Open Access Article
Open Access ArticleExportin-inspired artificial cell nuclear-exporting nanosystems†
Mitsuo
Inui
,
Yuta
Hamada
,
Nana
Sejima
,
Natsumi
Ueda
,
Tokitaka
Katayama
,
Kimika
Ono
and
Koji
Nagahama
 *
*
Department of Nanobiochemistry, Faculty of Frontiers of Innovative Research in Science and Technology (FIRST), Konan University, 7-1-20 Minatojima-minamimachi, Chuo-ku, Kobe 650-0047, Japan. E-mail: nagahama@konan-u.ac.jp
First published on 12th May 2022
Abstract
Inspired by the structural and chemical features of naturally occurring importin/exportin that allows them to pass through the nuclear pore complexes, we successfully developed an artificial nuclear-exporting nanosystem capable of eliminating compounds accumulated abnormally in the nucleus.
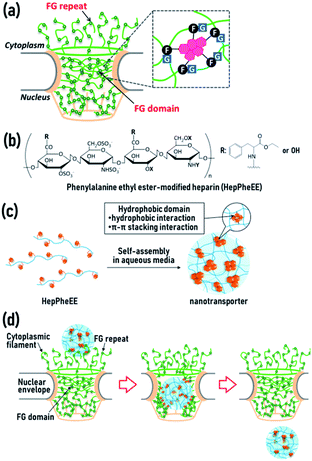
The nucleus is a crucial organelle in regulating the fate of cells, including reproduction, growth, metabolism, and death through the regulation of gene expression. Molecular transport between the cytoplasm and the nucleus is strictly controlled by a single type of gateway, the nuclear pore complex (NPC) (Fig. 1a and S1, ESI†).1,2 The central channel of NPCs contains disordered polypeptide chains with phenylalanine-glycine (FG) repeats,3,4 or a variation (glycine-leucine-phenylalanine-glycine, GLFG) (Fig. 1a).5,6 The polypeptide chains form three-dimensional hydrogel networks via intermolecular π–π stacking and hydrophobic interactions between phenyl groups of the phenylalanine residues, allowing the assembled FG domains to act as physical cross-linking points in the gel networks.7,8 The hydrogel networks play vital roles in the permeability barrier of the NPCs and permit passive diffusion of water, ions, and small molecules, whereas compounds larger than 4 nm rarely pass through NPCs.9–11
Artificial nuclear transport technologies capable of transporting bioactive molecules between the cytoplasm and the nucleus are promising for the development of nuclear-targeting medicines, diagnostics, live imaging, and drug delivery systems. Many researchers are currently focused on developing artificial nuclear-importing systems, and various types of nanomaterial-based nuclear-importing systems have been reported.12–16
In contrast, there are no reports focused on artificial nuclear-exporting technologies. Recent studies revealed that the abnormal accumulation of biomolecules, including proteins, RNA, and lipids in the nucleus, cause various types of diseases.17–23 Moreover, anticancer drugs such as doxorubicin (DOX) accumulated in the nucleus of non-target healthy cells cause severe side effects.24–27 These reports clearly demonstrate the need for studies aimed at developing artificial nuclear-exporting technologies as these would provide a promising approach to treatments for diseases and side effects caused by abnormal nuclear accumulation.
We recently developed a novel artificial nuclear nanotransporter inspired by importin, a naturally occurring nuclear transport machinery capable of importing cargo from the cytoplasm into the nucleus by passing through the NPCs.28 Importin contains characteristic multiple hydrophobic domains that transiently interact with the FG domains in the NPCs via reversible weak hydrophobic and π–π stacking interactions, leading to dissociation of the FG domains. This dissociation results in local disruption of the gel networks of the NPCs, allowing importin to easily penetrate the networks. The combined continuous local dissociation events finally enable importin to pass through the NPCs (Fig. S2, ESI†). We mimicked the essential structural and chemical features of importin for passage through the NPCs by generating a nuclear nanotransporter by self-assembly of amphiphilic polymers. These polymers contain several hydrophobic phenylalanine ethyl ester side-chains conjugated to a biocompatible hydrophilic heparin main-chain (HepPheEE, Fig. 1b). The self-assembled HepPheEE nanotransporters achieved remarkable rapid and highly efficient nuclear transport from the cytoplasm into the nucleus in cultured cells, tissue, organs, and living mice.28
Exportin is a naturally occurring nuclear transport machinery which transports cargo from the nucleus into the cytoplasm.29,30 Exportin also contains characteristic multiple hydrophobic domains capable of transiently interacting with FG domains in the NPCs. Although the direction of nuclear transport is opposite in importin and exportin, importin and exportin share the same mechanism for passage through the NPCs. Given this, we hypothesized that the importin-mimetic HepPheEE nanotransporter is adaptable for the development of artificial nuclear-exporting technologies. The aim of this study was to demonstrate proof-of-concept of this idea. To this end, we investigated the intracellular dynamics of the HepPheEE nanotransporters and the nuclear-export of DOX accumulated in the nucleus of healthy cells as a model drug cytotoxic to healthy cells. Moreover, we evaluated whether the nuclear-export of DOX by HepPheEE nanotransporters rescues healthy cells from the toxicity caused by DOX.
In our previous study, we used HepPheEE26, which contains an average of 26 PheEE side-chains conjugated with a heparin molecule. We previously found that relatively high numbers of PheEE residues interfere with nuclear importing capability,28 and thus here we designed HepPheEE with fewer PheEE side-chains. The synthesized HepPheEE (Scheme S1, ESI†) was characterized by 1H-NMR and FTIR analyses. The 1H-NMR (Fig. S3, ESI†) signal intensity of the anomeric protons of heparin (5.0 ppm, peaks a and b) and the aromatic protons of PheEE residues (7.2 ppm, peak k) were used to estimate that the average number of PheEE residues introduced into a heparin molecule was 10. FTIR analysis of HepPheEE10 (Fig. S4, ESI†) showed peaks at 702 and 814 cm−1 attributed to the –C–H bending vibration of the PheEE residues, while no peaks were detected at the same region for heparin. Furthermore, peaks at 1560 and 1733 cm−1 corresponding to –N–H bending vibration and –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration of the PheEE residues were detected in the spectrum of HepPheEE10, indicating the conjugation of PheEE to heparin through amide bonds. HepPheEE10 exhibited a critical aggregation concentration (CAC, 0.22 mg mL−1) in dilute aqueous solution by fluorescence assay using hydrophobic pyrene, whereas heparin did not (Fig. S5a, ESI†), indicating hydrophobic interaction-driven self-assembly of HepPheEE10. The average hydrodynamic diameter (Dh) of HepPheEE10 assemblies was 5.7 nm (Fig. S5b, ESI†), which is larger than heparin (4.3 nm, Fig. S6, ESI†). The zeta potential of HepPheEE10 nanoparticles was −56 mV. Characterization of the HepPheEE10 polymer and nanoparticle is summarized in Table 1. Combining these results, it appears that HepPheEE10 polymers self-assemble into nanoparticles with a heparin skeleton and multiple hydrophobic domains as physical cross-linking points to maintain the assembled structures (Fig. 1c).28
O stretching vibration of the PheEE residues were detected in the spectrum of HepPheEE10, indicating the conjugation of PheEE to heparin through amide bonds. HepPheEE10 exhibited a critical aggregation concentration (CAC, 0.22 mg mL−1) in dilute aqueous solution by fluorescence assay using hydrophobic pyrene, whereas heparin did not (Fig. S5a, ESI†), indicating hydrophobic interaction-driven self-assembly of HepPheEE10. The average hydrodynamic diameter (Dh) of HepPheEE10 assemblies was 5.7 nm (Fig. S5b, ESI†), which is larger than heparin (4.3 nm, Fig. S6, ESI†). The zeta potential of HepPheEE10 nanoparticles was −56 mV. Characterization of the HepPheEE10 polymer and nanoparticle is summarized in Table 1. Combining these results, it appears that HepPheEE10 polymers self-assemble into nanoparticles with a heparin skeleton and multiple hydrophobic domains as physical cross-linking points to maintain the assembled structures (Fig. 1c).28
| polymer | M w | # PheEE residues per heparin | CAC (mg mL−1) | D h (nm) | Zeta potential (mV) |
|---|---|---|---|---|---|
| a Molecular weight and numbers of amino acid derivatives per heparin were estimated by 1H-NMR analysis (DMSO-d6/D2O = 9/1). | |||||
| Heparin | 19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
— | — | 4.3 | −58 |
| HepPheEE10 | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 900 |
10 | 0.22 | 5.7 | −56 |
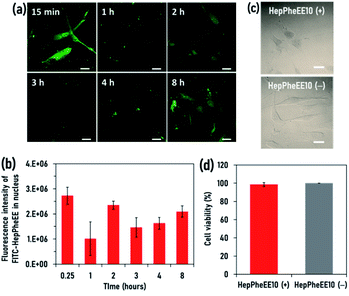
Human skeletal muscle satellite cells (SkmSCs) were used as a model healthy cell type to which DOX is cytotoxic as a side effect. FITC-HepPheEE10 nanoparticles introduced into the cytoplasm of SkmSCs showed rapid nuclear transport and most of the nanoparticles localized in the nucleus (Fig. S7, ESI†), indicating that HepPheEE10 nanoparticles pass through the NPCs from the cytoplasm into the nucleus via an importin-mimetic mechanism (Fig. 1d). Next, we evaluated the exportin-mimetic nuclear-exporting capability of HepPheEE10 nanoparticles (Fig. 2a). FITC-HepPheEE10 nanoparticles were allowed to accumulate in the nucleus for 15 min, then their intracellular dynamics was investigated. The fluorescence intensity of FITC-HepPheEE10 nanoparticles in the nucleus decreased up to 1 h (Fig. 2b), indicating nuclear export of the nanoparticles. Notably, the fluorescence intensity in the nucleus increased after 2 h, followed by an intensity decrease again after 3 h, suggesting that the nanoparticles travelled back and forth between the nucleus and the cytoplasm repeatedly. These results indicate that HepPheEE10 nanoparticles exhibit both importin-mimetic nuclear-importing and exportin-mimetic nuclear-exporting capabilities. Our molecular design (HepPheEE) appears to mimic the essential structural and chemical features of both importin and exportin.
To investigate the effects of the nuclear import/export of HepPheEE10 nanoparticles on cellular behaviour, the adhesion, shape, and proliferation of SkmSCs treated with HepPheEE10 nanoparticles were evaluated. SkmSCs treated with the nanoparticles adhered to the cell culture dish and spread normally (Fig. 2c). Importantly, the viability of SkmSCs treated with electroporation of the nanoparticles was almost 100%, similar to the viability of SkmSCs treated with electroporation of PBS (without HepPheEE nanoparticles) (Fig. 2d). These results indicate that the nuclear import/export capability of HepPheEE10 nanoparticles did not have a negative effect on fundamental cellular behaviour.
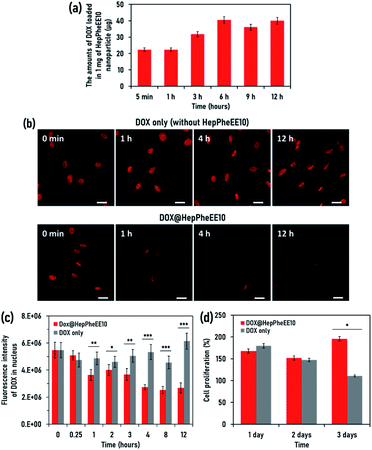
Next, we evaluated the potential cell rescue capability of HepPheEE10 nanoparticles from cytotoxicity caused by DOX. First, we evaluated the DOX-loading capability of HepPheEE10 nanoparticles. DOX contains hydrophobic regions composed of aromatic and aliphatic groups and HepPheEE10 nanoparticles possess multiple hydrophobic domains consisting of aromatic and aliphatic groups. Therefore, we anticipated that free DOX dissolved in solution can be loaded into the hydrophobic domains of HepPheEE10 nanoparticles via hydrophobic and π–π stacking interactions. HepPheEE10 nanoparticles were added to a solution of DOX and left for different times to allow DOX-loading, then the DOX-loaded nanoparticles were collected by centrifugation. Fig. 3a shows the amounts of DOX loaded in 1 mg of HepPheEE10 nanoparticles. As expected, the nanoparticles rapidly loaded DOX and the amounts increased over time to a maximum at 6 h. We performed DLS analysis of DOX-loading HepPheEE10 nanoparticles (Fig. S8, ESI†) and found that the DLS profile of HepPheEE10 nanoparticles changed after loading of DOX. In particular, the DOX-loading nanoparticles showed slightly large Dh value than that of HepPheEE10 nanoparticles, indicating the rearrangement of structures of HepPheEE10 nanoparticles in the presence of hydrophobic DOX. Considering this, we thought that it took long time period (6 h) to get equilibrium for DOX-loading process due to this rearrangement.
Next, we attempted to prepare a SkmSC model in which DOX is accumulated in the nucleus and causes sustained weak cytotoxicity for several days. We investigated the maximum amount of DOX accumulated in the nucleus by varying the amount of DOX added to the culture medium of SkmSCs and the length of exposure (Fig. S9, ESI†). This allowed optimization of the experimental conditions and generation of a SkmSC model, then the rescue capability of HepPheEE10 nanoparticles was investigated using the model. DOX was allowed to accumulate in the nucleus of SkmSCs, then the intracellular dynamics of DOX was investigated in the presence/absence of HepPheEE10 nanoparticles. The red fluorescence intensity of DOX in the nucleus did not decrease for 12 h in the absence of HepPheEE nanoparticles (DOX only) (Fig. 3b and S10, ESI†) whereas an obvious decrease in fluorescence intensity in the nucleus was observed for cells treated with the nanoparticles (Fig. 3b and S11, ESI†). Notably, the fluorescence intensity of DOX in the nucleus of cells treated with nanoparticles gradually decreased over time and the fluorescence intensity significantly decreased after 1 h compared to SkmSCs without nanoparticle treatment (Fig. 3c).
We investigated cellular dynamics of HepPheEE10 nanoparticles in the presence of DOX (Fig. S12, ESI†). The cellular dynamics of HepPheEE10 nanoparticles was slightly affected by the presence of DOX. In particular, nuclear-exporting capability of DOX-loading HepPheEE10 nanoparticles slightly decreased at the initial stage (15 min–2 h), and the nanoparticles tended to retain in the nucleus as compared to HepPheEE10 nanoparticles (Fig. 2b). Since hydrophobic PheEE moiety of HepPheEE10 nanoparticles share the binding sites with FG domains in the NPCs and DOX, the nuclear-exporting capability could be reduced. However, HepPheEE10 nanoparticles exhibited inherent nuclear-transporting capability and travelled back and forth between the nucleus and the cytoplasm. Notably, nuclear-exporting behaviour of DOX was consistent with nuclear-exporting behaviour of HepPheEE10 nanoparticles, indicating that the nanoparticles spontaneously loaded DOX accumulated in the nucleus and then exported the loaded DOX from the nucleus to the cytoplasm.
Importantly, the fluorescence intensity of HepPheEE10 nanoparticles in the whole cells also decreased up to 2 h (Fig. S12c, ESI†), indicating that HepPheEE10 nanoparticles disappeared from the cells. After that, the fluorescence intensity in the whole cells increased after 3 h, suggesting that the nanoparticles travelled back and forth between the extracellular space and the intracellular space. On the other hand, the red fluorescence intensity of DOX in both the nucleus and the whole cells continued to decrease over time in the presence of HepPheEE10 nanoparticles. Considering these results, it is likely that DOX could be mainly released from the nanoparticles at the extracellular spaces.
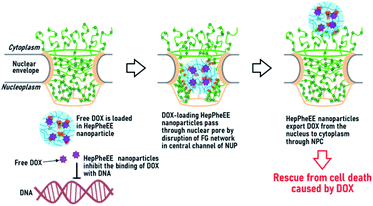
As a proof-of-concept, we evaluated the potential capability of HepPheEE10 nanoparticles to rescue SkmSCs from cytotoxicity caused by DOX. DOX-accumulated SkmSCs not treated with nanoparticles (DOX only) exhibited toxicity and the number of cells gradually decreased over 3 days (Fig. 3d). In contrast, DOX-accumulated SkmSCs treated with nanoparticles escaped cytotoxicity and proliferated, suggesting that HepPheEE10 nanoparticles inhibited binding between DOX and DNA in the nucleus by storing DOX within the hydrophobic domains of the nanoparticles and then exporting DOX from the nucleus to the cytoplasm (Fig. 4). Taking all these results together, we have demonstrated that HepPheEE10 nanoparticles successfully rescued SkmSCs from cytotoxicity caused by DOX. Thus, importin/exportin-mimetic HepPheEE10 nanoparticles could be the first example of an artificial nuclear-exporting system capable of eliminating compounds that accumulate abnormally in the nucleus.
In summary, inspired by the structural and chemical features of naturally occurring importin/exportin that allows them to pass through the NPCs, here we successfully developed an artificial nuclear-exporting nanosystem capable of eliminating compounds accumulated abnormally in the nucleus by adopting a simple molecular design and self-assembly of amphiphilic heparin-phenylalanine derivative conjugates. Given these promising results, we envision that artificial nuclear-exporting systems may not only provide new basic biological- and medical-technologies to study and treat diseases and side effects caused by abnormal nuclear accumulation, but also help better understand the nuclear export systems in living organisms.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported from JSPS KAKENHI Grant Number 17H04744.Notes and references
- L.-C. Tu, G. Fu, A. Zilman and S. M. Musser, EMBO J., 2013, 32, 3220–3230 CrossRef CAS PubMed.
- M. Rangl, A. Ebner, J. Yamada, C. Rankl, R. Tampé, H. J. Gruber, M. Rexach and P. Hinterdorfer, Angew. Chem., Int. Ed., 2013, 52, 10356–10359 CrossRef CAS PubMed.
- S. S. Patel, B. J. Belmont, J. M. Sante and M. F. Rexach, Cell, 2007, 129, 83 CrossRef CAS PubMed.
- P. Stelter, R. Kunza, J. Fischer and E. Hurt, J. Cell Biol., 2011, 195, 183–192 CrossRef CAS PubMed.
- S. Frey and D. Görlich, EMBO J., 2009, 28, 2554 CrossRef CAS PubMed.
- J. Fiserova, M. Spink, S. A. Richards, C. Saunter and M. W. Goldberg, J. Cell Sci., 2014, 127, 124–136 CAS.
- B. B. Hülsmann, A. A. Labokha and D. Görlich, Cell, 2012, 150, 738–751 CrossRef PubMed.
- M. Petri, S. Frey, A. Menzel, D. Görlich and S. Techert, Biomacromolecules, 2012, 13, 1882–1889 CrossRef CAS PubMed.
- T. Jovanovic-Talisman, J. Tetenbaum-Novatt, A. S. McKenney, A. Zilman, R. Peters, M. P. Rout and B. T. Chait, Nature, 2009, 457, 1023–1027 CrossRef CAS.
- J. Kim, A. Izadyar, N. Nioradze and S. Amemiya, J. Am. Chem. Soc., 2013, 135, 2321–2329 CrossRef CAS PubMed.
- R. Y. H. Lim, B. Fahrenkrog, J. Köser, K. Schwarz-Herion, J. Deng and U. Aebi, Science, 2007, 318, 640–643 CrossRef CAS PubMed.
- L. Pan, Q. He, J. Liu, Y. Chen, M. Ma, L. Zhang and J. Shi, J. Am. Chem. Soc., 2012, 134, 5722–5725 CrossRef CAS.
- J. Cheng, K. A. S. Fernando, L. M. Veca, Y.-P. Sun, A. I. Lamond, Y. W. Lam and S. H. Cheng, ACS Nano, 2008, 2, 2085–2094 CrossRef CAS.
- A. Arisaka, R. Mogaki, K. Okuro and T. Aida, J. Am. Chem. Soc., 2018, 140, 2687–2692 CrossRef CAS PubMed.
- R. Iwaura, M. Shirai, K. Yoshida and M. Ohnishi-Kameyama, Chem. Commun., 2014, 50, 9295–9297 RSC.
- D. M. Copolovici, K. Langel, E. Eriste and Ü. Langel, ACS Nano, 2014, 8, 1972–1994 CrossRef CAS PubMed.
- C. Almendáriz-Palacios, Z. E. Gillespie, M. Janzen, V. Martinez, J. M. Bridger, T. A. A. Harkness, D. D. Mousseau and C. H. Eskiw, Biomedicines, 2020, 8, 188 CrossRef PubMed.
- K.-L. Ma, L.-K. Song, Y.-H. Yuan, Y. Zhang, N. Han, K. Gao and N.-H. Chen, Neuropharmacology, 2014, 82, 132–142 CrossRef CAS PubMed.
- J. Li, J. Chen, C. L. Ricupero, R. P. Hart, M. S. Schwartz, A. Kusnecov and K. Herrup, Nat. Med., 2012, 18, 783–790 CrossRef PubMed.
- A. N. Coyne, V. Baskerville, B. L. Zaepfel, D. W. Dickson, F. Rigo, F. Bennett, C. P. Lusk and J. D. Rothstein, Sci. Transl. Med., 2021, 13, 604 Search PubMed.
- S. Sahadevan, K. M. Hembach, E. Tantardini, M. Pérez-Berlanga, M. Hruska-Plochan, S. Megat, J. Weber, P. Schwarz, L. Dupuis, M. D. Robinson, P. De Rossi and M. Polymenidou, Nat. Commun., 2021, 12, 3027 CrossRef PubMed.
- A. Pliss, S. M. Levchenko, L. Liu, X. Peng, T. Y. Ohulchanskyy, I. Roy, A. N. Kuzmin, J. Qu and P. N. Prasad, Nat. Commun., 2019, 10, 455 CrossRef CAS PubMed.
- M. Reyes, D. Peña-Oyarzún, P. Silva, S. Venegas, A. Criollo and V. A. Torres, FASEB J., 2020, 34, 4009–4025 CrossRef CAS PubMed.
- Y. C. Xiang, W. Shan and Y. A. Huang, Int. J. Pharm., 2018, 551, 14–22 CrossRef CAS PubMed.
- S. Shabalala, C. J. F. Muller, J. Louw and R. Johnson, Life Sci., 2017, 180, 160–170 CrossRef CAS PubMed.
- S. Wang, S. Kotamuraju, E. Konorev, S. Kalivendi, J. Joseph and B. Kalyabaraman, Biochem. J., 2002, 367, 729–740 CrossRef CAS PubMed.
- J.-G. Hou, B.-M. Jeon, Y.- Jin Yun, C.-H. Cui and S.-C. Kim, Int. J. Mol. Sci., 2019, 20, 1244 CrossRef CAS.
- K. Nagahama, Y. Sano, M. Inui, S. Aoyama, T. Katayama and K. Ono, Adv. Biosyst., 2020, 4, e1900189 CrossRef.
- K. Stade, C. S. Ford, C. Guthrie and K. Weis, Cell, 1997, 90, 1041–1050 CrossRef CAS.
- J. M. Wubben, S. C. Atkinson and N. A. Borg, Cells, 2020, 9, 2654 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2na00050d |
| This journal is © The Royal Society of Chemistry 2022 |