Novel anti-hyperuricemic hexapeptides derived from Apostichopus japonicus hydrolysate and their modulation effects on the gut microbiota and host microRNA profile†
Siqing
Fan‡
ab,
Yumeng
Huang‡
ac,
Guoding
Lu
d,
Na
Sun
d,
Rui
Wang
b,
Chenyang
Lu
 *ac,
Lijian
Ding
b,
Jiaojiao
Han
ac,
Jun
Zhou
ac,
Ye
Li
*ac,
Lijian
Ding
b,
Jiaojiao
Han
ac,
Jun
Zhou
ac,
Ye
Li
 ac,
Tinghong
Ming
*ac and
Xiurong
Su
ac
ac,
Tinghong
Ming
*ac and
Xiurong
Su
ac
aState Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Ningbo University, Ningbo, China. E-mail: luchenyang@nbu.edu.cn
bCollege of Food and Pharmaceutical Sciences, Ningbo University, Ningbo, China
cSchool of Marine Science, Ningbo University, Ningbo, China
dNingbo Green-Health Pharmaceutical Co., Ltd, Ningbo, China
First published on 4th March 2022
Abstract
Hyperuricemia (HUA) is the second most common metabolic disease nowadays, and is characterized by permanently increased concentrations of serum uric acid. In this study, two novel hexapeptides (GPAGPR and GPSGRP) were identified from Apostichopus japonicus hydrolysate and predicted to have xanthine oxidase (XOD) inhibitory activity by molecular docking. Their in vitro XOD inhibition rates reached 37.3% and 48.6%, respectively, at a concentration of 40 mg mL−1. Subsequently, in vivo experiments were carried out in a HUA mouse model, and we found that both peptides reduced the serum uric acid by inhibiting uric acid biosynthesis and reabsorption, as well as alleviated renal inflammation via suppressing the activation of the NLRP3 inflammasome. 16S rDNA sequencing indicated that both peptide treatments reduced the richness and diversity of the gut microbiota, altered the composition in the phylum and genus levels, but different change trends were observed in the phylum Verrucomicrobia and genera Akkermansia, Dubosiella, Alloprevotella, Clostridium unclassified and Alistipes. In addition, changes in the renal microRNA (miRNA) profiles induced by GPSGRP treatment were analyzed; 21 differentially expressed (DE) miRNAs were identified among groups, and KEGG pathway analysis indicated that their potential target genes were involved in pluripotency of stem cell regulation, mTOR signaling pathway and proteoglycans. Moreover, ten miRNAs involved in the HUA onset and alleviation were identified, which showed a high correlation with genera related to the metabolism of short-chain fatty acids, bile acids and tryptophan. This study delineated two hexapeptides as potential microbiota modulators and miRNA regulators that can ameliorate HUA.
Introduction
Hyperuricemia (HUA) is a progressive metabolic condition caused by permanently increased concentrations of serum uric acid and has gradually become the second most common metabolic disease after type 2 diabetes.1 Uric acid is mainly synthesized in the liver, and xanthine oxidase (XOD), one of the rate-limiting enzymes which is widely used as a HUA management target, catalyzes hypoxanthine to xanthine and then to uric acid.2 In addition, uric acid is excreted mainly via the kidneys mediated by reabsorption (GLUT9, URAT1, etc.) and secretion (MRP4, ABCG2, etc.).3 The overactive uric acid production in the liver that exceeds uric acid excretion by the kidneys and intestinal tract contributes to HUA, and long-term high uric acid levels and uric acid crystal deposition exacerbate oxidative stress, up-regulate the TLR4/MyD88/NF-κB signaling pathway to generate pro-IL-1β, and activate the NLRP3 inflammasome for the cleavage of IL-1β from pro-IL-1β, and subsequently leads to renal inflammation and injury.4 Moreover, HUA will increase the incidence of related diseases, including gout, kidney diseases, metabolic syndromes and cardiovascular disease.5 Due to the inefficiency of nonpharmacological therapies, such as diet management, in reducing uric acid in patients, pharmacological therapies are needed. There are three main categories of uric acid management drugs: uric acid production inhibitors (allopurinol and febuxostat), excretion promoters (probenecid and benzbromarone) and recombinant uricase (pegloticase). The first category is recommended as the first-line treatment for HUA in many countries, but various adverse effects have still been reported.6As previously reported, Apostichopus japonicus oligopeptide (AJOP), prepared with an edible protein, is a safer alternative that exerts anti-HUA effects by rebalancing uric acid metabolism and inhibiting the NLRP3 inflammasome and NF-κB signaling pathway activation.3,7 The various components and their multiple targets not only contribute to the comprehensive anti-HUA effects of AJOP but also hinder the further development of AJOP due to the unclear understanding of key peptides with anti-HUA effects. The purify-and-identify method has been widely used to screen peptides with distinct functions and structures via iterative rounds, but this method is one-sided, labor intensive and time-consuming.8 In a previous study of AJOP, in silico molecular docking instead of the purify-and-identify method was used to predict the XOD inhibition activity of the identified peptides with high abundance,7 but the anti-HUA effects and underlying mechanism of certain peptides need to be further explored both in vitro and in vivo.
The gut microbiota has become a potential target for understanding the onset and alleviation of HUA due to the correlation and causation relationship between them. Firstly, the absence of the gut microbiota in germ-free mice or after antibiotic treatment decreased joint inflammation induced by monosodium urate monohydrate crystal injection.9 Then, modulated gut microbiota composition and metabolism were observed in hyperuricemic individuals.10,11 In addition, the contributions of gut microbiota manipulations (probiotic and prebiotic) to HUA and renal inflammation alleviation have been reported by various studies,12,13 and the anti-hyperuricemic effects of AJOP were transferable via fecal microbiota transplantation.7 What's more, the microbiota produces thousands of diffusible metabolites, and the metabolites are reported to alleviate HUA by regulating the intestinal epithelial cell proliferation, ameliorating purine metabolism disorders, promoting uric acid excretion, and reducing inflammation.14 However, the underlying mechanism by which the microbiota remotely regulates the functions of central metabolic organs, such as the liver and kidneys, is largely undetermined.
MicroRNAs (miRNAs) are small (19–23 nt) and single-stranded noncoding RNA molecules that were once considered redundant; however, accumulating studies have shown the specific and global regulatory effects of miRNAs on individual genes and regulatory networks. As previously reported, miRNAs are differentially expressed in the renal tissues of HUA and healthy individuals,15 and several highly conserved miRNAs have been identified to be involved in the regulation of uric acid biosynthesis (miR-448, etc.), secretion (miR-34a, miR-143-3p, etc.), and immune response (miR-223, etc.);16–18 thus we proposed that miRNAs play critical roles in the control of HUA development. In addition, miRNAs were reported to be regulated by the metabolites produced by the gut microbiota, including short-chain fatty acids (SCFAs), tryptophan derivatives and bile acids (BAs).19–21 However, the modulation of miRNA profiles in HUA mice by treatment with distinct structural peptides remains unclear.
Herein, two peptides GPAGPR and GPSGRP were identified from Apostichopus japonicus hydrolysate and predicted to have XOD inhibitory activity by molecular docking. The anti-HUA effects of these two peptides were confirmed in vitro (XOD inhibition rate) and in vivo (HUA mice), and their modulation effects on the gut microbiota and host renal miRNA profile were explored.
Materials and methods
AJOP preparation and composition detection
Apostichopus japonicus (A. japonicus) was purchased from the Lulin Seafood Market (Ningbo, Zhejiang, China), and AJOP was prepared with 3% trypsin and alkaline protease in a ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 at 55 °C for 4 h as previously described.7 Then, the enzyme was inactivated, and the supernatant was ultrafiltered with a 1000 Da membrane (Millipore, Burlington, MA, USA) and freeze dried using a vacuum freeze-drying machine (LABCONCO Co., Ltd, Kansas City, MO, USA). Subsequently, the composition and abundance of peptides in AJOP were measured by MALDI-TOF/TOF (Applied Biosystems, Foster City, CA, USA).
1 at 55 °C for 4 h as previously described.7 Then, the enzyme was inactivated, and the supernatant was ultrafiltered with a 1000 Da membrane (Millipore, Burlington, MA, USA) and freeze dried using a vacuum freeze-drying machine (LABCONCO Co., Ltd, Kansas City, MO, USA). Subsequently, the composition and abundance of peptides in AJOP were measured by MALDI-TOF/TOF (Applied Biosystems, Foster City, CA, USA).
Prediction of XOD inhibitory activity by molecular docking
The thirteen major peptides (abundance >1%) detected by MALDI-TOF/TOF in this study were used as ligands in molecular docking studies to predict the in silico XOD inhibitory activity (Discovery Studio 2017, Beijing Chong Teng Technology Co., Ltd, Beijing, China), and the crystal structure of bovine milk XOD (PDB ID: 1FIQ) was obtained from the RCSB Protein Data Bank (http://www.rcsb.org/pdb) as the receptor. The CDOCKER program was used with the default parameters. The interaction between the ligand and receptor was characterized by -CDOCKER interaction energy (-CIE), and the highest -CIE value indicated the strongest interaction.Measurement of XOD inhibitory activity in vitro
The peptides GPAGPR and GPSGRP were synthesized by Shanghai Mujin Biotech Co., Ltd (Shanghai, China) and dissolved in 50 mM Tris-HCl buffer at pH 7.5. First, 40 μL of XOD solution (0.05 U mL−1) and 40 μL of peptide solutions (ranging from 10 to 60 mg mL−1) or allopurinol solutions (ranging from 3 to 18 μg mL−1) were mixed at 25 °C for 10 min. Subsequently, the reaction was started by adding 120 μL of xanthine solution (0.4 mM) into the mixture, the mixture was incubated at 25 °C, and the absorbance was measured at 290 nm every 30 s up to 25 min. Three replicates were set for each sample.At the end of the experiment, the reaction was terminated by adding 64 μL of HCl (1 M), and the production of uric acid was measured by HPLC (Agilent 1200 series, Agilent Technologies Inc., CA, USA) with an Agilent ZORBAX Eclipse XDB-Phenyl column (250 × 4.6 mm i.d., 5 μm, Santa Clara, CA, USA) as previously described.22 The mobile phase was composed of water with sodium 1-pentane sulfonate (0.52 mM) and monopotassium phosphate (0.20 M) (pH 4.0, solvent A) and acetonitrile (solvent B) in a ratio of 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 (v/v). The detection wavelength was 290 nm and the flow rate was 1.0 mL min−1 at room temperature. The percentage of inhibition rate was calculated as follows: XOD inhibition rate (%) = [1 − (sample reaction UA production/control reaction UA production)] × 100%.
15 (v/v). The detection wavelength was 290 nm and the flow rate was 1.0 mL min−1 at room temperature. The percentage of inhibition rate was calculated as follows: XOD inhibition rate (%) = [1 − (sample reaction UA production/control reaction UA production)] × 100%.
Animal experiment
The present study, referring to the laboratory animal care, was approved by the Ethical Committee of Experimental Animal Care at Ningbo University under permit no. 10525.Twenty male C57BL/6 mice (five week old, 22.44 ± 1.37 g) purchased from Zhejiang Laboratory Animal Center (Hangzhou, Zhejiang, China) were housed in a specific pathogen free (SPF) room (23–25 °C, 60–80% relative humidity and a 12 h light/12 h dark cycle) with free access to water and food. After one week of acclimatization, twenty mice were randomly divided into four groups, namely, the control, model, GPAGPR and GPSGRP groups, with 5 mice per group. At 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 00, the mice in the control group received 200 μL of saline by oral gavage, and the mice in the remaining three other groups received 200 μL of HUA solution (containing 200 mg kg−1 d−1 of hypoxanthine, 30 mg kg−1 d−1 of yeast extract and 250 mg kg−1 d−1 of potassium oxonate) every day for 12 weeks as previously described.23 At 15
00, the mice in the control group received 200 μL of saline by oral gavage, and the mice in the remaining three other groups received 200 μL of HUA solution (containing 200 mg kg−1 d−1 of hypoxanthine, 30 mg kg−1 d−1 of yeast extract and 250 mg kg−1 d−1 of potassium oxonate) every day for 12 weeks as previously described.23 At 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 00, the mice in the peptide groups were administered 200 μL of GPAGPR or GPSGRP (10 mg kg−1 d−1) by oral gavage for 12 weeks, while the mice in the control and model groups were administered 200 μL of saline by oral gavage. The dosages of GPAGPR and GPSGRP were selected according to the previous results reported by our group8 and are equivalent to 49.8 mg d−1 in humans.24
00, the mice in the peptide groups were administered 200 μL of GPAGPR or GPSGRP (10 mg kg−1 d−1) by oral gavage for 12 weeks, while the mice in the control and model groups were administered 200 μL of saline by oral gavage. The dosages of GPAGPR and GPSGRP were selected according to the previous results reported by our group8 and are equivalent to 49.8 mg d−1 in humans.24
At the end of 12 weeks, urine was collected from each mouse and stored at −80 °C. After anaesthetizing the animals, the serum was separated, and the liver, kidney and cecal contents were collected and stored at −80 °C for further analysis. The uric acid and blood urea nitrogen (BUN) in the serum and urine were quantified using commercial kits (No. C012-2-1 and C013-2-1) purchased from Nanjing Jiancheng Co., Ltd (Nanjing, Jiangsu, China).
Measurement of mRNA and protein expression
mRNA and protein expressions were measured by qRT-PCR and western blot analysis. For qRT-PCR, total RNA was extracted from the liver and kidneys using the TransZol Up Plus RNA kit (TransGen Biotech, Beijing, China), and cDNA was synthesized using mRNA as a template with TransScript® All-in-One First-Strand cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). mRNA levels were measured with the primers described in Table S1† using an ABI 7500 Real Time Detection System (Applied Biosystems, Bedford, MA, USA) following the default program in a 20 μL reaction volume. Three replicates were set for each sample, and the mRNA level was normalized to the β-actin level and calculated using the 2−ΔΔCT method.As previously reported,25 the liver and kidneys were lysed in RIPA buffer (Sangon Biotech Co., Ltd, Shanghai, China), the homogenate was centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 min, the supernatant was collected as total protein, and the protein concentration was measured via a BCA protein assay kit (Beijing ComWin Biotech Co., Ltd, Beijing, China) according to the manufacturer's protocol. Denatured proteins were separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes (Millipore Corp, Billerica, MA, USA). Nonspecific binding to the membranes was blocked with 5% nonfat milk powder for 1 h at room temperature, incubated overnight with primary antibodies against Glut9 (1
000g for 15 min, the supernatant was collected as total protein, and the protein concentration was measured via a BCA protein assay kit (Beijing ComWin Biotech Co., Ltd, Beijing, China) according to the manufacturer's protocol. Denatured proteins were separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes (Millipore Corp, Billerica, MA, USA). Nonspecific binding to the membranes was blocked with 5% nonfat milk powder for 1 h at room temperature, incubated overnight with primary antibodies against Glut9 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800, D162759, 54 kDa), ADA (1
800, D162759, 54 kDa), ADA (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700, D221495, 41 kDa), ABCG2 (1
700, D221495, 41 kDa), ABCG2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500, D155255, 72 kDa), IL-1β (1
500, D155255, 72 kDa), IL-1β (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, #63124, 17 kDA), ASC (1
1000, #63124, 17 kDA), ASC (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800, #67824, 22 kDa), cleaved caspase-1 (1
800, #67824, 22 kDa), cleaved caspase-1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900, #89332, 22 kDa) and caspase-1 (1
900, #89332, 22 kDa) and caspase-1 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000, #24232, 48 kDa) at 4 °C, and then with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. The first three primary antibodies were purchased from Sangon Biotech Co., Ltd (Shanghai, China) and the rest were purchased from Cell Signaling Technology (Shanghai, China). Finally, the bands were visualized using an ECL kit (Advansta Inc, San Jose, CA, USA) with a ChemiDoc XRS+ Imaging System (Bio-Rad Laboratories Inc, Hercules, CA, USA).
1000, #24232, 48 kDa) at 4 °C, and then with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. The first three primary antibodies were purchased from Sangon Biotech Co., Ltd (Shanghai, China) and the rest were purchased from Cell Signaling Technology (Shanghai, China). Finally, the bands were visualized using an ECL kit (Advansta Inc, San Jose, CA, USA) with a ChemiDoc XRS+ Imaging System (Bio-Rad Laboratories Inc, Hercules, CA, USA).
16S rDNA amplicon sequencing and data analysis
As previously reported,26 total bacterial genomic DNA was extracted from the cecal contents with an E.Z.N.A.® Stool DNA kit (Omega, Norcross, GA, USA) and quantified with a NanoDrop 2000c spectrophotometer (Thermo-Fisher Scientific, Waltham, MA, USA). The V3–V4 region of 16S rDNA was amplified with barcoded primers 319F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) with the default program in a 25 μL reaction volume, containing 12.5 μL of Premix Ex Taq™ Hot Start Version (Takara Biotechnology Co. Ltd, Dalian, China), 0.1 μM each primer, and 20 ng of template. Amplification was initiated at 98 °C for 30 s, followed by 35 cycles of denaturation at 98 °C for 10 s, primer annealing at 54 °C for 30 s, extension at 72 °C for 45 s, and final extension for 10 min. The presence of amplicons was confirmed by gel electrophoresis, and the purified PCR products were used to prepare a PCR product library. Subsequently, the amplicon sequencing was performed in LC-Bio (Hangzhou, Zhejiang, China) with an Illumina MiSeq platform (Illumina, San Diego, CA, USA) using 2 × 300 bp PE sequencing and multiple sequencing runs. The raw data were filtered via QIIME 2 (https://qiime2.org/) and classified with the RDP database. In addition, α diversity analysis was calculated using Mothur,27 and β diversity analysis was calculated using QIIME2. The differentially abundant genera were identified via the Kruskal–Wallis test. Spearman's correlation analysis between the abundance of differentially abundant genera and phenotypes of HUA was calculated using SPSS (version 19.0, Chicago, IL, USA), and correlations were defined as significant when p < 0.05 and FDR < 0.25.miRNA profile sequencing and analysis
Total RNA was extracted from the kidneys using a commercial kit following the manufacturer's recommendations, and the concentration and quality of total RNA were detected using a NanoDrop 2000c spectrophotometer. The RNA was purified using 15% agarose gels and enriched by ethanol precipitation. A TruSeq Small RNA Sample Prep kit (Illumina, San Diego, CA, USA) was used to construct the library, which was subsequently sequenced using the Illumina HiSeq 2000/2500 platform at 1 × 50 bp.Raw reads were submitted to ACGT101-miR (LC Sciences, Houston, TX, USA) to remove adapter dimers, junk, low complexity sequences, common RNA families and repeats. Subsequently, known and novel miRNAs were identified by mapping the unique sequences (19–26 nt) to specific species precursors in miRbase (version 22.0) using BLAST. In this study, differentially expressed (DE) miRNAs between two groups based on normalized deep-sequencing counts were analyzed using Student's t test, while DE-miRNAs among the three groups were analyzed using ANOVA, and the significance threshold was set to 0.05. In addition, both TargetScan (version 5.0) and Miranda (version 3.3a) were used to predict the target genes of certain DE-miRNAs, and the overlapping genes predicted by both software programs were used for further analysis. Moreover, the target genes were annotated by GO and KEGG pathway analyses.
Statistical analysis
All data are presented as mean ± standard deviation (SD). The differences among groups were calculated by one-way analysis of variance (ANOVA) followed by Tukey's post hoc tests via MATLAB R2012a (MathWorks, Natick, MA, USA), and p < 0.05 was defined as the statistical significance.Accession number
The sequences have been deposited in the Read Archive Database under PRJNA752999 (16S rDNA sequencing) and PRJNA753202 (miRNA sequencing).Results
Potential interactions between XOD and the major peptides in AJOP
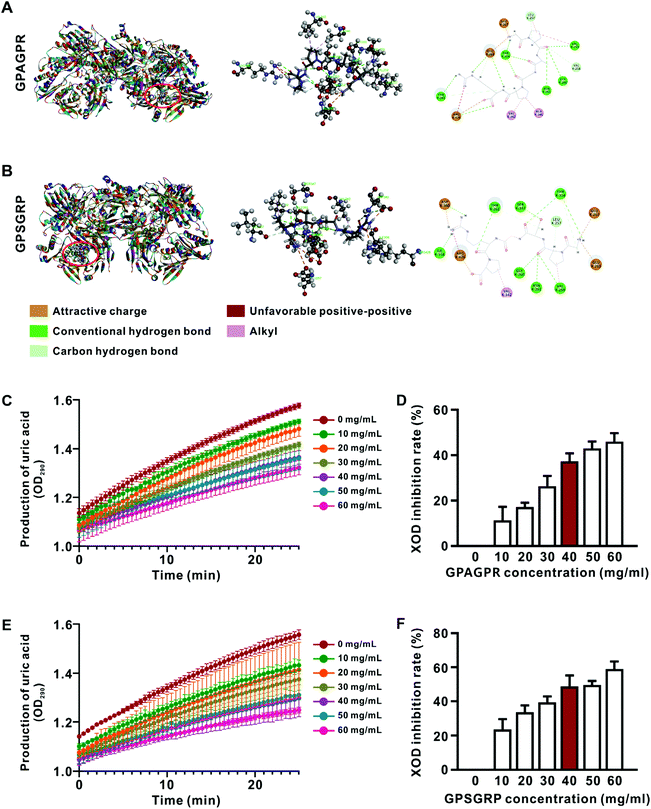
The peptide profiles in AJOP were measured by MALDI-TOF/TOF-MS as previously described,7 and 13 peptides with relatively high abundance (>1%) were identified (Table S2†). Subsequently, the potential interactions between XOD and peptides were predicted via molecular docking, and two hexapeptides with similar structures, GPAGPR and GPSGRP, showed high binding ability with XOD characterized by lower CIE (Fig. 1A, B and Table S3†). Therefore, these two similar peptides were predicted to have XOD inhibitory activities and are involved in HUA alleviation.Inhibitory effects of GPAGPR and GPSGRP on XOD activity in vitro
The peptides GPAGPR and GPSGRP were synthesized, and their inhibitory activities on XOD were subsequently measured and compared in vitro, and allopurinol (an XOD inhibitor used in clinic) was used as the positive control. The production of uric acid gradually increased in the presence of both peptides at all assessed concentrations from 0 to 25 min (Fig. 1C and E). The inhibition rate of XOD reached 41.2% in the presence of allopurinol at a concentration of 3 μg mL−1 (Fig. S1†), while GPAGPR and GPSGRP showed similar inhibition rates (37.3% and 48.6%) at a concentration of 40 mg mL−1 (Fig. 1D and F). This experiment confirmed the XOD inhibitory activities of GPAGPR and GPSGRP in vitro, even though they are relatively far lower than that of allopurinol. Then, we investigated their anti-HUA effects in vivo via a high-purine-diet (HPD)-induced HUA mouse model.GPAGPR and GPSGRP treatments attenuated HUA in mice
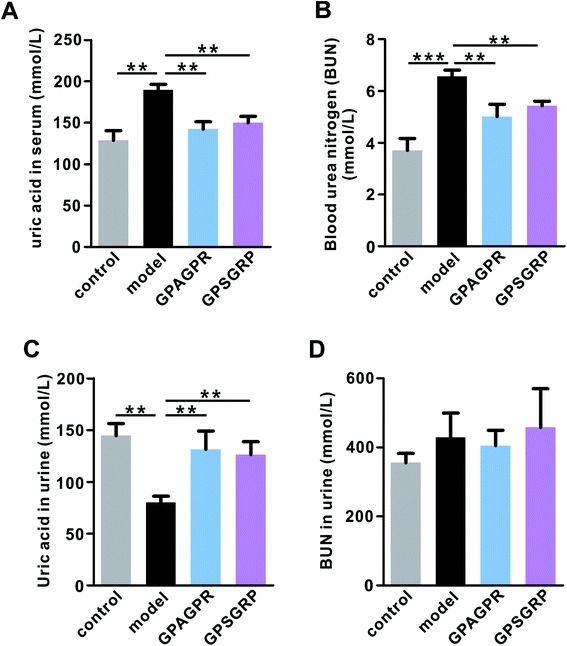
The body weights of mice in all groups gradually increased during the 12-week experiment. Both peptides significantly restored the increased body weight gain induced by the HPD treatment (p < 0.01), but relatively lower body weight gain was observed in the GPSGRP group (Fig. S2†). In addition, treatment with both peptides attenuated the phenotypes of HUA in mice, characterized by reduced uric acid (p < 0.01) and urea nitrogen in serum (p < 0.01) (Fig. 2A and B), and enhanced uric acid in the urine (p < 0.01) (Fig. 2C), and no significant difference was observed between the GPAGPR and GPSGRP groups. Moreover, neither HPD nor peptide treatments affected the urea nitrogen level of urine (Fig. 2D).GPAGPR and GPSGRP treatments rebalanced uric acid metabolism in HUA mice
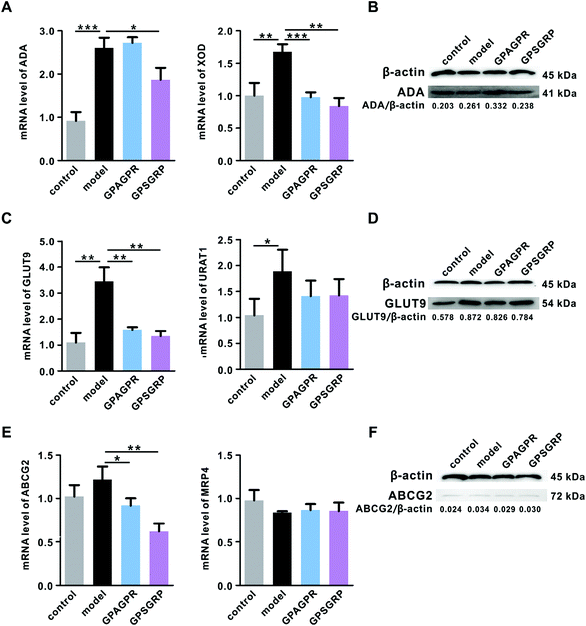
As expected, the mRNA level of XOD induced by HPD in the liver was significantly downregulated by the treatment with peptides (p < 0.001 and p < 0.01), while ADA, another key enzyme involved in the uric acid biosynthesis, was upregulated by GPAGPR (p > 0.05) and downregulated by GPSGRP (p < 0.05) in both mRNA and protein levels (Fig. 3A and B). In addition, the peptides inhibited uric acid reabsorption and secretion in the kidneys by the corresponding regulation of GLUT9 (p < 0.01) and ABCG2 (p < 0.05 and p < 0.01) at the mRNA and protein levels, respectively, but showed no significant effects on URAT1 and MRP4 (Fig. 3C–F).GPAGPR and GPSGRP treatments regulated cytokine release in HUA mice
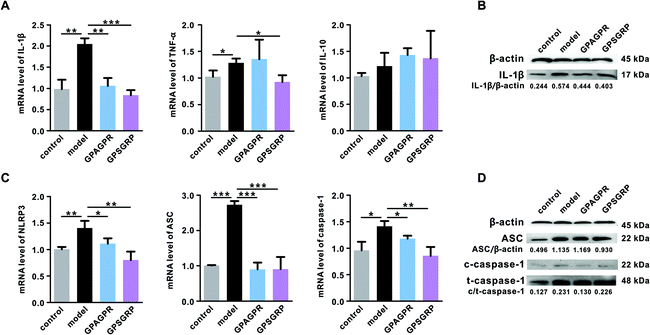
Long-term high uric acid in serum will lead to potential inflammation of the kidneys. Therefore, the mRNA and protein levels of the representative inflammatory cytokines, including IL-1β, TNF-α and IL-10, in the kidneys were measured by qRT-PCR and western blot analysis. Both peptides significantly reduced the mRNA and protein levels of IL-1β (p < 0.01 and p < 0.001). GPSGRP significantly downregulated the mRNA level of TNF-α (p < 0.05), and GPAGPR treatment further aggravated the enhanced mRNA level of TNF-α induced by HPD (p > 0.05), although not reaching a significant level. In addition, neither HPD nor peptide significantly affected IL-10 at the mRNA level (p > 0.05) (Fig. 4A and B).Considering the important roles of the NLRP3 inflammasome and TLR4/MyD88/NF-κB pathway in the release of cytokines, particularly IL-1β, the expression levels and activation of related proteins were measured. As expected, NLRP3 inflammasome activation was induced by HPD, characterized by enhanced NLRP3 (p < 0.01), ASC (p < 0.001) and caspase-1 (p < 0.05) in mRNA levels, and this activation was significantly inhibited by both peptides (Fig. 4C). In addition, the ratio of cleaved caspase-1 (c-caspase-1) to total caspase-1 (t-caspase-1) was decreased by the peptide treatments (Fig. 4D). Otherwise, neither HPD nor the peptide treatments affected the mRNA level of TLR4, even though it plays a vital role in innate immunity (Fig. S3A†). However, the increased mRNA level of MyD88 (p < 0.05), one of the key components related to TLR4 signal transduction, was further aggravated by the GPAGPR treatment (p < 0.01) but reduced by GPSGRP (p < 0.05) (Fig. S3B†). The mRNA level of iNOS, a factor downstream of the MyD88 signaling pathway, presented the same trend as the HUA phenotypes: HPD significantly upregulated the expression level of iNOS (p < 0.001), and the peptide treatments restored it to the control level (p < 0.01 and p < 0.05) (Fig. S3D†). However, constant mRNA levels of TRAF6 and COX-2 were observed among the groups (Fig. S3C and S3E†).
GPAGPR and GPSGRP treatments modulated the gut microbiota in HUA mice
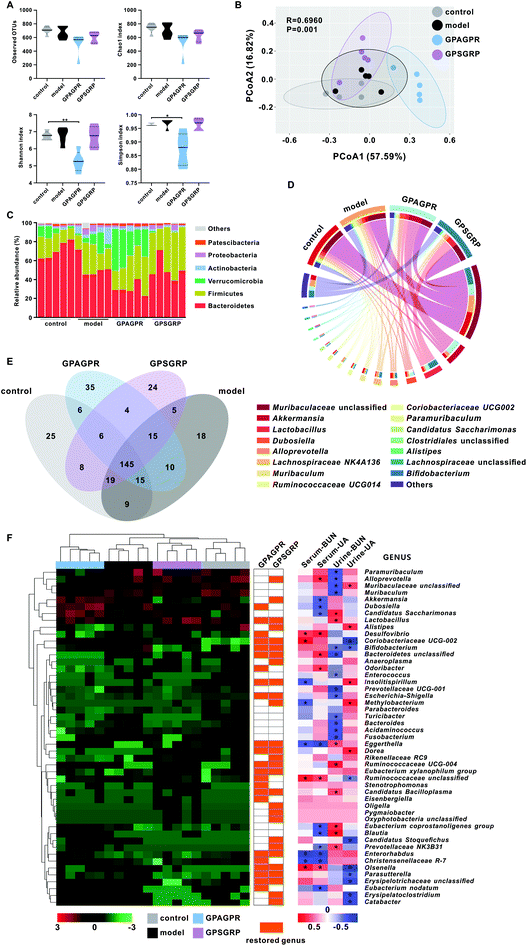
After 12 weeks of treatment, cecal contents were collected and the extracted DNAs were sequenced to show the effects of HPD and peptide treatment on the gut microbiota. The reduced abundance (observed OTUs and Chao 1 index) of the gut microbiota induced by HPD was further aggravated by both peptide treatments, while the increased diversity (Shannon and Simpson indices) was restored (Fig. 5A). Weighted UniFrac PCoA showed that HPD shifted the overall structure of the gut microbiota away from the control group but the microbiota structure was subsequently altered by the peptide treatments. In addition, the GPAGPR and GPSGRP groups showed different structures, showing the different modulatory effects of these two peptides on the gut microbiota (Fig. 5B).Six dominant phyla with more than 1% relative abundance were identified in this study, including Bacteroidetes, Firmicutes, Verrucomicrobia, Actinobacteria, Proteobacteria and Patescibacteria. HPD enhanced the abundance of the last five phyla and reduced the abundance of only Bacteroidetes. Both the GPAGPR and GPSGRP treatments aggravated the changes in the abundance of Bacteroidetes, Firmicutes and Patescibacteria and restored the abundance of Actinobacteria and Proteobacteria to the control levels, but Verrucomicrobia was restored by only GPSGRP (Fig. 5C and Table S4†). In addition, 15 genera with high abundance (>1% in at least one group) were shown in Fig. 5D and Table S5,† including Akkermansia, Lactobacillus and Bifidobacterium, which were well-known probiotics. Among these 15 genera, 8 and 9 genera were restored to control levels by GPAGPR and GPSGRP treatments, respectively, while only five genera were restored by both peptides. In addition, different change trends were observed in five genera, including Akkermansia, Dubosiella, Alloprevotella, Clostridiales unclassified and Alistipes. At the species level, 145 species were detected in all groups, while 25, 18, 35 and 24 species were identified in only the control, model, GPAGPR and GPSGRP groups, accounting for 10.7%, 7.6%, 14.8% and 10.6% of the total species in the four groups, respectively (Fig. 5E).
Forty-nine differentially abundant genera were identified by the Kruskal–Wallis test in this study (Fig. 5F and Table S6†). Among them, 23 and 28 genera were restored to the control level after GPAGPR and GPSGRP treatments, respectively, while 15 genera were restored by both peptides, which belonged to the phyla Actinobacteria (n = 5), Firmicutes (n = 5), Proteobacteria (n = 3), Tenericutes (n = 1) and Bacteroidetes (n = 1). In addition, Spearman's correlation analysis was carried out for these genera with phenotypes of HUA in serum and urine. Among them, 40 out of 49 genera were significantly correlated with at least one index, while only three genera, Eggerthella, Ruminococcaceae unclassified, and Olsenella, were significantly correlated with three indices.
GPAGPR and GPSGRP treatments altered the miRNA profile of the kidneys in HUA mice
Due to the relatively better XOD inhibition and HUA alleviation activity of GPSGRP, the miRNA profile of kidneys in HUA mice that received GPSGRP treatment was assessed. In total, 9 small RNA libraries were constructed, and 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 718
718![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 484 ± 1
484 ± 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 271
271![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 019, 2
019, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 369
369![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 893 ± 174
893 ± 174![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 885 and 2
885 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800
800![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 077 ± 689
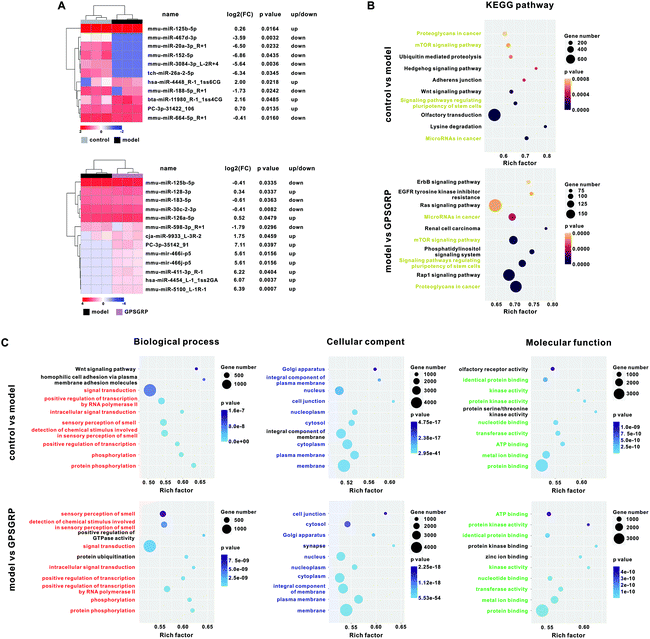
077 ± 689![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 950 clean reads were generated from the control, model and GPSGRP groups, respectively (Table S7†). In total, 11 differentially expressed (DE) miRNAs (four upregulated and seven downregulated miRNAs) were identified in the model group compared with that of the control group, while 13 DE-miRNAs (nine upregulated and four downregulated miRNAs) were identified in the peptide group compared with that of the model group (Fig. 6A). Twenty-one nonrepetitive and DE miRNAs were identified among the three groups in this study and the heatmap is shown in Fig. S4 and Table S8.† We sought to explore the role of these DE-miRNAs in regulating the expression of target genes and related functions. The putative target mRNAs of the DE-miRNAs were predicted and used for KEGG pathway analysis, and the top 10 significantly enriched KEGG pathways with the lowest p values were identified in the control and peptide groups when compared with those of the model group. Among them, four significantly enriched KEGG pathways were identified in both pairwise comparisons: microRNAs in cancer; pluripotency of stem cell regulation; the mTOR signaling pathway; and proteoglycans in cancer (Fig. 6B). In addition, the top five significantly enriched GO terms of the three categories in this study are shown in Fig. 6C. Among them, protein phosphorylation, and membrane and protein binding were the categories with the highest rich factor and lowest p value in terms of biological processes, cellular component and molecular function, respectively, in both pairwise comparisons.
950 clean reads were generated from the control, model and GPSGRP groups, respectively (Table S7†). In total, 11 differentially expressed (DE) miRNAs (four upregulated and seven downregulated miRNAs) were identified in the model group compared with that of the control group, while 13 DE-miRNAs (nine upregulated and four downregulated miRNAs) were identified in the peptide group compared with that of the model group (Fig. 6A). Twenty-one nonrepetitive and DE miRNAs were identified among the three groups in this study and the heatmap is shown in Fig. S4 and Table S8.† We sought to explore the role of these DE-miRNAs in regulating the expression of target genes and related functions. The putative target mRNAs of the DE-miRNAs were predicted and used for KEGG pathway analysis, and the top 10 significantly enriched KEGG pathways with the lowest p values were identified in the control and peptide groups when compared with those of the model group. Among them, four significantly enriched KEGG pathways were identified in both pairwise comparisons: microRNAs in cancer; pluripotency of stem cell regulation; the mTOR signaling pathway; and proteoglycans in cancer (Fig. 6B). In addition, the top five significantly enriched GO terms of the three categories in this study are shown in Fig. 6C. Among them, protein phosphorylation, and membrane and protein binding were the categories with the highest rich factor and lowest p value in terms of biological processes, cellular component and molecular function, respectively, in both pairwise comparisons.
In addition, 10 miRNAs (miR-143-3p, miR-34a, miR-223, miR-17-5p, miR-186, miR-125b-5p, miR-152-5p, miR-188-5p, miR-128-3p and miR-30b) were identified to be involved in uric acid metabolism, NLRP3 inflammasome, inflammation related signaling pathways and renal injury, and four of them (miR-125b-5p, miR-152-5p, miR-188-5p and miR-128-3p) were differentially expressed among groups (Tables S8 and S9†). Subsequently, spearman's correlation analysis was carried out for these 10 miRNAs with the abundance of 49 differentially abundant genera. Among them, genus Rikenellaceae RC9 was positively correlated (coefficient >0.95) with five miRNAs, while genera Lactobacillus, Ruminococcaceae UCG-004, Eubacterium, Candidatus, Oligella, Pygmaiobacter, Oxyphotobacteria unclassified, Anaeroplasma, Escherichia, Shigella and Parasutterella were correlated (coefficient >0.95 or <−0.95) with four miRNAs. On the other hand, miR-188-5p and miR-128-3p were correlated with 16 and 18 genera, respectively (Table S10†).
Discussion
Identification of peptides with distinct structures and functions and their subsequent use as biomarkers will contribute to the clarification of the underlying mechanism of peptides, the optimization of hydrolysis strategies and the improvement of hydrolysate product quality control. The AJOP mixture was previously reported to show beneficial effects on HUA.7 In this study, two dominant peptides, GPAGPR and GPRGRP, were identified and investigated for their XOD inhibition and anti-HUA activities by the combination of in silico, in vitro and in vivo experiments, and their modulation effects on gene expression, gut microbiota and host miRNA profile were also explored.The biological activities of peptides are related to their amino acid residue profiles. The XOD inhibitory activities of peptides are partly attributed to tryptophan (Trp) residues due to the similar C6 and C5 ring structures of Trp, allopurinol and xanthine,22,28 while treatment with arginine (Arg) alone suppresses uric acid generation.29 However, GPAGPR and GPSGRP contain one Arg each and no Trp but still showed XOD inhibitory activities (Fig. 1C–F). Both GPAGPR and GPSGRP are rich in Gly and Pro, but the presence of Gly and Pro was reported to increase the availability of peptides that act as antioxidants instead of XOD inhibitors.30 In fact, although XOD inhibition and antioxidation are different and independent processes, XOD inhibition is beneficial in most pathophysiological states due to the decrease in reactive oxygen species (ROS), which partly explains the antioxidant activity of allopurinol.31 Therefore, we proposed that the amino acid residues related to antioxidant properties mainly contributed to the XOD inhibitory activity in these two peptides identified in this study.
GPAGPR and GPSGRP were predicted as XOD inhibitors via molecular docking and reduced the expression levels of XOD and inhibited uric acid biosynthesis in HUA mice, as expected (Fig. 3A and B). However, inhibited uric acid reabsorption, characterized by reduced mRNA and protein levels of GLUT9, was also observed in the peptide treatment group (Fig. 3C and D) although no interaction was predicted between GLUT9 and peptides. In this study, we proposed that the regulation of GLUT9 is mediated via miRNAs. miRNAs are noncoding RNA molecules involved in the posttranscriptional regulation of gene expression by regulating mRNA degradation and/or binding to the 3′-UTR of target mRNAs to block protein translation, and they target more than 60% of protein coding genes in humans. As previously reported, miR-143-3p directly targets GLUT9 to reduce uric acid reabsorption at the cellular level,16 and in this study, compared with the model group, the peptide treatment group showed increased miR-143-3p expression and decreased GLUT9 expression (Table S9†). Furthermore, miRNAs were reported to regulate multiple genes involved in inflammation. Studies have shown that the selective inhibition of the NF-κB pathway and NLRP3 inflammasome, which plays a vital role in generating pro-IL-1β and modifying pro-IL-1β to IL-1β, respectively, has renal protective effects.4 In this study, the activation of the NLRP3 inflammasome and NF-κB pathway was inhibited by the peptide treatments, along with reduced IL-1β mRNA and protein expression levels (Fig. 3 and Fig. S1†). Notably, miRNA-223 directly targets the NLRP3 inflammasome,18 while miR-17-5p regulates it indirectly by binding and degrading the mRNA encoding thioredoxin-interacting protein.32 Additionally, miR-186 showed inhibitory activity against the TLR4/NF-κB pathway in a mouse model.33 In addition to regulating the related pathways, miR-488 and miR-920 were reported to directly target IL-1β.34 All the related miRNAs except miR-488 and miR-920 were detected in this study, and the expected changing trend was found for miR-17-5p, which were downregulated in the model group and subsequently upregulated by the peptide treatments, while the expressions of miR-223 and miR-186 were only increased in the GPSGRP group compared to that of the model group (Table S9†). Additionally, most of the DE-miRNAs identified in this study were reported to be closely related to renal injury. Among them, miRNA-125b-5p and miR-152-5p were associated with renal injury and used as diagnostic biomarkers.35,36 Furthermore, the interaction between miR-188-5p and PTEN was shown to regulate the PI3K/AKT signaling pathway and mediate diabetic kidney disease alleviation. MiR-128-3p directly targeted SIRT1 to induce kidney damage, and miR-30b participated in the onset of HUA by targeting the IL-6 receptor in mice.37–39 However, the detected expression trends of some of the above-mentioned DE-miRNAs were not as expected (Table S9†), which might be due to the complicated network between miRNAs and mRNAs, and other target molecules of these DE-miRNAs might be involved in the alleviation of HUA and renal injury.
Previous studies reported the vital roles of the gut microbiota in HUA alleviation, and clarified that the anti-HUA effects of specific peptides or mixtures were mediated by the gut microbiota.7,40,41 Firstly, genera Escherichia and Proteus were reported to promote uric acid formation by secreting XOD, while genera Lactobacillus and Pseudomonas inhibited purine absorption and degraded uric acid in the intestine.14 In this study, the expected decrease in Escherichia and increase in Lactobacillus were detected in the two peptide treatment groups, with an extremely high abundance of the genus Lactobacillus in the GPSGRP group (21.49%), and both HPD and the peptide treatments showed no effect on the genus Pseudomonas (Table S6†). In addition, genera Lactobacillus, Clostridium, Streptococcus, Akkermansia and Faecalibaculum were known as SCFA producers,42 and SCFAs were reported to provide energy for intestinal wall cells to excrete uric acid and alleviate HUA.43 Among them, Lactobacillus and Streptococcus were increased by both peptide treatments, while the abundance of genus Akkermansia, also known as a potential next-generation probiotic, was increased by the HPD treatment (8.49%), further aggravated by GPAGPR (30.72%) but reduced by the GPSGRP treatment (0.36%) (Table S6†). Moreover, allopurinol, one of the most important clinical XOD inhibitors, was reported to modulate the gut microbiota by specifically decreasing the abundance of the genera Adlercreutzia, Anaerostipes, Bilophila, Morganella and Desulfovibrio.44 Except for the undetected genus Morganella in this study, the GPAGPR and GPSGRP treatments decreased the abundance of two (Bilophila and Desulfovibrio) and three genera (Bilophila, Adlercreutzia and Anaerostipes) that allopurinol did, respectively. However, it is not very clear whether these genera are closely related to the XOD inhibitory activity and anti-HUA effects.
In addition, metabolites derived from the gut microbiota are known as the potential regulators of miRNA expression. For example, tryptophan-derived metabolites (indole or its derivatives) were shown to negatively regulate miR-181 expression in adipocytes and alleviate obesity and insulin resistance; SCFAs were shown to control miRNA expression by regulating the DNA methylation or histone deacetylation of promoters and subsequently alleviate obesity and autoimmunity in the host; and bile acids were shown to increase the miR-92a-1-5p expression involved in the generation of an intestinal metaplasia phenotype from gastric cells by the FOXD1/NF-κB/CDX2 regulatory axis.19–21,45 Among the 49 differentially abundant genera identified in this study, genera Lactobacillus, Bifidobacterium and Enterococcus catalyze the hydrolysis of conjugated BAs to preserve the metabolic balance of BA,46 genera Lactobacillus and Bifidobacterium converted tryptophan to indole and its derivatives,47 and genera Lactobacillus, Akkermansia, Eubacterium and Blautia generated SCFAs by the degradation of dietary fiber.33 Spearman's correlation analysis indicated that both Lactobacillus and Eubacterium correlated with four miRNAs, and genera related to the metabolism of SCFAs, tryptophan and BA were totally correlated with 17, 7 and 12 miRNAs (Table S10†), proposing the vital roles of microbiota metabolites between the gut microbiota and miRNAs.
In conclusion, the peptides GPAGPR and GPSGRP screened from AJOP in this study exhibited XOD inhibitory activities and anti-HUA effects in vitro and in vivo. In addition, these peptides modulated the gut microbiota composition and altered the renal miRNA expression in the host, and we proposed that the microbiota-derived metabolites, including SCFAs, BAs and tryptophan derivatives, might act as links between miRNA and the gut microbiota, but the underlying mechanism needs to be further explored.
Author contributions
CYL and XRS designed this study. SQF, YMH, GDL, NS and RW carried out the experiments. SQF, YMH, NS, JJH, JZ, YL and CYL performed the data analysis. SQF, GDL, CYL, THM and LJD prepared the manuscript.Conflicts of interest
All authors declare that they have no conflict of interest.Acknowledgements
This work was sponsored by the National Key R&D Program of China (2018YFD0901102), China Postdoctoral Science Foundation (2021M691677), Public Welfare Project of Ningbo City (2019C10064), Postdoctoral Science Foundation of Zhejiang Province (ZJ2020006), Fundamental Research Funds for the Provincial Universities of Zhejiang (SJLY2021015), Fund of State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products, Ningbo University (ZS20190105), and K.C. Wong Magna Fund in Ningbo University.References
- S. Hao, C. Zhang and H. Song, Natural products improving hyperuricemia with hepatorenal dual effects, J. Evidence-Based Complementary Altern. Med., 2016, 2016, 7390504 Search PubMed.
- R. Harrison, Structure and function of xanthine oxidoreductase: where are we now?, Free Radical Biol. Med., 2002, 33, 774–797 CrossRef CAS PubMed.
- H. Wan, J. Han, S. Tang, W. Bao, C. Lu, J. Zhou, T. Ming, Y. Li and X. Su, Comparisons of protective effects between two sea cucumber hydrolysates against diet induced hyperuricemia and renal inflammation in mice, Food Funct., 2020, 11, 1074–1086 RSC.
- J. Tan, L. Wan, X. Chen, X. Li, X. Hao, J. Li and H. Ding, Conjugated linoleic acid ameliorates high fructose-induced hyperuricemia and renal inflammation in rats via NLRP3 inflammasome and TLR4 signaling pathway, Mol. Nutr. Food Res., 2019, 63, e1801402 CrossRef PubMed.
- C. E. Ekpenyong and N. Daniel, Roles of diets and dietary factors in the pathogenesis, management and prevention of abnormal serum uric acid levels, PharmaNutrition, 2015, 3, 29–45 CrossRef CAS.
- M. M. Ansari-Ramandi, M. Maleki, A. Alizadehasl, A. Amin, S. Taghavi, M. J. Alemzadeh-Ansari, A. Kazem Moussavi and N. Naderi, Safety and effect of high dose allopurinol in patients with severe left ventricular systolic dysfunction, J. Cardiovasc. Thorac. Res., 2017, 9, 102–107 CrossRef PubMed.
- C. Lu, S. Tang, J. Han, S. Fan, Y. Huang, Z. Zhang, J. Zhou, T. Ming, Y. Li and X. Su, Apostichopus japonicus oligopeptide induced heterogeneity in the gastrointestinal tract microbiota and alleviated hyperuricemia in a microbiota-dependent manner, Mol. Nutr. Food Res., 2021, 65, e2100147 CrossRef PubMed.
- J. Han, Z. Huang, S. Tang, C. Lu, H. Wan, J. Zhou, Y. Li, T. Ming, Z. Jim Wang and X. Su, The novel peptides ICRD and LCGEC screened from tuna roe show antioxidative activity via Keap1/Nrf2-ARE pathway regulation and gut microbiota modulation, Food Chem., 2020, 327, 127094 CrossRef CAS PubMed.
- A. T. Vieira, L. Macia, I. Galvão, F. S. Martins, M. C. Canesso, F. A. Amaral, C. C. Garcia, K. M. Maslowski, E. De Leon, D. Shim, J. R. Nicoli, J. L. Harper, M. M. Teixeira and C. R. Mackay, A role for gut microbiota and the metabolite-sensing receptor GPR43 in a murine model of gout, Arthritis Rheumatol., 2015, 67, 1646–1656 CrossRef CAS PubMed.
- Z. Guo, J. Zhang, Z. Wang, K. Y. Ang, S. Huang, Q. Hou, X. Su, J. Qiao, Y. Zheng, L. Wang, E. Koh, H. Danliang, J. Xu, Y. K. Lee and H. Zhang, Intestinal microbiota distinguish gout patients from healthy humans, Sci. Rep., 2016, 6, 20602 CrossRef CAS PubMed.
- T. Shao, L. Shao, H. Li, Z. Xie, Z. He and C. Wen, Combined signature of the fecal microbiome and metabolome in patients with gout, Front. Microbiol., 2017, 8, 268 Search PubMed.
- T. Koguchi and T. Tadokoro, Beneficial effect of dietary fiber on hyperuricemia in rats and humans: a review, Int. J. Vitam. Nutr. Res., 2019, 89, 89–108 CrossRef CAS PubMed.
- H. Wang, L. Mei, Y. Deng, Y. Liu, X. Wei, M. Liu, J. Zhou, H. Ma, P. Zheng, J. Yuan and M. Li, Lactobacillus brevis DM9218 ameliorates fructose-induced hyperuricemia through inosine degradation and manipulation of intestinal dysbiosis, Nutrition, 2019, 62, 63–73 CrossRef CAS PubMed.
- J. Wang, Y. Chen, H. Zhong, F. Chen, J. Regenstein, X. Hu, L. Cai and F. Feng, The gut microbiota as a target to control hyperuricemia pathogenesis: Potential mechanisms and therapeutic strategies, Crit. Rev. Food Sci. Nutr., 2021, 1–11, DOI:10.1080/10408398.2021.1874287.
- Y. T. Xu, Y. R. Leng, M. M. Liu, R. F. Dong, J. Bian, L. L. Yuan, J. G. Zhang, Y. Z. Xia and L. Y. Kong, MicroRNA and long noncoding RNA involvement in gout and prospects for treatment, Int. Immunopharmacol., 2020, 87, 106842 CrossRef CAS PubMed.
- Z. Zhou, Y. Dong, H. Zhou, J. Liu and W. Zhao, MiR-143-3p directly targets GLUT9 to reduce uric acid reabsorption and inflammatory response of renal tubular epithelial cells, Biochem. Biophys. Res. Commun., 2019, 517, 413–420 CrossRef CAS PubMed.
- W. F. Sun, M. M. Zhu, J. Li, X. X. Zhang, Y. W. Liu, X. R. Wu and Z. G. Liu, Effects of Xie-Zhuo-Chu-Bi-Fang on miR-34a and URAT1 and their relationship in hyperuricemic mice, J. Ethnopharmacol., 2015, 161, 163–169 CrossRef PubMed.
- Q. B. Zhang, D. Zhu, F. Dai, Y. Q. Huang, J. X. Zheng, Y. P. Tang, Z. R. Dong, X. Liao and Y. F. Qing, MicroRNA-223 suppresses IL-1β and TNF-α production in gouty inflammation by targeting the NLRP3 inflammasome, Front. Pharmacol., 2021, 12, 637415 CrossRef CAS PubMed.
- H. N. Sanchez, J. B. Moroney, H. Gan, T. Shen, J. L. Im, T. Li, J. R. Taylor, H. Zan and P. Casali, B cell-intrinsic epigenetic modulation of antibody responses by dietary fiber-derived short-chain fatty acids, Nat. Commun., 2020, 11, 60 CrossRef CAS PubMed.
- A. T. Virtue, S. J. McCright, J. M. Wright, M. T. Jimenez, W. K. Mowel, J. J. Kotzin, L. Joannas, M. G. Basavappa, S. P. Spencer, M. L. Clark, S. H. Eisennagel, A. Williams, M. Levy, S. Manne, S. E. Henrickson, E. J. Wherry, C. A. Thaiss, E. Elinav and J. Henao-Mejia, The gut microbiota regulates white adipose tissue inflammation and obesity via a family of microRNAs, Sci. Transl. Med., 2019, 11(496), eaav1892, DOI:10.1126/scitranslmed.aav1892.
- T. Li, H. Guo, H. Li, Y. Jiang, K. Zhuang, C. Lei, J. Wu, H. Zhou, R. Zhu, X. Zhao, Y. Lu, C. Shi, Y. Nie, K. Wu, Z. Yuan, D. M. Fan and Y. Shi, MicroRNA-92a-1–5p increases CDX2 by targeting FOXD1 in bile acids-induced gastric intestinal metaplasia, Gut, 2019, 68, 1751–1763 CrossRef CAS PubMed.
- C. Hou, D. Liu, M. Wang, C. Gong, Y. Li, L. Yang, M. Yao, E. Yuan and J. Ren, Novel xanthine oxidase-based cell model using HK-2 cell for screening antihyperuricemic functional compounds, Free Radical Biol. Med., 2019, 136, 135–145 CrossRef CAS PubMed.
- N. Liu, Y. Wang, M. Yang, W. Bian, L. Zeng, S. Yin, Z. Xiong, Y. Hu, S. Wang, B. Meng, J. Sun and X. Yang, New rice-derived short peptide potently alleviated hyperuricemia induced by potassium oxonate in rats, J. Agric. Food Chem., 2019, 67, 220–228 CrossRef CAS PubMed.
- S. Reagan-Shaw, M. Nihal and N. Ahmad, Dose translation from animal to human studies revisited, FASEB J., 2008, 22, 659–661 CrossRef CAS PubMed.
- C. C. Wang, L. Du, H. H. Shi, L. Ding, T. Yanagita, C. H. Xue, Y. M. Wang and T. T. Zhang, Dietary EPA-enriched phospholipids alleviate chronic stress and LPS-induced depression- and anxiety-like behavior by regulating immunity and neuroinflammation, Mol. Nutr. Food Res., 2021, 65, e2100009 CrossRef PubMed.
- C. Lu, T. Sun, Y. Li, D. Zhang, J. Zhou and X. Su, Modulation of the gut microbiota by krill oil in mice fed a high-sugar high-fat diet, Front. Microbiol., 2017, 8, 905 CrossRef PubMed.
- P. D. Schloss, S. L. Westcott, T. Ryabin, J. R. Hall, M. Hartmann, E. B. Hollister, R. A. Lesniewski, B. B. Oakley, D. H. Parks, C. J. Robinson, J. W. Sahl, B. Stres, G. G. Thallinger, D. J. Van Horn and C. F. Weber, Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities, Appl. Environ. Microbiol., 2009, 75, 7537–7541 CrossRef CAS PubMed.
- A. B. Nongonierma and R. J. Fitzgerald, Tryptophan-containing milk protein-derived dipeptides inhibit xanthine oxidase, Peptides, 2012, 37, 263–272 CrossRef CAS PubMed.
- W. A. Saka, R. E. Akhigbe, A. O. Abidoye, O. S. Dare and A. O. Adekunle, Suppression of uric acid generation and blockade of glutathione dysregulation by L-arginine ameliorates dichlorvos-induced oxidative hepatorenal damage in rats, Biomed. Pharmacother., 2021, 138, 111443 CrossRef CAS PubMed.
- A. Alemán, B. Giménez, E. Pérez-Santin, M. C. Gomez-Guillen and P. Montero, Contribution of Leu and Hyp residues to antioxidant and ACE-inhibitory activities of peptide sequences isolated from squid gelatin hydrolysate, Food Chem., 2012, 125, 334–341 CrossRef.
- J. Luo, D. Yan, S. Li, S. Liu, F. Zeng, C. W. Cheung, H. Liu, M. G. Irwin, H. Huang and Z. Xia, Allopurinol reduces oxidative stress and activates Nrf2/p62 to attenuate diabetic cardiomyopathy in rats, J. Cell. Mol. Med., 2020, 24, 1760–1773 CrossRef CAS PubMed.
- D. Chen, B. J. Dixon, D. M. Doycheva, B. Li, Y. Zhang, Q. Hu, Y. He, Z. Guo, D. Nowrangi, J. Flores, V. Filippov, J. H. Zhang and J. Tang, IRE1α inhibition decreased TXNIP/NLRP3 inflammasome activation through miR-17–5p after neonatal hypoxic-ischemic brain injury in rats, J. Neuroinflammation, 2018, 15, 32 CrossRef PubMed.
- Y. Zhang, L. Ma, E. Lu and W. Huang, Atorvastatin upregulates microRNA-186 and inhibits the TLR4-mediated MAPKs/NF-κB pathway to relieve steroid-induced avascular necrosis of the femoral head, Front. Pharmacol., 2021, 12, 583975 CrossRef CAS PubMed.
- W. Zhou, Y. Wang, R. Wu, Y. He, Q. Su and G. Shi, MicroRNA-488 and -920 regulate the production of proinflammatory cytokines in acute gouty arthritis, Arthritis Res. Ther., 2017, 19, 203 CrossRef PubMed.
- A. Duan, L. Liu, Y. Lou, D. Zhang, H. Li, Y. Chen, W. Cui and L. Miao, Diagnostic Value of Urinary miR-152–5p in Patients with IgA Nephropathy with Elevated Proteinuria Levels., Clin. Lab., 2019, 65(7) DOI:10.7754/Clin.Lab.2019.190111.
- S. Wang, L. Wu, L. Du, H. Lu, B. Chen and Y. Bai, Reduction in miRNA-125b-5p levels is associated with obstructive renal injury, Biomed. Rep., 2017, 6, 449–454 CrossRef CAS PubMed.
- M. Xue, Y. Cheng, F. Han, Y. Chang, Y. Yang, X. Li, L. Chen, Y. Lu and B. Sun, Triptolide attenuates renal tubular epithelial-mesenchymal transition via the MiR-188–5p-mediated PI3K/AKT pathway in diabetic kidney disease, Int. J. Biol. Sci., 2018, 14, 1545–1557 CrossRef CAS PubMed.
- T. Ye, X. Yang, H. Liu, P. Lv, H. Lu, K. Jiang, E. Peng, Z. Ye, Z. Chen and K. Tang, Theaflavin protects against oxalate calcium-induced kidney oxidative stress injury via upregulation of SIRT1, Int. J. Biol. Sci., 2021, 17, 1050–1060 CrossRef CAS PubMed.
- X. Zhong, Y. Chen, C. Yao, L. Xu, Y. Peng, Q. Yang, M. Zhao and X. Guo, MicroRNA-30b participates in the pathological process of hyperuricemia by regulating interleukin-6 receptor, Nucleosides, Nucleotides Nucleic Acids, 2020, 39, 1162–1178 CrossRef CAS PubMed.
- J. Han, X. Wang, S. Tang, C. Lu, H. Wan, J. Zhou, Y. Li, T. Ming, Z. J. Wang and X. Su, Protective effects of tuna meat oligopeptides (TMOP) supplementation on hyperuricemia and associated renal inflammation mediated by gut microbiota, FASEB J., 2020, 34, 5061–5076 CrossRef CAS PubMed.
- J. Han, Z. Wang, C. Lu, J. Zhou, Y. Li, T. Ming, Z. Zhang, Z. J. Wang and X. Su, The gut microbiota mediates the protective effects of anserine supplementation on hyperuricaemia and associated renal inflammation, Food Funct., 2021, 12, 9030–9042 RSC.
- L. Zhao, F. Zhang, X. Ding, G. Wu, Y. Y. Lam, X. Wang, H. Fu, X. Xue, C. Lu, J. Ma, L. Yu, C. Xu, Z. Ren, Y. Xu, S. Xu, H. Shen, X. Zhu, Y. Shi, Q. Shen, W. Dong, R. Liu, Y. Ling, Y. Zeng, Q. Zhang, J. Wang, L. Wang, Y. Wu, B. Zeng, H. Wei, M. Zhang, Y. Peng and C. Zhang, Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes, Science, 2018, 359, 1151–1156 CrossRef CAS PubMed.
- M. Nieuwdorp, P. W. Gilijamse, N. Pai and L. M. Kaplan, Role of the microbiome in energy regulation and metabolism, Gastroenterology, 2014, 146, 1525–1533 CrossRef CAS PubMed.
- Y. Yu, Q. Liu, H. Li, C. Wen and Z. He, Alterations of the gut microbiome associated with the treatment of hyperuricaemia in male rats, Front. Microbiol., 2018, 9, 2233 CrossRef PubMed.
- J. Du, P. Zhang, J. Luo, L. Shen, S. Zhang, H. Gu, J. He, L. Wang, X. Zhao, M. Gan, L. Yang, L. Niu, Y. Zhao, Q. Tang, G. Tang, D. Jiang, Y. Jiang, M. Li, A. Jiang, L. Jin, J. Ma, S. Shuai, L. Bai, J. Wang, B. Zeng, D. Wu, X. Li and L. Zhu, Dietary betaine prevents obesity through gut microbiota-derived microRNA-378a family, Gut Microbes, 2021, 13, 1–19 Search PubMed.
- Z. Song, Y. Cai, X. Lao, X. Wang, X. Lin, Y. Cui, P. K. Kalavagunta, J. Liao, L. Jin, J. Shang and J. Li, Taxonomic profiling and populational patterns of bacterial bile salt hydrolase (BSH) genes based on worldwide human gut microbiome, Microbiome, 2019, 7, 9 CrossRef PubMed.
- H. M. Roager and T. R. Licht, Microbial tryptophan catabolites in health and disease, Nat. Commun., 2018, 9, 3294 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1fo03981d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |