Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
In silico characterisation of the complete Ly6 protein family in Fasciola gigantica supported through transcriptomics of the newly-excysted juveniles†
Sarah D.
Davey
 a,
Iain W.
Chalmers
a,
Iain W.
Chalmers
 *a,
Narcis
Fernandez-Fuentes
a,
Martin T.
Swain
a,
Dan
Smith
b,
Syed M.
Abbas Abidi
c,
Mohammad K.
Saifullah
c,
Muthusamy
Raman
d,
Gopalakrishnan
Ravikumar
d,
Paul
McVeigh
e,
Aaron G.
Maule
e,
Peter M.
Brophy
a and
Russell M.
Morphew
*a,
Narcis
Fernandez-Fuentes
a,
Martin T.
Swain
a,
Dan
Smith
b,
Syed M.
Abbas Abidi
c,
Mohammad K.
Saifullah
c,
Muthusamy
Raman
d,
Gopalakrishnan
Ravikumar
d,
Paul
McVeigh
e,
Aaron G.
Maule
e,
Peter M.
Brophy
a and
Russell M.
Morphew
 a
a
aInstitute of Biological, Environmental & Rural Sciences (IBERS), Aberystwyth, Ceredigion, UK. E-mail: iwc@aber.ac.uk
bComputational and Analytical Sciences, Rothamsted Research, Harpenden, Hertfordshire, UK
cDepartment of Zoology, Aligarh Muslim University, Aligarh, Uttar Pradesh 202001, India
dTamil Nadu Veterinary and Animal Sciences University, Chennai, Tamil Nadu 600051, India
eSchool of Biological Sciences, Queen's University Belfast, Belfast, UK
First published on 8th November 2021
Abstract
Fasciola gigantica is one of the aetiological trematodes associated with fascioliasis, which heavily impacts food-production systems and human and animal welfare on a global scale. In the absence of a vaccine, fascioliasis control and treatment is restricted to pasture management, such as clean grazing, and a limited array of chemotherapies, to which signs of resistance are beginning to appear. Research into novel control strategies is therefore urgently required and the advent of ‘omics technologies presents considerable opportunity for novel drug and vaccine target discovery. Here, interrogation of the first available F. gigantica newly excysted juvenile (NEJ) transcriptome revealed several protein families of current interest to parasitic flatworm vaccine research, including orthologues of mammalian complement regulator CD59 of the Ly6 family. Ly6 proteins have previously been identified on the tegument of Schistosoma mansoni and induced protective immunity in vaccination trials. Incorporating the recently available F. gigantica genome, the current work revealed 20 novel Ly6 family members in F. gigantica and, in parallel, significantly extended the F. hepatica complement from 3 to 18 members. Phylogenetic analysis revealed several distinct clades within the family, some of which are unique to Fasciola spp. trematodes. Analysis of available proteomic databases also revealed three of the newly discovered FhLy6s were present in extracellular vesicles, which have previously been prioritised in studying the host-parasite interface. The presentation of this new transcriptomic resource, in addition to the Ly6 family proteins here identified, represents a wealth of opportunity for future vaccine research.
Introduction
In recent decades, the advent of high-throughput sequencing and bioinformatics-based computational processing has allowed parasitologists to access a wide array of genetic data.1 Publication of this data has been vital in developing a better understanding of the molecular processes, key genes and proteins involved in parasitism, as well as clarifying evolutionary relationships between the species.2,3An area of research that could benefit from the use of ‘omics data is the study of Fasciola spp. trematodes, which are aetiological agents in the zoonotic disease, fascioliasis.4,5Fasciola spp. trematodes have complex life cycles and undergo development in the environment, molluscan intermediate host and definitive mammalian hosts. The juvenile stages are particularly relevant to the pathology of this disease due to the migration of the parasite through the host abdominal cavity to the liver, resulting in significant mechanical and inflammatory damage to the host.6 Despite the global importance of this disease, relatively little genomic or bioinformatic data has been historically available for these parasites, thus there is substantial opportunity to expand on the understanding of fascioliasis’ molecular underpinning.7
Fasciola gigantica lacked complete genomic representation until very recently, with the independent publication of two new draft genomes.8,9 Consequently, research into F. gigantica molecular biology has been reliant on limited transcriptomic data. Whole-organism transcriptomes are currently available for the egg, miracidial, redial, cercarial, later juvenile (42 and 70 days post infection) and adult stages, in addition to smaller datasets which focus specifically on the host-parasite interface and pathogenicity.10–14 The newly excysted juvenile (NEJ) has been entirely neglected, with no transcriptomic datasets publicly available and most studies focusing on small subsets of data, such as specific genes for RNAi interference.15 Considering the importance of this stage to host invasion and disease progression, it is essential that this gap in knowledge be closed.16
The mechanisms in which Fasciola spp. parasites interface with their hosts has been an area of intense scientific scrutiny in recent decades, particularly focussing on excretory-secretory (ES) proteins, extracellular vesicles (EVs) and the tegument.17–23 The tegument has previously been investigated in F. hepatica given its proposed involvement in immunological processes of early infection and in-host resolution via antibody-dependent cellular cytotoxicity.18 Components of the tegument have also demonstrated antigenic properties and therefore represent an attractive target for vaccine research.24
Yet to be explored for F. gigantica are the CD59-like proteins of the Ly6 family, named for their homology to human CD59 (UniProt: P13987), a complement cascade inhibitor which regulates membrane attack complex (MAC) mediated cytotoxicity of the human complement cascade.25 Homologous proteins have been identified on the teguments of a range of parasitic helminths, including Schistosoma mansoni, Fasciola hepatica and Opisthorchis viverrini.26–28 These proteins exhibit conserved structures: uPAR-like domains, ten cysteines in conserved positions and a three-fingered tertiary structure which approximates to that of human CD59.28,29 Despite the similarity of these sequences to human CD59, it is unknown whether their functions within helminths are comparable, namely proposed dysregulation of host complement via interruption of MAC formation as a form of protective immunomodulation, or entirely unique.26 Considering that preliminary immunization trials using S. mansoni Ly6 family proteins demonstrate promising effects on host antibodies, complete characterisation of these proteins within F. gigantica has the potential to reveal a novel vaccine candidate for future research.29–31
To date, characterisation of the Ly6 proteins in Fasciola spp. has been limited to a single study in which three Ly6 proteins, FhCD59-1 to 3, with FhCD59-1 represented by seven isoforms, were identified.27 To the authors’ knowledge, no attempts have been made to characterise the Ly6 proteins of F. gigantica. Alongside the recent genome, transcriptomic data from a newly-excysted juvenile (NEJ) described herein represents a unique opportunity to investigate Ly6 proteins, given that NEJs are strongly associated with pathogenesis, and thus represent the ideal phase for intervention via vaccination.32 This study aimed to exploit the recent expansion of F. gigantica nucleotide databases to deliver the most comprehensive characterisation of Fasciola Ly6 proteins to date.
Materials and methods
Excystment of Fasciola gigantica metacercariae
F. gigantica metacercariae were obtained from naturally infected wild snails collected in Aligarh, Uttar Pradesh, India, by researchers at Aligarh Muslim University. Excystment was performed as reported by McVeigh et al.33 Briefly, the outer cyst walls were removed by incubation in a solution of 1% w/v pepsin, 4 mM HCl, for 90 min at 37 °C. Post pepsin incubation, cysts were washed several times in distilled water. Excystment was initiated by incubation of the cysts in 0.6% w/v sodium bicarbonate, 0.45% w/v sodium chloride, 0.4% w/v sodium tauroglycocholate, 0.025 M HCl and 0.4% w/v L-Cysteine, for up to 4 h at 37 °C. After approximately 75 min, at 10–15 min intervals, NEJs were transferred to DMEM (Life Technologies), in which they were maintained at 37 °C overnight for which excystment was completed within 18 h. NEJs were centrifuged at 400×g for 1 min and the DMEM removed. NEJs were washed in warmed DMEM, centrifuged at 400×g for 1 min and the DMEM removed. F. gigantica NEJs were then flash frozen in liquid nitrogen and stored at −80 °C for RNA extraction.RNA extraction and normalised cDNA production
In total 30 mg of F. gigantica NEJs were used for total RNA isolation. Total RNA (tRNA) was purified using the Qiagen RNeasy kit as per the manufacturer's instructions. NEJs were homogenised using Eppendorf micropestles and supported by passing the homogenate through a 21 gauge (0.8 μm) needle. tRNA was quantified using NanoDrop (ThermoFisher, UK). 4.5 μg of tRNA was normalised to prevent over representation of abundant transcripts and used for complimentary DNA synthesis following the Evrogen (Russia) CS010-1C protocol using the SMART approach.34 cDNA library production, GsuI digestion and 454 sequencing was performed at the Centre for Genomic Research at the University of Liverpool (UK) using one plate of the 454 GS FLX platform. All 454 sequence data representing the F. gigantica NEJ transcriptome is available through NCBI Transcriptome Shotgun Assembly (TSA) under the accession number GJHP01000000.Transcriptome bioinformatics
Assembly statistics were generated using the ‘assembly-stats’ JavaScript repository.35 Mapping of the NEJ transcriptome against the publically available F. gigantica genome (GenBank accession GCA_006461475.1) was performed using EasyBuild BLAST+ (version 2.11.0) with an E-value cut-off of 1 × 10−10 and a minimum bit score of 70.8 Gene ontology (GO) term and InterPro annotations were then produced for the complete F. gigantica NEJ transcriptome using the Functional Annotation workflow within OmicsBox.36Gene ontology (GO) term enrichment of the Fasciola gigantica newly-excysted juvenile transcriptome against the genome
GO annotations for the draft genome predicted protein data (GenBank GCA_006461475.1) were generated using PANNZER2 for comparison with GO outputs previously generated for the transcriptome.8,37 Gene identifiers were mapped between the two datasets using reciprocal BLASTs with an E-value threshold of 0.1 using in-house scripts. GO term enrichment analysis was then performed using GOAtools (version 0.5.9) script in Python using paired t-tests and FDR corrected P-values.38 Child GO terms were not propagated to parents as an attempt to reduce inheritance over-representation.39 Significant enrichment was measured at two levels: significant, q < 0.05 and highly significant, q < 0.001.Bioinformatic characterisation of Fasciola gigantica and Fasciola hepatica Ly6s
Sequences for Ly6 members identified previously by Shi et al. and Chalmers et al. were retrieved for F. hepatica and S. mansoni members, respectively.27,28 Recently identified SmLy6 members Smp_202630 and Smp_064430 were also included (putatively identified by Chalmers et al.).28 Accession numbers for the S. mansoni proteins were updated from Chalmers et al. to the latest versions using WormBase ParaSite (version WBPS14) and the S. mansoni Meta RNAseq library (V7).27tBLASTn searches were performed using the reference sequences against the F. gigantica newly excysted juvenile (NEJ) transcriptome, using the local database function within BioEdit (version 7.2.5.).40 Sequences with E-values smaller than the designated 1 × 10−5 cut-off were subsequently retrieved and translated into proteins using ExPASy translate.41 BLASTp searches of an adult F. gigantica predicted protein library and draft genome were subsequently performed using the same reference sequences, in addition to putatively identified Ly6 proteins from the NEJ transcriptome, using the same E-value cut-off.8,12
Ly6 proteins of F. hepatica were subsequently characterised in the same manner as those found in F. gigantica, with the exception that the previously characterised F. gigantica orthologues were also incorporated into the query sequence list during the BLAST searches of the transcriptomes. Complete life-stage specific characterisation was performed using a F. hepatica adult predicted protein library, as well as six stage-specific assemblies (egg; metacercaria; newly excysted juveniles at 1, 3 and 24 hours; adult) provided in curated format by Queens University, Belfast.7,42 Characterised Ly6 proteins from F. hepatica were also identified in the Liverpool F. hepatica genome (accession PRJEB25283) in WormBase BLASTp using standard parameters and an E-value cut-off of 1 × 10−5. Interspecies, F. gigantica to F. hepatica, orthologues were defined as >90% identity over 100 continuous residues.
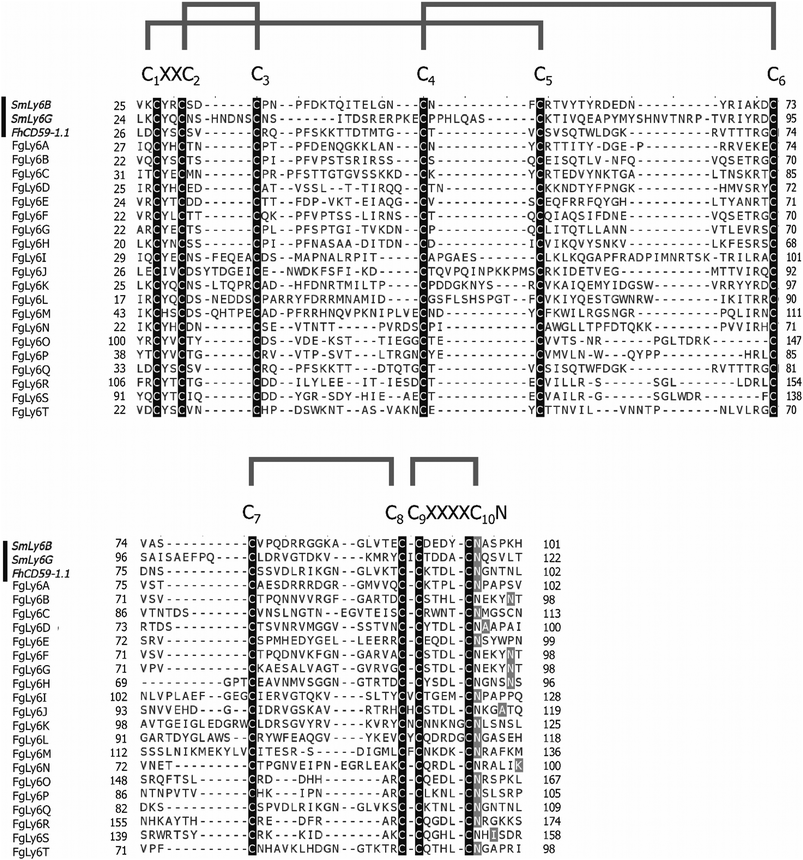
Sequences were screened for uPAR-domain features; namely the presence of at least ten cysteine residues within 120 amino acids, a C1–XX–C2 N-terminal motif (where X represents any amino acid) and a C10–N motif within the C-terminal (Pfam accession PF00021). Additional canonical Ly6 protein features were also identified in accordance with the literature.27,28 This included a signal peptide identified with Signal-P 5.0 and a GPI-anchor identified using the big-PI GPI modification site predictor.44,45
De novo tertiary structural predictions were generated for the domain region of the mature proteins (C-terminal pre-C1XXC2 motif and N-terminal post-GPI anchor removed) as previously described.28 Briefly, structural modelling was done using Rosetta following a de novo protocol generating 1000 full-atom models.46,47 Structural decoys were clustered and top scoring models as per Rosetta scores were visualised and analysed using PyMol (version 2.3.3) (Schrödinger, LLC).
Phylogenetics analysis
Phylogenetic analysis using domain region protein sequences from the characterised Ly6 sequences from F. gigantica, F. hepatica and S. mansoni was performed using MEGAX (version 10.0.5).48 To aid in the isolation of fasciolid-specific clades, proteins with Ly6 domain features from Clonorchis sinensis (PRJDA7281), Opisthorchis felineus (PRJNA413383) and Opisthorchis viverrini (PRJNA222628) were also added via BLASTp FgLy6 homology at 1 × 10−5. Maximum likelihood reconstruction was performed with 2000 bootstraps, JTT model substitution and otherwise default parameters. The resulting distance-scaled tree was separated into clades according to a minimum ancestral root support value of 40. Life stage expression data, extrapolated from sequence presence or absence in each of the transcriptomic and genomic databases searched, was also added.Proteomic annotation
Querying of available F. hepatica proteomic databases was performed to retrieve representative EV and tegument expression data for each of the newly identified proteins.18,22 Expression data for the S. mansoni orthologues was also retrieved for comparison between orthologues.28Results
Fasciola gigantica newly excysted juvenile transcriptomic profile
Assembly statistics for the F. gigantica NEJ transcriptome are summarised in Table 1. The NEJ transcriptome contained a total of 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 551 transcripts, of which 4,031 could not be classified by BLASTx, InterPro, or GO mapping/annotation; 2097 produced BLAST/InterPro hits only; 960 produced were successfully mapped and 9463 reached completion with full GO annotation. A direct count of GO terms identified the top five annotation terms for each class to be as follows: Biological Process (BP) - protein phosphorylation, oxidation–reduction processes, translation, proteolysis and regulation of transcription (DNA-templated); Cellular Component (CC) – integral component of membrane, nucleus, membrane, cytoplasm and ribosome; and Molecular Function (MF) – ATP binding, metal ion binding, protein binding, nucleic acid binding and RNA binding. When mapped against the available F. gigantica genome, 97% of the 16551 assembled transcripts produced strong hits, with 495 additional transcripts revealed in the NEJ (supplementary file 1, ESI†).
551 transcripts, of which 4,031 could not be classified by BLASTx, InterPro, or GO mapping/annotation; 2097 produced BLAST/InterPro hits only; 960 produced were successfully mapped and 9463 reached completion with full GO annotation. A direct count of GO terms identified the top five annotation terms for each class to be as follows: Biological Process (BP) - protein phosphorylation, oxidation–reduction processes, translation, proteolysis and regulation of transcription (DNA-templated); Cellular Component (CC) – integral component of membrane, nucleus, membrane, cytoplasm and ribosome; and Molecular Function (MF) – ATP binding, metal ion binding, protein binding, nucleic acid binding and RNA binding. When mapped against the available F. gigantica genome, 97% of the 16551 assembled transcripts produced strong hits, with 495 additional transcripts revealed in the NEJ (supplementary file 1, ESI†).
| Metric | F. gigantica NEJ transcriptome. |
|---|---|
| Total Sequences (Contigs) | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 551 551 |
| Mean sequence length (bp) | 799 |
| Longest transcript (bp) | 4239 |
| Contig N50 length (bp) | 988 |
| Contig N90 length (bp) | 442 |
| GC content (%) | 45.14 |
| AT content (%) | 54.86 |
| N content (Gapped) (%) | 0.00 |
Gene ontology (GO) term enrichment
To determine any functional representation in the F. gigantica NEJ transcriptome compared to the newly produced draft genome a GO-term enrichment analysis was performed. A total of 376 GO-terms were compared across the datasets (supplementary file 2), with biological processes (BP) most heavily represented (n = 203), followed by cellular components (CC, n = 89) and molecular function (MF, n = 84). 13 terms were found to be significantly enriched in the NEJ (p < 0.05) with 7 terms reaching significance at p < 0.001. Enrichment was observed mostly in CC terms (n = 9), followed equally by BP (n = 2) and MF (n = 2). The most significantly enriched terms (p < 0.001) included translation (GO:0006412), mRNA splicing via spliceosome (GO:0000398), ribosome (GO:0005840), mitochondrion (GO:0005739), mitochondrial matrix (GO:0005759), RNA binding (GO:0003723) and structural component of ribosome (GO:0003735).Conversely, 24 terms were found to be significantly enriched in the genome (p < 0.05), with 14 of these reaching significance at p < 0.001. Genome enriched terms were more evenly distributed between BP, CC and MF, with 9, 5 and 10 terms respectively, and included potassium ion transmembrane transport (GO:0071805), multicellular organismal development (GO:0007275), endopeptidase activity (GO:0004175), cysteine-type peptidase activity (GO:0008234), dynein intermediate light chain binding (GO:0051959) and structural constituent of cytoskeleton (GO:0005200).
Protein domain analysis by transcript frequency
Transcript domain analysis was performed on the F. gigantica NEJ transcriptome to determine the presence of commonly targeted domains in helminth development research. Domains were ranked according to frequency (measured in number of annotated transcripts) and the top 20 domains were selected. A total of 1,563 domains with unique InterPro handles were identified, with a sequence frequency ranging from 1 to 109 across all identified domains. Several domains of relevance to vaccine research were included in the top 20 (Table 2).| InterPro ID (IPR-). | Domain. | Number of NEJ transcripts. | Example vaccines or drug candidates |
|---|---|---|---|
| 002048 | EF-hand | 109 | Tegument-allergen like (TAL) proteins.49 Calpains50 |
| 000504 | RNA recognition motif | 102 | — |
| 000719 | Protein kinase | 98 | ePKs (conventional protein kinases)51 |
| 017986 | WD40-repeat-containing | 73 | — |
| 005225 | Small GTP-binding protein | 63 | — |
| 001841 | Zinc finger, RING-type | 45 | — |
| 013087 | Zinc finger, C2H2-type | 44 | — |
| 020683 | Ankyrin repeat-containing | 36 | — |
| 013026 | Tetratricopeptide repeat-containing | 33 | — |
| 001650 | Helicase, C terminal | 32 | — |
| 008139 | Saposin B type | 31 | Saposins.52 |
| 000477 | Reverse transcriptase | 30 | — |
| 000608 | Ubiquitin-conjugating enzyme E2 | 29 | — |
| 001452 | SH3 | 29 | — |
| 014001 | Helicase superfamily 1/2, ATP binding | 29 | — |
| 017452 | GPCR, rhodopsin-like, 7TM | 28 | G-coupled protein receptors53 |
| 001781 | Zinc finger, LIM-type | 28 | — |
| 001623 | DnaJ | 28 | — |
| 004088 | K homology, type 1 | 27 | — |
| 001478 | PDZ | 26 | Syntenin54 |
Characterisation of Ly6 proteins of Fasciola gigantica
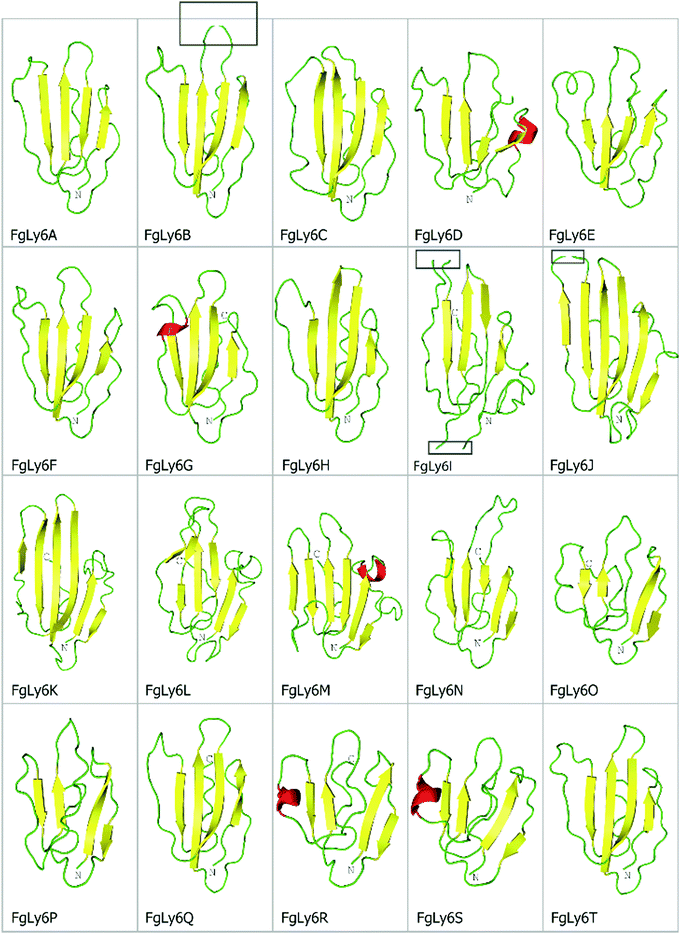
As Ly6 proteins are unlikely to be identified during a domain search, additional homology BLAST (tBLASTn or BLASTp) searches were performed using known Ly6 proteins from F. hepatica and S. mansoni against the NEJ transcriptome, the adult F. gigantica transcriptome and an F. gigantica genome.8 Twenty hit sequences displayed uPAR-domain features in approximately conserved locations when aligned with F. hepatica and S. mansoni Ly6 reference sequences (Fig. 1). These sequences were arbitrarily designated as FgLy6-A through T (Table 3). Of these, five (FgLy6-C, -E, -F, -G and -H) were only present in the NEJ. Three additional FgLy6s, -Q, -R and -S, were found in the adult and genome datasets only, with no NEJ equivalent at 1 × 10−5. Six additional sequences met the 1 × 10−5 threshold but contained no domain features and were removed from further analysis. Following uPAR-domain characterisation, the putative FgLy6 sequences were screened for secondary Ly6 features, including a signal peptide and a GPI anchor. Of the twenty FgLy6s, eighteen had signal peptides that could be confidently predicted by Signal P 5.0 and all twenty sequences had GPI-anchors present in their C-terminals. All predictions also fell in approximately conserved locations within the FgLy6 sequences when aligned with F. hepatica and S. mansoni reference sequences (Fig. 1). Finally, tertiary structural predictions for the twenty putatively identified FgLy6s were produced using de novo modelling. All twenty FgLy6s converged into the conserved three-finger domain fold and demonstrate highly conserved core structures, as shown in Fig. 2.| Ly6 | GenBank accession | Sequence length (amino acids) | Signal peptide | C-terminal transmembrane domain | NEJ F. gigantica transcript ID. | Transcriptomic support in adult F. gigantica. | F. gigantica genomic support. | F. hepatica orthologue identified. Δ |
|---|---|---|---|---|---|---|---|---|
| (+) = Present (−) = absent. Δ > 90% sequence identity with F. gigantica sequence at protein level, minimum 100 continuous amino acids. * Indicates sequences which may have orthologues but were below cut-off. a – Present in Young et al. (2010) adult F. hepatica transcriptome. b – Present in at least one of the Cwiklinski et al. (2015) F. hepatica multi-stage transcriptomes. | ||||||||
| A | TPP67930 | 127 | + | + | 01777 | + | + | +b |
| B | TPP57504 | 122 | + | + | 05428 | + | + | − |
| C | TPP61687 | 132 | + | + | 04026 | − | + | +b |
| D | TPP58533 | 121 | + | + | 11947 | + | + | +ab |
| E | — | 109 | + | − | 11274 | − | − | −* |
| F | TPP58767 | 122 | + | + | 05429 | − | + | +b |
| G | TPP66235 | 122 | + | + | 11065 | − | + | +b |
| H | TPP66236 | 120 | + | + | 14360 | − | + | +b |
| I | TPP67438 | 150 | + | + | 10885 | + | + | +ab |
| J | TPP56171 | 143 | + | + | 10181 | + | + | +ab |
| K | TPP58682 | 152 | + | + | 11200 | + | + | +ab |
| L | TPP59839 | 138 | + | + | 01142 | + | + | +ab |
| M | TPP61345 | 153 | + | + | 09765 | + | + | +ab |
| N | — | 117 | + | + | 02813 | + | + | +ab |
| O | TPP67399 | 189 | + | + | 06228 | + | + | +ab |
| P | TPP57978 | 127 | − | + | 02241 | + | + | − |
| Q | TPP67483 | 129 | + | + | − | + | + | +b |
| R | TPP67398 | 197 | + | + | − | + | + | −* |
| S | TPP57976 | 166 | − | − | − | + | + | +a |
| T | TPP67484 | 121 | + | + | − | − | + | −* |
Expansion of the Ly6 protein family of Fasciola hepatica
Following the complete in silico characterisation of the FgLy6s leading to a considerable expansion on known Fasciola species Ly6 members, a follow up analysis was performed to determine if orthologous F. hepatica proteins could be identified. Orthologous proteins were defined as those with > 90% identity over at least 100 continuous amino acids. Of the twenty FgLy6s, fifteen had F. hepatica orthologues in at least one transcriptomic dataset (Table 3). FgLy6-E, -R and -T also matched to F. hepatica orthologous sequences on direct alignment but were marginally below the cut-off threshold during a BLAST search (85% ID over 109 residues, 90% ID over 81 residues and 86% ID over 192 residues for E, R and T respectively). FhLy6-A and FhLy6-Q represented previously characterised F. hepatica Ly6 proteins (FhLy6-2 and FhLy6-1.1 respectively).27In addition to the sixteen FgLy6-orthologous proteins, three additional proteins exclusive to F. hepatica were also identified (FhLy6 -U, -V and -W) using transcriptomic data. Of these, one had already been identified in previous works (FhCD59-3, given here as FhLy6-V) and was the only F. hepatica specific protein with genomic support.27 The remaining two had not previously been identified.
Phylogenetic analysis
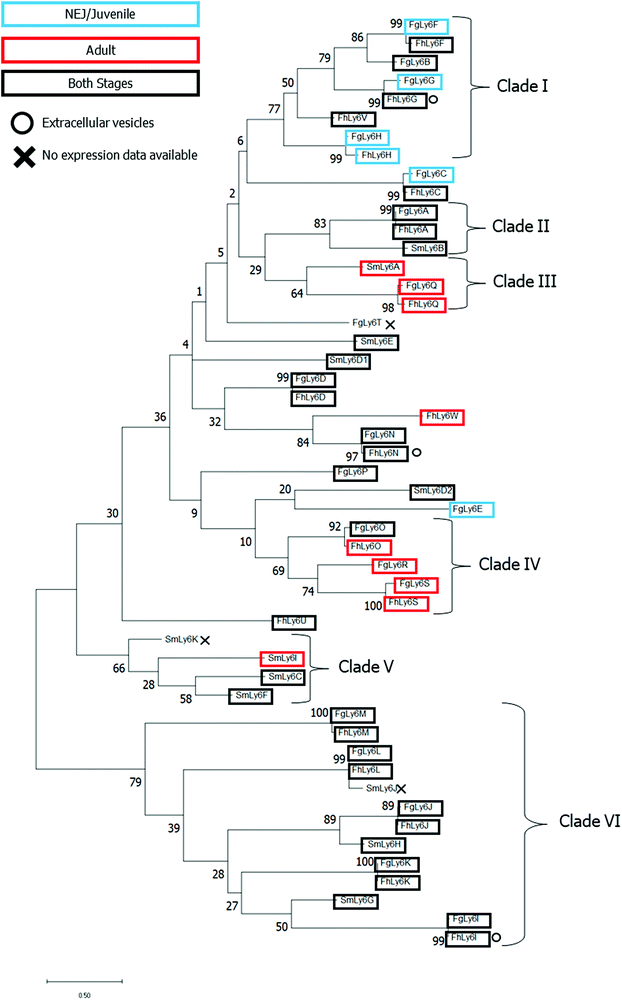
A maximum-likelihood phylogenetics analysis with 2000 bootstraps was performed on domain-region Ly6 proteins from F. gigantica, F. hepatica and S. mansoni. Existing F. hepatica sequences were renamed to correspond with the F. gigantica alphabetic nomenclature. The phylogenetic relationships between all three species’ Ly6 proteins and associated transcriptomic and proteomic expression data are provided in Fig. 3.Several clades, defined by an ancestral root support value ≥40%, were resolved, including two clades which were unique to the two Fasciola spp. (clades I and IV, clade root support value of 77% and 69%, respectively). Clade I was also the only defined clade to contain juvenile-specific FgLy6s (F, G and H). Six FgLy6s could not be grouped into a clade and appeared to be relatively distinct from both S. mansoni proteins and each other (FgLy6-C, -D, -E, -N, -P and -T), with the only closely related proteins being their FhLy6 orthologues, when present. Repeated phylogenetic analysis to include putatively identified Opisthorchis spp. and C. sinensis Ly6 proteins revealed that FgLy6-B, -F, -G, -H and -V remain from S. mansoni when contextualised by orthologues from other trematodes (root support value 62%, supplementary file 4, ESI†).
Querying of published proteomic databases for F. hepatica was also performed to attempt to localise the Ly6 members within the parasite. FhLy6s which matched to accessions in the Davis et al. extracellular vesicle proteome were distributed throughout the phylogenetic tree, with three FhLy6s, -G, - I and -N, found to be expressed in at least one experimental replicate.22 All three were found to be expressed in both life stages, with two of the three belonging to distinct clades (I and VI for G and I, respectively). Querying of the Wilson et al. tegumental transcriptome returned no hits for any of the FhLy6 members.18
Discussion
F. gigantica, despite being a zoonotic trematode of considerable global socio-economic importance, has thus far been largely neglected in respect to molecular characterisation, especially in comparison to its temperate relative, F. hepatica.12 The 18 h NEJ transcriptome described here represents the first to be produced for this ontogenetic stage of F. gigantica and provides an exciting opportunity to explore the stage-specific expression of key target proteins. To date, novel proteins have already been revealed through the analysis of this important life stage transcriptome including a Zeta class glutathione transferase (GST) and a novel second Sigma class GST isoform.55 Interestingly, several of the domains highlighted within the top twenty analysis indicate that further expansion of potential vaccine-candidates may also be possible.For example, the top domain hit, EF-hand domain proteins, include the tegument-allergen-like (TAL) proteins, which have previously been identified as potential vaccine candidates due to their association with host immune responses, specifically IgE-based.49 In S. mansoni, thirteen TAL proteins have been identified and characterised in total.49,56 Though TAL-family proteins have also been studied in both F. gigantica (FgCaBPs) and F. hepatica (FhCaBPs), far fewer are known at present (four in F. gigantica, three in F. hepatica). Also represented among the EF-hand domain proteins are the calpains, a family of cysteine proteases which have been highlighted as potential vaccine candidates in schistosomes.50,57 Two calpains of F. hepatica have previously been identified using ‘omics data, while F. gigantica orthologues remain formally unidentified.18 While EF-hand domains are not exclusive to these two families, putative annotation of the 109 EF-hand transcripts revealed the presence of 13 tegument antigens and 5 calpains, indicating the potential for family expansion and isoform discovery in F. gigantica using this transcriptome.58,59
In addition to the presence of several domains of interest, GO term enrichment analysis indicated several significantly enriched terms in the 18 h NEJ transcriptome compared to genomic data. The results of this study were comparable to previous work by Zhang et al. (2019), wherein GO enrichment analysis was performed between various F. gigantica life stages, including juvenile vs adult.13 Though there were no overlapping GO terms between the two studies, this is likely an artefact of the difference in term propagation depth between analyses, as well as the age of the juvenile studied, 18 h NEJ vs. 70 days post infection juveniles. Several common ‘themes’ were observed in the descriptive terms, including signalling-, microtubule- and cytoskeletal-associated terms. This likely reflects the considerable development required to transition into definitive host from the environmental stages, as well as responses to requirements for immune-interfacing and migration.13,60,61
During this study, the Ly6 family proteins were targeted for further analysis, as it was unlikely that they would be picked up by a domain-level query. This proved to be the case in the NEJ transcriptome, as only a single uPAR-like domain protein was annotated during the domain analysis; a contrast to the twenty subsequently characterised. Further investigation also noted that this sequence did not have all the features required to be classified as a uPAR-domain protein (at least ten cysteine residues, C1–XX–C2 N-terminal motif, C10–N motif; isotig 18073, annotated with InterPro accession IPR016054) and was not a partial sequence, therefore was classified as a mis-annotation. This demonstrates the need for alternative discovery pipelines for Ly6 family proteins aside from domain identification.
The considerable expansion of the Ly6 protein family described herein is extremely positive for the progression of vaccine research in Fasciola spp. trematodes. Characterisation of the S. mansoni Ly6 proteins revealed several candidates able to stimulate a protective immune response, notably SmLy6D.28 Preliminary vaccine trials using SmLy6D demonstrated a reduction in adult worm burden of >50% in a mouse model, with significant reductions in egg production and clinical signs of inflammation also observed.30 Though no direct homologue to SmLy6D was found in this study, the high level of diversity observed in the Fasciola spp. Ly6 proteins is encouraging for the discovery of an equivalent candidate. Additionally, the high level of orthology demonstrated between F. gigantica and F. hepatica Ly6 proteins (15 of 20 FgLy6s with orthologues present in F. hepatica at > 90% sequence identity over 100 amino acids) is also encouraging for the cross-species coverage of any vaccine candidates progressed from this research.
On complete characterisation of the Fasciola spp. Ly6 proteins, phylogenetic analysis was performed on all sequences, in addition to the S. mansoni homologues, to determine if the sequences could be clustered into distinct Ly6 groups. Results demonstrated the presence of several distinct clades, with the previously defined life-stage expression patterns from the FhLy6s reflected by their newly characterised FgLy6 orthologues.7,43 This support of stage-specific expression by distinct phylogenetic clades has previously been observed in other vaccine candidate families, such as the cathepsin L proteases.10,62,63 Ly6 proteins S, Q and H were found to be expressed only in the adult (S and Q) or juvenile stages (H) across both fasciolids, which indicates the potential for stage-targeted intervention strategies. FgLy6-F, -G and -C were also noted to be juvenile specific, however, this juvenile specific cluster conflicted with their FhLy6 orthologues, which were expressed in both stages. This is likely an artifact of improved transcriptomic quality in the newer NEJ transcriptome in comparison to the older adult F. gigantica transcriptome.12 Therefore, these isotypes could also be expressed in both stages in F. gigantica. Considering the extent to which the S. mansoni Ly6s have been studied and the expansive available ‘omic data in this parasite, it is unlikely that there are undiscovered SmLy6s which would conform to the Fasciola spp. distinct clades and thus it is likely that these eight family members, B, F, G, H, O, R, S and V, are distinct within the platyhelminthes.28,64 Notably, no orthologues of the primary S. mansoni vaccine candidate, SmLy6D were identified in either of the Fasciola spp.
Although S. mansoni Ly6s B, D and F have previously been identified as major antigens of the tegument, little is known about the functions of tegumental Ly6 proteins.28 FhLy6 proteins have been found not to contain essential active sites to fulfil roles as complement inhibitors, however the presence of a GTPase may indicate that Ly6 proteins are capable of signalling.18,65 None of the newly identified FhLy6s appeared to be present in the tegumental proteome. However, additional proteomic data with increased proteomic sensitivity is required to confirm this, as Ly6 retrieval appears to be highly dependent on the techniques observed.27,66 Two of the S. mansoni Ly6 proteins which have previously been identified in the tegument have closely related Fasciola spp. homologues (Q with SmLy6A, support value 61, and A with SmLy6B, support value 81, respectively) and may therefore be expected to be expressed in the tegument.28
Whilst additional transcriptomic and proteomic resources are now available for both F. gigantica and F. hepatica, published data is frequently orphaned from raw sequence files and instead rely on indexing using historic BN-numbers, which refer to an outdated, previous version of the PRJEB25283 F. hepatica genome.13,22,23,67 In light of the newly published F. gigantica genomes, and the transcriptome described herein, reflection upon existing resources to incorporate these updates would be highly valuable, as the current form of indexing precludes any further analysis. Thus, there is a real opportunity to reanalyse historic datasets, such as the study by Zhang et al., to support future analyses on key target protein families.13
Conclusions
The considerable expansion of the Ly6 protein family described in this study demonstrates the value of novel transcriptomic resources, especially in the invasive stages. Whilst additional confirmatory research (for example subcloning and RNAi) would ideally need to be performed, leveraging ‘omics resources in parasite research represents a relatively low-cost method for the identification of novel anthelmintic targets, as well as allowing the characterisation of a range of key biological and molecular functions. Preliminary exploration of the NEJ transcriptome described herein indicates there are still many protein families of interest that could be explored and thus this transcriptome represents an attractive resource for future therapy and diagnostic research.Data availability
The 18 h NEJ F. gigantica Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL/GenBank under the accession GJHP00000000. The version described in this paper is the first version, GJHP01000000. In addition, The NEJ F. gigantica transcriptome is available to interrogate viahttps://sequenceserver.ibers.aber.ac.uk/.Author contributions
Conceptualization – P. M. B., A. G. M., I. W. C. and R. M. M.; data curation – S. D. D.; formal analysis – S. D. D., N. F. F., R. M. M., M. S. and D. S.; funding acquisition – P. M. B., A. G. M., S. M. A. A., K. S., M. R., G. R., P. M., R. O. M. and N. F. F.; investigation – S. M. A. A., K. S., M. R. and G. R.; methodology – R. M. M., I. W. C., N. F. F. and M. S.; supervision – R. M. M., P. M. B., A. G. M. and I. W. C.; writing – original draft – S. D. D., R. M. M. and I. W. C.; writing – review & editing – S. D. D., R. M. M., I. W. C., N. F. F., M. S. and P. M. B.Conflicts of interest
The authors declare there are no conflicts of interest.Acknowledgements
This work was supported by the Biotechnology and Biological Science Research Council (BBSRC): BB/H009256/1: Developing a “validation portfolio” to exploit key virulence proteins in Fasciola species for parasite control and BBS/E/W/0012843D: Grasslands Gogerddan. Super Computing Wales (https://www.supercomputing.wales) provided computing resources to carry out de-novo 3D structural modelling. Many thanks to Professor Karl Hoffmann for providing feedback on a preprint version of this article.References
- P. McVeigh, Parasitology, 2020, 147, 835–840 CrossRef.
- W. M. Degrave, S. Melville, A. Ivens and M. Aslett, Int. J. Parasitol., 2001, 31, 532–536 CrossRef CAS PubMed.
- C. Cantacessi, B. E. Campbell, A. R. Jex, N. D. Young, R. S. Hall, S. Ranganathan and R. B. Gasser, Parasite Immunol., 2011, 34, 265–275 CrossRef.
- K. Ashrafi, M. D. Bargues, S. O’Neill and S. Mas-Coma, Travel Medicine and Infectious Disease, 2014, 12, 636–649 CrossRef PubMed.
- K. Cwiklinski and J. P. Dalton, Int. J. Parasitol., 2018, 48, 321–331 CrossRef CAS PubMed.
- M. Moazeni and A. Ahmadi, Exp. Parasitol., 2016, 169, 81–89 CrossRef PubMed.
- N. D. Young, R. S. Hall, A. R. Jew, C. Cantacessi and R. B. Gasser, Biotechnol. Adv., 2010, 28, 222–231 CrossRef CAS PubMed.
- Y. Choi, S. Fontenla, P. U. Fischer, T. H. Le, A. Costábile, D. Blair, P. J. Brindley, J. F. Tort, M. M. Cabada and M. Mitreva, Mol. Biol. Evol., 2020, 37, 84–99 CrossRef CAS.
- T. Pandey, A. Ghosh, V. N. Todur, V. Rajendran, P. Kalita, J. Kalita, R. Shukla, P. B. Chetri, H. Shukla, A. Sonkar, D. L. Lyngdoh, R. Singh, H. Khan, J. Nongkhlaw, K. C. Das and T. Tripathi, ACS Omega, 2020, 5, 11084–11091 CrossRef CAS PubMed.
- M. W. Robinson, R. Menon, S. M. Donnelly, J. P. Dalton and S. Ranganathan, Mol. Cell. Proteomics, 2009, 8, 1891–1907 CrossRef CAS PubMed.
- M. Xu, L. Ai, J. Fu, A. J. Nisbet, Q. Liu, M. Chen, D. Zhou and X. Zhu, PLoS One, 2012, 7, e53387 CrossRef CAS PubMed.
- N. D. Young, A. R. Jew, C. Cantacessi, R. S. Hall, B. E. Campbell, T. W. Spithill, S. Tangkawattana, P. Tangkawattana, T. Laha and R. B. Gasser, PLoS Neglected Trop. Dis., 2011, 5, e1004 CrossRef CAS PubMed.
- X. Zhang, K. Cwiklinski, R. Hu, W. Zheng, Z. Sheng, F. Zhang, H. M. Elsheikha, J. P. Dalton and X. Zhu, BMC Genomics, 2019, 20, 729 CrossRef PubMed.
- X. Zhang, W. Cong, H. M. Elsheikha, G. Liu, J. Ma, W. Huang, Q. Zhao and X. Zhu, Infect., Genet. Evol., 2017, 51, 33–40 CrossRef CAS PubMed.
- A. Anandanarayanan, O. K. Raina, H. Lalrinkima, A. Rialch, M. Sankar and A. Varghese, PLoS Neglected Trop. Dis., 2017, 11, e0006109 CrossRef.
- S. A. Beckham, D. Piedrafita, P. I. Phillips, N. Samarawickrema, R. H. P. Law, P. M. Smooker, N. S. Quinsey, J. A. Irving, D. Greenwood, S. H. L. Verhelst, M. Bogyo, B. Turk, T. H. Coetzer, L. C. Wijeyewickrema, T. W. Spithill and R. N. Pike, Int. J. Biochem. Cell Biol., 2009, 41, 1601–1612 CrossRef CAS PubMed.
- C. M. Hamilton, D. J. Dowling, C. E. Loscher, R. M. Morphew, P. M. Brophy and S. M. O’Neill, Infect. Immun., 2009, 77, 2488–2498 CrossRef CAS PubMed.
- R. A. Wilson, J. M. Wright, W. de Castro-Borges, S. J. Parker-Manuel, A. A. Dowle, P. D. Ashton, N. D. Young, R. B. Gasser and T. W. Spithill, Int. J. Parasitol., 2011, 41, 1347–1359 CrossRef CAS PubMed.
- L. Guasconi, L. S. Chiapello and D. T. Masih, Immunobiology, 2015, 220, 934–939 CrossRef CAS.
- A. M. van der Ree and F. Mutapi, Exp. Parasitol., 2015, 157, 48–58 CrossRef PubMed.
- K. Cwiklinski, E. de la Torre-Escudero, M. Trelis, D. Bernal, P. J. Dufresne, G. P. Brennan, S. O’Neill, J. Tort, S. Paterson, A. Marcilla, J. P. Dalton and M. W. Robinson, Mol. Cell. Proteomics, 2015, 14, 3258–3273 CrossRef CAS PubMed.
- C. Davis, H. Phillips, J. J. Tomes, M. T. Swain, T. J. Wilkinson, P. M. Brophy and R. M. Morphew, PLoS Neglected Trop. Dis., 2019, 13, e0007191 CrossRef CAS PubMed.
- E. de la Torre-Escudero, J. Q. Gerlach, A. P. S. Bennett, K. Cwiklinski, H. L. Jewhurst, K. M. HUson, L. Joshi, M. Kilcoyne, S. O’Neill, J. P. Dalton and M. W. Robinson, PLoS Neglected Trop. Dis., 2019, 13, e0007087 CrossRef PubMed.
- N. S. Degheidy and H. A. Shalaby, Res. J. Parasitol., 2010, 5, 79–89 CrossRef CAS.
- A. Davies, D. L. Simmons, G. Hale, R. A. Harrison, H. Tighe, P. J. Lachmann and H. Waldmann, J. Exp. Med., 1989, 170, 637–654 CrossRef CAS PubMed.
- J. Mulvenna, B. Sripa, P. J. Brindley, J. Gorman, M. K. Jones, M. L. Colgrave, A. Jones, S. Nawaratna, T. Laha, S. Suttiprapa, M. J. Smout and A. Loukas, Proteomics and Systems Biology, 2010, 10, 1063–1078 CAS.
- Y. Shi, H. Toet, V. Rathinasamy, N. D. Young, R. B. Gasser, T. Beddoe, W. Huang and T. W. Spithill, Exp. Parasitol., 2014, 144, 57–64 CrossRef CAS PubMed.
- I. W. Chalmers, C. M. Fitzsimmons, M. Brown, C. Pierrot, F. M. Jones, J. M. Wawrzyniak, N. Fernandez-Fuentes, E. M. Tukahebwa, D. W. Dunne, J. Khalife and K. F. Hoffmann, PLoS Neglected Trop. Dis., 2015, 9, e0003920 CrossRef PubMed.
- L. P. Farias, G. Krautz-Peterson, C. A. Tararam, B. O. Araujo-Montoya, T. R. Fraga, H. K. Rofatto, F. P. Silva Jr., L. Isaac, A. A. Da’dara, R. A. Wilson, C. B. Shoemaker and L. C. C. Leite, PLoS Neglected Trop. Dis., 2013, 7, e2482 CrossRef.
- F. C. Cardoso, G. C. Macedo, E. Gava, G. T. Kitten, V. L. Mati, A. L. de Melo, M. V. Caliari, G. T. Almeida, T. M. Venancio, G. Verjovski-Almeida and S. C. Oliveira, PLoS Neglected Trop. Dis., 2008, 2, e308 CrossRef.
- L. P. Farias, C. A. Tararam, P. A. Miyasato, M. Y. Nishiyama Jr., K. C. Oliveira, T. Kawano, S. Verjoyski-Almeida and L. C. de Cerqueira Leite, Parasitol. Res., 2011, 108, 125–135 CrossRef.
- D. Piedrafita, J. C. Parsons, R. M. Sandemann, P. R. Wood, S. E. Estuningsih, S. Partoutomo and T. W. Spithill, Parasite Immunol., 2011, 23, 473–482 CrossRef.
- P. McVeigh, E. M. McCammick, P. McCusker, R. M. Morphew, A. Mousley, A. Abidi, K. M. Saifullah, R. Muthusamy, R. Gopalakrishnan, T. W. Spithill, J. P. Dalton, P. M. Brophy, N. J. Marks and A. G. Maule, PLoS Neglected Trop. Dis., 2014, 8, e3185 CrossRef.
- Y. Y. Zhu, E. M. Machleder, A. Chenchik, R. Li and P. D. Siebert, Biotechniques, 2001, 30, 892–897 CrossRef CAS PubMed.
- R. Challis, ‘assembly-stats’ Source Code Version 17.02, 2017, used under an MIT license, accessed via GitHub, DOI: 10.5281/zendo.322347.
- BioBam Bioinformatics, ‘OmicsBox – bioinformatics made easy.’ Version 1.2.4., 2019, accessed viahttps://www.biobam.com/omicsbox.
- P. Törönen, A. Medlar and L. Holm, Nucleic Acids Res., 2018, 46, W84–W88 CrossRef PubMed.
- D. V. Klopfenstein, L. Zhang, B. S. Pedersen, F. Ramírez, A. W. Vesztrocy, A. Naldi, C. J. Mungall, J. M. Yunes, O. Botvinnik, M. Weigel, W. Dampier, C. Dessimoz, P. Flick and H. Tang, Sci. Rep., 2018, 8, 10872 CrossRef CAS PubMed.
- S. Grossmann, S. Bauer, P. N. Robinson and M. Vingron, Bioinformatics, 2007, 23, 3024–3031 CrossRef CAS PubMed.
- T. A. Hall, Nucleic Acids Symp. Ser., 1999, 41, 95–98 CAS.
- P. Artimo, M. Jonnalagedda, K. Arnold, D. Baratin, G. Csardi, E. de Castro, S. Devaud, V. Flegel, A. Fortier, E. Gasteiger, A. Grosdidier, C. Hernandez, V. Ioannidis, D. Kuznetsov, R. Liechti, S. Moretti, K. Mostaguir, N. Redaschi, G. Rossier, I. Xenarios and H. Stockinger, Nucleic Acids Res., 2012, 40, W597–W603 CrossRef CAS PubMed.
- K. Cwiklinski, J. P. Dalton, P. J. Dufresne, J. La Course, D. J. L. Williams, J. Hodgkinson and S. Paterson, Genome Biol., 2015, 16, 71 CrossRef PubMed.
- K. Cwiklinski, H. Jewhurst, P. McVeigh, T. Barbour, A. G. Maule, J. Tort, S. M. O’Neill, M. W. Robinson, S. Donnelly and J. P. Dalton, Mol. Cell. Proteomics, 2018, 17, 792–809 CrossRef CAS PubMed.
- J. J. A. Armenteros, K. D. Tsirigos, C. K. Sønderby, T. N. Petersen, O. Winther, S. Brunak, G. von Heijne and H. Nielsen, Nat. Biotechnol., 2019, 37(420), 423 Search PubMed.
- B. Eisenhaber, P. Bork and F. Eisenhaber, J. Mol. Biol., 1999, 292, 741–758 CrossRef CAS PubMed.
- A. Leaver-Fay, M. Tyka, S. M. Lewis, O. F. Lange, J. Thompson, R. Jacak, K. W. Kaufman, P. D. Renfrew, C. A. Smith, W. Sheffler, I. W. Davis, S. Cooper, A. Treuille, D. J. Mandell, F. Richter, Y. A. Ban, S. J. Fleishman, J. E. Corn, D. E. Kim, S. Lyskov, M. Berrondo, S. Mentzer, Z. Popović, J. J. Havranek, J. Karanicolas, R. Das, J. Meiler, T. Kortemme, J. J. Gray, B. Kuhlman, D. Baker and P. Bradley, Methods Enzymol., 2011, 487, 545–574 CAS.
- S. Raman, R. Vernon, J. Thompson, M. Tyka, R. Sadreyey, J. Pei, D. Kim, E. Kellogg, F. DiMaio, O. Lange, L. Kinch, W. Sheffler, B. Kim, R. Das, N. V. Grishin and D. Baker, Proteins: Struct., Funct., Bioinf., 2009, 77, 89–99 CrossRef CAS PubMed.
- S. Kumar, G. Stecher, M. Li, C. Knyaz and K. Tamura, Molecular Biology and Evolution, 2018, 35, 1547–1549 CrossRef CAS.
- C. M. Fitzsimmons, F. M. Jones, A. Stearn, I. W. Chalmers, K. F. Hoffmann and J. Wawrzyniak, PLoS Neglected Trop. Dis., 2012, 6, e1593 CrossRef CAS.
- N. Ohta, T. Kumagai, H. Maruyama, A. Yoshida, Y. He and R. Zhang, Parasitol. Int., 2004, 53, 175–181 CrossRef CAS PubMed.
- L. F. Andrade, L. A. Nahum, L. G. A. Avelar, L. L. Silva, A. Zerlotini, J. C. Ruiz and G. Oliveira, BMC Genomics, 2011, 12, 215 CrossRef CAS PubMed.
- T. A. Don, J. M. Bethony and A. Loukas, Int. J. Infect. Dis., 2008, 12, e39–e47 CrossRef PubMed.
- S. Hahnel, N. Wheeler, Z. Lu, A. Wangwiwatsin, P. McVeigh, A. Maule, M. Berriman, T. Day, P. Ribeiro and C. G. Grevelding, PLoS Pathogens, 2018, 14, e1006718 CrossRef.
- B. C. Figueiredo, N. R. G. Assis, S. B. Morais, N. D. Ricci, C. S. Pinheiro, V. P. Martins, R. M. Bicalho, A. A. Da-dara, P. J. Skelly and S. C. Oliveira, PLoS Neglected Trop. Dis., 2014, 8, e3107 CrossRef PubMed.
- R. B. Stuart, S. Zwaanswijk, N. D. MacKintosh, B. Witikornkul, P. M. Brophy and R. M. Morphew, Parasitol. Res., 2021, 120, 979–991 CrossRef PubMed.
- J. Carson, C. M. Thomas, A. McGinty, G. Takata and D. J. Timson, Mol. Biochem. Parasitol., 2018, 221, 14–22 CrossRef CAS PubMed.
- S. Chaimon, Y. Limpanont, O. Reamtong, S. Ampawong, O. Phyphisut, P. Chusongsang, J. Ruangsittichai, U. Boonyuen, D. Watthanakulpanich, A. J. O’Donoghue, C. R. Caffrey and P. Adisakwattana, Parasites Vectors, 2019, 12, 383 CrossRef PubMed.
- P. Subpipattana, R. Grams and S. Vichasri-Grams, Exp. Parasitol., 2012, 130, 364–373 CrossRef CAS PubMed.
- S. Banford, O. Drysdale, E. M. Hoey, A. Trudgett and D. J. Timson, Biochimie, 2013, 95, 751–758 CrossRef CAS PubMed.
- T. Tansatit, S. Sahaphong, S. Riengrojpitak, V. Vivanant and P. Sobhon, Vet. Parasitol., 2006, 135, 269–278 CrossRef CAS PubMed.
- R. E. B. Hanna, D. Moffett, M. W. Robinson, W. G. Z. O. Jura, G. P. Brennan, I. Fairweather and L. T. Threadgold, Parasitology, 2019, 274, 108923 CAS.
- J. P. Dalton, S. O’Neill, C. Stack, P. Collins, A. Walshe, M. Sekiya, S. Doyle, G. Mulcahy, D. Hoyle, E. Khaznadji, N. Moiré, G. Brennan, A. Mousley, N. Kreshchenko, A. G. Maule and S. M. Donnelly, Int. J. Parasitol., 2003, 33, 1173–1181 CrossRef CAS.
- R. M. Morphew, H. A. Wright, E. J. LaCourse, J. Porter, J. Barrett, D. J. Woods and P. M. Brophy, PLoS Neglected Trop. Dis., 2011, 5, e937 CrossRef CAS PubMed.
- Z. Han, P. J. Brindley, S. Wang and Z. Chen, Genomics Hum. Genet., 2009, 10, 211–240 CrossRef CAS PubMed.
- H. Toet, D. M. Piedrafita and T. W. Spithill, Int. J. Parasitol., 2014, 44, 915–927 CrossRef CAS PubMed.
- W. Castro-Borges, A. Dowle, R. S. Curwen, J. Thomas-Oates and R. A. Wilson, PLoS Neglected Trop. Dis., 2011, 5, e993 CrossRef CAS PubMed.
- S. Huang, D. Yue, J. Hou, X. Zhang, F. Zhang, C. Wang and X. Zhu, Parasitol. Res., 2018, 118, 453–460 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1mo00254f |
| This journal is © The Royal Society of Chemistry 2022 |