Open Access Article
Open Access ArticleA preliminary investigation comparing high-volume and low-volume air samplers for measurement of PAHs, NPAHs and airborne bacterial communities in atmospheric particulate matter
Egide
Kalisa
ab,
Vincent
Kuuire
a and
Matthew
Adams
 *a
*a
aUniversity of Toronto Mississauga, Department of Geography, Geomatics and Environment, Mississauga, ON L5L 1C6, Ontario, Canada. E-mail: md.adams@utoronto.ca
bUniversity of Rwanda, Department of Biology, College of Science and Technology, P.O. Box 3900, Kigali, Rwanda
First published on 28th July 2022
Abstract
Exposure to atmospheric particulate matter (PM) constitutes a severe public health threat in African countries' urban areas. However, monitoring is scarce to non-existent in large parts of Africa due to the lack of resources to acquire, operate and maintain the expensive reference monitors used to measure PM and its composition. In this study, PM10 (particulate matter, <10 μm) was collected simultaneously using high-volume and low-volume air samplers (an HVAS and an LVAS) at an urban site in Rwanda. Polycyclic aromatic hydrocarbons (PAHs), nitro-PAHs (NPAHs) and bacterial community structures (estimated using 16 rRNA gene sequences) were characterized, and results were compared for the two sampler types. The 24 h mean PM10 concentrations were higher in the HVAS than in the LVAS, but both exceeded WHO guidelines. Fewer PAH and NPAH compounds were identified using the LVAS, suggesting that the LVAS collected an insufficient mass of PM10 to reach detection limits. Species diversity of airborne bacteria was lower in the LVASs; however, in contrast, the LVASs yielded a higher average DNA concentration. Principal coordination analysis (PCoA) results indicated that bacterial communities were distinctly different between the HVAS and LVAS samplers. Both sampling instruments have potential benefits; however, their samples should not be directly compared without a comprehensive performance evaluation in the area of study.
Environmental significanceAir pollution in Africa is high, and data are limited due to few reference monitoring stations, which are very expensive and require consistent maintenance. This study provides the first preliminary investigation in Rwanda comparing high-volume and low-volume air sampling systems; low-volume systems are more affordable and require less infrastructure than high-volume reference monitors. Polycyclic aromatic hydrocarbons (PAHs), nitro-PAHs (NPAHs) and bacterial community structures associated with airborne particulate matter were characterized. Both samplers have potential benefits; however, air samples exhibited high variability in particulate composition and should not be directly compared without a comprehensive performance evaluation in the study area. The low-volume sampler is a significantly more affordable option and could be a reasonable intermediary solution and educational tool for collecting air quality information in Africa. |
1. Introduction
PM10 (particulate matter less than 10 μm in aerodynamic diameter) is often the focus of air quality monitoring in Africa and is used as a general air quality indicator.1 The WHO has recognized PM10 as a health hazard and has set a 24 hours guideline of 45 μg m−3.2 Atmospheric PM is a mixture of several chemical species, including polycyclic aromatic hydrocarbons (PAHs), nitro-PAHs (NPAHs), trace metals, dust, minerals, soot, and biological matter, including viruses, bacteria and fungi.3,4 These PM-associated chemical and biological components have been reported to cause several diseases in humans, including chronic obstructive pulmonary disease (COPD), asthma, and lung cancer.4 Several studies on PM's chemical and biological composition have been performed in developed countries,5 but information about aerosol composition, which is the first step towards addressing air pollution, is scarce to non-existent in sub-Saharan Africa.PAHs and NPAHs are ubiquitous environmental pollutants primarily generated during the incomplete combustion of organic materials.6 They are released into the urban atmosphere from anthropogenic sources (e.g., diesel vehicles, gasoline, industrial processes, cool and wood combustion) and natural sources (e.g., volcanic eruptions and forest fires).7 In the atmosphere, PAHs (two or three rings) exist in the vapour phase, whereas multi-ringed, PAHs (five rings or more) are found to be bound with particles.7 Four-ring PAHs may occur in the vapour or particle phase. Given the toxicity of PAHs and their nitro-derivatives, the US Environmental Protection Agency (EPA) has identified 16 PAH priority compounds.8 The PAH compounds such as benzo(a)pyrene have received great attention in air pollution and epidemiological studies due to their carcinogenic and mutagenic effects.6 The PAH (fluoranthene) and NPAH (1-nitropyrene, 2-nitropyrene, and 2-nitrofluoranthene) compounds have been reported to be the most dominant PAHs and NPAH in atmospheric particulate matter, and these compounds are mainly emitted by automobile exhaust.9 Studies indicated that the NPAHs such as 1-nitropyrene exhibit higher carcinogenicity and mutagenicity effects on humans than their parent PAHs.10
Air sampling by filtration, where the air is drawn through a filter using a sampling pump, is widely used. High-volume air samplers (HVASs) and low-volume air samplers (LVASs) have been commonly used to collect samples of PM for chemical and biological characterization.11–14 The difference between an HVAS and an LVAS is the flow rate. For PAH analysis, studies using HVASs to collect PM onto filters have collected volumes of air of 3224 m3 (at a flow rate of > 1 m3 min−1),15 1440 m3 (flow rate = 1 m3 min−1)16,17 and 960 m3 (flow rate = 0.666 m3 min−1).18 Others have used LVAS and medium-volume air samplers (MVASs), collecting total volumes of air < 200 m3.19,20 An advantage of using an HVAS to collect PM samples for chemical and biological characterization is a larger quantity of particles for laboratory analysis. However, LVASs have also been used to measure particle-bound PAH concentrations in high-income regions, including a study in Birmingham, UK.21 LVASs are small devices that are low-cost and easy to maintain. They have small filter area sizes that require smaller solvent volumes during the extraction process, while HVASs, with larger filter area sizes, involves the use of large volumes of toxic organic solvents and long extraction times, which is more costly and more environmentally damaging.22 Over the past few decades, interest in bioaerosols research has proliferated due to the discovery of the significant impacts of bioaerosols on human health and in atmospheric events such as cloud formation, precipitation, and atmospheric chemistry.23,24 The challenges in the characterization of bioaerosols in airborne particulates are the low density of microorganisms in the air;25 the variability in microorganisms in the atmosphere; DNA sequencing-related challenges;26 and a lack of standardized methodologies for the collection of samples and extraction of their DNA.27 In addition, due to the very low biomass concentrations in bioaerosols, several researchers have used air samplers with flow rates ranging from 200 to 1130 litres per minute to collect enough biomass from the air for microbiological analyses.20,28
In this study, we compare HVAS and LVAS samplers in measuring PAH concentrations, NPAH concentrations, and airborne bacterial communities' species diversity. PAHs, NPAHs and airborne bacteria associated with PM10 samples were determined from samples collected at an urban site in Rwanda using an HVAS and an LVAS simultaneously. PAHs and NPAHs were selected because they are ubiquitous pollutants in urban areas, with carcinogenic and mutagenic effects on humans,29 and information on their prevalence in Africa is sparse. Further, PAHs and NPAHs are linked to cancer and asthma even at very low concentrations,30 but governments in Africa do not commonly measure them due to the lack of specialized techniques and the skilled personnel required. Therefore, this study does not solely intend to recommend the performance of samplers based on their performance but to investigate whether LVASs, which are more cost-efficient than HVAS, can be used in Rwanda as an option to characterize PAHs, NPAHs and airborne bacteria associated with PM10 and fill the existing research gap in the atmospheric aerosol composition.
2. Materials and methods
2.1 Particulate matter collection
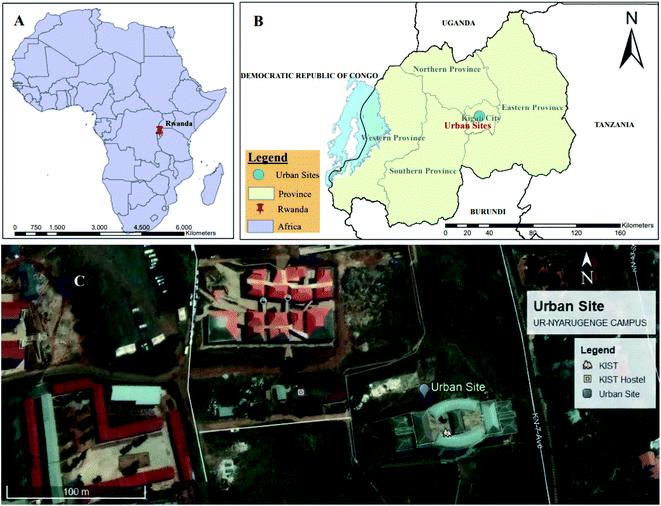
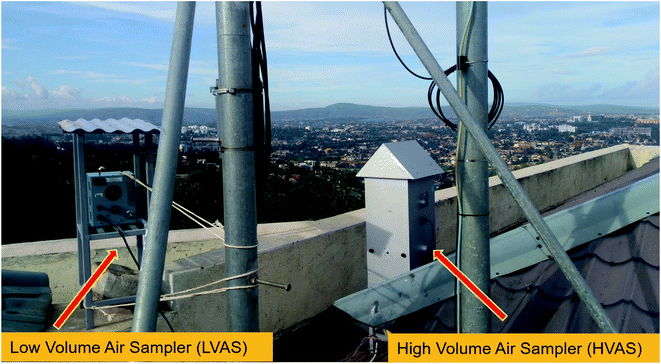
In this study, samples were collected simultaneously using an HVAS and an LVAS on the rooftop of the Kigali Institute of Science – Building Four at the University of Rwanda (∼10 m above ground level), which is an urban location (1.9616°S, 30.0640°E). In Fig. 1, we present the sampling site at its relative location in Africa and satellite imagery of the local geography.Our sampling periods were 24 hours for each day (from 12:00 am to 12:00 am of the following day, local time) and we collected seven consecutive paired samples between April 1 and April 7, 2018 when there was a stable climate (little variation in meteorological conditions) with no precipitation. HVAS samples were collected on a 126 × 166 mm Whatman glass fibre filter (GFF) (pore size: 1 μm) using a Sibata HVAS (HVS-RW-1000F, Japan) operating at a flow rate of 1000 L min−1. In addition, we collected the LVAS samples on 47 mm Whatman GFFs (pore size: 0.8 μm) using a Hi-Q Environmental Products Company CF-901 air sampler operating at a flow rate of 80 L min−1. The distance between HVAS and LVAS was 1.5 m and the rooftop collocated samplers are illustrated in Fig. 2. GFFs were chosen for this study as being ideal for the analysis of both the chemical and biological composition of air particulate matter because they are robust and inert for DNA extraction and provide consistency in the capture of low levels of organic black carbon.12,31
Gravimetric analysis was performed for all samples at the School of Science laboratory at the University of Rwanda, following the procedures previously described in detail by Kalisa et al.32 PM10 filters were weighed before sampling using an internally calibrated electronic microbalance (KERN, Balingen, Germany, readability 0.1 μg). After gravimetric analysis, each PM filter was cut into two equal parts using sterilized scissors; one was analyzed for PAHs and NPAHs, while the other was analyzed for airborne bacteria. The two filters (HVAS and LVAS) from each sampling day were analyzed for 15 PAHs, 7 NPAHs and airborne bacterial communities. Fig. 3 shows the general workflow for the experimental design.
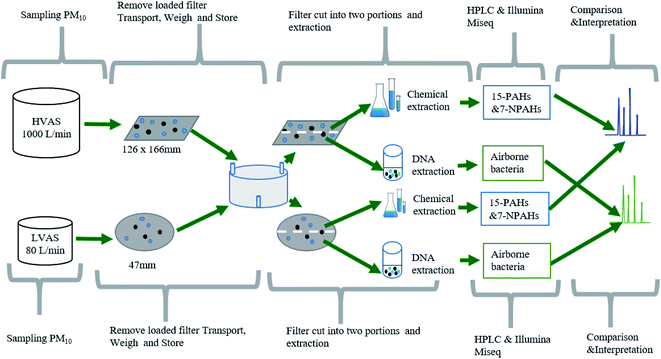 | ||
| Fig. 3 General workflow for the experimental design, showing how PM10 samples were collected from both HVAS and LVAS, subdivided and analyzed for PAHs, NPAHs and airborne bacteria. | ||
2.2 Meteorological conditions
Daily average air temperature (°C), wind speed (WS) and relative humidity (%) during the sampling period were obtained from a Rwandan meteorological weather station (1.9562°S, 30.0566°E) situated 2 km from our sampling site in similar land use conditions (Table 1).| Sampling days | Temperature | Relative humidity | Wind speed |
|---|---|---|---|
| Day 1 | 17.0 °C | 87.0% | 0–1 m s−1 |
| Day 2 | 18.0 °C | 85.9% | 1.1–2 m s−1 |
| Day 3 | 23.8 °C | 61.6% | 2.1–3 m s−1 |
| Day 4 | 21.5 °C | 69.6% | >3 m s−1 |
| Day 5 | 22.0 °C | 66.7% | 0–1 m s−1 |
| Day 6 | 20.9 °C | 71.0% | 1.1–2 m s−1 |
| Day 7 | 17.7 °C | 89.7% | 2.1–3 m s−1 |
2.3 Analysis of PAHs and NPAHs
The filter extraction process, instrumental analyses, and reagents used in this study were described previously. Briefly, each sample was analyzed for 15 PAHs and 7 NPAHs. The concentrations of NPAHs (2-NFR and 2-NP) were grouped (2-NFR + 2-NP) because they could not be separated in the chromatographic system.32 First, one-half of each of the PM10 sample filters from each sampler was divided into small pieces (0.5 cm2) and placed in a flask. Then, five deuterated PAHs (naphthalene-d8 (NaP-d8), acenaphthylene-d10 (Ace-d10), phenanthrene-d10 (Phe-d10), chrysene-d12 (Chr-d12) and perylene-d12 (Per-d12)) and an NPAH (2-fluoro-7-nitrofluorene) were added as internal standards. Next, the PM10 filter samples from each sampler were extracted (using ultrasonic extraction) in benzene–ethanol (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v).
1, v/v).
Fifteen PAHs including naphthalene (NaP), acenaphthene (Ace), fluorene (Fle), phenanthrene (Phe), anthracene (Ant), fluoranthene (Fle), pyrene (Pyr), benz(a)anthracene (BaA), chrysene (Chr), benzo(b)fluoranthene (BbF), benz(k)fluoranthene (BkF), dibenz(a,h)anthracene (DBA), benz(g,h,i)perylene (BPe) and indeno(1,2,3-cd)pyrene (IDP) were identified using an HPLC (High-Performance Liquid Chromatograph, Shimadzu Inc., Japan), equipped with a fluorescence detector based on the absorption and subsequent emission of light.33 The fluorescence detector was set at the optimum excitation (Ex) and emission (Em) wavelengths for each PAH compound as previously described by.33 The HPLC system consisted of a pump, an integrator, a degasser, an auto sample injector, a column oven, a guard column, and an analytical column (Inertial Series column-C18 (4.6 i.d. × 250 mm), GL Sciences, Tokyo, Japan) and the mobile phase was operated under a gradient concentration using a mixture of acetonitrile and water (1 mL min−1).
Seven NPAHs including 9-nitroanthracene (9-NA), 2-nitropyrene (2-NP), 2-nitrofluoranthene (2-NFR), 1-nitroperylene (1-NP), 7-nitrobenz(a)anthracene (7-NBaA), 6-nitrobenz(a)pyrene (6-NBaP), 1,3-dinitropyrenes (1,3-DNP) and 1,8-dinitropyrenes (1,8-DNP) – were analysed using an HPLC with a chemiluminescence detector (HPLC-CLD, Shimadzu). The reagents for the mobile phase for NPAH analysis consisted of an imidazole buffer and an acetonitrile solution containing bis(2,4,6-trichlorophenyl) oxalate and hydrogen peroxide with flow rate of 1 mL min−1.33 The NPAH column (Cosmosil 4.6 i.d. × (250 + 150) mm, Nacalai Tesque Inc., Kyoto, Japan) was made with Pt/Rh (reverse-phase). An HPLC was used for aerosol monitoring due to its sensitivity and ability to identify and quantify individual PAH and NPAH compounds, compared to other analytical tools.34
2.4 Quality control (QC) and quality assurance (QA)
To remove organic contaminants, all filters used in our study were pre-heated at a high temperature between 550–600 °C for 240 min before use.32 After sample collection, each filter was removed from the sampler, covered, inverted, and wrapped in aluminum foil inside a plastic bag. All filters were stored in containers packed with dry ice during transportation from the sampling site to the laboratory for gravimetric analysis; then, filters were stored at −20 °C before filter extraction and analysis. The samplers (filter holders) were sterilized with 75% ethanol before sampling.35 All inside surfaces of the HVAS and LVAS samplers were maintained sterile until sampling. The gravimetric analysis was carried out for each PM sample in a weighing room where the temperature ranged between 20 °C and 25 °C and relative humidity ranged between 30 and 45%.36 Two field blanks from each sampler and solvent blank experiments were conducted, and no target PAH and NPAH compounds were found in any of the blank samples for both samplers. The recovery of internal standard deuterated PAHs and NPAHs was quantitatively measured following the methodology described by:36 surrogate recoveries ranged from 60% to 102%.2.5 Microbial community analysis (bacteria)
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) of chloroform and isoamyl alcohol was added to the solution to prevent emulsification and to reduce foaming; (5) 10 M ammonium acetate to a final concentration of 2.5 M was added to the removed aqueous phase in order to precipitate out the proteins associated with the DNA from the solution; (6) 0.5 times the recorded volume of isopropanol was added to the aqueous layer in a new tube to precipitate DNA. After extraction of DNA, the concentration was calculated per volume of extracted DNA (re-suspended in 20 μg). The genomic DNA detected in PM10 samples obtained from both HVAS and LVAS samplers was quantified using a Qubit Fluorometer (Invitrogen).
1) of chloroform and isoamyl alcohol was added to the solution to prevent emulsification and to reduce foaming; (5) 10 M ammonium acetate to a final concentration of 2.5 M was added to the removed aqueous phase in order to precipitate out the proteins associated with the DNA from the solution; (6) 0.5 times the recorded volume of isopropanol was added to the aqueous layer in a new tube to precipitate DNA. After extraction of DNA, the concentration was calculated per volume of extracted DNA (re-suspended in 20 μg). The genomic DNA detected in PM10 samples obtained from both HVAS and LVAS samplers was quantified using a Qubit Fluorometer (Invitrogen).
Amplicon libraries were constructed following the Illumina MiSeq protocol.38 The 16S rRNA V3–V4 region was used for the 16S rRNA gene in bacteria (341F-CCTACGGGAGGCAGCAG and 907R CCGTCAATTCMTTTGAGTTT).38 The PCR thermo-cycle used was as follows: 30 cycles (1) at 95 °C for 60 s (denaturing stage); (2) 55 °C for 60 s, 72 °C for 60 s (annealing stage); (3) and the last stage at 72 °C for 10 min (extending stage). AMPure XP beads and Illumina MiSeq protocol were used to purify PCR products and index the amplicons, respectively.38
3. Results
3.1 Concentrations of PM10 from the HVAS and the LVAS
The 24 h mean concentrations of measured PM10 were higher with the HVAS (75.77 ± 17.03 μg m−3) than the LVAS (51.82 ± 12.89 μg m−3) (Table 2). The Wilcoxon–Mann–Whitney test indicated that the mean concentration of PM10 measured in the HVAS was significantly higher than the mean concentration of PM10 in the LVAS (p = 0.0387). The measured 24 h mean concentration of PM10 using both the HVAS and LVAS at the site exceeded the 24 hours mean in the WHO's guidelines for PM10 (45 μg m−3).2| Analytes | HVAS (n = 7) | LVAS (n = 7) | p-Value |
|---|---|---|---|
| PM10 [μg m−3] | 75.77 ± 18.39 | 51.82 ± 13.92 | <0.0001 |
| PAH [ng m−3] | 22.90 ± 8.09 | 20.67 ± 15.02 | 0.7077 |
| NPAH [pg m−3] | 305.12 ± 301.90 | 282.66 ± 174.31 | 0.6178 |
Meteorological parameters recorded (daily) during each sampling period showed stable weather conditions at our sampling site, suggesting that weather conditions should have minimal or no influence on the PM10 concentrations measured during the sampling period. The mean temperature was 20.2 ± 2.4 °C, the wind speed was 2.4 ± 1.9 m s−1, and relative humidity was 75.8 ± 10.3%. Rwanda is a landlocked country situated at a high altitude, with a stable tropical climate, exhibiting minor temperature variations and slow wind speeds.
3.2 Concentrations of total PAHs and NPAHs
The mean concentrations of the ∑15 PAHs in PM10 were 22.90 ± 8.9 ng m−3 and 20.6 ± 15.02 ng m−3 for the HVAS and LVAS, respectively. The mean of ∑7 NPAHs was 305.11 ± 301.90 pg m−3 with the HVAS and 282.66 ± 174.31 pg m−3 with the LVAS (Table 2).The individual PAHs (BPe, Flu and BbF) and NPAHs (9-NA, 2-NP + 2-NFR and 1,8-DNP) were the most dominant compounds detected in both samplers. Concentrations are provided in Table 3.
| Compounds | HVAS | LVAS | |
|---|---|---|---|
| a ND: not detected. | |||
| PAH (ng m −3 ) | |||
| NaP | 0.29 ± 0.07 | 0.58 ± 0.24 | |
| Ace | 0.03 ± 0.00 | 0.02 ± 0.02 | |
| Fle | 0.13 ± 0.07 | 0.08 ± 0.04 | |
| Phe | 1.42 ± 0.70 | 1.86 ± 2.75 | |
| Ant | 0.03 ± 0.01 | 0.03 ± 0.01 | |
| Flu | 4.61 ± 2.89 | 3.72 ± 3.22 | |
| Pyr | 0.29 ± 0.06 | 0.35 ± 0.24 | |
| BaA | 0.52 ± 0.25 | 0.9 ± 0.65 | |
| Chr | 0.67 ± 0.32 | 1.18 ± 0.78 | |
| BbF | 2.71 ± 0.82 | 2.49 ± 1.45 | |
| BkF | 1.28 ± 0.32 | 1.15 ± 0.67 | |
| BaP | 2.64 ± 0.83 | 1.56 ± 1.04 | |
| DBA | 0.06 ± 0.02 | 0.02 ± 0.02 | |
| BPe | 5.92 ± 1.73 | 4.74 ± 2.74 | |
| IDP | 2.31 ± 0.70 | 1.98 ± 1.11 | |
| Total PAHs (ng m−3) | 23.00 ± 8.90 | 20.67 ± 15.02 | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||
| NPAH (pg m −3 ) | |||
| 1,8-DNP | 50.48 ± 92.79 | 10.6 ± 4.11 | |
| 1,3-DNP | 0.42 ± 0.40 | 0.28 ± 0.29 | |
| 9-NA | 145.79 ± 111.28 | 145.73 ± 126.36 | |
| 2-NP + 2-NFR | 91.68 ± 41.48 | 84.37 ± 42.35 | |
| 1-NP | 1.33 ± 1.29 | 3.22 ± 1.17 | |
| 7-NBaA | 15.42 ± 14.64 | ND | |
| Total NPAHs (pg m−3) | 305.12 ± 301.90 | 282.66 ± 174.31 | |
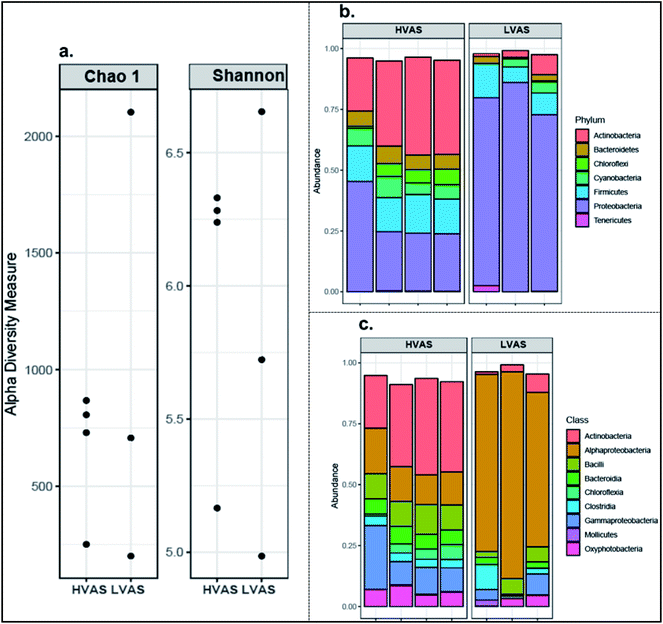
3.3 DNA and microbial community composition
The variations in DNA concentrations detected in PM10 samples using the two sampler types are presented in Fig. 4. The results indicated that the LVAS yielded a higher average DNA concentration (1.8 ng μL−1) than the HVAS (0.2 ng μL−1). In addition, the DNA concentration generally increased with increasing concentrations of PM10 (Fig. 4).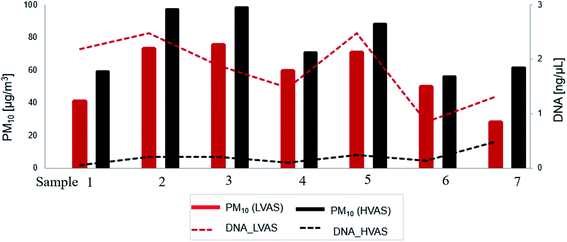 | ||
| Fig. 4 Variation in DNA concentrations (ng μL−1) and PM10 concentrations (μg m−3) detected with a high-volume air sampler (HVAS) and a low-volume air sampler (LVAS) at an urban site in Rwanda. | ||
3.4 Community diversity and microbial community composition
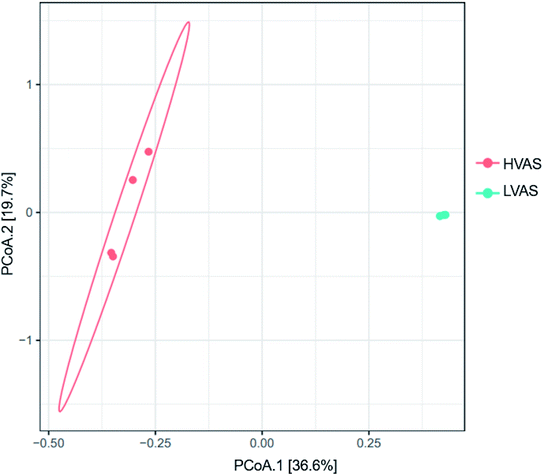
Airborne bacterial community diversity was higher in samples from the HVAS (6.00 ± 0.50) than the LVAS (5.70 ± 0.83) (as estimated by the mean of the Shannon index). The Shannon index value indicated that bacterial communities were more diverse in the HVAS than in the LVAS (Fig. 5). The bacterial species richness (as estimated from the mean of the Chao1 index) was higher in LVAS (1003 ± 985) than in HVAS samples (663 ± 280) (Fig. 5). Due to the low density of microorganisms in the air, four out of seven samples were successfully quantifiable from the HVAS compared to three from the LVAS (Fig. 5). However, the results of the t-test showed no statistically significant differences in species richness (p-value = 0.6146) or diversity (p-value = 0.7212). These results suggest that the difference in flow rate had little effect on species richness and diversity when both samplers were run simultaneously.The dominant bacterial phyla were proteobacteria, followed by Actinobacteria, Firmicutes, Chloroflexi, Bacteroidetes, and Cyanobacteria (Fig. 5). The most common bacteria in the two air samplers were grouped into the classes of Alphaproteobacteria, Actinobacteria, Bacilli and Bacteroidia. Proteobacteria were more abundant in the LVAS while Actinobacteria were more abundant in the HVAS. In the Proteobacteria, Alphaproteobacteria were the most abundant class identified in the LVAS. Principal coordination analysis (PCoA) results indicated that there were distinct bacterial communities in the HVAS and LVAS samples (PERMANOVA, p < 0.033) (Fig. 6).
4. Discussion
The present investigation characterized PAHs, NPAHs and airborne microorganisms associated with PM10 in Rwanda, the first study in the region. We demonstrated that high-volume air samplers (HVASs) and low-volume monitors (LVASs) could measure PAHs, NPAHs and airborne microorganisms associated with particulate air pollution. However, our analysis of HVAS samples showed more diversity in PAHs and NPAHs compounds and microbial communities. Thus, there are differences between the two instruments, and they are not comparable.The discrepancies between the HVAS and the LVAS for PAHs compounds results are consistent with a previous study44 that conducted a 1 year study to compare HVAS and LVAS results for levoglucosan, ions, elements and PAH analysis. Comparable results were obtained for both samplers except for PAHs, which were more variable due to the molecular complexity of the PAH with high molecular weight.44 The mean concentrations of PAHs found in this study for both the HVAS (23.0 ng m−3) and the LVAS (20.6 ng m−3) were in the same range as a previous study conducted at the same site over the same wet season in a Rwandan urban background location (21.8 ng m−3), using an HVAS, from March to July 2017. The PAH concentrations measured in Rwanda using both samplers were also generally lower than in previous studies that collected particulates using HVASs in East African countries such as Uganda (74 ng m−3)45 and Kenya (201 ng m−3).46 The high mean concentrations in Kenya and Uganda were expected due to a large number of diesel-fueled vehicles/motorcycles, many industries, and larger population growth compared to Rwanda.36
The LVAS measured more of the lighter PAHs (2.57 ng m−3) than the HVAS (1.9 ng m−3). In contrast, a higher sampling efficiency was observed for high molecular weight PAHs with the HVAS (21.01 ng m−3) than the LVAS (18.09 ng m−3) in a side-by-side comparison. A possible explanation for this disagreement may involve volatilization and flow rate (volume of air collected).47 found that compounds with lower molecular weights were more volatile and thus likely to show significant losses during long sampling periods; however, the sampling times were equal in our study. This can be described as a negative artifact (the loss of semivolatile organic compounds (SVOCs) collected on large filters using HVASs).48,49 These last authors showed that an LVAS modified with a polyurethane foam (PUF) sorbent measured more of the lighter (smaller molecular weight) SVOCs in a side-by-side comparison with an HVAS with PUF,36 suggesting that higher flow rates through the HVAS may have decreased retention of the lighter SVOCs, which may be what we are observing with our data. Several studies have recommended adsorbents such as PUF or styrene/divinylbenzene (XAD) for sampling PAHs in vapour phases.50 In contrast,51 showed that adsorbents such as PUF or XAD for PAH analysis may cause contamination between PAH and PUF and is also a time-consuming approach.
The source of PAHs and NPAHs in cities depends on the type of energy used since many cities in developing countries use wood, oil and coal for heating and cooking. In Rwanda, the primary source of PAHs and NPAHs are old diesel vehicle emissions and biomass burning.36 The 2 and 3-ring PAHs (Nap, Ant, Phe, Fle, Phe) are mainly emitted from biomass burning (wood burning) while PAHs of 4 rings (Flu, Pyr, BaA, Chr, BbF) 5-rings (BbF, BkF, BaP) and 6 rings (DBA, BPe, IDP) are mainly emitted from diesel engines and they are used as marker of vehicular exhaust emissions.7 The PAHs (Phe, Flue and Pyr) have been associated with salt particles.7 The NPAHs detected in this study in Rwanda originated from different sources such as diesel exhaust (1-NP, 7-NBaA, and 1, 3-, 1,8-DNPs), secondary formation (2-NP and 2-NFR) and from both primary emissions (vehicle and wood-burning) and secondary formation (9-NA). In this study, we found discrepancies in the levels of these PAH and NPAH compounds detected in both samplers.36
For instance, the dominant PAHs in both samplers were BPe, Flu, and BbF, while 9-NA, 2-NP and 2-NFR dominated the NPAHs. These compounds have been previously detected in Kenya, Uganda and Japan in areas with high vehicle emissions.16,52–54 The mean concentrations of all carcinogenic PAHs compounds described by the International Agency for Research on Cancer (IARC)55 were higher in HVAS than in LVAS samples. Benzo(a)pyrene (BaP) is used as a marker for carcinogenic risk levels (group 1) due to its recognized properties as an epidemiological health hazard.56 The atmospheric standard for BaP is set in Europe at 1 ng/m3 (ref. 57) and in New Zealand at 0.3 ng/m3 (ref. 58) as an annual guideline. The value of BaP in this study was 2.64 ng m−3 in the HVAS and 1.56 ng m−3 in the LVAS. The concentration of BaP measured in this study for HVAS was comparable to that found in our previous study at the same site but more than four times lower than that measured at an urban roadside in Kigali.36 Research has shown that the volatilization of particulate PAHs collected on filters represents a considerable loss, especially PAHs containing less than five rings.50 These PAHs (vapour-phase PAHs) volatilize during sampling. The small filter area size in LVAS could cause the possible reaction of collected organics with other airborne chemicals such as ozone, which could cause degradation of PAHs on the filter during sampling compared with an HVAS.50,59 Losses of PAHs from small filter areas and flow rates may be due to their reaction with other environmental pollutants during long sampling periods. For example, BaP deposited on a filter was reported to undergo chemical reactions with ozone and nitric acid, with losses as high as 85%,60 suggesting that the small filter size is more likely to enhance large reactions in an LVAS than in an HVAS.
Only three publications available studying Africa have analyzed NPAH in air particulate matter in Rwanda,36 Egypt61 and Algeria.62 The mean concentrations of 1-NP detected in both the HVAS and the LVAS in Rwanda were higher than those reported in Algeria62 and Egypt.61 1-NP is emitted from vehicles, especially diesel engines.34 There is a high density of ageing diesel vehicles (high emission rates) in Rwanda (often very old and poorly maintained), suggesting that vehicle exhausts were a major contributor to these NPAHs. Nitrated PAHs (NPAHs) are formed through nitration reactions of PAHs, and DNP (1,3-, 1,6-, or 1,8-DNP) is produced by the nitration reaction of 1-NP. Therefore, the atmospheric concentration of 1-NP is always higher than that of DNP. In this study, the mean concentration of 1,8-DNP was significantly higher than 1-NP.34 A possible reason for this discrepancy is the misidentification of fragments produced from coexisting substances as the m/z value of DNP, which is considered at the trace level of quantification. A previous study conducted at the same site in Rwanda also found that the 1,8-DNP concentration was higher than that of 1-NP,36 suggesting an influence of location, season, and climate, which results in spatio-temporal variations in the composition, concentration, and toxicity of these compounds. This strongly suggests that contributors other than automobiles likely account for the 1,8-DNP in Rwanda. The highest mean concentrations of 1, 8-DNP and 7-NBaA were detected in the HVAS rather than the LVAS. These NPAHs, which exist at concentrations orders of magnitude lower than PAHs, are challenging to determine due to their very low concentrations compared to other NPAHs. Thus, it is expected that the high flow rate of an HVAS (1000 L min−1) is likely to capture a significantly larger amount of aerosolized particulate (PAHs) on its large filter than an LVAS. The 24 hours of sampling with the LVAS provided sufficient material to investigate most PAH and NPAH levels in Rwanda. However, in developed countries such as New Zealand, with relatively clean air and situated far from other polluted land masses, the detection of these PAHs and NPAHs collected and analysed using same methodology as our current study requires long sampling periods (7 days) to achieve adequate analytical sensitivity.32
This study demonstrated that HVASs are more effective than LVASs for airborne microbial analyses associated with particulate matter. A higher concentration of genomic DNA was detected from the LVAS than from the HVAS. Studies have indicated that the diversity and richness of airborne microbial communities depend on high-quality DNA for downstream analysis.63 High levels of DNA observed in the LVAS suggested that particulates sampled using an LVAS are uniformly distributed over the filter and can reach equilibrium (saturation); thus, a small filter area size can be extracted and still reflect the whole filter.64 It was found that the HVAS was better suited to detect airborne microbial communities than the LVAS65 compared HVAS and LVAS systems for viral aerosol sampling during an emergency response and suggested that HVAS be considered in conducting a health risk assessment. However, if low concentrations are expected (clean environments with low concentrations of particulates), the highest volume of air or longest sampling duration are preferred to capture representative samples for downstream airborne microbial analysis.65 The community diversity of airborne bacteria estimated as a Shannon index value was higher in the HVAS than in the LVAS. These results suggest that the flow rate affected measured community diversity when both samplers were run simultaneously. Previous studies did not find significant differences in bacterial diversity and richness between different air pollution levels in China,66,67 in which similar techniques (culture-independent methods) were also employed.68 indicated that the number of total viable bacteria compared to total microbes increased with an increasing air quality index (AQI) in Qingdao, China. However, the association between airborne microorganisms and an increase or decrease in air pollution remains unclear.69 Using culture-independent techniques, the airborne bacterial communities sampled in Rwanda using an HVAS and an LVAS were similar to those identified in other ambient air samples. In this study, the phyla and classes detected with both samplers are commonly found in most atmospheric studies of outdoor air in developed countries.66,70–72 The bacterial phyla Actinobacteria, Firmicutes, Chloroflexi, Bacteroidetes, and Cyanobacteria were detected in both samplers, which are previously found to contain species pathogenic to humans and plants.28,70,73–75 A greater abundance of Actinobacteria, usually found in soil and dust samples, was most dominant in the HVAS samples. Rwanda has dirt roads, and the high flowrate and large filter size of the HVAS could lead to the collection of more air volume with atmospheric dust-associated bacteria on the large filter area size compared to the LVAS with a low-flow rate.
Proteobacteria and Alphaproteobacteria dominated the LVAS, while Firmicutes were dominant in the HVAS. Our pore filter size for the LVAS was 0.8 μm compared to 1.0 μm for the HVAS and Proteobacteria have been previously found to be more abundant in PM2.5 than in PM10. This was also supported by,28 who found a higher abundance of Proteobacteria in PM2.5 than PM10 in China. The tendency of Firmicutes (soil- and plant-associated bacteria) to form aggregates was thought to contribute to their association with large particle fractions.76 The high abundance of Firmicutes in the HVAS can be attributed to the high flow rate that collected a larger volume of air (and thus dust-associated bacteria) than the LVAS. Bacterial communities collected from the HVAS and the LVAS showed clear clustering of samples, suggesting that the samplers had different and distinctive communities. However, the HVAS communities resembled each other more than the LVAS samples. These results were in good agreement with a previous study that compared the performance of HVASs and LVASs and found that certain species were different from one sampler to the other in polluted environments such as subways.63
Our study used PCoA to elucidate the differences in bacteria communities from PM10 fractions by sampler types. The results found in this study show a significant difference between the two samples despite being co-allocated in close locations. These discrepancies (distinct bacterial communities) in the HVAS and LVAS samples can be explained by different factors such as sampler type, flowrate, and filter type and pore size. HVAS is in a housing designed to face the prevailing winds. Thus, PM10 impaction inlet ensures wind-direction insensitivity compared to LVAS. Both samplers were from different manufacturers, and this difference in design may also affect collection efficiency, as demonstrated in a previous study.77 Additionally, meteorological factors such as wind can influence the diversity and richness of airborne microorganisms.78 The large filter area size for HVAS with a high-flow rate, which was more than 10 times that of LVAS, could lead to the collection of more soil and plant bacteria communities attached to PM than LVAS with a low-flow rate and small filter size. A large filter area size can lead to more formation of bacteria. In contrast, the small filter area size in LVAS could cause the possible reaction of collected microorganisms with other airborne pollutants, which could cause the degradation of bacteria. Further, the level of PAHs and NPAHs found in this study were higher in HVAS than in LVAS. According to,79 PM air pollution usually contains organic compounds, soot and toxic metals, which are harmful to bioaerosols. Other studies found that the large filter samples collected with high flowrate sampler result in the uneven distribution of biomass across the filter, further complicating DNA extractions and downstream analysis.80 A large filter area size can also allow the bacteria to quickly disperse in the atmosphere compared to a small filter area. Previous study showed that the flow rate and pore size could affect the collection efficiency of airborne microorganisms.81 They showed that the relative survival of microbial communities became lower with a high flow rate and a filter with a smaller pore size usually has a higher resistance to a high flow rate.
This study demonstrated that commercially available HVASs were more efficient in detecting airborne bacteria communities in air particulate matter than LVAS. The bacterial community's genera, including Acinetobacter, Pseudomonas, Methylobacterium and Micrococcus, detected in this study, have been found to degrade PAHs, especially those with lower molecular weights (2 to 4 rings) as their sole carbon source.82 In the present study, PAHs with lower molecular weight were high in the LVAS while the airborne bacterial community diversity was higher in samples from the HVAS; these could have degraded PAHs, especially those with lower molecular weight, but further investigation is required.
5. Limitations of this study
This study focused only on PAHs and NPAHs as model chemicals and airborne bacteria as model biologicals to compare two sampling devices. Due to the logistical complexity of sample collection and the extensive variety of analytical techniques, the study period was limited and lacked the long-term monitoring component necessary for extensive regulatory review. However, the information provided in this paper can be used as a baseline for further long-term analysis in Africa, where there is a lack of information on the chemical and biological composition of PM. This study analyzed the difference between an HVAS and an LVAS for PAHs and NPAHs and conducted a bacterial analysis using a single sample duration (24 hours) at a single site (urban) in one season (wet) using one collection medium (glass fibre filters). While these factors could have affected the samplers' performance, time and resources did not allow the investigation of other parameters such as different sampling durations, different filter media, meteorological factors, and analysis of other chemicals (trace metals, dust, minerals, soot, or smoke), and biological compositions (airborne virus and fungi) of particulate matter. Therefore, further studies are required and should consider the effect of all the mentioned factors, with long-term monitoring to understand the association between HVASs and LVASs and the recovery of chemical and biological materials associated with airborne particulates.6. Conclusion
This study was the first investigation in Rwanda to compare HVAS and LVAS sampling systems operating in conjunction to investigate chemical (PAHs and NPAHs) and biological (bacterial) abundance and speciation associated with airborne PM10. We collected PM10 data with two sampling devices simultaneously (side-by-side) in the same season and location, suggesting that meteorological factors affected both sampling devices in the same manner. The 24 h mean PM10 concentrations were higher in the HVAS than the LVAS and exceeded WHO guidelines for both samplers. Both samplers allowed the successful determination of particle-bound PAH and NPAH concentrations and bacterial communities. However, the HVAS enabled the detection of important PAH and NPAH compounds, and microbial communities present in air samples that are known to impact human health. There was a performance difference between the two samplers, and they are not directly comparable. HVASs are very expensive and require consistent maintenance, thus limiting air quality information on aerosol composition in Africa. Commercial companies should design more affordable and easier-to-use samplers that can be used as an alternative to collect air quality information in Africa, where emissions are high, and data are lacking.Author contributions
E. K. and M. A. conceived and designed the study, E. K. collected and analysed the data, and E. K. and M. A. interpreted the data, wrote the manuscript. E. K., M. A. and V. K. contributed equally to the analysis and discussion of the results and edit the final version of the manuscript.Conflicts of interest
We declare no competing financial interests.Acknowledgements
We thank the University of Rwanda and Rwanda Environmental Management Authority for providing permits and access to the sampling site. In addition, we would like to thank K. Hayakawa (Kanazawa University, Japan), S. Pointing (Yale-NUS College, Singapore), K. Lee, S. Archer, and D. Lacap-Bugler (Auckland University of Technology, New Zealand) for providing support in the study design. E. K. wishes to thank the financial support from the University of Toronto postdoctoral fellowship award.References
- E. P. Petkova, D. W. Jack, N. H. Volavka-Close and P. L. Kinney, Air Qual., Atmos. Health, 2013, 6, 603–614 CrossRef CAS.
- World Health Organization, WHO Global Air Quality Guidelines. Particulate matter (PM2.5 and PM10), ozone, nitrogen dioxide, sulfur dioxide and carbon monoxide, 2021 Search PubMed.
- D. S. Jyethi, P. S. Khillare and S. Sarkar, Environ. Sci. Pollut. Res. Int., 2014, 21, 366–378 CrossRef CAS PubMed.
- O. M. Morakinyo, M. I. Mokgobu, M. S. Mukhola and R. P. Hunter, Int. J. Environ. Res. Public Health, 2016, 13, 1–22 Search PubMed.
- Y. Zhai, X. Li, T. Wang, B. Wang, C. Li and G. Zeng, Environ. Int., 2018, 113, 74–90 CrossRef PubMed.
- K.-H. Kim, S. A. Jahan, E. Kabir and R. J. C. Brown, Environ. Int., 2013, 60, 71–80 CrossRef CAS PubMed.
- K. Ravindra, R. Sokhi and R. Van Grieken, Atmos. Environ., 2008, 42, 2895–2921 CrossRef CAS.
- L. H. Keith, Polycyclic Aromat. Compd., 2015, 35, 147–160 CrossRef CAS.
- I. J. Keyte, A. Albinet and R. M. Harrison, Sci. Total Environ., 2016, 566–567, 1131–1142 CrossRef CAS PubMed.
- S. Lara, F. Villanueva, P. Martín, S. Salgado, A. Moreno and P. Sánchez-Verdú, Chemosphere, 2022, 294, 133745 CrossRef CAS PubMed.
- J. M. Delgado-Saborit, C. Stark and R. M. Harrison, Environ. Int., 2011, 37, 383–392 CrossRef CAS PubMed.
- M. A. Alghamdi, M. Shamy, M. A. Redal, M. Khoder, A. H. Awad and S. Elserougy, Sci. Total Environ., 2014, 479–480, 109–116 CrossRef CAS PubMed.
- M. S. Happo, O. Sippula, P. I. Jalava, H. Rintala, A. Leskinen, M. Komppula, K. Kuuspalo, S. Mikkonen, K. Lehtinen, J. Jokiniemi and M. Hirvonen, Part. Fibre Toxicol., 2014, 11, 60 CrossRef PubMed.
- D. Yan, T. Zhang, J. Su, L.-L. Zhao, H. Wang, X.-M. Fang, Y.-Q. Zhang, H.-Y. Liu and L.-Y. Yu, Front. Microbiol., 2016, 1–12 Search PubMed.
- N. Yousef, M. J. Omar, M. R. Bin, K. Aziz and N. Mohd, Atmos. Environ., 2002, 36, 247–254 CrossRef.
- K. Hayakawa, Polycyclic Aromatic Hydrocarbons : Environmental Behavior and Toxicity in East Asia, Springer, Singapore, 2018 Search PubMed.
- P. Castells, F. J. Santos and M. T. Galceran, J. Chromatogr. A, 2003, 1010, 141–151 CrossRef CAS PubMed.
- M. Tsapakis and E. G. Stephanou, Environ. Pollut., 2005, 133, 147–156 CrossRef CAS PubMed.
- S. K. Pandey, K.-H. Kim and R. J. C. Brown, TrAC, Trends Anal. Chem., 2011, 30, 1716–1739 CrossRef CAS.
- G. L. Andersen, W. J. Wilson, T. Z. DeSantis, J. L. Radosevich and J. H. Shinn, Lett. Appl. Microbiol., 2003, 34, 162–167 Search PubMed.
- J. M. Delgado-Saborit, N. Aquilina, S. Baker, S. Harrad, C. Meddings and R. M. Harrison, Anal. Methods, 2010, 2, 231 RSC.
- H. Giergielewicz-Mozajska, L. Dabrowski and J. Namiesnik, Crit. Rev. Anal. Chem., 2001, 31, 149–165 CrossRef CAS.
- V. R. Després, J. Alex Huffman, S. M. Burrows, C. Hoose, A. S. Safatov, G. Buryak, J. Fröhlich-Nowoisky, W. Elbert, M. O. Andreae, U. Pöschl and R. Jaenicke, Tellus B: Chem. Phys. Meteorol., 2012, 64(1), 15598 CrossRef.
- J. Fröhlich-Nowoisky, D. a Pickersgill, V. R. Després and U. Pöschl, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 12814–12819 CrossRef PubMed.
- H. Behzad, T. Gojobori and K. Mineta, Genome Biol. Evol., 2015, 7, 1216–1226 CrossRef CAS PubMed.
- W. Jiang, P. Liang, B. Wang, J. Fang, J. Lang, G. Tian, J. Jiang and T. F. Zhu, Nat. Protoc., 2015, 10, 768–779 CrossRef CAS PubMed.
- I. Luhung, B. Cao, D. Miller, C. K. Ng, V. W.-C. Chang and Y. Wu, PLoS One, 2015, 10, e0141158 CrossRef PubMed.
- C. Cao, W. Jiang, B. Wang, J. Fang, J. Lang, G. Tian, J. Jiang and T. F. Zhu, Environ. Sci. Technol., 2014, 1499–1507 CrossRef CAS PubMed.
- J. L. Durant, W. F. Busby, A. L. Lafleur, B. W. Penman and C. L. Crespi, Mutat. Res., Genet. Toxicol., 1996, 371, 123–157 CrossRef CAS.
- J. J. West, A. Cohen, F. Dentener, B. Brunekreef, T. Zhu, B. Armstrong, M. L. Bell, M. Brauer, G. Carmichael, D. L. Costa, D. W. Dockery, M. Kleeman, M. Krzyzanowski, N. Künzli, C. Liousse, S. C. C. Lung, R. V. Martin, U. Pöschl, C. A. Pope, J. M. Roberts, A. G. Russell and C. Wiedinmyer, Environ. Sci. Technol., 2016, 50, 4895–4904 CrossRef CAS PubMed.
- K. Peltonen and T. Kuljukka, J. Chromatogr., 1995, 710, 93–108 CrossRef CAS.
- E. Kalisa, E. Nagato, E. Bizuru, K. Lee, N. Tang, S. Pointing, K. Hayakawa, S. Archer and D. Lacap-Bugler, Atmos. Pollut. Res., 2019, 10, 1396–1403 CrossRef CAS.
- K. Hayakawa, N. Tang, E. G. Nagato, A. Toriba, S. Sakai, F. Kano, S. Goto, O. Endo, K. ichi Arashidani and H. Kakimoto, Environ. Pollut., 2018, 233, 474–482 CrossRef CAS PubMed.
- K. Hayakawa, Chem. Pharm. Bull., 2016, 64, 83–94 CrossRef CAS PubMed.
- M. Gao, R. Jia, T. Qiu, M. Han, Y. Song and X. Wang, Atmos. Environ., 2015, 118, 203–210 CrossRef CAS.
- E. Kalisa, E. G. Nagato, E. Bizuru, K. C. Lee, N. Tang, S. B. Pointing, K. Hayakawa, S. D. J. Archer and D. C. Lacap-bugler, Environ. Sci. Technol., 2018, 52, 12179–12187 CrossRef CAS PubMed.
- S. D. J. Archer, I. R. McDonald, C. W. Herbold, C. K. Lee and C. S. Cary, Front. Microbiol., 2015, 6, 1–11 Search PubMed.
- S. D. J. Archer, K. C. Lee, T. Caruso, T. Maki, C. K. Lee, S. C. Cary, D. A. Cowan, F. T. Maestre and S. B. Pointing, Nat. Microbiol., 2019, 4(6), 925–932 CrossRef CAS PubMed.
- B. J. Callahan, P. J. McMurdie, M. J. Rosen, A. W. Han, A. J. A. Johnson and S. P. Holmes, Nat. Methods, 2016, 13, 581–583 CrossRef CAS PubMed.
- T. Maki, K. Hara, F. Kobayashi, Y. Kurosaki, M. Kakikawa, A. Matsuki, B. Chen, G. Shi, H. Hasegawa and Y. Iwasaka, Atmos. Environ., 2015, 282–293 CrossRef CAS.
- J. Oksanen, F. G. Blanchet, R. Kindt, P. Legendre, P. R. Minchin, et al., Community Ecol. Package R Package Version 23-1 Search PubMed.
- K. C. Lee, S. D. J. Archer, T. Caruso, L. Gillman, M. C. Lau, S. C. Cary, C. K. Lee and S. B. Pointing, Front. Microbiol., 2018, 9, 2619 CrossRef PubMed.
- M. J. Anderson, K. E. Ellingsen and B. H. McArdle, Ecol. Lett., 2006, 9, 683–693 CrossRef PubMed.
- K. Sugita, Y. Kin, M. Yagishita, F. Ikemori, K. Kumagai, T. Ohara, M. Kinoshita, K. Nishimura, Y. Takagi and D. Nakajima, Genes Environ., 2019, 41, 1–11 CrossRef PubMed.
- K. Arinaitwe, B. T. Kiremire, D. C. G. Muir, P. Fellin, H. Li, C. Teixeira and D. N. Mubiru, Environ. Sci. Technol., 2012, 46, 11524–11531 CrossRef CAS PubMed.
- M. Muendo, Y. Hanai, Y. Kameda and S. Masunaga, Atmos. Res., 2006, 7(2), 147–157 CAS.
- G. P. Schwartz, J. M. Daisey and P. J. Lioy, Am. Ind. Hyg. Assoc. J., 1981, 42, 258–263 CrossRef CAS.
- H. Tang, E. A. Lewis, D. J. Eatough, R. M. Burton and R. J. Farber, Atmos. Environ., 1994, 28, 939–947 CrossRef CAS.
- D. J. Eatough, H. Tang, W. Cui and J. Machir, Inhalation Toxicol., 1995, 7, 691–710 CrossRef CAS.
- C. S. Davis, P. Fellin and R. Otson, J. Air Pollut. Control Assoc., 1987, 37, 1397–1408 CrossRef CAS PubMed.
- V. Paolini, E. Guerriero, A. Bacaloni, M. Rotatori, P. Benedetti and S. Mosca, Aerosol Air Qual. Res., 2016, 16, 175–183 CrossRef CAS.
- X.-Y. Yang, Y. Okada, N. Tang, S. Matsunaga, K. Tamura, J.-M. Lin, T. Kameda, A. Toriba and K. Hayakawa, Atmos. Environ., 2007, 41, 2710–2718 CrossRef CAS.
- K. Arinaitwe, B. T. Kiremire, D. C. G. Muir, P. Fellin, H. Li, C. Teixeira and D. N. Mubiru, Environ. Sci. Technol., 2012, 46, 11524–11531 CrossRef CAS PubMed.
- M. Muendo, Y. Hanai, Y. Kameda and S. Masunaga, Environ. Forensics, 2006, 7, 147–157 CrossRef CAS.
- IARC-International Agency for Research on Cancer, The Carcinogenicity of Outdoor Air Pollution, Lyon, 2013, vol. 140 Search PubMed.
- H. I. Abdel-Shafy and M. S. M. Mansour, Egypt. J. Pet., 2015, 25, 107–123 CrossRef.
- European Environmental Agency, Air Quality in Europe – 2020 Report, 2020 Search PubMed.
- Ministry for the Environment and Statistics New Zealand, Environment Aotearoa 2015, 2015 Search PubMed.
- K. Liu, F. Duan, K. He, Y. Ma and Y. Cheng, 2014, 8(2), 284–292.
- K. Peltonen and T. Kuljukka, J. Chromatogr. A, 1995, 710, 93–108 CrossRef CAS.
- H. F. Nassar, N. Tang, T. Kameda, A. Toriba, M. I. Khoder and K. Hayakawa, Atmos. Environ., 2011, 45, 7352–7359 CrossRef CAS.
- R. Ladji, N. Yassaa, C. Balducci, A. Cecinato and B. Youcef, Atmos. Res., 2009, 92(2), 258–269 CrossRef CAS.
- K. O. Bøifot, J. Gohli, G. Skogan and M. Dybwad, Environ. Microbiome, 2020, 15, 1–16 CrossRef PubMed.
- R. Subramanian, A. Y. Khlystov, J. C. Cabada and A. L. Robinson, Aerosol Sci. Technol., 2004, 38, 27–48 CrossRef CAS.
- C. Cooper, J. Slagley, J. Lohaus Jr, E. Escamilla, C. Bliss, D. Semler, D. Felker, D. Smith and D. Ott, J. Emerg. Manag., 2014, 12, 161 CrossRef PubMed.
- A. C. Woo, M. S. Brar, Y. Chan, M. C. Y. Y. Lau, F. C. C. C. Leung, J. A. Scott, L. L. P. P. Vrijmoed, P. Zawar-Reza and S. B. Pointing, Atmos. Environ., 2013, 74, 291–300 CrossRef CAS.
- Y. Wang, Q. Zhang, Y. Zhang, H. Zhao, F. Tan, X. Wu and J. Chen, Chemosphere, 2019, 216, 516–523 CrossRef CAS PubMed.
- J. Gong, J. Qi, B. E, Y. Yin and D. Gao, Environ. Pollut., 2020, 257, 113485 CrossRef CAS PubMed.
- M. Shammi, M. M. Rahman and S. M. Tareq, Front. Environ. Sci., 2021, 9, 1–15 Search PubMed.
- R. M. Bowers, N. Clements, J. B. Emerson, C. Wiedinmyer, M. P. Hannigan and N. Fierer, Environ. Sci. Technol., 2013, 47(21), 12097–12106 CrossRef CAS PubMed.
- K. Dannemiller, N. Yamamoto, N. Burshtein, J. Peccia, O. Yarden and Y. Rudich, Atmos. Chem. Phys., 2012, 2681–2690 Search PubMed.
- D. Haas, H. Galler, J. Luxner, G. Zarfel, W. Buzina, H. Friedl, E. Marth, J. Habib and F. F. Reinthaler, Atmos. Environ., 2013, 65, 215–222 CrossRef CAS.
- S.-H. Lee, H.-J. Lee, S.-J. Kim, H. M. Lee, H. Kang and Y. P. Kim, Sci. Total Environ., 2010, 408, 1349–1357 CrossRef CAS PubMed.
- R. M. Bowers, S. McLetchie, R. Knight and N. Fierer, ISME J., 2011, 5, 601–612 CrossRef CAS PubMed.
- J. F. Gao, X. Y. Fan, H. Y. Li and K. L. Pan, Aerosol Air Qual. Res., 2017, 17, 788–798 CrossRef.
- H. Gou, J. Lu, S. Li, Y. Tong, C. Xie and X. Zheng, Environ. Pollut., 2016, 202–210 CrossRef CAS PubMed.
- W. G. Lindsley, F. M. Blachere, D. H. Beezhold, R. E. Thewlis, B. Noorbakhsh, S. Othumpangat, W. T. Goldsmith, C. M. McMillen, M. E. Andrew, C. N. Burrell and J. D. Noti, Influenza Other Respir , Viruses, 2016, 10(5), 404–413 Search PubMed.
- R. Tignat-Perrier, A. Dommergue, A. Thollot, C. Keuschnig, O. Magand, T. M. Vogel and C. Larose, Sci. Rep., 2019, 9, 14441 CrossRef PubMed.
- M. Gao, X. Yan, T. Qiu, M. Han and X. Wang, Atmos. Environ., 2016, 128, 10–19 CrossRef CAS.
- N. Lang-Yona, Y. Mazar, M. Pardo and Y. Rudich, J. Visualized Exp., 2016, 1–11 Search PubMed.
- S. A. Grinshpun, M. P. Buttner, G. Mainelis and K. Willeke, in Manual of Environmental Microbiology, John Wiley & Sons, Ltd, 2016, pp. 3.2.2-1–3.2.2-17 Search PubMed.
- D. Ghosal, S. Ghosh, T. K. Dutta and Y. Ahn, Front. Microbiol., 2016, 7 Search PubMed.
| This journal is © The Royal Society of Chemistry 2022 |