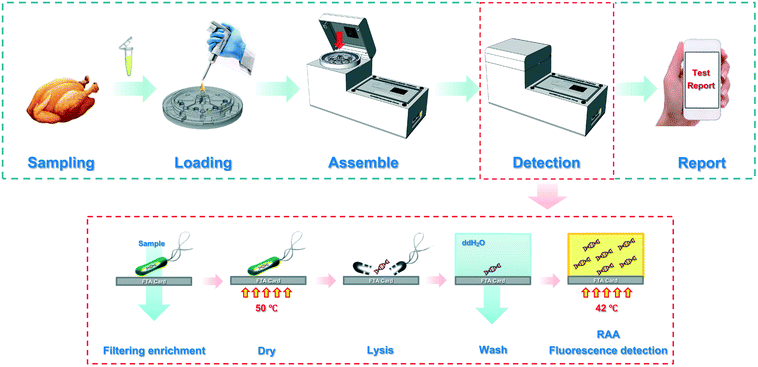
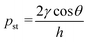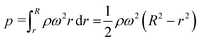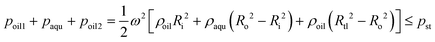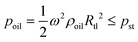Automatic and multi-channel detection of bacteria on a slidable centrifugal disc based on FTA card nucleic acid extraction and recombinase aided amplification†
Siyuan
Wang
ab,
Gaozhe
Cai
c,
Hong
Duan
a,
Wuzhen
Qi
b and
Jianhan
Lin
 *ab
*ab
aKey Laboratory of Agricultural Information Acquisition Technology, Ministry of Agriculture and Rural Affairs, China Agricultural University, Beijing 100083, China. E-mail: jianhan@cau.edu.cn
bKey Laboratory of Modern Precision Agriculture System Integration Research, Ministry of Education, China Agricultural University, Beijing 100083, China
cState Key Laboratory of Transducer Technology, Shanghai Institute of Microsystem and Information Technology, Chinese Academy of Sciences, Shanghai, 200050, China
First published on 11th November 2021
Abstract
Rapid screening of foodborne pathogens is key to preventing food poisoning. In this study, a slidable centrifugal disc was developed for automatic and multi-channel detection of Salmonella typhimurium using Flinders Technology Associates (FTA) cards for nucleic acid extraction and recombinase aided amplification (RAA) for nucleic acid detection. The slidable FTA switching and centrifugal fluidic control were elaborately combined to achieve fully automatic operations, including centrifugation of the bacterial sample to obtain the concentrated bacteria, heating and drying of the FTA card to extract the nucleic acids, washing of the FTA card to remove the impurities, and RAA detection of the extracted DNA to determine the concentration. Under the optimal conditions, this slidable centrifugal disc was able to detect 10 CFU mL−1 in a spiked chicken meat supernatant in 1 h with an average recovery of 101.8% and an average standard deviation of 6.5%. This disc has been demonstrated as an alternative for sample-in-result-out detection of Salmonella and has shown potential for simultaneous detection of multiple bacteria.
1. Introduction
In recent years, food safety has become an increasing global public health concern. Pathogenic bacteria, as the primary cause of foodborne diseases, bring serious harm to human beings.1,2 Among these pathogenic bacteria, Salmonella is the leading cause of foodborne diseases worldwide.3 According to the World Health Organization,4 600 million cases of foodborne illnesses are reported every year and 420![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people have died. In China, Salmonella also ranks number one in microbial food poisoning.5 For bacterial detection, the traditional culture is the gold standard method, but it takes as long as 2–3 days.6 Other recommended methods, such as the polymerase chain reaction (PCR)7 and enzyme-linked immunosorbent assay,8 are often subject to cross contamination and the need for expensive equipment and well-trained technicians. Therefore, there is an urgent demand for rapid, sensitive and in-field detection methods of foodborne pathogens to ensure food safety.
000 people have died. In China, Salmonella also ranks number one in microbial food poisoning.5 For bacterial detection, the traditional culture is the gold standard method, but it takes as long as 2–3 days.6 Other recommended methods, such as the polymerase chain reaction (PCR)7 and enzyme-linked immunosorbent assay,8 are often subject to cross contamination and the need for expensive equipment and well-trained technicians. Therefore, there is an urgent demand for rapid, sensitive and in-field detection methods of foodborne pathogens to ensure food safety.
In the past few decades, various isothermal nucleic acid amplification technologies, including strand exchange amplification (SEA),9 loop-mediated isothermal amplification (LAMP),10 recombinase polymerase amplification (RPA)11 and recombinase aided amplification (RAA),12 have been widely reported for bacterial detection due to their high sensitivity, simple operation and low cost.9,11,13 Recently, RAA received increasing attention since it could be completed at a constant temperature from 37 °C to 42 °C and was able to detect Salmonella as low as 102 copies per reaction in 20 min when it was combined with fluorescent probes.14 Besides, RAA was also reported for detection of viable Staphylococcus aureus by combining with propidium monoazide and had a lower detection limit of 102 CFU mL−1 under 3 h enrichment and 101 CFU mL−1 under 6 h enrichment in artificially contaminated milk.15
The Flinders Technology Associates (FTA, GE Healthcare, Kent, England) card is a kind of special filter paper with strong denatured and chelating agents to lyse the captured bacteria and immobilize their genomic DNA on its matrix.16,17 To date, the FTA card has been often reported to extract nucleic acids from various bacterial samples and combined with real-time PCR and LAMP for Salmonella detection due to its simple operation, fast purification and wide applicability.18,19 Recently, an interesting study was reported for in-field detection of Salmonella by integrating the FTA card onto a microdevice for extraction and purification of genomic DNA from bacterial cells, which was then combined with colorimetric fuchsin-based LAMP for DNA detection.20 This microdevice was able to detect Gram-negative bacteria (Salmonella spp. and Escherichia coli O157:H7) as low as 3.0 × 101 CFU per sample and Gram-positive bacteria (Staphylococcus aureus) as low as 3.0 × 102 CFU per sample in 75 min.
Since the slidable chip (slipchip) was first reported in 2009, it has received great attention due to its simple fabrication, easy operation and in-field applicability.21–24 More importantly, slidable chips do not depend on external pumps and/or valves, thus greatly improving their portability and disposability.25,26 In recent years, slidable chips have been often used for various biological detection methods, including RPA,27 colorimetric biosensing28 and volumetric biosensing,29,30 to achieve rapid detection of biological targets. Besides, centrifugal discs or chips were often reported31,32 and combined with molecular biological and immunological assays for automatic and rapid detection of various bacteria, including Mycobacterium tuberculosis,33Escherichia coli O157:H7, Salmonella typhimurium, and Vibrio parahaemolyticus.34 An important challenge for centrifugal discs is their controllable sequential operation on different reagents. Some interesting attempts have been made to automatically and successively control the reagents based on the differences of channel size,35 chamber location36 or slope valve angle,37etc. using their corresponding centrifugal speed. However, these methods still needed complex structures and careful operation. Thus, the combination of a centrifugal disc and a slidable chip might be a promising way to automatically achieve sample-in-result-out detection of foodborne pathogens.
In this study, a slidable centrifugal disc was elaborately developed for automatic and multi-channel detection of Salmonella typhimurium using FTA cards for nucleic acid extraction and recombinase aided amplification for nucleic acid detection. As shown in Scheme 1, eight bacterial samples were first loaded onto their respective sample chambers at the disc's octagonal center and centrifuged to flow through their respective FTA cards, resulting in capture and concentration of target bacteria. After a mini heater was used to dry the cards for 15 min, the captured target bacteria were lysed with a lysis agent pre-chelated on the cards, resulting in the release of the genomic nucleic acids from the bacterial cells and the immobilization of the nucleic acids on the cards. Then, the FTA cards were slidably switched to their washing chambers and incubated for 15 min, followed by second centrifugation to remove the wastes. Finally, the FTA cards were slidably switched to their RAA detection chambers, and the nucleic acids on the FTA cards were isothermally amplified at a constant temperature of 47.5 °C respectively, followed by real-time monitoring of their fluorescence signals to quantitatively determine the target bacteria and wireless transmission of the results from the prototype to a smartphone via Bluetooth.
2. Materials and methods
2.1 Materials and reagents
Salmonella typhimurium (ATCC 14028) was used as the target bacteria, and Listeria monocytogenes (ATCC 13932), E. coli O157:H7 (ATCC 43888), Vibrio parahaemolyticus (ATCC 17802) and Bacillus cereus (ATCC 11778) were used as non-target bacteria. The FTA cards and a 4 mm puncher were purchased from Whatman (Little Chalfont, Buckinghamshire, UK). The washing buffer with 10 mM Tris-HCl and 0.1 mM EDTA was purchased from Thermo Fisher (Waltham, MA, USA). The RAA kit, including a recombinase, single-stranded binding protein, DNA polymerase, dNTP mixture, reaction buffer and magnesium acetate, was purchased from Ampu Biotech (Weifang, China). The RAA forward primer, reverse primer and fluorescent probe were previously reported by the Hangzhou Airport Office of the Zhejiang Entry-Exit Inspection and Quarantine Bureau (see Table S1†) and synthesized by Sangon Biotech (Shanghai, China). Polydimethylsilicone oil from HiBase (Beijing, China) was used for coating and binding different layers. The silicone elastomer kit (Sylgard 184) from Dow Corning (Midland, MI, USA) was used to fabricate the polydimethylsiloxane (PDMS) channel. Brain heart infusion (BHI) medium from Remel (Lenexa, KS, USA) and Luria–Bertani medium (Aoboxing Biotech, Beijing, China) were used for bacterial culture.2.2 Design and fabrication of the slidable centrifugal disc
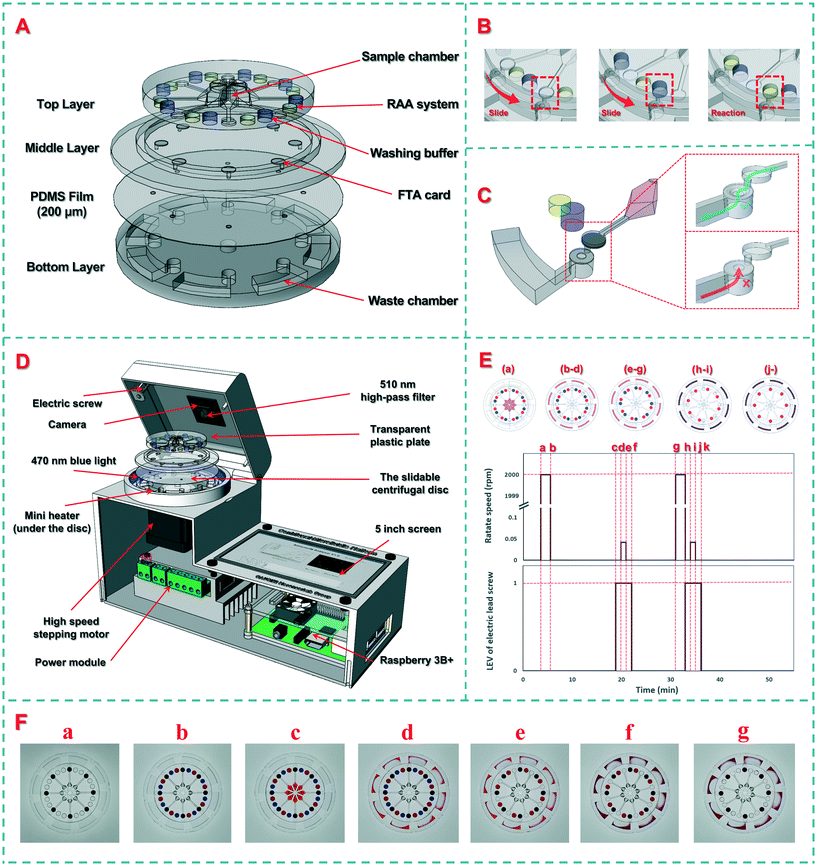
The slidable centrifugal disc is the most important component. As shown in Scheme 2(A), the disc consists of four layers: (1) the top layer (diameter: 50 mm, thickness: 5 mm) with eight sample chambers (volume: 100 μL) at the octagonal center to house eight bacterial samples and eight groups of total 24 cylindrical chambers (diameter: 4 mm, distance to the disc center: 20 mm) at the periphery to house eight FTA cards in their respective centrifugation chambers, eight washing buffers in their respective buffer chambers and eight RAA reaction reagents in their respective detection chambers, (2) the middle layer (diameter: 70 mm, thickness: 2 mm) with eight circular slots (diameter: 4 mm, distance to the disc center: 20 mm) for installing eight FTA cards in the centrifugation chambers, (3) a PDMS film (diameter: 70 mm, thickness: 200 μm) with eight holes (diameter: 1 mm, distance to the disc center: 21.5 mm) for acting as the check valves, and (4) the bottom layer (diameter: 70 mm, thickness: 5 mm) with eight waste chambers (volume: 250 μL) for collecting the wastes. In the top layer, 100 μL of bacterial sample was preloaded onto a sample chamber at the octagonal center, which was connected to its respective centrifugation chamber through a straight channel (width and height: 300 μm). Besides, 50 μL of washing buffer and 25 μL of RAA reaction reagent were preloaded onto a washing buffer chamber and detection chamber, respectively, through two small holes (diameter: 0.6 mm) punched at the top of each chamber, which allowed the washing buffer and RAA reagent to be injected into and to fill in both chambers before the holes were sealed with tape. In the bottom layer, each curved waste chamber was filled with an absorbent cotton to collect more wastes. The sliding scheme of the disc and the structure of the check valve are shown in Scheme 2(B) and (C).The molds of these layers were designed using the SolidWorks software (Concord, MA, USA) and printed using an Object30 3D printer (Vero Whiteplus RGD835, Stratasys, Eden Prairie, MN, USA). After the prepolymer and the curing agent were fully mixed at a ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and cast from the molds after heat curing to obtain these layers, the middle layer, PDMS film and bottom layer were bonded through surface oxygen plasma (Harrick Plasma, Ithaca, NY, USA), while the top layer and the middle layer were bonded through a silicone oil layer. First, 1 mL of polydimethylsilicone oil with a viscosity of 5000 cSt was added onto the center of a glass plate. After spinning at 4000 rpm for 30 s, a uniform oil layer with a thickness of ∼20 μm was formed. Then, the top layer was slowly placed on the glass plate to coat its surface with a thin layer of silicone oil and carefully peeled off using tweezers. Finally, the top layer was placed onto the middle layer, followed by static standing for 30 min to allow the silicone oil to gradually distribute between these two layers via capillary force.
1 and cast from the molds after heat curing to obtain these layers, the middle layer, PDMS film and bottom layer were bonded through surface oxygen plasma (Harrick Plasma, Ithaca, NY, USA), while the top layer and the middle layer were bonded through a silicone oil layer. First, 1 mL of polydimethylsilicone oil with a viscosity of 5000 cSt was added onto the center of a glass plate. After spinning at 4000 rpm for 30 s, a uniform oil layer with a thickness of ∼20 μm was formed. Then, the top layer was slowly placed on the glass plate to coat its surface with a thin layer of silicone oil and carefully peeled off using tweezers. Finally, the top layer was placed onto the middle layer, followed by static standing for 30 min to allow the silicone oil to gradually distribute between these two layers via capillary force.
2.3 Development of the sample-in-result-out device
To achieve sample-in-result-out detection of the target bacteria, a supporting device was developed for slidable switching of the reagents, centrifugal filtration of the samples, automatic control of the temperature and light, and real-time monitoring of the fluorescence signal. As shown in Scheme 2(D), this device consists of an actuator, a detector, and software. The actuator included a high-speed stepping motor for rotating the disc and a vertically movable transparent plastic plate for applying pressure on the top layer. The stepping motor was programmed to rotate at a high speed of 3000 rpm for centrifugation and at a low speed of 0.042 rpm (15 degrees per minute) for slidable switching. The detector consisted of a circular mini heater for heating the disc, a circular LED array for providing the excitation light, and a high-resolution camera with a 510 nm high-pass filter for collecting the fluorescence signals. The mini heater (Youpusi Electronics, Beijing, China) was placed below the disc to provide a constant temperature for drying the FTA cards and activating the RAA detection. The tailor-made LED array (power: 10 W, wavelength: 470 nm, Jiadeng, Hangzhou, China) was mounted at a distance of 50 mm from the center to excite the dissociative FAM (5-carboxyfluorescein) groups in the detection chamber. The camera (WeiXin Optics, Chengdu, China) was mounted at the center of the top cover, and a high-pass filter with a wavelength of 510 nm from Hoda Optics (Shanghai, China) was installed in front of the camera to filter the scattered excitation light. The fluorescence images of the disc were collected using the camera for real-time analysis. The embedded software was developed using Python and the PyQt framework on a Raspberry Pi 3B+ to automatically perform the procedures and analyze the images. Besides, an Android smartphone app was developed to communicate with this supporting device via Bluetooth for wireless transmission of the detection results to the food safety monitoring cloud platform. The interfaces of the device and smartphone are shown in Fig. S1 and S2.†2.4 Preparation of the bacterial cultures
Prior to culture, both the target bacterial strain and the non-target bacterial strains were stored with glycerol at −20 °C. For bacteria culture, 50 μL of each bacterial strain was first added into 5 mL of the LB liquid medium and shaken at 180 rpm at 37 °C for 12–16 h. Then, each culture was serially diluted with sterile PBS to obtain the bacteria at the final concentrations from 1.0 × 101 to 1.0 × 106 CFU mL−1. For bacteria enumeration, the bacterial sample was serially diluted with sterile PBS, and 100 μL of the dilutions were surface plated on LB agar plates, followed by incubation at 37 °C for 22–24 h. The visible colonies were counted to obtain the concentration of the bacteria.2.5 RAA detection of the nucleic acids
The RAA detection was performed according to the manufacturer's instructions with slight modifications. The RAA detection system with a total volume of 50 μL included 14.5 μL of buffer A (recombinase, single-stranded binding protein, DNA polymerase and dNTP mixture), 1 μL of forward primers (10 μM), 1 μL of reverse primers (10 μM), 0.5 μL of fluorescent probes (10 μM), 1.5 μL of buffer B with Mg(CH3COO)2, and 6.5 μL of deionized water. The nucleic acids immobilized on the FTA cards were used as the DNA templates. The circular LED array was used to excite the dissociative FAM groups in the detection chamber, and the high-resolution camera was used to collect the fluorescence images every 10 s. A total of 120 fluorescence images were obtained during the whole RAA detection process. Since a small air bubble was inevitable to occur at each RAA detection chamber, the Laplace edge detection algorithm was used to process each image and thus avoid the interference of this air bubble. The formation of this air bubble was due to the gathering of air in the FTA card after it was washed with washing buffer, switched to the RAA detection chamber and detected by RAA. The average G/(R + G + B) value for each processed image was calculated to obtain the time for the fluorescence signal to reach the threshold, which was defined as the sum of the average G/(R + G + B) values of the 3rd image to the 20th image and 10 times of their standard deviation. The threshold time was used to determine the bacterial concentration.2.6 Detection of the target bacteria using this disc and supporting device
To establish the calibration curve for this device, different concentrations of Salmonella typhimurium ranging from 1.0 × 101 to 10 × 106 CFU mL−1 were detected. Prior to detection, the washing buffer and RAA reagent were preloaded into the disc. The procedure is shown in Scheme 2(E), while the pictures of the disc at different stages are shown in Scheme 2(F). The first 3 minutes were used to load the bacterial samples. First, the disc was centrifuged at 3000 rpm for 2 min (from moment a, 3rd min, to moment b, 5th min), allowing the bacterial samples to flow through their FTA cards. After the FTA cards were heated at 65 °C for 15 min (from moment b, 5th min, to moment d, 20th min) to lyse the bacterial cells and immobilize the nucleic acids on the cards, the transparent plastic plate was extruded with an electric screw to fix the top layer, and the bottom layer was rotated 15 degrees using the stepping motor to slidably switch the FTA cards to the washing chambers (from moment c, 19th min, to moment f, 22nd min), followed by incubation for 10 min (from moment e, 21st min to moment g, 31st min) and centrifugation at 3000 rpm for 2 min to remove the impurities (from moment g, 31st min, to moment h, 33rd min). Finally, the FTA cards were slidably switched to the RAA detection chambers (from moment h, 33rd min, to moment k, 36th min), and the extracted nucleic acids were isothermally amplified at the optimal temperature for 20 min (from moment k, 36th min), followed by real-time monitoring of the fluorescence signal under the excitation of the blue LED array to obtain the time threshold for quantitative determination of the bacterial concentration. A demonstration video of the automatic disc fluid control was recorded as Video S1† and a time-lapse video was recorded as Video S2† to show the monitoring of the fluorescence signals in the detection chambers.2.7 Detection of Salmonella typhimurium in the spiked chicken samples
To verify the applicability of this slidable centrifugal disc for detection of Salmonella typhimurium in real food samples, 25 g of chicken meats were first purchased from a local supermarket and added into 225 mL of sterile PBS according to China's food safety national standards, followed by homogenization for 4 min using a stomacher (BagMixer CC, InterScience, Paris, France) to obtain the supernatant. Then, different concentrations of Salmonella typhimurium were added into the supernatant to prepare the spiked chicken samples with bacterial concentrations from 1.0 × 101 to 1.0 × 106 CFU mL−1. Finally, the spiked samples were detected using the disc and supporting device.3. Results and discussion
3.1 The principle of the centrifugal disc
In this study, the combination of the slipchip and centrifugal disc is the core to achieve automatic detection of Salmonella, which mainly depends on the controllable delivery of the solutions (bacterial sample and washing buffer) using high-speed centrifugation and slidable switching of the FTA card to the corresponding reagents using the silicone oil. Since the centrifugation has been theoretically proved and experimentally demonstrated with the capability of manipulating the solutions, the feasibility of the slidable switching using the silicone oil was analyzed by establishing the physical model. During the high-speed centrifugation, the silicone oil layer has to maintain no relative movement to ensure the preloaded washing buffer and RAA reagent in their respective chambers, so the surface tension on the silicone oil should be larger than the centrifugal force.According to the Young–Laplace equation, the pressure resulting from the surface tension of the silicone oil can be expressed as:
where r and R are the distances between the center and the near-end and far-end of the solution, respectively; ρ is the density of the liquid; ω is the angular velocity.
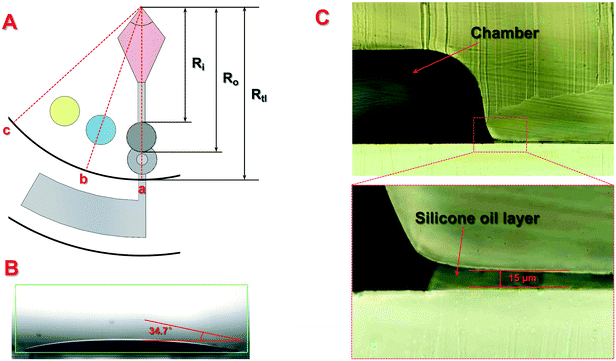
According to the distribution of the liquid and silicone oil on the disc during centrifugation, the force exerted on the silicone oil at the edge of the top layer can be divided into three typical situations, where the distance from the center to the front or end of the chamber on the top layer is defined as Ri and Ro, respectively, while the radius of the top layer is defined as Rtl. as shown in Fig. 1(A):
(1). For point a, the centrifugal force on the edge of the top layer was contributed by the sample being centrifuged and the silicone oil from the end of the centrifugal chamber to the edge of the top layer. In order to maintain no relative movement of the silicone oil, ω should meet the following requirements:
(2). For point b, the centrifugal force on the edge of the top layer was contributed by the silicone oil from the center to the front of the chamber, the silicone oil from the end of the chamber to the edge of the top layer and the liquid in the chamber. In order to maintain no relative movement of the silicone oil, ω should meet the following requirements:
(3). For point c, the centrifugal force on the edge of the top layer was completely contributed by the silicone oil from the edge to the center of the top layer. In order to maintain no relative movement of the silicone oil, ω should meet the following requirements:
By solving the range of ω respectively from the above cases, it can be concluded that there will be no relative movement of the silicone oil layer when the rotation speed is lower than ∼26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, which is much higher than the rotation speed needed in this study. To further verify the theoretical analysis, rotation speeds from 0 rpm to 5000 rpm were used for the centrifugation of the disc. Results showed that the disc can effectively seal the liquid inside the chambers at different speeds, indicating that the scheme of the slipchip and centrifugal disc can be combined together for bacteria detection.
000 rpm, which is much higher than the rotation speed needed in this study. To further verify the theoretical analysis, rotation speeds from 0 rpm to 5000 rpm were used for the centrifugation of the disc. Results showed that the disc can effectively seal the liquid inside the chambers at different speeds, indicating that the scheme of the slipchip and centrifugal disc can be combined together for bacteria detection.
3.2 Extraction of nucleic acids using the FTA cards
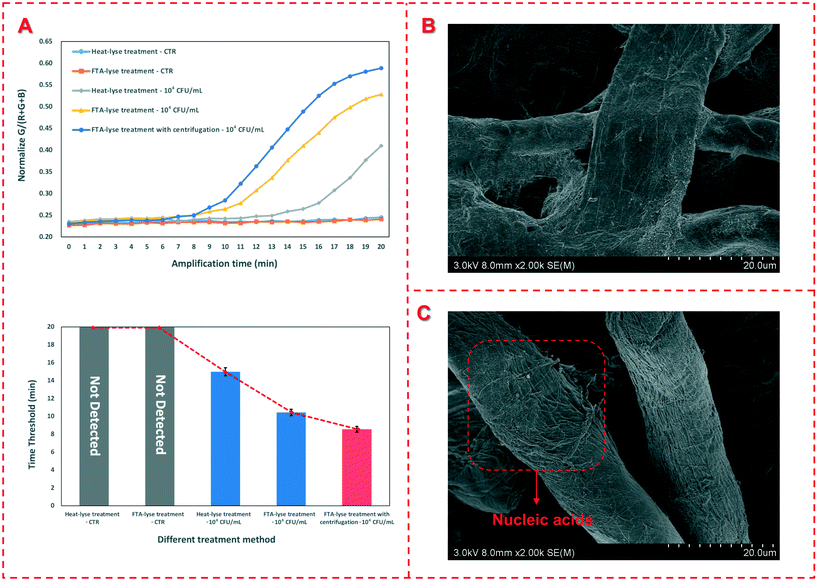
In general, the bacterial sample was first applied onto the FTA card and completely dried, and a small disk was then punched from the card for downstream DNA detection, resulting in a reduced amount of DNA templates and thus loss of sensitivity of bacterial detection. In this study, more volume of the bacterial sample was centrifuged to flow through the FTA card for enrichment of the target bacteria and extraction of their nucleic acids, which were subsequently detected using RAA. To verify the enrichment of the target bacteria using the FTA card, target bacterial samples with a concentration of 1.0 × 104 CFU mL−1 were prepared and pretreated in parallel using three different methods, including the traditional boiling method, the common FTA card method and this centrifugal FTA card method. Besides, negative samples (PBS) pretreated with the traditional boiling method and the common FTA card method were set as the control. The results are shown in Fig. 2(A). No fluorescence signal was produced after the templates extracted from the negative samples by the traditional boiling method and the common FTA card were detected using RAA. The fluorescence signal occurred at 15.0 min and 10.4 min for the target bacterial sample pretreated with the traditional boiling method and the common FTA card respectively, which were longer than that for the centrifugal FTA card method (8.5 min), indicating that more bacteria were captured through centrifugation and more DNA templates were extracted so that the sensitivity of this method was improved. In addition, TEM images were taken to characterize the FTA cards before and after nucleic acid extraction. As shown in Fig. 2(B) and (C), many filamentous nucleic acids are found on the surface of the FTA fiber after extraction, indicating that the nucleic acids were successfully extracted and immobilized on the FTA card.3.3 Optimization of the device
In this study, bacterial samples were centrifuged to flow through FTA cards for nucleic acid extraction, so the centrifugal speed was investigated to check if it has an impact on the nucleic acid extraction. Thus, different speeds ranging from 2000 rpm to 4000 rpm were applied for centrifugation of the bacterial sample at the concentration of 1.0 × 104 CFU mL−1 and the time thresholds were obtained. As shown in Fig. 3(A), when the speed is below 2000 rpm, the sample could not be centrifuged to flow through the FTA card since the centrifugal force is insufficient to drive the sample. When the speed increases from 2500 rpm to 4000 rpm, the time threshold remains around 9.3 min with on obvious change, indicating that the centrifugal speed has little effect on the nucleic acid extraction.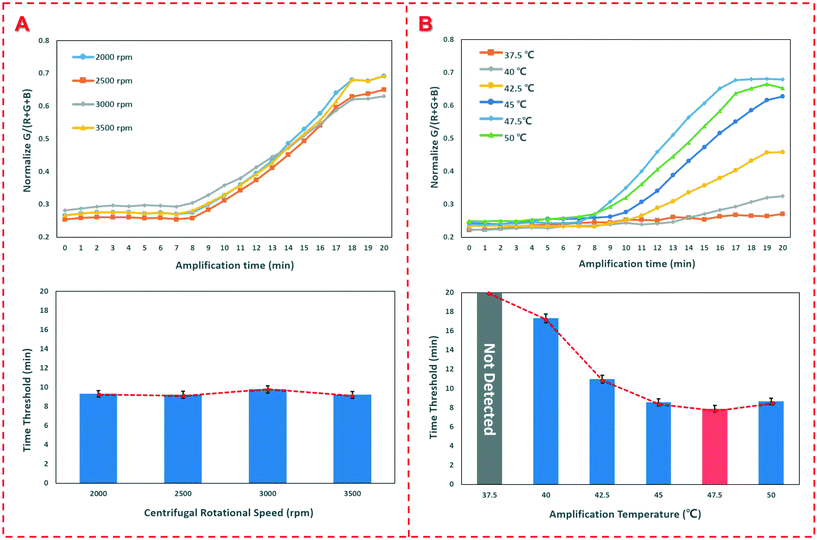 | ||
| Fig. 3 Optimization of the device. (A) Optimization of the centrifugal speed; (B) optimization of the reaction temperature. | ||
The reaction temperature is particularly crucial for RAA detection of the extracted nucleic acids. Thus, different heating temperatures from 37.5 °C to 50.0 °C were applied for the RAA reaction. The results are shown in Fig. 3(C). When the heating temperature was set as 37.5 °C, no obvious fluorescence signal was produced. When the temperature increases from 40.0 °C to 47.5 °C, the time threshold decreases from 17.3 min to 7.8 min. The further increase in the temperature to 50 °C does not result in an obvious decrease in the time threshold. Therefore, the optimal temperature of 47.5 °C was used in this study, which was slightly higher than those of other reports12,15 since PDMS has a poor thermal conductivity and only the bottom was heated for the RAA detection.
3.4 Calibration model of the device
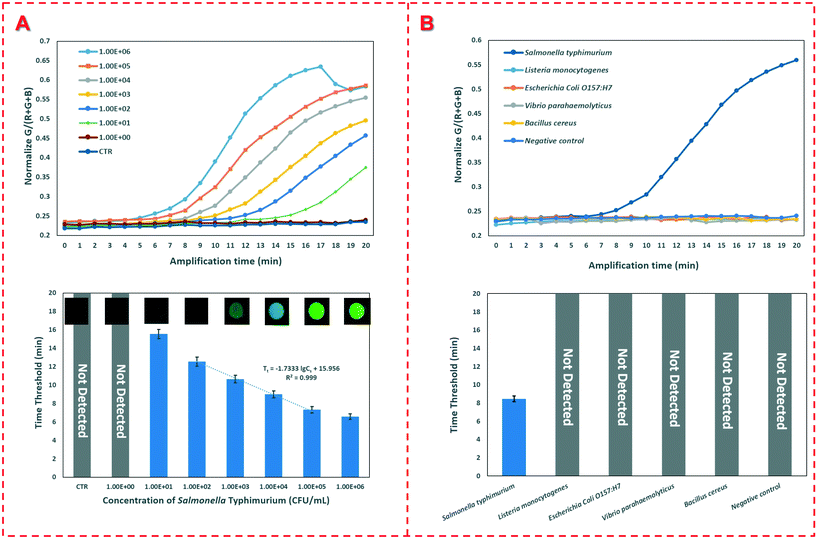
To detect an unknown concentration of Salmonella typhimurium, the calibration curve was established using the device to detect different concentrations of the target bacteria. Under optimal conditions, different concentrations of Salmonella typhimurium ranging from 1.0 × 101 CFU mL−1 to 1.0 × 106 CFU mL−1 were detected using this device. As shown in Fig. 4(A), the time threshold decreases from 15.5 min to 6.6 min as the concentration of Salmonella typhimurium increases from 1.0 × 101 CFU mL−1 to 1.0 × 106 CFU mL−1. A good linear relationship between the time threshold (Tt) and the bacterial concentration (Cs) ranging from 1.0 × 102 CFU mL−1 to 1.0×105 CFU mL−1 is found and can be expressed as:Tt = −1.733 × lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cs + 15.956 (R2 = 0.999) Cs + 15.956 (R2 = 0.999) |
To evaluate the sensitivity of the device, 20 negative controls were detected and the low detection limit of this device was determined to be 1.0 × 101 CFU mL−1. The excellent sensitivity of this device could be attributed to the following aspects: (1) the target bacteria were captured and enriched on the FTA cards; (2) the nucleic acids were effectively extracted from the bacterial cells and concentrated on the FTA cards; (3) the extracted genomic DNA templates were accurately detected using the real-time fluorescence RAA; and (4) the whole bacterial detection procedure was automatically performed without manual intervention.
To further evaluate the specificity of this device, Listeria monocytogenes, E. coli O157:H7, Vibrio parahaemolyticus and Bacillus cereus with a concentration of 1.0 × 106 CFU mL−1 and a negative control were detected. As shown in Fig. 4(B), only the target Salmonella typhimurium with the same concentration has a fluorescence signal, while all the non-target bacteria and three negative controls do not have any signals, indicating that this device has good specificity.
3.5 Applicability of the device for food sample testing
To further verify the applicability of the device for detection of Salmonella typhimurium in real food samples, chicken meats were pretreated to prepare the spiked samples at different concentrations. Triple measurements at each concentration were performed using this device under the optimal conditions. The recovery of the spiked Salmonella typhimurium was calculated as the ratio of the detected concentration to the spiked concentration. As shown in Table 1, the recovery for different concentrations of spiked bacteria ranges from 91.65% to 112.57%, and the average recovery is 101.48%, indicating that this device is capable of practical detection of Salmonella typhimurium in chicken meats.4. Conclusions
In this study, a slidable centrifugal disc was successfully developed for automatic and multi-channel detection of Salmonella typhimurium using FTA cards for nucleic acid extraction and recombinase aided amplification for nucleic acid detection. The combination of the slidable chip with the centrifugal disc was demonstrated to achieve complete automation of the bacterial detection procedure. Besides, the FTA card was verified with the capability of continuous-flow separation and efficient concentration of target bacteria, and effective extraction and stable immobilization of nucleic acids. Under the optimal conditions, this slidable centrifugal disc was able to detect Salmonella as low as 10 CFU mL−1 in a spiked chicken meat supernatant in 1 h with an average recovery of 101.8% and an average CV of 6.5%. Although this slidable centrifugal disc has been demonstrated to achieve sample-in-result-out detection of Salmonella, it still faces an important challenge that the temperature for RAA detection needs to be precisely controlled using a mini heater and might suffer from environmental conditions to some degree. Besides, this disc with eight independent channels could be used for simultaneous detection of eight samples and might be further improved for high-throughput detection of multiple bacteria.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by the National Natural Scientific Foundation of China (32071899) and Walmart Foundation (SA1703161).References
- F. Asgharpour, R. Rajabnia, E. F. Shahandashti, M. A. Marashi and Z. Moulana, Jundishapur J. Microbiol., 2014, 7, e10019 CrossRef.
- M. R. Kabir, M. A. Hossain, S. K. Paul, C. Mahmud and N. Kobayashi, Mymensingh Med. J., 2012, 21, 618–623 CAS.
- M. T. Brandi, C. E. Cox and M. Teplitski, Phytopathology, 2013, 103, 316–325 CrossRef PubMed.
- WHO, https://www.who.int/news/item/21-04-2021-infosan-quarterly-summary-2021-1, 2021.
- J. F. Yang, Y. H. Li, J. Zhao, P. F. Li, C. Zhu, Y. H. Song, L. Y. Zhang and B. W. Zhu, Food Sci. Biotechnol., 2015, 24, 23–30 CrossRef CAS.
- A. Rohde, J. A. Hammerl, I. Boone, W. Jansen, S. Fohler, G. Klein, R. Dieckmann and S. Al Dahouk, Trends Food Sci. Technol., 2017, 62, 113–118 CrossRef CAS.
- C. Huang, B. Y. Mahboubat, Y. Ding, Q. Yang, J. Wang, M. Zhou and X. Wang, LWT--Food Sci. Technol., 2021, 142, 111075 CrossRef CAS.
- T. Di Febo, M. Schirone, P. Visciano, O. Portanti, G. Armillotta, T. Persiani, E. Di Giannatale, M. Tittarelli and M. Luciani, Food Anal. Methods, 2018, 12, 322–330 CrossRef.
- M. Li, A. Ge, M. Liu, B. Ma, C. Ma and C. Shi, Talanta, 2020, 219, 121221 CrossRef CAS.
- Y. Shang, Q. Ye, S. Cai, Q. Wu, R. Pang, S. Yang, X. Xiang, C. Wang, F. Zha, Y. Ding, Y. Zhang, J. Wang, X. Sun and J. Zhang, LWT--Food Sci. Technol., 2021, 142, 110999 CrossRef CAS.
- J. Li, B. Ma, J. Fang, A. Zhi, E. Chen, Y. Xu, X. Yu, C. Sun and M. Zhang, Foods, 2019, 9 Search PubMed.
- Z. Qin, X. Xiang, L. Xue, W. Cai, J. Gao, J. Yang, Y. Liang, L. Wang, M. Chen, R. Pang, Y. Li, J. Zhang, Y. Hu and Q. Wu, Microchem. J., 2021, 164, 106050 CrossRef CAS.
- S. Wang, N. Liu, L. Zheng, G. Cai and J. Lin, Lab Chip, 2020, 20, 2296–2305 RSC.
- X. Zhang, L. Guo, R. Ma, L. Cong, Z. Wu, Y. Wei, S. Xue, W. Zheng and S. Tang, J. Microbiol. Methods, 2017, 139, 202–204 CrossRef CAS PubMed.
- G. Xie, D. Zhou, G. Zhao, X. Feng, Z. P. Aguilar and H. Xu, LWT--Food Sci. Technol., 2021, 135, 110249 CrossRef CAS.
- D. Rajendram, R. Ayenza, F. M. Holder, B. Moran, T. Long and H. N. Shah, J. Microbiol. Methods, 2006, 67, 582–592 CrossRef CAS PubMed.
- K. T. L. Trinh, R. A. Stabler and N. Y. Lee, Sens. Actuators, B, 2020, 314, 128057 CrossRef CAS.
- T. Lalani, M. D. Tisdale, J. Liu, I. Mitra, C. Philip, E. Odundo, F. Reyes, M. P. Simons, J. A. Fraser, E. Hutley, P. Connor, B. E. Swierczewski, E. Houpt, D. R. Tribble and M. S. Riddle, PLoS One, 2018, 13, e0202178 CrossRef.
- E. Pepey, T. Taukhid, N. Keck, A. Lusiastuti, J. C. Avarre, G. Sundari, S. Sarter and D. Caruso, J. Fish Dis., 2021, 44, 505–512 CrossRef CAS.
- K. T. L. Trinh, T. N. D. Trinh and N. Y. Lee, Biosens. Bioelectron., 2019, 135, 120–128 CrossRef CAS PubMed.
- W. Lyu, M. Yu, H. Qu, Z. Yu, W. Du and F. Shen, Biomicrofluidics, 2019, 13, 041502 CrossRef PubMed.
- Y. Xia, Z. Liu, S. Yan, F. Yin, X. Feng and B.-F. Liu, Sens. Actuators, B, 2016, 228, 491–499 CrossRef CAS.
- J. Yang, X. Liu, Y. Pan, J. Yang, B. He, Y. Fu and Y. Song, Sens. Actuators, B, 2019, 291, 192–199 CrossRef CAS.
- D. V. Zhukov, E. M. Khorosheva, T. Khazaei, W. Du, D. A. Selck, A. A. Shishkin and R. F. Ismagilov, Lab Chip, 2019, 19, 3200–3211 RSC.
- H. Peng, M. Zhu, Z. Gao, C. Liao, C. Jia, H. Wang, H. Zhou and J. Zhao, Biomed. Microdevices, 2020, 22, 18 CrossRef PubMed.
- Y. Wang, S. Liu, T. Zhang, H. Cong, Y. Wei, J. Xu, Y. P. Ho, S. K. Kong and H. P. Ho, Lab Chip, 2019, 19, 3870–3879 RSC.
- F. Shen, E. K. Davydova, W. Du, J. E. Kreutz, O. Piepenburg and R. F. Ismagilov, Anal. Chem., 2011, 83, 3533–3540 CrossRef CAS PubMed.
- G. Cai, W. Wu, S. Feng and Y. Liu, Analyst, 2021, 146, 4622–4629 RSC.
- Y. Song, Y. Zhang, P. E. Bernard, J. M. Reuben, N. T. Ueno, R. B. Arlinghaus, Y. Zu and L. Qin, Nat. Commun., 2012, 3, 1283 CrossRef PubMed.
- Y. Song, Y. Wang and L. Qin, J. Am. Chem. Soc., 2013, 135, 16785–16788 CrossRef CAS PubMed.
- H. V. Nguyen, V. D. Nguyen, H. Q. Nguyen, T. H. T. Chau, E. Y. Lee and T. S. Seo, Biosens. Bioelectron., 2019, 141, 111466 CrossRef CAS PubMed.
- H. Van Nguyen and T. S. Seo, Biosens. Bioelectron., 2021, 181, 113161 CrossRef CAS.
- A. R. Homann, L. Niebling, S. Zehnle, M. Beutler, L. Delamotte, M. C. Rothmund, D. Czurratis, K. D. Beller, R. Zengerle, H. Hoffmann and N. Paust, Lab Chip, 2021, 21, 1540–1548 RSC.
- H. V. Nguyen, V. D. Nguyen, E. Y. Lee and T. S. Seo, Biosens. Bioelectron., 2019, 136, 132–139 CrossRef CAS.
- B. H. Park, J. H. Jung, H. Zhang, N. Y. Lee and T. S. Seo, Lab Chip, 2012, 12, 3875–3881 RSC.
- H. V. Nguyen, K. Y. Kim, H. Nam, S. Y. Lee, T. Yu and T. S. Seo, Lab Chip, 2020, 20, 3293–3301 RSC.
- D. H. Kang, N. K. Kim, S. W. Park, W. Lee and H. W. Kang, Lab Chip, 2020, 20, 4433–4441 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1lc00915j |
| This journal is © The Royal Society of Chemistry 2022 |