Open Access Article
Open Access ArticleMicrowave-assisted green synthesis of Desmodium triquetrum-mediated silver nanoparticles: enhanced antibacterial, antibiofilm, and cytotoxicity activities against human breast cancer cell lines
Faiza
Maryani
 *a and
Abdi Wira
Septama
b
*a and
Abdi Wira
Septama
b
aResearch Centre for Chemistry, National Research and Innovation Agency (BRIN), South Tangerang, Indonesia. E-mail: marynfaiza@gmail.com; faiza.maryani@brin.go.id
bResearch Centre for Pharmaceutical Ingredients and Traditional Medicine, National Research and Innovation Agency (BRIN), South Tangerang, Indonesia
First published on 16th September 2022
Abstract
Green synthesis of nanoparticles using plant extracts has attracted great interest due to it being a low cost and environment-friendly method. In this study, a simple, fast, and green process for the synthesis of silver nanoparticles by microwave irradiation using aqueous Desmodium triquetrum extract (DTE) was investigated. The plant extract contained polyphenols that were observed by liquid chromatography-tandem mass spectrometry (LCMS/MS) analysis. The biosynthesized silver nanoparticles showed a maximum peak at 430 nm in the UV-Vis spectrum. The hydrodynamic size (Z-average size), polydispersity index (PI), and zeta potential of the AgNPs were assessed by dynamic light scattering (DLS) analysis showing the values of 88.8 nm, 0.355, and −27.4 mV, respectively. Spherical silver nanoparticles were observed in the FE-SEM image with an average diameter of 21.63 ± 4.85 nm. The X-ray diffraction (XRD) result showed a crystalline face-centred cubic structure of the silver nanoparticles. The FTIR spectra revealed phytochemicals involved in the synthesis of AgNPs. Determination of the antibacterial and antibiofilm activities of the AgNPs was conducted against four pathogens, namely B. subitilis, P. aeruginosa, E. coli and K. pneumoniae. These AgNPs exhibited strong antibacterial activity against B. subitilis (MIC value of 3.9 μg mL−1) and P. aeruginosa (7.8 μg mL−1) and moderate antibacterial activity against E. coli (15.1 μg mL−1) and K. pneumoniae (31.25 μg mL−1). In addition, the silver nanoparticles showed potent inhibition of biofilm formation at supra-MIC (2 MIC) for all tested pathogens and significant cytotoxicity against human breast cancer cell lines (MCF-7). These results suggested that Desmodium triquetrum could be an effective natural resource for the synthesis of silver nanoparticles, which have possible applications as antibacterial, antibiofilm, and cytotoxic agents against breast cancer cell lines.
Introduction
Nanoparticles have attracted massive attention for their applications in various fields including electronics, energy, catalysis, cosmetics, electrochemistry, biomedicine, food technology, the environment, etc. Nanoparticles possess sizes from 1–100 nm and a high surface area generating special chemical and physical features compared to those of bulk molecules.1 One of the noble metallic nanoparticles is silver nanoparticles, which have achieved the highest-level commercialization and attracted the most attention due to their unique features, such as non-toxic nature, attractive physical and chemical functionalities, and therapeutic capability.2 Silver nanoparticles have shown a great potential in biomedical applications, including anti-microbial, anti-cancer, anti-viral, anti-fungal, tissue scaffolding, wound dressings, and drug delivery.3–5In conventional methods, physical and chemical processes are generally used to prepare silver nanoparticles. These processes using hazardous solvents and producing toxic by-products are harmful to the environment.6,7 To reduce these drawbacks, a biological approach that focuses on replacing harmful chemicals could be a good alternative for synthesising silver nanoparticles. Green synthesis involves biological sources, such as bacteria,8 yeasts,9 actinomycetes,10 viruses,11 fungi,12 and plants.13 However, using microorganisms to synthesize silver nanoparticles requires more complex processes compared to using plants, including maintenance of a sterile environment, microbial culture and isolation.14 Plant-mediated synthesis provides major benefits, including its ease of monitoring, low time consumption, low cost, and effective replacement of chemicals in the reduction process, and is the safest choice to produce silver nanoparticles.5,15,16
Recent studies have reported that parts of the plant used for biosynthesized silver nanoparticles are roots,17 leaves,18–20 seeds21,22 and bark.23 Plant extracts contain many bioactive compounds (e.g. flavonoids, phenols, terpenoids, alkaloids, proteins, sugars, enzymes and co-enzymes) that act as reducing agents and capping agents in the synthesis of nanoparticles.24,25 As reducing agents, they can reduce silver nitrate (Ag+) into metallic silver nanoparticles (Ag0). The presence of potent reducing agents in the plant extract stimulates a fast reaction time and promotes the arrangement of smaller nanoparticles.24 In addition, as a capping agent, the bioactive compounds play an important role in preventing nanoparticle aggregation and decreasing the toxicity, which leads to the enhancement of the biocompatibility.26 According to previous studies, flavonoids are one of the most generally declared compounds that have the potential for the biosynthesis of silver nanoparticles.27
Desmodium triquetrum, a medicinal plant from Fabaceae (family) and Faboideae (subfamily), is distributed in East Asia, South Asia, and Southeast Asia.28 Previous studies have reported that Desmodium triquetrum exhibits antidiabetic, hypoglycemic, anti-inflammatory, antioxidant, and hepatoprotective activities.29,30 This plant contains bioactive compounds including phenylpropanoids, saponins, triterpenoids, and polyphenols.29,31
While other researchers reported the synthesis and antibacterial activity of silver nanoparticles using a methanolic extract of the stem of Desmodium gangeticum32 and a chloroform-methanol extract of the whole plant of Desmodium adscendens,33 this study investigated an aqueous extract of the leaves of Desmodium triquetrum (DTE) as a reducing and capping agent for the first time for the synthesis of silver nanoparticles by microwave irradiation. Microwave irradiation was applied to improve the rate of kinetics of the reaction and decrease the reaction time. The biosynthesized silver nanoparticles (DTE-AgNPs) were tested for antibacterial and antibiofilm activities against B. subitilis, P. aeruginosa, E. coli and K. pneumoniae and their cytotoxicity against MCF-7 cell lines.
Experimental
Materials
Dried leaves of Desmodium triquetrum DC were purchased from Research Centre for Pharmaceutical Ingredients and Traditional Medicine in Tawangmangu, Central Java (Indonesia). Silver nitrate, crystal violet, and NaCl were purchased from Merck. Fetal bovine serum (FBS) was used as received from Gibco. Broth heart infusion (BHI), McFarland standard, and agar were obtained from Himedia Mumbai India. Penicillin streptomycin (PS), tetracycline, Dulbecco's modified Eagle's medium (DMEM), and thiazolyl blue tetrazolium bromide (MTT) were obtained from Sigma. All of the bacteria were purchased from Indonesia Marine Education and Research Organisation, Bali.Preparation of plant extract
The dried leaves of Desmodium triquetrum DC collected from Research Institute for Spices and Medicinal Plants in Tawangmangu, Central Java (Indonesia) were ground to obtain a powder. The powder (10 g) in 100 mL of deionized water was heated at 65 °C for 30 minutes. Subsequently, the mixture was cooled and separated using Whatman filter paper. Then, the filtrate was kept at 4 °C for further use.Determination of chemical compounds by liquid chromatography with tandem mass spectrometry (LCMS-MS)
Identification of chemical constituents was accomplished using QTOF LCMS/MS (quadrupole time of flight liquid chromatography-mass spectrometry) at Research Centre for Chemistry, BRIN. The column used in this measurement was an ACQUITY UPLC BEH C8 1.7 μm 2.1 × 100 mm. Solvent A (0.1% formic acid in H2O) and solvent B (0.1% formic acid in acetonitrile) were applied as binary mobile phases. The flow rate was 0.3 mL min−1. Data were determined by UNIFI software.Biosynthesis of silver nanoparticles
Silver nitrate solution (2 mM) was prepared by dissolving AgNO3 (0.0017 g) in 50 mL of deionized water. This solution was mixed with 1 mL of the aqueous plant extract. The mixture was irradiated using a domestic microwave oven (Modena MG 2516) operating at 900 Watt for 1.5 minutes. A UV-Vis spectrometer was applied to monitor the reaction mixture. The centrifugation was conducted at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min to separate the supernatant and precipitate silver nanoparticles. The precipitate was collected and then washed thrice with deionized water to remove impurities. The purified sample was dried in an oven at 70 °C to obtain black powder silver nanoparticles.
000 rpm for 30 min to separate the supernatant and precipitate silver nanoparticles. The precipitate was collected and then washed thrice with deionized water to remove impurities. The purified sample was dried in an oven at 70 °C to obtain black powder silver nanoparticles.
Characterization of silver nanoparticles
UV-Vis spectral analysis was conducted using an Agilent Technologies Cary 60 UV-Vis spectrophotometer. The FTIR spectra were obtained using a Brucker Tensor II with an ATR attachment. Particle size, heterogeneity, and zeta potential were measured using a Horiba SZ-100 instrument. The morphology and elemental mapping of the nanoparticles were determined using a JEOL JIB-4610F field emission scanning electron microscope by energy dispersive X-ray spectroscopy (FESEM-EDX). The XRD analysis was recorded using a Rigaku Smartlab instrument with monochromatic radiation, Cu Kα radiation (θ = 1.5417 Å), at a voltage of 40 kV and a current of 30 mA.Determination of minimum inhibitory concentrations
The broth microdilution method according to standard protocols, slightly modified (NCCLS, 2008), was applied to investigate the minimum inhibitory concentration (MIC). In brief, the overnight cultured selected bacteria were prepared in sterile 0.85% NaCl and adjusted to standard 0.5 McFarland, which is equal to 1 × 108 CFU mL−1. Then, sterile 0.85% NaCl was utilized to dilute the suspension and afford 1 × 106 CFU mL−1. The bacterial suspension was mixed with two-fold dilutions of each sample in a 96-well plate and incubated at 37 °C for 24 h. The MIC was expressed as the lowest concentration that generated suppression of visible growth. The minimal bactericidal concentration (MBC) was reflected as the lowest concentration that enables the killing of the bacteria.Biofilm formation assay
The effect of AgNPs on biofilm formation of the tested bacteria was studied using a 96-well plate as previously described.34 Briefly, 100 μL of the bacterial suspension in sterile 0.85% NaCl (1 × 106 CFU mL−1) was added to a 96-well plate that contained serial concentrations of AgNPs (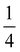 MIC,
MIC, 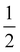 MIC, MIC and 2 MIC) in BHI. The untreated cells were applied as a negative control. After 24 h of incubation at 37 °C, the bacteria in the wells were removed and rinsed twice with sterile PBS. The wells were stained with 200 μL of 0.3% crystal violet solution, followed by incubation for 30 min at room temperature. Subsequently, the plate was rinsed with sterile PBS and 200 μL of DMSO was put into the well to solubilize the stained biofilm. Absorbance was evaluated at 595 nm which represents the biofilm formation.
MIC, MIC and 2 MIC) in BHI. The untreated cells were applied as a negative control. After 24 h of incubation at 37 °C, the bacteria in the wells were removed and rinsed twice with sterile PBS. The wells were stained with 200 μL of 0.3% crystal violet solution, followed by incubation for 30 min at room temperature. Subsequently, the plate was rinsed with sterile PBS and 200 μL of DMSO was put into the well to solubilize the stained biofilm. Absorbance was evaluated at 595 nm which represents the biofilm formation.
Cytotoxicity activity assay
An MTT ((3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-tetrazolium bromide) assay35,36 was conducted to demonstrate cytotoxicity activity against the MCF7 (breast cancer) cell line. Cancer cells in DMEM, FBS 10%, and PS 1% were put into a 96-well microplate at the density of 7000 cells per well. The cells were incubated in a CO2 incubator for 24 h. Then, the cells were treated with Desmodium triquetrum extract (DTE) and AgNPs at varying concentrations for 24 h. After incubation, each well was washed with 100 μg mL−1 PBS and 10 μl of MTT solution (5 mg mL−1) and 90 μL of serum-free medium were sequentially added. The microplate was incubated for 3 h. The solution in the microplate was discarded and DMSO (100 μl) was added to the well plate to dissolve formazan crystals. A multi-well spectrophotometer (Varioskan Flash, Thermo Fisher Scientific) was used to determine the absorbance of the samples at 570 nm. The cytotoxicity activity of DTE and AgNPs was demonstrated as a percentage of cell viability compared to the control using the following formula: % of cell viability = mean OD/control × 100.Results and discussion
Phytochemical profile of Desmodium triquetrum extract (DTE)
LCMS/MS quadrupolemeasurement was carried out to identify organic compounds in the plant extract. According to the result (Table 1), there are 6′′-O-p-coumaroylaloesin and polyphenols including 3-O-[β-D-glucopyra-nosyl-(1 → 2)]-β-D-glucopyranosyl-7-O-α-L-glucopyranosyl-kaempferol, kaempferol-3-O-rutinoside, quercetin, kaempferol-3-O-β-D-glucopyranoside, and quercimeritrin in the plant extract. These organic compounds may be significant for the biosynthesis of silver nanoparticles due to the fact that they could reduce silver ions to silver nanoparticles, contribute to the stability of silver nanoparticles, and avoid aggregation of the nanoparticles.37Visual observation and UV-visible spectroscopy analysis
Desmodium triquetrum extract (DTE) assists in the reduction of silver ions (Ag+) to silver atoms followed by clusters leading to the formation of colloidal silver nanoparticles. The colour alteration of the mixture proving the presence of silver nanoparticles was observed after microwave irradiation (Fig. 1). The colour changed from pale yellow to yellowish-brown, which appeared as a result of the excitation of the surface plasmon resonance (SPR) band in the silver nanoparticles, as seen in the UV-Vis region.38Fig. 2 presents the UV absorbance spectra of DTE and the biosynthesized silver nanoparticles (DTE-AgNPs) with various reaction times from 10 to 90 seconds. The peak absorbance of DTE-AgNPs was monitored at 430–440 nm. The SPR band is slightly increased between zero and 30 s, since the transformation of silver ions to silver nanoparticles is slow. Enhancing the reaction time generates an excellent SPR band due to a lot of silver ions converting to silver nanoparticles. The longest-time reaction exhibited the sharpest and strongest absorption, which was observed at 430 nm (Fig. 2). | ||
| Fig. 1 Photographs of the aqueous solution of silver nitrate (AgNO3) with the addition of plant extract (A) before microwave irradiation (0 minute) and (B) after microwave irradiation for 90 s. | ||
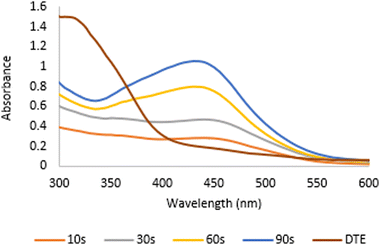 | ||
| Fig. 2 The UV absorbance spectra of DTE (Desmodium triquetrum extract) and the biosynthesized AgNPs (DTE-AgNPs) with various reaction times from 10 to 90 seconds. | ||
Dynamic light scattering (DLS) analysis
DLS measurements were accomplished to obtain the hydrodynamic particle size, zeta potential, and polydispersity index (PI) of the biosynthesized AgNPs. As shown in Fig. 3, the average particle size (Z-average) of the biosynthesized AgNPs (Fig. 3A) was 88.8 nm with the PI value of 0.355 (Fig. 3A) indicating that the nanoparticles were relatively monodispersed and the zeta potential showed negative charge with a value of −27.4 mV (Fig. 3B). The obtained particle size is the hydrodynamic size determined by the accumulation of the metal core size, the size of the capping agents or biomolecules attached to the silver nanoparticles, and the electrical double layer between particles.39 Therefore, the average size of the biosynthesized AgNPs based on DLS analysis is larger than that by macroscopic analysis, such as SEM or TEM. Nanoparticles with a size below 150 nm and a PI value of 0.3 are capable of cellular uptake.40 The negative zeta potential value reveals that the negatively charged functional groups of the plant extract play important roles in the stability of the colloidal silver nanoparticles.41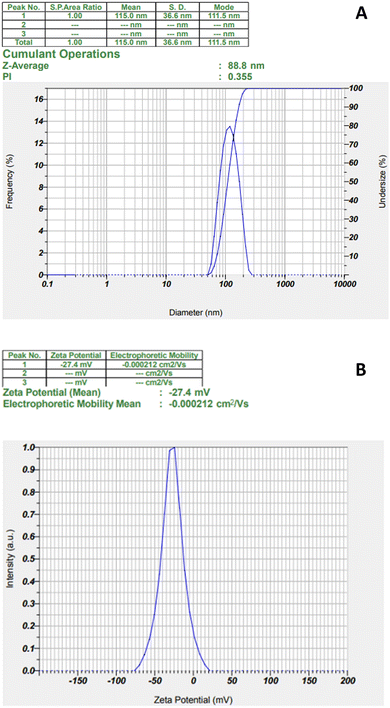 | ||
| Fig. 3 The particle size and PI analysis (A) and zeta potential (B) of biosynthesized AgNPs from Desmodium triquetrum extract. | ||
FE-SEM and EDX analysis
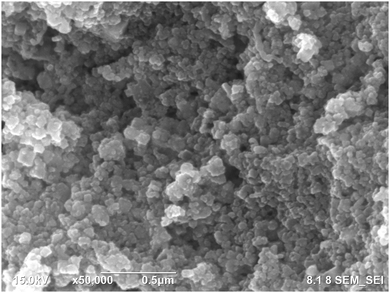
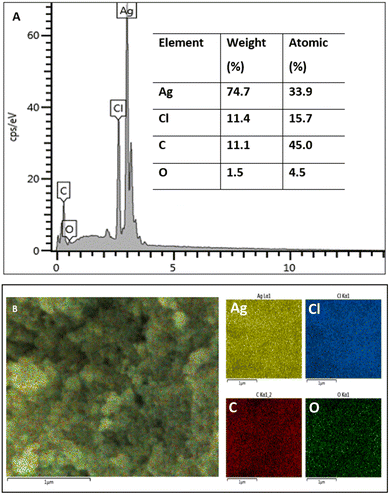
FE SEM and EDX measurements were conducted to obtain the morphology and elemental composition of the synthesized silver nanoparticles. The spherical silver nanoparticles are observed in Fig. 4. Image J software was used to calculate the average size of the silver nanoparticles, which was 21.63 ± 4.85 nm. The size and shape of the nanoparticles affect their biological activity. Smaller nanoparticles can more easily penetrate cells leading to them being more cytotoxic against cancer cells and more efficient against microbial pathogens in comparison with larger particles.42,43 Moreover, the spherical structure of nanoparticles has a larger specific surface area generating more damage to the bacterial cell as compared to nanorods and nanowires with smaller surface area.44The elemental analysis is presented in Fig. 5. The EDX spectrum (Fig. 5A) showed a strong signal peak, correlated to high emission energy at 3 keV revealing the existence of silver.45,46 The quantitative analysis showed that the weight % of silver nanoparticles was 74.7% and the atomic % was 33.9%. Another element that showed an intense peak is chloride (Cl). Previous studies also reported the intense peaks attributed to the presence of silver (Ag) and chloride (Cl) in the elemental analysis of biosynthesized silver nanoparticles using aqueous plant extracts.47,48 Chloride (Cl−) anions are macronutrients that significantly exist in plants contributing to maintaining photosynthesis and homeostatis.49 Generally, other elements observed by EDX analysis including carbon, oxygen, and chloride were demonstrated as capping agents of the biosynthesized silver nanoparticles in agreement with other studies.50,51 The elemental mapping of the biosynthesized AgNPs is presented in Fig. 5B, exhibiting that all elements are homogeneously dispersed.
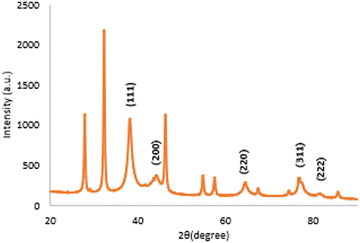
X-ray diffraction (XRD) analysis
The XRD pattern of the biosynthesized silver nanoparticles is exhibited in Fig. 6. The five distinct diffraction peaks of 2θ values were observed at 38.11°, 44.25°, 64.41°, 77.45°, and 81.33° corresponding to the (111), (200), (220), (311), and (222) planes. A similar result was reported by Jayaprakash et al., who demonstrated that the synthesized silver nanoparticles have a face-centred cubic (FCC) structure.52 The d spacing values presented at 2.3590 Å, 2.0453 Å, 1.4454 Å, 1.2313 Å, and 1.1821 Å are matched with the standard silver values.53 The additional peaks observed in Fig. 6 were due to the presence of organic compounds from the extract as reducing and stabilizing agents.54 The average crystalline size calculated using the Scherrer formula was observed to be 7.428 nm.Fourier transform infrared (FTIR)
Functional groups of the samples were identified using FTIR analysis. DTE showed peaks at 3352, 2931, 1600, 1515, 1444, 1391, 1203, 1063 and 819 cm−1 (Fig. 7A). O–H stretching in polyphenols was observed at 3352 cm−1, C–H stretching was presented at 2931 cm−1, and a strong peak at 1600 cm−1 was related to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching. The peak presented at 1515 cm−1 corresponded to N–H bending vibration in amide linkages of the peptides in the plant extract. The other peaks at 1444 and 1391 cm−1 corresponded to O–H bending, 1203 and 1063 cm−1 were related to C–O stretching, and 819 and 793 cm−1 corresponded to C
O stretching. The peak presented at 1515 cm−1 corresponded to N–H bending vibration in amide linkages of the peptides in the plant extract. The other peaks at 1444 and 1391 cm−1 corresponded to O–H bending, 1203 and 1063 cm−1 were related to C–O stretching, and 819 and 793 cm−1 corresponded to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bending. The biosynthesized AgNPs (Fig. 7B) revealed peaks at 3727 and 3601 cm−1 (O–H stretching), 1993 and 1896 cm−1 (C–H bending), 1594 cm−1 (C
C bending. The biosynthesized AgNPs (Fig. 7B) revealed peaks at 3727 and 3601 cm−1 (O–H stretching), 1993 and 1896 cm−1 (C–H bending), 1594 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching), and 1512 cm−1 (N–H bending vibration). O–H bending was presented at 1440 and 1358 cm−1. Peaks at 1221 and 1051 cm−1 can be assigned to C–O stretching and 819 and 773 cm−1 can be attributed to C
C stretching), and 1512 cm−1 (N–H bending vibration). O–H bending was presented at 1440 and 1358 cm−1. Peaks at 1221 and 1051 cm−1 can be assigned to C–O stretching and 819 and 773 cm−1 can be attributed to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bending. The spectrum of the AgNPs showed a reduction in intensity and slight shifts in the position of peaks indicating polyphenols as the main components in the extract assisting in the synthesis and encapsulating of silver nanoparticles.55
C bending. The spectrum of the AgNPs showed a reduction in intensity and slight shifts in the position of peaks indicating polyphenols as the main components in the extract assisting in the synthesis and encapsulating of silver nanoparticles.55
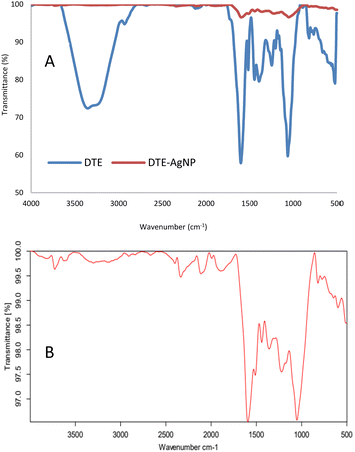 | ||
| Fig. 7 The FTIR spectra of desmodium triquetrum extract (DTE) and biosynthesized AgNPs (DTE-AgNPs) (A) and the magnified spectrum of the biosynthesized AgNPs (DTE-AgNPS) (B) from 500 to 4000 cm−1. | ||
Antibacterial activity
Several clinical isolates (B. subtillis, P. rugosa, E. coli, and K. pneumonia) were selected to determine the effect of AgNPs on pathogens. On the basis of the broth microdilution method, AgNPs demonstrated potent antibacterial activity against B. subitilis and P. aeruginosa with MIC values of 3.9 and 7.8 μg mL−1, respectively (Table 2). These AgNPs produced a moderate antibacterial effect against E. coli (an MIC value of 15.1 μg mL−1) and K. pneumoniae (31.2 μg mL−1). In contrast, DTE showed weak activity against all these selected pathogens with MIC values in the range of 250 to 500 μg mL−1. It has been reported that AgNPs display a potent antibacterial effect against both Gram-negative and the Gram-positive bacteria.56 However, the antibacterial mechanism of AgNPs is still unclear. The antibacterial action of AgNPs against the Gram-negative bacteria that have a thin peptidoglycan layer may be due to the capability of nanoparticles to penetrate the membrane of the cells. It is reported that AgNPs may induce membrane cell damage that allows nanoparticles to penetrate the cell and react with intracellular material including DNA and finally lead to cell damage.57 In addition, AgNPs also enabled the generation of reactive oxygen species (ROS) production, which causes cell damage and protein leakage.58 In the present study, a better antibacterial effect was found in the Gram-positive bacteria (B. subtillis,an MIC value of 3.9 μg mL−1), which probably resulted from synergistic interaction between the silver ion and secondary metabolites from the plants. It has been reported that the synthesised AgNPs using Eucalyptus citriodora, which contained several phytochemical compounds including flavonoids, displayed strong antibacterial activity against various pathogens such as Staphylococcus aureus, Acinetobacter baumannii, and Enterococcus faecalis.59Biofilm formation assay
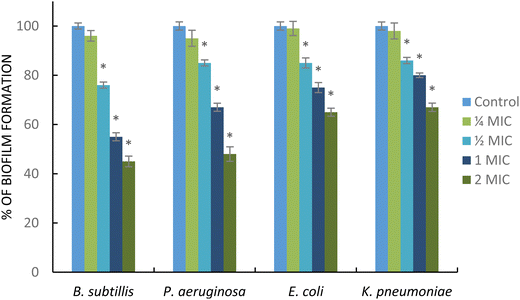
Biofilm formation plays important roles in the initial step of antimicrobial resistance. Different pathogenic bacteria, such as Gram-negative and Gram-positive bacteria, are capable of producing a biofilm. In addition, the development of the biofilm also generates the failure of antibiotic therapy.60 In this study, the AgNPs showed antibiofilm activity against the tested bacteria. As shown in Fig. 8, AgNPs at sub-MIC ( MIC) significantly decreased the biofilm formation in B. subtillis, P. eruginosa, K. pneumoniae, and E. coli. Interestingly, these nanoparticles at supra-MIC (2 MIC) strongly inhibit the formation of biofilms for all tested pathogens. The AgNPs displayed strong antibiofilm activity particularly in B. subtillis, and P. eruginosa. This result was supported by a previous report; Ali et al. reported that AgNPs obtained from the green synthesis method using Eucalyptus globulus as a capping and reducing agent demonstrated strong activity in inhibiting biofilm formation in P. aeruginosa and E. coli.61 Additionally, AgNPs synthesized using Styrax benzoin possessed significant antibiofilm activity against E. coli.62 The mechanism of action of AgNPs inhibiting biofilm formation is still unclear. However, the antibiofilm activity may be associated with the release of silver ions in high concentration in the film biological system enabling the killing of the planktonic bacterial cells.63
MIC) significantly decreased the biofilm formation in B. subtillis, P. eruginosa, K. pneumoniae, and E. coli. Interestingly, these nanoparticles at supra-MIC (2 MIC) strongly inhibit the formation of biofilms for all tested pathogens. The AgNPs displayed strong antibiofilm activity particularly in B. subtillis, and P. eruginosa. This result was supported by a previous report; Ali et al. reported that AgNPs obtained from the green synthesis method using Eucalyptus globulus as a capping and reducing agent demonstrated strong activity in inhibiting biofilm formation in P. aeruginosa and E. coli.61 Additionally, AgNPs synthesized using Styrax benzoin possessed significant antibiofilm activity against E. coli.62 The mechanism of action of AgNPs inhibiting biofilm formation is still unclear. However, the antibiofilm activity may be associated with the release of silver ions in high concentration in the film biological system enabling the killing of the planktonic bacterial cells.63
In vitro cytotoxic activity of the biosynthesized silver nanoparticles against breast cancer cell lines (MCF7)
An MTT assay was carried out to demonstrate the in vitro cytotoxicity activity of DTE and the biosynthesized AgNPs towards MCF7 (breast cancer cell line). Fig. 9 presents the percentage of cell viability over a range of concentrations (1.5625–12 μg mL−1). The viability of MCF7 cells was observed from 86% to 100% (for all tested concentrations of DTE) and from 6% to 45% (for all tested concentrations of AgNPs). Cell viability is enhanced by reducing the concentrations of the samples, showing dose-dependent cytotoxicity. Cell viability of DTE above 70% indicated non-cytotoxicity towards the MCF7 cell line in this range of concentrations, whereas the biosynthesized AgNPs showed a cytotoxic effect with cell viability below 70%.64 The strong cytotoxic effect of the biosynthesized silver nanoparticles occurs due to high cellular uptake of silver nanoparticles possessing a large surface-to-volume ratio and cancer cells obtaining high proliferation rate and irregular metabolism becoming more accessible to attack by silver nanoparticles.65 Silver nanoparticles are capable of generating cell cycle arrest, DNA fragmentation, and potent apoptosis in cancer cells.66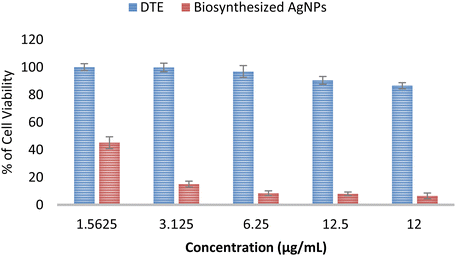 | ||
| Fig. 9 Cytotoxic activity of Desmodium triquetrum extract (DTE) and the biosynthesized AgNPs against human breast cancer cell lines (MCF-7). | ||
Conclusions
A simple and rapid synthesis of silver nanoparticles using Desmodium triquetrum extract (DTE) as an alternative green method was accomplished. DTE contained bioactive compounds, such as polyphenols, which assisted in the reduction of nitrate ions to form silver nanoparticles and stabilization of silver nanoparticles. The results of UV-Vis spectrophotometry, FTIR, DLS, FE-SEM, EDX, and XRD analyses exhibited the existence of biosynthesized silver nanoparticles (DTE-AgNPs). The DTE-AgNPs demonstrated potent antibacterial and antibiofilm properties against the tested pathogens, and strong cytotoxicity against human breast cancer cell lines (MCF-7).Author contributions
FM: project administration, conceptualization, investigation, methodology, writing of the original draft, and writing-review & editing. AWS: investigation, methodology, and writing of the original draft. FM is the main contributor for this manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work did not receive any specific grant from funding agencies. The authors acknowledge the facilities and technical support from Characterization Laboratories at Research Centre for Chemistry and Research Centre for Physic, National Research and Innovation Agency (BRIN), Republic of Indonesia.References
- I. Khan, K. Saeed and I. Khan, Arabian J. Chem., 2019, 12, 908–931 CrossRef CAS.
- A. Naganthran, G. Verasoundarapandian, F. E. Khalid, M. J. Masarudin, A. Zulkharnain, N. M. Nawawi, M. Karim, C. A. C. Abdullah and S. A. Ahmad, Materials, 2022, 15, 427 CrossRef CAS PubMed.
- A. Almatroudi, Open Life Sci., 2020, 15(1), 819–839 CAS.
- C. Vanlaveni, S. Lallianrawna, A. Biswas, M. Selvaraj, B. Changmai and S. L. Rokhum, RSC Adv., 2021, 11, 2804 RSC.
- A. N. Maranciuc, D. Chicea and M. Chicea, Mol. Sci., 2022, 23, 5778 CrossRef PubMed.
- M. S. Samuel, S. Jose, E. Selvarajan, T. Mathimani and A. Pugazhendhi, J. Photochem. Photobiol., B, 2020, 202, 111642 CrossRef CAS PubMed.
- S. Pirtarighat, M. Ghannadnia and S. Baghshashi, J. Nanostruct. Chem., 2019, 9, 1–9 CrossRef CAS.
- J. H. Jo, P. Singh, Y. J. Kim, C. Wang, R. Mathiyalagan, C.-G. Jin and D. C. Yang, Artif. Cells, Nanomed., Biotechnol., 2016, 44, 1576–1581 CrossRef CAS PubMed.
- M. Shu, F. He, Z. Li, X. Zhu, Y. Ma, Z. Zhou, Z. Yang, F. Gao and M. Zeng, Nanoscale Res. Lett., 2020, 15, 14 CrossRef CAS PubMed.
- S. Kumari, T. Nimisha, G. Anjum and V. Hooda, Inorg. Nano-Met. Chem., 2021, 51, 1386–1395 CAS.
- P. Velusamy, G. V. Kumar, V. Jeyanthi, J. Das and R. Pachaiappan, Toxicol. Res., 2016, 32, 95–102 CrossRef CAS PubMed.
- M. G. Casagrande and R. D. Lima, Front. Bioeng. Biotechnol., 2019, 7, 287 CrossRef PubMed.
- C. Hano and B. H. Abbasi, Biomolecules, 2021, 12, 31 CrossRef.
- A. Rautela, J. Rani and M. Debnath, J. Anal. Sci. Technol., 2019, 10, 5 CrossRef.
- H. Hemlata, P. R. Meena, A. P. Singh and K. K. Tejavath, ACS Omega, 2020, 5(10), 5520–5528 CrossRef.
- S. M. Rakib Uz Zaman, et al. , Challenges, 2022, 13, 18, DOI:10.3390/challe13010018.
- S. Dangi, A. Gupta, D. K. Gupta, S. Singh and N. Parajuli, Chem. Data Collect., 2020, 28, 100411, DOI:10.1016/j.cdc.2020.100411.
- H. A. Widatalla, L. F. Yassin, A. A. Alrasheid, S. A. R. Ahmed, M. O. Widdatallah, S. H. Eltilib and A. A. Mohamed, Nanoscale Adv., 2022, 4, 911–915 RSC.
- L. Muthulakshmi, K. Suganya, M. Murugan, J. Annaraj, V. Duraipandiyan, D. A. Al Farraj, M. S. Elshikh, A. Juliet, M. Pasupuleti and J. Arockiaraj, J. King Saud Univ., Sci., 2022, 34(5), 102083 CrossRef.
- P. Rani, B. Ahmed, J. Singh, J. Kaur, M. Rawat, N. Kaur, A. S. Matharu, M. Alkahtani, E. A. H. Alhomaidi and J. Lee, Saudi J. Biol. Sci., 2022, 29, 103296 CrossRef CAS PubMed.
- S. Shobana, S. Veena, S. S. M. Sameer, K. Swarnalakshmi and L. A. Vishal, Curr. Pharm. Biotechnol., 2020, 21, 980–989 CAS.
- C. Thummaneni, D. V. S. Prakash, R. Golli and M. Vangalapati, Mater. Today: Proc., 2022, 62(6), 4001–4005 CAS.
- A. D. Mahapatra, C. Patra, J. Mondal, C. Sinha, P. C. Sadhukhan and D. Chattopadhyay, ChemistrySelect, 2020, 5, 4770–4777 CrossRef.
- A. Roy, O. Bulut, S. Some, A. K. Mandal and M. D. Yilmaz, RSC Adv., 2019, 9, 2673–2702 RSC.
- N. Tarannum, D. Divya and Y. K. Gautam, RSC Adv., 2019, 9, 34926–34948 RSC.
- R. K. Selvakesavan and G. Franklin, Nanotechnol., Sci. Appl., 2021, 14, 179–195 CrossRef PubMed.
- T. Mustapha, N. Misni, N. R. Ithnin, A. M. Daskum and N. Z. Unyah, Int. J. Environ. Res. Public Health, 2022, 19, 674 CrossRef CAS PubMed.
- S. P. Vedpal, P. DHanabal, M. V. Dhamodaran, N. L. Chaitnya, B. Duraiswamy, U. Jayaram and S. Neha, J. Chem. Pharm. Res., 2016, 8, 91–97 CAS.
- X. Lin, X. Zhou, W. Sun, L. Zhang, C. Zhang and X. Zang, Trop. J. Pharm. Res., 2020, 19, 829–835 CrossRef CAS.
- V. Vedpal, S. Jupudi, S. Jubie, N. P. Deepika and S. P. Dhanabal, Nat. Prod. Res., 2021, 35, 413–420 CrossRef.
- G. Zhang, Y. Chen, K. Tariq, Z. An, S. Wang, F. Q. Memon, W. Zhang and H. Si, Czech J. Food Sci., 2020, 38, 115–122 CrossRef CAS.
- B. Azad and A. Banerjee, J. Pharm. Innov., 2014, 3(7), 77–81 Search PubMed.
- J. R. Lakkakula, D. T. Ndinteh, S. F. V. Vuuren, D. K. Olivier and R. W. M. Krause, IET Nanobiotechnol., 2017, 11(8), 1017–1026 CrossRef.
- A. W. Septama, A. N. Tasfiyati, R. Kristiana and A. Jaisi, S. Afr. J. Bot., 2022, 146, 728–734 CrossRef CAS.
- K. Ali, Q. Saquib, B. Ahmed, M. A. Siddiqui, J. Ahmed, M. A. Shaeri, A. A. Al-khedhairy and J. Musarrat, Process Biochem., 2020, 91, 387–397 CrossRef CAS.
- T. Ernawati, M. Minarti, G. Widiyarti, M. Megawati and F. Maryani, J. Appl. Pharm. Sci., 2022, 12(04), 096–107 CrossRef CAS.
- W. M. Melkamu and L. T. Bitew, Heliyon, 2021, 7, e08459 CrossRef PubMed.
- S. Das, L. Langbang, M. Haque, V. K. Belwal, K. Aguan and A. S. Roy, J. Pharm. Anal., 2021, 11, 422–434 CrossRef.
- M. S. Alwhibi, A. S. Dina, A. A. Manal, A. B. Alangery, H. A. Dehaish and Y. A. Alwasel, Green Process Synth., 2021, 1, 412–420 CrossRef.
- B. Cervantes, L. Arana, S. M. Cuesta, M. Bruno, I. Alkorta and I. V. Nieto, Jpn. Clin. Med., 2019, 8, 1464 CrossRef CAS PubMed.
- E. Y. Ahn, H. Jin and Y. Park, Mater. Sci. Eng., 2019, 101, 204–216 CrossRef CAS PubMed.
- H. Arshad, M. A. Sami, S. Sadaf and U. Hassan, Sci. Rep., 2021, 11, 5996 CrossRef CAS.
- W. Liu, et al. , Nanotoxicology, 2010, 4(3), 319–330 CrossRef CAS.
- D. Acharya, K. M. Singha, P. Pandey, B. Mohanta, J. Rajkumari and L. P. Singha, Sci. Rep., 2018, 8, 201 CrossRef PubMed.
- V. Lakkim, M. C. Reddy, R. R. Pallavali, K. R. Reddy, C. V. Reddy, I. Inamuddin, A. L. Bilgrami and D. Lomada, Antibiotics, 2020, 9, 902 CrossRef CAS PubMed.
- S. A. Gaddam, V. S. Kotakadi, G. K. Subramanyam, J. Penchalaneni, V. N. Challagundla, S. G. Dvr and V. R. Pasupuleti, Sci. Rep., 2021, 11, 21969 CrossRef CAS.
- M. T. Yassin, A. A. F. Mostafa, A. A. Al-Askar and F. O. Al-Otibi, Crystals, 2022, 12, 603 CrossRef CAS.
- K. Okaiyeto, M. O. Ojemaye, H. Hoppe, L. V. Mabinya and A. I. Okoh, Molecules, 2019, 24, 4382 CrossRef CAS PubMed.
- A. Sidorowicz, T. Szymanski and J. D. Rybka, Biology, 2021, 10, 784 CrossRef CAS PubMed.
- S. Singla, A. Jana, R. Thakur, C. Kumari, S. Goyal and J. Pradhan, OpenNano, 2022, 7, 100047 CrossRef.
- Z. Umar, U. R. Bello, A. Mathur, G. Tailor, J. Chaudhary and S. Singh, Curr. Res. Green Sustainable Chem., 2021, 4, 100203 CrossRef CAS.
- N. Jayaprakash, J. J. Vijaya, L. J. Kennedy, K. Priadharsini and P. Palani, Mater. Lett., 2014, 137, 358–361 CrossRef CAS.
- S. H. Chai, Y. Wang, Y. Qiao, P. Wang, Q. Li, C. Xia and M. Ju, J. Photochem. Photobiol., B, 2018, 178, 457–462 CrossRef.
- A. Rautela, J. Rani and M. Debnath, J. Anal. Sci. Technol., 2019, 10, 5 CrossRef.
- K. N. Yusof, S. S. Alias, Z. Harun, H. Basri and F. H. Azhar, ChemistrySelect, 2018, 3, 8881–8885 CrossRef CAS.
- S. Paosen, S. Wunnoo, S. Bilhman, S. Lethongkam, A. W. Septama and S. P. Voravuthikunchai, Int. J. Food. Sci., 2021, 56, 5881–5892 CrossRef CAS.
- M. Hossain, S. A. Polash and M. Takikawa, Front. Bioeng. Biotechnol., 2019, 7, 239 CrossRef PubMed.
- B. Ahmed, A. Hashmi, M. S. Khan and J. Musarrat, Adv. Powder Technol., 2018, 29, 1601–1616 CrossRef CAS.
- S. Paosen, J. Saising, A. W. Septama and S. P. Voravuthikunchai, Mater. Lett., 2017, 209, 201–206 CrossRef CAS.
- F. G. V. Lopez, J. Reifman and A. Wallqvist, PLoS Comput. Biol., 2015, 11, e1004452 CrossRef PubMed.
- K. Ali, B. Ahmed, S. Dwivedi, Q. Saquib, A. A. Al-Khedhairy and J. Musarrat, PLoS One, 2015, 10, e0131178 CrossRef PubMed.
- J. Du, H. Singh and T. H. Yi, Bioprocess Biosyst. Eng., 2016, 39, 1923–1931 CrossRef CAS PubMed.
- L. Huang, Y. Lou, D. Zhang, L. Ma, H. Qian, Y. Hu, P. Ju, D. Xu and X. Li, Chem. Eng. J., 2020, 381, 122662 CrossRef CAS.
- International Organization for Standardization. ISO 10993-5:2009. 2009.
- S. Khorrami, A. Zarrabi, M. Khaleghi, M. Danaei and N. R. Mozafari, Int. J. Nanomed., 2018, 13, 8013–8024 CrossRef CAS.
- A. Bandyopadhyay, B. Roy, P. Shaw, P. Mondal, M. K. Mondal, P. Chowdhury, S. Bhattacharya and A. Chattopadhyay, Nucleus, 2020, 63, 191–202 CrossRef.
| This journal is © The Royal Society of Chemistry 2022 |