-editing systems due to the preferable accumulation of these platforms in that organ. To broaden the in vivo applications of mRNA-based gene editing, Cheng et al. developed a SORT NP platform for gene editing in different organs (Fig. 9E).3 Multiple classes of SORT NPs engineered by adding a supplemental SORT molecule specifically delivered Cas9 mRNA/sgRNA to lung, spleen, and liver, and selectively edited various cell types including hepatocytes, epithelial cells, endothelial cells, and immune cells (T cells and B cells). These SORT NPs represent the first platform that is able to selectively deliver Cas9 mRNA and sgRNA to different organs, holding great promise for the treatment of disease-causing mutations with different cellular origins. Previous studies on mRNA-based gene editing have mainly focused on the validation of systems in cells that carry reporter genes or genetically engineered animal models. Therapeutic efficacy studies of mRNA-based gene editing systems can be more challenging, because high editing efficiency is normally required to generate a therapeutic effect. To validate the therapeutic effect of mRNA-based gene editing, a cationic LNP platform was developed for the targeted delivery of Cas9 mRNA/sgRNA to macrophages.48 The treatment of macrophages with NPs loading Cas9 mRNA and sgRNA-targeting NLRP3 (sgNLRP3), a target in the treatment of various inflammatory diseases, disrupted NLRP3 of macrophages and subsequently suppressed the activation of the NLRP3 inflammasome in the presence of multiple stimuli. In the LPS-induced septic shock and monosodium urate crystal-induced peritonitis mouse model, intravenous administration of NPs containing Cas9 mRNA/sgNLRP3 effectively ameliorated acute inflammation. Several methods can identify novel platforms for the delivery of Cas9 mRNA/sgRNA or other functional mRNAs to specific cell types. For example, a high-throughput screening method named Fast Identification of Nanoparticle Delivery (FIND) was proposed to identify NP platforms for mRNA-based gene editing in endothelial cells.49
4. Clinical status of mRNA-based therapeutics
To date, there are more than 1000 clinical trials of mRNA-based therapeutics, including the FDA-approved covid vaccines BNT 162 and Spikevax. Among them, LNPs account for a large part. Representative records are listed in Table 1, and most are in the clinical phase I and II. LNPs are served not only as vaccines against SARS-CoV-2 and other infections such as Chikungunya virus and Zika virus, but also as cancer therapeutics and drugs for metabolic diseases.| Name | Delivery platform | Disease | Administration route | Clinicaltrials.gov identifier | Phase |
|---|---|---|---|---|---|
| BNT-162 | LNP | SARS-CoV-2 | i.m. injection | FDA-approved | |
| Spikevax | LNP | SARS-CoV-2 | i.m. injection | FDA-approved | |
| mRNA-1944 | LNP | Chikungunya virus | i.v. injection | NCT03829384 | Phase I |
| mRNA-3745 | LNP | Glycogen Storage Disease | i.v. injection | NCT05095727 | Phase I |
| mRNA-1647 | LNP | Cytomegalovirus Infection | i.m. injection | NCT05085366 | Phase III |
| mRNA-1893 | LNP | Zika Virus | i.m. injection | NCT04917861 | Phase II |
| mRNA-2752 | LNP | Relapsed/refractory solid tumor malignancies or lymphoma | Intratumoral injection | NCT03739931 | Phase I |
| mRNA-4157 | LNP | Melanoma | NCT03897881 | Phase II | |
| BD111 | Lentivirus | Refractory herpetic viral keratitis | Corneal injection | NCT04560790 | Phase I/II |
| LCs | Langerhans-type dendritic cells | Melanoma | i.v. injection | NCT01456104 | Phase I |
| W_pro1/BNT112 | Liposomes | Prostate cancer | i.v. injection | NCT04382898 | Phase I/II |
| BNT141 | CLDN18.2-positive tumors | i.v. injection | NCT04683939 | Phase I/II |
LNPs hold the advantages of easy formulation, uniform size, modularity, high mRNA loading and transfection efficiency, and good biocompatibility. However, structural changes might occur during long-term storage, and the stability of LNPs may need to be further improved. Polymer-based carriers could show better stability. The strong chemical designability in polymer molecules also makes polymer an efficient and promising material for mRNA delivery. However, the polymer chemists may still need to pay attention to making the molecule weight and polydispersity of polymers be controllable, thus avoiding variation between batches. Hybrid carriers may combine the advantages of both LNPs and polymers such as easy formulation, stability and functionality, while bulk screening of effective formulations is needed. Protein-based carriers are highly biocompatible for mRNA delivery. However, considering proteins are inherently complex molecules, which may be unstable and sensitive to temperature, pH, ionic concentration and other formulation properties.
Although all types of platforms could have their corresponding advantages and disadvantages, they are all promising delivery systems that may generate effective and new-generation mRNA medicine. We also expect that with increasing efforts from worldwide scientists in different fields, these current disadvantages could be finally overcome and the potential of mRNA medicine will be maximally fulfilled in the future.
5. Conclusions and outlook
The rapid development, improvement, and application of mRNA technology are inseparable from the cross fields of biology, medicine, pharmacy, materials science, and chemistry. Unlike DNA, mRNA does not need to be delivered to the nucleus and does not interfere with the human genome. However, the large molecular weight and single-stranded structure of mRNA make it unstable. Therefore, chemical modification of mRNA is required to ensure its function. This tutorial review began from the viewpoint of chemistry, through representative examples from the design and development of delivery platforms, to the biomedical application of mRNA therapeutics. It offers a substantial summary of the currently “hot” but still infant mRNA field, with some challenges still to overcome. We also point out the following factors that may promote the development of mRNA technology.First of all, although the current storage requirements of mRNA vaccines have been improved, the commonly used low temperature method of storing mRNA vaccine still restrict applications in low-resource areas. Thus, further chemical modification of mRNA or the development of a better delivery platform to improve stability will be potential research directions.
Secondly, although the side effects of the current mRNA vaccines are acceptable, further reducing the side effects of mRNA and avoiding unnecessary immune responses are also among future research directions. Further modification of mRNA on the 5′ cap and 3′ Poly(A) tail to reduce immunogenicity is required. At present, it could be helpful to combine ionizable lipid materials with exosomes and cell membranes derived from the human body to deliver mRNA, which may reduce side effects and improve therapeutic effects. Most importantly, the principles for developing more suitable delivery platforms should also be fully understood.
In addition, mRNA NPs tend to accumulate in the liver rather than other organs when administrated via intravenous injection, which limits the applications of mRNA in the treatment of other organ-related diseases. For example, some studies have focused on adjusting the ratio of delivery platform components to achieve selective organ targeting or used inhalation methods to treat lung-related diseases.3,29 However, there is little research on the delivery of mRNA to the brain, where crossing the blood–brain barrier must be considered. Studies have shown that a certain intensity of ultrasound can open the blood–brain barrier, so it may be possible to combine ultrasound and mRNA technology in the treatment of brain diseases.
For mRNA vaccines, how to better target immune cells is also a research direction. Perhaps research into albumin “free-ride” methods22 will guide the way to improving the immune system-targeting effect of mRNA vaccines.
Further, although there is some work that combines mRNA imaging and treatment, such efforts need to be strengthened. Perhaps PET imaging can be integrated into the mRNA delivery process, or the effect of the treatment can be observed through imaging methods and adjust the treatment strategy according to the feedback.
Of course, the research and development of delivery platforms have always been the focus of chemists and material scientists. Thus, we have also illustrated how different chemical structures, components, and ratios affect the efficiency of mRNA delivery through various representative examples. As for the synthesis of lipid materials, directions for future chemists will likely include high channels, rapid synthesis, fewer steps, no protection/deprotection of the reaction, no catalysts, preferably no solvents, and no purification, which benefit the manufacture and clinical translation. Furthermore, design multifunctional LNP platforms are of great interest. All the four components can be further designed and modified to not only improve the in vivo transfection efficiency, but also endow LNPs with desirable functions in one platform. In addition, exploring new polymers capable of delivering mRNA is also one important direction. Hyperbranched polymers and dendrimers with plenty of amine groups could be promising. Block copolymers with both functional block and positive-charged block or zwitterionic block may also be constructed to perform therapeutic functions. As for inorganic materials like MOF and COF, improving the size uniformity and stability in physiological environment by novel synthesis method and surface modification might also be considered. Moreover, the delivery platforms can be further developed. Not limited by NP solution, gel, patch, capsule, microneedle, and even wearable devices can be employed for specific diseases with biosensing ability to monitor the progress of diseases.50
We anticipate that this review could benefit the researchers in various fields including biology, medicine, pharmacy, materials science, chemistry, etc., and promote the vigorous development of the mRNA field. With the development of emerging mRNA technologies, we also expect that mRNA medicine could be greatly expanded to various biomedical applications (not limited to the areas we discussed above). The COVID-19 mRNA vaccine is likely just a start, and we may be at the beginning of a new Era in mRNA medicine.
Author contributions
Y. X., N. K., and W. T. created the outline of this review. Y. X., Z. T., and X. H. wrote the manuscript and designed the figures. All authors discussed, edited, and reviewed the manuscript before submission.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by the National Natural Science Foundation of China (No. 82122076, N. Kong), Harvard Medical School/Brigham and Women's Hospital Department of Anesthesiology-Basic Scientist Grant (No. 2420 BPA075, W. Tao), Center for Nanomedicine Research Fund (No. 2019A014810, W. Tao), Nanotechnology Foundation (No. 2022A002721, W. Tao.), Farokhzad Family Distinguished Chair Foundation (No. 018129, W. Tao), and the US METAvivor Early Career Investigator Award (No. 2018A020560, W. Tao). W. Tao is also a recipient of the Khoury Innovation Award (No. 2020A003219), The Gillian Reny Stepping Strong Center for Trauma Innovation Breakthrough Innovator Award (No. 113548), and the American Heart Association (AHA) Collaborative Sciences Award (No. 2018A004190).References
- M. Cobb, Curr. Biol., 2015, 25, R526 CrossRef CAS PubMed.
- Z. Tang, N. Kong, X. Zhang, Y. Liu, P. Hu, S. Mou, P. Liljeström, J. Shi, W. Tan, J. S. Kim, Y. Cao, R. Langer, K. W. Leong, O. C. Farokhzad and W. Tao, Nat. Rev. Mater., 2020, 5, 847 CrossRef CAS PubMed.
- Q. Cheng, T. Wei, L. Farbiak, L. T. Johnson, S. A. Dilliard and D. J. Siegwart, Nat. Nanotechnol., 2020, 15, 313 CrossRef CAS PubMed.
- N. Kong, W. Tao, X. Ling, J. Wang, Y. Xiao, S. Shi, X. Ji, A. Shajii, S. T. Gan, N. Y. Kim, D. G. Duda, T. Xie, O. C. Farokhzad and J. Shi, Sci. Transl. Med., 2019, 11, eaaw1565 CrossRef CAS PubMed.
- M. A. Islam, Y. Xu, W. Tao, J. M. Ubellacker, M. Lim, D. Aum, G. Y. Lee, K. Zhou, H. Zope, M. Yu, W. Cao, J. T. Oswald, M. Dinarvand, M. Mahmoudi, R. Langer, P. W. Kantoff, O. C. Farokhzad, B. R. Zetter and J. Shi, Nat. Biomed. Eng., 2018, 2, 850 CrossRef CAS PubMed.
- A. August, H. Z. Attarwala, S. Himansu, S. Kalidindi, S. Lu, R. Pajon, S. Han, J.-M. Lecerf, J. E. Tomassini, M. Hard, L. M. Ptaszek, J. E. Crowe and T. Zaks, Nat. Med., 2021, 27, 2224 CrossRef CAS PubMed.
- C. J. McKinlay, N. L. Benner, O. A. Haabeth, R. M. Waymouth and P. A. Wender, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E5859 CrossRef PubMed.
- U. Capasso Palmiero, J. C. Kaczmarek, O. S. Fenton and D. G. Anderson, Adv. Healthcare Mater., 2018, 7, 1800249 CrossRef PubMed.
- Y. Wang, J. Tang, Y. Yang, H. Song, J. Fu, Z. Gu and C. Yu, Angew. Chem., Int. Ed., 2020, 59, 2695 CrossRef CAS PubMed.
- N. Kong, R. Zhang, G. Wu, X. Sui, J. Wang, N. Y. Kim, S. Blake, D. De, T. Xie, Y. Cao and W. Tao, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2112696119 CrossRef CAS PubMed.
- K. J. Kauffman, J. R. Dorkin, J. H. Yang, M. W. Heartlein, F. DeRosa, F. F. Mir, O. S. Fenton and D. G. Anderson, Nano Lett., 2015, 15, 7300 CrossRef CAS PubMed.
- L. Miao, L. Li, Y. Huang, D. Delcassian, J. Chahal, J. Han, Y. Shi, K. Sadtler, W. Gao, J. Lin, J. C. Doloff, R. Langer and D. G. Anderson, Nat. Biotechnol., 2019, 37, 1174 CrossRef CAS PubMed.
- J. M. Richner, S. Himansu, K. A. Dowd, S. L. Butler, V. Salazar, J. M. Fox, J. G. Julander, W. W. Tang, S. Shresta, T. C. Pierson, G. Ciaramella and M. S. Diamond, Cell, 2017, 168, 1114 CrossRef CAS PubMed.
- J. B. Miller, S. Zhang, P. Kos, H. Xiong, K. Zhou, S. S. Perelman, H. Zhu and D. J. Siegwart, Angew. Chem., Int. Ed., 2017, 56, 1059 CrossRef CAS PubMed.
- S. Kreiter, M. Diken, A. Selmi, J. Diekmann, S. Attig, Y. Hüsemann, M. Koslowski, C. Huber, Ö. Türeci and U. Sahin, Cancer Res., 2011, 71, 6132 CrossRef CAS PubMed.
- K. K. L. Phua, K. W. Leong and S. K. Nair, J. Controlled Release, 2013, 166, 227 CrossRef CAS PubMed.
- M. Fotin-Mleczek, K. M. Duchardt, C. Lorenz, R. Pfeiffer, S. Ojkic-Zrna, J. Probst and K.-J. Kallen, J. Immunother., 2011, 34, 1 CrossRef CAS PubMed.
- B. Weide, S. Pascolo, B. Scheel, E. Derhovanessian, A. Pflugfelder, T. K. Eigentler, G. Pawelec, I. Hoerr, H.-G. Rammensee and C. Garbe, J. Immunother., 2009, 32, 498 CrossRef CAS PubMed.
- M. Segel, B. Lash, J. Song, A. Ladha, C. C. Liu, X. Jin, S. L. Mekhedov, R. K. Macrae, E. V. Koonin and F. Zhang, Science, 2021, 373, 882 CrossRef CAS PubMed.
- Q. Cheng, T. Wei, Y. Jia, L. Farbiak, K. Zhou, S. Zhang, Y. Wei, H. Zhu and D. J. Siegwart, Adv. Mater., 2018, 30, 1805308 CrossRef PubMed.
- X. Zhang, W. Zhao, G. N. Nguyen, C. Zhang, C. Zeng, J. Yan, S. Du, X. Hou, W. Li, J. Jiang, B. Deng, D. W. McComb, R. Dorkin, A. Shah, L. Barrera, F. Gregoire, M. Singh, D. Chen, D. E. Sabatino and Y. Dong, Sci. Adv., 2020, 6, eabc2315 CrossRef CAS PubMed.
- L. Miao, J. Lin, Y. Huang, L. Li, D. Delcassian, Y. Ge, Y. Shi and D. G. Anderson, Nat. Commun., 2020, 11, 2424 CrossRef CAS PubMed.
- S. M. Lee, Q. Cheng, X. Yu, S. Liu, L. T. Johnson and D. J. Siegwart, Angew. Chem., Int. Ed., 2021, 60, 5848 CrossRef CAS PubMed.
- X. Hou, X. Zhang, W. Zhao, C. Zeng, B. Deng, D. W. McComb, S. Du, C. Zhang, W. Li and Y. Dong, Nat. Nanotechnol., 2020, 15, 41 CrossRef CAS PubMed.
- Y. Li, R. Jarvis, K. Zhu, Z. Glass, R. Ogurlu, P. Gao, P. Li, J. Chen, Y. Yu, Y. Yang and Q. Xu, Angew. Chem., Int. Ed., 2020, 59, 14957 CrossRef CAS PubMed.
- X. Yu, S. Liu, Q. Cheng, T. Wei, S. Lee, D. Zhang and D. J. Siegwart, Adv. Healthcare Mater., 2020, 9, 1901487 CrossRef CAS PubMed.
- S. Liu, Q. Cheng, T. Wei, X. Yu, L. T. Johnson, L. Farbiak and D. J. Siegwart, Nat. Mater., 2021, 20, 701 CrossRef CAS PubMed.
- N. L. Benner, R. L. McClellan, C. R. Turlington, O. A. W. Haabeth, R. M. Waymouth and P. A. Wender, J. Am. Chem. Soc., 2019, 141, 8416 CrossRef CAS PubMed.
- A. K. Patel, J. C. Kaczmarek, S. Bose, K. J. Kauffman, F. Mir, M. W. Heartlein, F. DeRosa, R. Langer and D. G. Anderson, Adv. Mater., 2019, 31, 1805116 CrossRef PubMed.
- Y. Jiang, Q. Lu, Y. Wang, E. Xu, A. Ho, P. Singh, Y. Wang, Z. Jiang, F. Yang, G. T. Tietjen, P. Cresswell and W. M. Saltzman, Nano Lett., 2020, 20, 1117 CrossRef CAS PubMed.
- C. J. McKinlay, J. R. Vargas, T. R. Blake, J. W. Hardy, M. Kanada, C. H. Contag, P. A. Wender and R. M. Waymouth, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E448 CrossRef CAS PubMed.
- C. Fornaguera, M. Guerra-Rebollo, M. Ángel Lázaro, C. Castells-Sala, O. Meca-Cortés, V. Ramos-Pérez, A. Cascante, N. Rubio, J. Blanco and S. Borrós, Adv. Healthcare Mater., 2018, 7, 1800335 CrossRef PubMed.
- D. Zhang, E. N. Atochina-Vasserman, D. S. Maurya, N. Huang, Q. Xiao, N. Ona, M. Liu, H. Shahnawaz, H. Ni, K. Kim, M. M. Billingsley, D. J. Pochan, M. J. Mitchell, D. Weissman and V. Percec, J. Am. Chem. Soc., 2021, 143, 12315 CrossRef CAS PubMed.
- H. Y. Choi, T.-J. Lee, G.-M. Yang, J. Oh, J. Won, J. Han, G.-J. Jeong, J. Kim, J.-H. Kim, B.-S. Kim and S.-G. Cho, J. Controlled Release, 2016, 235, 222 CrossRef CAS PubMed.
- P. Sun, Z. Li, J. Wang, H. Gao, X. Yang, S. Wu, D. Liu and Q. Chen, Chem. Commun., 2018, 54, 11304 RSC.
- W. Chen, M. Schilperoort, Y. Cao, J. Shi, I. Tabas and W. Tao, Nat. Rev. Cardiol., 2022, 19, 228 CrossRef PubMed.
- J. Liu, J. Chang, Y. Jiang, X. Meng, T. Sun, L. Mao, Q. Xu and M. Wang, Adv. Mater., 2019, 31, 1902575 CrossRef PubMed.
- Y. Fukushima, S. Uchida, H. Imai, H. Nakatomi, K. Kataoka, N. Saito and K. Itaka, Biomaterials, 2021, 270, 120681 CrossRef CAS PubMed.
- M. L. Fahrni, I. A.-N. Ismail, D. M. Refi, A. Almeman, N. C. Yaakob, K. M. Saman, N. F. Mansor, N. Noordin and Z.-U.-D. Babar, J. Pharm. Policy Pract., 2022, 15, 16 CrossRef PubMed.
- M. Alberer, U. Gnad-Vogt, H. S. Hong, K. T. Mehr, L. Backert, G. Finak, R. Gottardo, M. A. Bica, A. Garofano, S. D. Koch, M. Fotin-Mleczek, I. Hoerr, R. Clemens and F. von Sonnenburg, Lancet, 2017, 390, 1511 CrossRef CAS.
- R. A. Feldman, R. Fuhr, I. Smolenov, A. Ribeiro, L. Panther, M. Watson, J. J. Senn, M. Smith, Ö. Almarsson, H. S. Pujar, M. E. Laska, J. Thompson, T. Zaks and G. Ciaramella, Vaccine, 2019, 37, 3326 CrossRef CAS PubMed.
- N. Kose, M. Fox Julie, G. Sapparapu, R. Bombardi, N. Tennekoon Rashika, A. D. de Silva, M. Elbashir Sayda, A. Theisen Matthew, E. Humphris-Narayanan, G. Ciaramella, S. Himansu, S. Diamond Michael and E. Crowe James, Sci. Immunol., 2019, 4, eaaw6647 CrossRef CAS PubMed.
- M. A. Oberli, A. M. Reichmuth, J. R. Dorkin, M. J. Mitchell, O. S. Fenton, A. Jaklenec, D. G. Anderson, R. Langer and D. Blankschtein, Nano Lett., 2017, 17, 1326 CrossRef CAS PubMed.
- C. R. Stadler, H. Bähr-Mahmud, L. Celik, B. Hebich, A. S. Roth, R. P. Roth, K. Karikó, Ö. Türeci and U. Sahin, Nat. Med., 2017, 23, 815 CrossRef CAS PubMed.
- S. L. Hewitt, A. Bai, D. Bailey, K. Ichikawa, J. Zielinski, R. Karp, A. Apte, K. Arnold, S. J. Zacharek, M. S. Iliou, K. Bhatt, M. Garnaas, F. Musenge, A. Davis, N. Khatwani, S. V. Su, G. MacLean, S. J. Farlow, K. Burke and J. P. Frederick, Sci. Transl. Med., 2019, 11, eaat9143 CrossRef CAS PubMed.
- Y. Wang, K. Tiruthani, S. Li, M. Hu, G. Zhong, Y. Tang, S. Roy, L. Zhang, J. Tan, C. Liao and R. Liu, Adv. Mater., 2021, 33, 2007603 CrossRef CAS PubMed.
- X. Huang, R. Zheng, F. Ding, J. Yang, M. Xie, X. Liu, J. Li, J. Feng, X. Zhu and C. Zhang, ACS Mater. Lett., 2020, 2, 1509 CrossRef CAS.
- C. Xu, Z. Lu, Y. Luo, Y. Liu, Z. Cao, S. Shen, H. Li, J. Liu, K. Chen, Z. Chen, X. Yang, Z. Gu and J. Wang, Nat. Commun., 2018, 9, 4092 CrossRef PubMed.
- C. D. Sago, M. P. Lokugamage, K. Paunovska, D. A. Vanover, C. M. Monaco, N. N. Shah, M. Gamboa Castro, S. E. Anderson, T. G. Rudoltz, G. N. Lando, P. Munnilal Tiwari, J. L. Kirschman, N. Willett, Y. C. Jang, P. J. Santangelo, A. V. Bryksin and J. E. Dahlman, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E9944 CrossRef CAS PubMed.
- Y. Fang, X. Zhao, T. Tat, X. Xiao, G. Chen, J. Xu and J. Chen, Matter, 2021, 4, 1102 CrossRef CAS.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |