Non-peptide secondary metabolites from poisonous mushrooms: overview of chemistry, bioactivity, and biosynthesis
Seulah
Lee†
ab,
Jae Sik
Yu†
a,
Seoung Rak
Lee†
ac and
Ki Hyun
Kim
 *a
*a
aSchool of Pharmacy, Sungkyunkwan University, Suwon 16419, Republic of Korea. E-mail: khkim83@skku.edu
bDivision of Life Sciences, Korea Polar Research Institute, KIOST, Incheon 21990, Republic of Korea
cDepartment of Chemistry, Princeton University, New Jersey 08544, USA
First published on 5th October 2021
Abstract
Covering: up to June 2021
A wide variety of mushrooms have traditionally been recognized as edible fungi with high nutritional value and low calories, and abundantly produce structurally diverse and bioactive secondary metabolites. However, accidental ingestion of poisonous mushrooms can result in serious illnesses and even death. Chemically, mushroom poisoning is associated with secondary metabolites produced in poisonous mushrooms, causing specific toxicity. However, many poisonous mushrooms have not been fully investigated for their secondary metabolites, and the secondary metabolites of poisonous mushrooms have not been systematically summarized for details such as chemical composition and biosynthetic mechanisms. The isolation and identification of secondary metabolites from poisonous mushrooms have great research value since these compounds could be lethal toxins that contribute to the toxicity of mushrooms or could provide lead compounds with remarkable biological activities that can promote advances in other related disciplines, such as biochemistry and pharmacology. In this review, we summarize the structures and biological activities of secondary metabolites identified from poisonous mushrooms and provide an overview of the current information on these metabolites, focusing on their chemistry, bioactivity, and biosynthesis.
1 Introduction
Fungi are ubiquitous in nature. Approximately 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 species have been identified in terrestrial, freshwater, and marine environments, but several millions are expected to exist,1 indicating that fungi are one of the most well-adjusted organisms. Fungi produce a large number of structurally diverse secondary metabolites. In general, although the ecological role of most fungal secondary metabolites remains to be clarified, these compounds are considered to be produced to provide the producer organism a survival advantage.2 Natural product chemists have been interested in the isolation and structural elucidation of fungal secondary metabolites. Fungal secondary metabolites have long been investigated,3 and the existence of bioactive metabolites derived from fungi is well-recognized. The discovery of the antibiotic, penicillin from Penicillium rubens by Fleming4 in 1928 stimulated an intensive search for new antibiotics and bioactive secondary metabolites from fungi.5 The fungal secondary metabolites from the subsequent natural product research have been useful as lead structures for the development of new drugs.6–8 In addition to the beneficial properties, the deleterious aspects of fungi have been known and utilized for a long time. For example, mycotoxins derived from fungi growing on food or crops, such as ergot fungi of the Claviceps genus, caused mass poisoning of people in the Middle Ages in Europe.9 However, the pharmacological property of ergot to induce strong contractions of the uterus was known and used by midwives as early as 1582.9 Ergometrine, discovered in ergot in 1932, became a medication in obstetrics to facilitate delivery of the placenta and prevent bleeding after childbirth.
000 species have been identified in terrestrial, freshwater, and marine environments, but several millions are expected to exist,1 indicating that fungi are one of the most well-adjusted organisms. Fungi produce a large number of structurally diverse secondary metabolites. In general, although the ecological role of most fungal secondary metabolites remains to be clarified, these compounds are considered to be produced to provide the producer organism a survival advantage.2 Natural product chemists have been interested in the isolation and structural elucidation of fungal secondary metabolites. Fungal secondary metabolites have long been investigated,3 and the existence of bioactive metabolites derived from fungi is well-recognized. The discovery of the antibiotic, penicillin from Penicillium rubens by Fleming4 in 1928 stimulated an intensive search for new antibiotics and bioactive secondary metabolites from fungi.5 The fungal secondary metabolites from the subsequent natural product research have been useful as lead structures for the development of new drugs.6–8 In addition to the beneficial properties, the deleterious aspects of fungi have been known and utilized for a long time. For example, mycotoxins derived from fungi growing on food or crops, such as ergot fungi of the Claviceps genus, caused mass poisoning of people in the Middle Ages in Europe.9 However, the pharmacological property of ergot to induce strong contractions of the uterus was known and used by midwives as early as 1582.9 Ergometrine, discovered in ergot in 1932, became a medication in obstetrics to facilitate delivery of the placenta and prevent bleeding after childbirth.
Mushrooms are fungi belonging to the higher phyla Ascomycota and Basidiomycota that produce fleshy, spore-bearing fruiting bodies.10 Natural product chemists have considered mushrooms rich sources of structurally diverse and unique secondary metabolites with a remarkable range of bioactivities and potential applications in humans for various diseases.10–12 In practice, the bioactive secondary metabolites from mushrooms have proven to be invaluable in drug discovery and development by providing core template structures and pharmacophores of pharmaceutically and commercially successful products, such as the antibiotic retapamulin13 or strobilurin-derived fungicides for crop plant protection.6,14 However, some mushrooms, referred to as toxic or poisonous mushrooms, can sometimes cause massive and fatal toxicity. Out of 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 species of mushrooms reported, 70–80 are known to be toxic. The representative secondary metabolites responsible for the toxicity are amanitins, the toxins of the death cap (Amanita phalloides) and other species,15,16 and orellanine (see Section 2.7.2.) produced by Cortinarius orellanus and related webcaps.17 Since fruiting bodies of mushrooms produce spores for their sexual reproduction, most fruiting bodies retain either toxins or bitter and/or pungent metabolites as a chemical defense mechanism to prevent fungivores from feeding.2 These metabolites may also be toxic or lethal to humans, but simultaneously, can also be applied beneficially to humans, as in the case of ergometrine derived from ergot. This review intends to switch the perspective that the toxins or secondary metabolites of poisonous mushrooms are always harmful and to provide an overview of the current knowledge on the chemistry, bioactivity, and biosynthesis of secondary metabolites of poisonous mushrooms.
000 species of mushrooms reported, 70–80 are known to be toxic. The representative secondary metabolites responsible for the toxicity are amanitins, the toxins of the death cap (Amanita phalloides) and other species,15,16 and orellanine (see Section 2.7.2.) produced by Cortinarius orellanus and related webcaps.17 Since fruiting bodies of mushrooms produce spores for their sexual reproduction, most fruiting bodies retain either toxins or bitter and/or pungent metabolites as a chemical defense mechanism to prevent fungivores from feeding.2 These metabolites may also be toxic or lethal to humans, but simultaneously, can also be applied beneficially to humans, as in the case of ergometrine derived from ergot. This review intends to switch the perspective that the toxins or secondary metabolites of poisonous mushrooms are always harmful and to provide an overview of the current knowledge on the chemistry, bioactivity, and biosynthesis of secondary metabolites of poisonous mushrooms.
1.1 Poisonous mushrooms
Mushrooms can be found extensively in a variety of natural environments, and the visual identification of mushroom species with stalks and caps is well established. Picking mushrooms from the wild for food is an age-long practice, and edible mushrooms have been used in human diet, not only as a delicious low-calorie food item but also as dietary supplements/mushroom nutraceuticals owing to their nutritional and therapeutic properties.18 However, the growing popularity of wild mushroom consumption has led to the increased incidence of mushroom poisoning—the harmful effects caused by the ingestion of toxic substances present in mushrooms. Mushroom poisoning is usually the result of intake of wild mushrooms by misunderstanding of a toxic mushroom as an edible species owing to their close resemblance in terms of color and general morphology.19In 2019, mushroom poisoning was proposed to be classified into six major groups based on the key clinical features: (1) cytotoxic mushroom poisoning, (2) gastrointestinal irritant mushroom poisoning, (3) mycotoxic mushroom poisoning, (4) endocrine, metabolic, and related toxicity mushroom poisoning, (5) neurotoxic mushroom poisoning, and (6) miscellaneous adverse reactions to mushrooms.20 This system reformed the major classification published in 1994 because the existing classifications of mushroom poisoning did not include more recently described syndromes. Mushroom poisoning is a significant and increasing form of a toxin-induced disease. Poisonous mushrooms contain a variety of toxins whose potency can be influenced by many intrinsic and extrinsic factors present around the mushroom. The symptoms of toxicity change over time, but generally begin with the gastrointestinal tract and then progress to organ damage and even death. Chemically, mushroom poisoning is associated with exceptionally powerful toxins, which can be defined as secondary metabolites produced via specific biochemical pathways in the fungal cells of poisonous mushrooms.
Intensive chemical investigations into toxic secondary metabolites produced by poisonous mushrooms have been conducted since the 1950s. To date, >100 mushroom toxins have been identified and reported; however, there are still many lethal mushrooms that need to be fully investigated. Deaths due to mushroom poisoning have been reported worldwide, and new poisonous mushrooms are continually being discovered and reported. However, little is known about the mechanisms of these toxins beyond their structures. The reason is probably that the fruiting bodies of mushrooms are usually short-lived entities, which cannot even be cultivated. The true ecological reasons behind the evolution of toxic metabolite–synthetic pathways in poisonous mushrooms remain unknown in most cases. However, it is interesting to note that poisonous mushrooms produce structurally diverse and unique secondary metabolites, which cause toxic or lethal effects in humans, but simultaneously can show pharmaceutical bioactivities from a different angle. Previous investigations into natural products of poisonous mushrooms have substantially enhanced our understanding of secondary metabolites from poisonous mushrooms and revealed their structural diversity. It is now time to accept the fact that poisonous mushrooms are not something to be avoided.
1.2 Scope of the review
This review covers the structurally characterized secondary metabolites of poisonous mushrooms, primarily non-peptide secondary metabolites, with a focus on the period up to June 2021. It summarizes the structures of the newly discovered compounds, identified biological activities, potential utilities in medicine, and known toxicities relevant to mushroom poisoning, as well as an overview of their biosynthetic mechanism. Several previous reviews have covered poisonous mushrooms,21–23 focusing either on the mushrooms or their toxicity, but the chemistry (structure and biosynthesis) of the secondary metabolites of these poisonous mushrooms has rarely been reviewed systematically. There are an estimated 900−1000 species in the genus Amanita, including some deadly poisonous species such as A. phalloides, A. verna, and A. virosa, which are the most intensively studied resources among the poisonous mushrooms.24 The lethal Amanita species have attracted the attention of chemists because of the cyclopeptides, as the major lethal toxins. Toxic cyclopeptides, composed mainly of three groups, namely phallotoxins, amatoxins, and virotoxins, have been specifically studied for their structure, total synthesis, biosynthesis, mechanism of toxicity, and potential use as anticancer agents for the past 70 years.15,24–29 Previous reviews have thoroughly summarized the chemistry, biosynthesis mechanisms, and toxicity mechanisms of the cyclopeptides produced by the lethal Amanita species.30–33 Thus, in this review, the peptides reported from the Amanita species are not included. A recent review focused on the chemistry (structure and synthesis), toxicology, and bioprospecting of mushroom toxins, and summarized the chemical information and toxicology of 124 known mushroom toxins by source.33 Thus, in the present review, we cover only non-peptide secondary metabolites from poisonous mushrooms, including those that have not been mentioned in the recent review of mushroom toxins (2019)33 and summarize their chemistry by structural classification, bioactivity, and biosynthesis, which have not been covered in the recent review,33 as well as discussing future research directions.2 Non-peptide secondary metabolites from poisonous mushrooms
2.1 Fatty acids
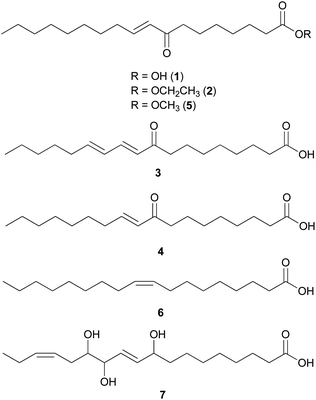
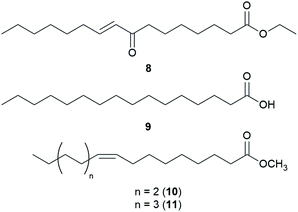
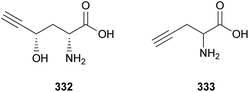
Fatty acids are principal elements of biological membranes, which serve as storage substances and act as second messengers regulating protein localization.34 They also act as crucial compounds in establishing pathogenic plant–fungal interactions.35 Formed from iterative reaction cycles of acetyl-CoA and malonyl-CoA, fatty acids naturally occur as an unbranched chain comprising an even number of carbon chains, from 4 to 28.36 Fatty acid synthesis in fungi is carried out by large architecturally diverse multifunctional synthases, fungal fatty acid synthases (FAS), which constitute a 2.6-megadalton α6β6 heterododecameric complex.37–41 FAS promotes the loading of the acetyl primer and malonyl elongation substrates from CoA to an acyl carrier protein (ACP), via monofunctional acetyl transferase and bifunctional malonyl/palmitoyl transferase (MPT), and their condensation to acetoacetyl-ACP under decarboxylation by ketoacyl synthase.34,37,42 Subsequently, the β-carbon groups are processed by ketoacyl reductase, dehydratase, and enoyl reductase to produce fully saturated acyl-ACP that can serve as a primer for the next condensation reaction. The growing acyl chain is elongated by two carbon units in each reaction cycle until C16 or C18 fatty acids are back-transferred to CoA by MPT, and the end products released are palmitate or stearate, which are precursors of diverse long-chain fatty acids.34,37,42,43 Type I fungal FAS integrates all of these enzymatic activities into unique protein assemblies catalyzing more than 40 reaction steps.1,37Palmitic acid (9), a widely known hexadecanoic acid (C16:0) that serves as the precursor of diverse long-chain fatty acids, such as stearic acid, oleic acid, and palmitoleic acid,50 was identified in the toxic mushroom A. spissacea.48 It was evaluated for cytotoxicity against human lung cancer cells, A549, H1264, H1299, and Calu-6, but did not exhibit any significant cytotoxicity against any of the tested cell lines. Compound 9 is the most common saturated fatty acid, accounting for 20–30% of total fatty acids in the human body.51,52
Boletus pseudocalopus is an inedible mushroom that is known to cause gastrointestinal irritation.53 Methyl palmitoleate (10) and methyl 11(Z)-eicosenoate (11) were isolated from B. pseudocalopus and evaluated for antiproliferative activity against human cancer cell lines A549, Hs756T, and H3122, but were inactive against all tested cell lines (IC50 >20 μM).54
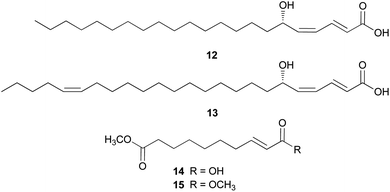
Amanita pantherina is a toxic mushroom that causes muscle cramps, insanity, and audiovisual disorders; ibotenic acid, muscimol, stizolobic acid, and stizolobinic acid are its main toxic metabolites (see Section 2.7.1.).55 Pantheric acids A–C (12–14) are new fatty acid derivatives with various carbon chain lengths corresponding to C22 (12), C24 (13), and C10 (14) identified from this mushroom, along with 1,10-dimethyl ester-2-decenedioic acid (15), a known fatty acid ester with a C10 chain.55 Based on the finding that dietary fatty acids are essential metabolites in lipid metabolism of adipocytes,56,57 the isolated compounds (12–15) were evaluated for their effects on lipid metabolism. Compounds 12–14 were found to regulate lipid metabolism, which reduced the mRNA expression of lipolytic genes ATGL and MCAD at a concentration of 20 μM during adipocyte maturation, while elevating the transcription of lipogenic genes FASN and SREBP1.55
2.2 Polyacetylenes
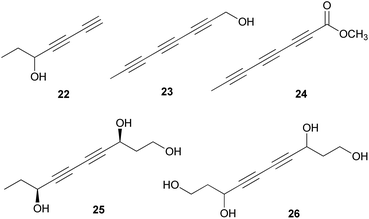
Polyacetylenes are also referred to as acetylenic compounds, and contain alkynyl functional groups.59 Fatty acids, which are the building blocks for the biosynthesis of polyacetylenes, are derivatized by various enzymes and result in the oxidation of a preexisting double bond to a triple bond.60 There are two hypotheses regarding the biogenesis of acetylenic bonds: the first states that the formation of these bonds involves catalysis by NAD+ or NADP+ and desaturation via an iron-catalyzed dehydrogenation step with molecular oxygen,61 and the second states that the elimination reaction of an activated enol carboxylate intermediate, thermodynamically driven by carbon anhydride formation, results in the formation of acetylenic bonds.62 These pathways result in the production of monoacetylenic intermediates, which are precursors of a broad variety of natural polyacetylenes.60Although it has been reported that C18 fatty acids serve as the core of most acetylenic compounds,59 those identified from toxic mushrooms have short carbon chains ranging from C7 to C10. As part of the research on the polyacetylene content of Basidiomycetes, Hearn et al. examined the culture fluids of species belonging to families not previously reported as polyacetylene producers and characterized novel natural C7–C10 polyacetylenes.63 (−)-Hepta-1,3-diyn-5-ol (22) was identified as the main constituent in the poisonous mushrooms G. spectabilis and G. hybridus, but its absolute configuration was not determined. Triynol (23) and triyne acid methyl ester (24) were found to be the main polyacetylenes of toxic mushrooms Psilocybe merdaria, Kuehneromyces mutabilis, and Ramaria flava. 4,6-Decadiyne-1,3,8-triol (25) was isolated from G. spectabilis in 1986 by Kusano et al., and only its planar structure was reported.64 In 1989, compound 25 was isolated again from the same source by Hashimoto et al., and the absolute configuration was determined as (3S,8S).65 It was evaluated for antioxidant activity, but did not exhibit any significant scavenging activity against the ABTS (2,2′-azino-bis(3-ethylbenzothiazolin-6-sulfonic acid) and DPPH radical cations and superoxide radical anion up to 200 μM.66 4,6-Decadiyne-1,3,8,10-tetraol (26), a novel polyisoprenol, was also isolated from G. spectabilis, but its stereochemistry remains to be deduced.67
2.3 Sphingolipids
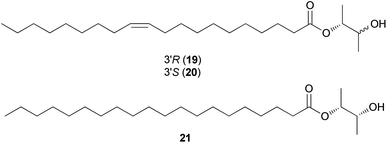
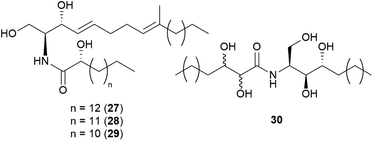
Sphingolipids are membrane lipids that are important in cell regulation, signal transduction, heat stress response, vesicle trafficking, and endocytosis.68–72 In fungi, sphingolipid biosynthesis starts with the condensation of serine and palmitoyl-CoA by serine palmitoyltransferase (SPT) to produce 3-ketosphinganine, which is then reduced by 3-ketosphinganine reductase (3KSR) in an NADPH-dependent reaction to form dihydrosphingosine.73 The long-chain base (known as (2S,3R,4E)-2-amino-octadec-4-ene-1,3-diol) is the building block of all common sphingolipids including ceramides and glycosphingolipids.73Ceramides play key roles in apoptotic signaling and are found in fungi at relatively low levels.74 Novel ceramides, (2S,2′R,3R,4E,8E)-N-2′-hydroxypentadecanoyl-2-amino-9-methyl-4,8-octadecadiene-1,3-diol (28) and (2S,2′R,3R,4E,8E)-N-2′-hydroxytetradecanoyl-2-amino-9-methyl-4,8-octadecadiene-1,3-diol (29), were identified from the toxic mushroom A. pantherina, along with a known synthetic compound, (2S,2′R,3R,4E,8E)-N-2′-hydroxyhexadecanoyl-2-amino-9-methyl-4,8-octadecadiene-1,3-diol (27).75 A new phytosphingosine-type ceramide, paxillamide (30), was isolated from the fruiting bodies of the toxic mushroom Paxillus panuoides.76 The biosynthesis of sphingolipid scaffolds of 27–29 (9-methyl trans-4-trans-8-sphingadienine, also known as 4-t,8-t-9-methylceramide) and 30 (phytoceramide) has been previously described.74
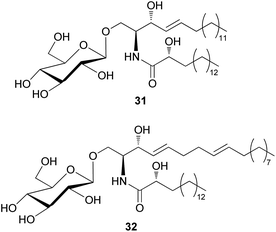
The amount of glycoceramides is higher than that of regular ceramides in fungi. The change in glucosylceramide level is known to have a severe impact on biological properties of fungi under different physiological conditions.74 Despite their abundance in fungi, only two glucosylceramides have been detected in toxic mushrooms; 8,9-dihydrosoyacerebroside I (31) and soyacerebroside I (32) have been isolated from Amanita subjunquillea, also known as “the East Asian death cap”, which exerts toxic effects, such as delayed gastrointestinal symptoms, hepatotoxicity, and mortality.77 Compounds 31 and 32 were evaluated for cytotoxic activity against A549, SK-OV-3, SK-MEL-2, and HCT15 human tumor cell lines. Compound 31 did not exhibit significant cytotoxicity against any of the tested cell lines (IC50 >30 μM), while 32 exhibited cytotoxicity against SK-OV-3 (IC50 = 24.17 μM) and HCT-15 cells (IC50 = 28.61 μM) and was inactive against the other two cell lines (IC50 >30 μM).77 Glucosylceramides are formed through the enzymatic reaction of glucosyltransferase on ceramide scaffolds.74
2.4 Phenolics
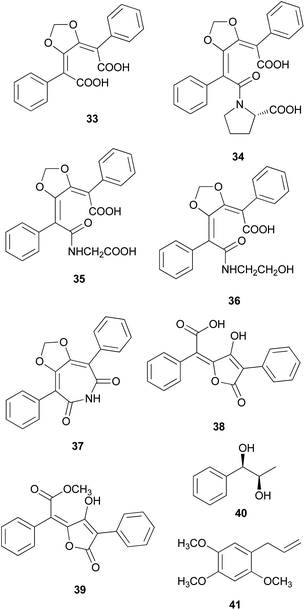
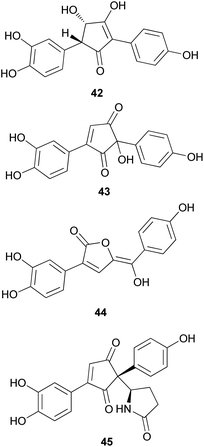
Phenolic compounds, typically isolated from mushrooms, consist of a common aromatic ring with one or more hydroxyl groups. These metabolites are normally synthesized through the shikimate and phenylpropanoid pathways.78 Phosphoenolpyruvate reacts with erhtyrose-4-phosphate to yield chorismic acid,79 which serves as a precursor for the aromatic amino acids, L-tyrosine and L-phenylalanine, via the activity of chorismate mutase. Alternatively, chorismic acid can follow an alternative anabolic route catalyzed by the enzyme, anthranilate synthase, to form L-tryptophan.80L-tyrosine and L-phenylalanine can undergo enzymatic oxydeamination to form arylpyruvic acids, and they can be dimerized to form terphenylquinones, which can be derivatized into p-terphenyl compounds.81,82 Further oxidative-ring cleavage and decarboxylation from terphenylquinones form the pulvinone skeleton that is commonly found in fungi, and these terphenylquinones act as starting compounds in the biogenesis of terphenyls.81 Aromatic amino acids also act as a precursor for C6–C3 phenylpropanoids and can be broadly classified into simple phenolics, which have a low molecular weight and single aromatic ring, and complex phenolics with a high molecular weight.Structurally similar to ustalic acid analogs, diphenyl cyclopenteneone derivatives (42–45) have been isolated from the toxic mushroom, Paxillus involutus, also known as Poison Pax, which is a member of the division Basidiomycetes and distributed across Europe, South Africa, and New Zealand.85 The consumption of these species causes vomiting and high fever, and in severe cases, it can lead to hemolytic symptoms, which have been observed in both laboratory animals and human beings; however, the specific antigen of the metabolite responsible for this severe toxicity remains unknown.85–91 From the fruiting bodies of P. involutus, involutin (42), involutone (43), (5Z)-3-(3,4-dihydroxyphenyl)-5-[hydroxy(4-hydroxyphenyl) methylene]-2(5H)-furanone (44), and 4-(3,4-dihydroxyphenyl)-2-(4-hydroxyphenyl)-2-(2-pyrrolidon-5-yl)-4-cyclopentene-1,3-dione (45) have been isolated as new metabolites.92–95 Absolute configurations of compounds 43 and 45 are not confirmed and compounds 42–45 do not display any significant cytotoxic activities.
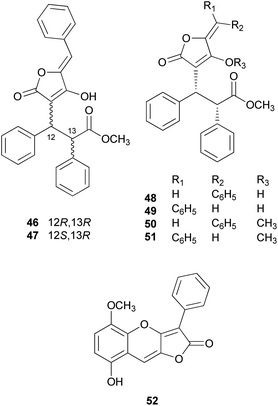
There are polymer types of phenolic compounds that can vary in molecular weight. When phenylalanine hydroxylase is reduced, phenylalanine is converted into phenylpyruvic acid, and its dimerization can lead to terphenylquinone formation. Compound 39, isolated from T. ustale, is considered a secondary metabolite derived from terphenylquinones. It has also been isolated from another inedible mushroom, Pulveroboletus ravaenlii, commonly known as Ravenel's bolete or powdery sulfur bolete owing to the color of its fruiting bodies. Compound 39 has been found to reduce the viability of human cancer cells, including four lung cancer cell lines (NCI-H1264, NCI-H1299, A549, and Calu-6), two pancreatic cancer cell lines (MIA PaCa-2 and PANC-1), and one liver cancer cell line (HepG2), by inducing apoptosis; the IC50 values of compound 39 range from 21.65 to 146.17 μM.96 This compound is also involved in cell differentiation, promoting the osteogenesis of mesenchymal stem cells and decreasing the proportion of hypertrophic adipocytes.97 Furthermore, related phenolic analogs, compounds 46–52, have been isolated from the fruiting bodies of P. ravenelii.98–100 Among these compounds, pulveraven A (46), pulveraven B (47), isoravenelone (48), ravenelone (49), and pulverolide (52) were isolated as new fungal butenolide-type compounds from P. ravenelii, while the biosynthesis of 52 was proposed from 39 (Scheme 1).100 Unlike compound 39, no study has investigated the cytotoxic activity of compounds 46–52. Instead, a previous study tested compounds 39, 46, and 47 for their potential to inhibit cyclooxygenase (COX) activity and carcinogen-induced preneoplastic lesion formation using mouse mammary organ culture (MMOC) assays. Compound 39 weakly inhibited COX-1 activity, with IC50 values ranging from 192 to 218 μM, while it was inactive as a COX-2 inhibitor, with IC50 values of >100 μg mL−1. Compound 47 was inactive in both COX-1 and COX-2 inhibition assays, with IC50 values of >100 μg mL−1, and 46 was not tested due to insufficient material. In the MMOC assay, 39 was inactive, with IC50 values of >10 μg mL−1, while 46 and 47 showed suitable inhibitory activities, with IC50 values of 6.0 and 0.8 μM, respectively.98
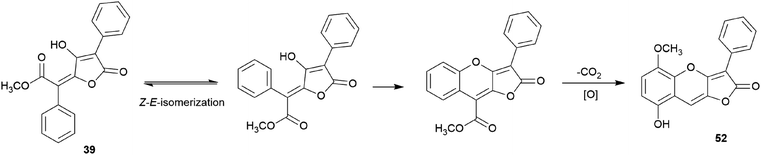 | ||
| Scheme 1 Plausible biosynthetic pathway of pulverolide (52). Adapted from Zhang et al.100 | ||
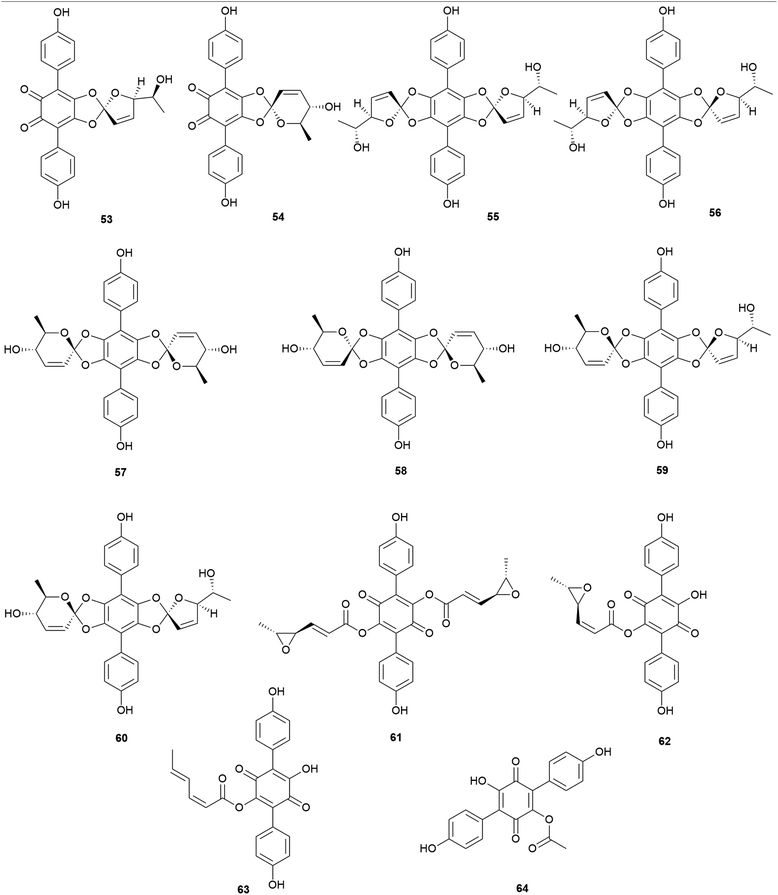
Other examples of terphenylquinones are spiromentins, which are commonly found in the toxic mushroom Paxillus atrotomentosus. These structures consist of a spirostructural moiety that is derived from terphenylquinones, featuring unique 4,5-dihydroxy-1,2-benzoquinone.101 Compounds 53–60 have been isolated from the fruiting bodies of P. atrotomentosus as spriromentins B (53), C (54), and E–J (55–60).102 Compounds 55–60 were identified for the first time from this mushroom, but only the relative configuration has been reported. Because of its bitter taste, this mushroom is not considered edible, and there have been cases of poisoning by this species in Europe, but not much information is available.88,103 Interestingly, this species is no longer called P. atrotomentosus but is known as Tapinella atrotomentosa, owing to the physical difference in its stipe from those of the members of the genus Paxillus, and the genetic analysis showed that T. atrotomentosa is only distantly related.104 In addition to spiromentins, further derived products, flavomentins A–C (61–63) and 2-O-acetylatromentin (64) have also been isolated from this mushroom.102 These isolated compounds 53–64 are known to be fungal pigments, and the particular bioactivities of compounds 53–64 have not been tested.
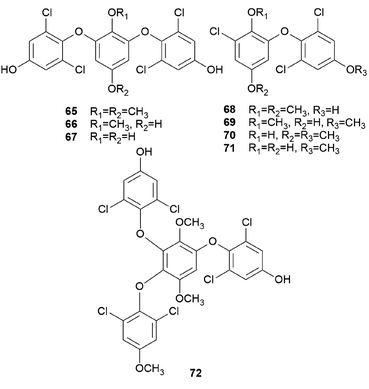
Interestingly, some phenolic compounds from toxic mushrooms consist of halogen groups. Secondary metabolites from Russula subnigricans Hongo, known to be a toxic mushroom that is predominantly found in East Asia, are an example of such phenolic compounds. The ingestion of R. subnigricans causes vomiting and diarrhea, followed by severe upper body ache and hematuria.105 In severe cases, it causes speech impediment, cardiac damage, loss of consciousness, and acute rhabdomyolysis.106,107 Distinctively, this mushroom produces chlorinated phenyl ethers, russuphelins A–G (65–71), and a chlorohydroquinone tetramer russuphelol (72).108–110
Compound 65 has been tested for its cytotoxicity against various tumor cells. It showed cytotoxic activities against the mouse lymphocytic leukemia cell line L1210 (IC50 = 2.47 μg mL−1), mouse colon adenocarcinoma cell line Colon 26 (IC50 = 10.16 μg mL−1), mouse leukemia cell line P388 (IC50 = 4.64 μg mL−1), mouse leukemia cell line P388/ADM (IC50 = 2.96 μg mL−1), human mouth epidermal carcinoma cell line KB (IC50 = 2.09 μg mL−1), non-small-cell lung carcinoma cell line A549 (IC50 = 0.49 μg mL−1), murine melanoma cell line B16 (IC50 = 6.28 μg mL−1), chronic myeloid leukemia cell line K562 (IC50 = 0.16 μg mL−1), human colorectal adenocarcinoma cell line DLD-1 (IC50 = 0.79 μg mL−1), and human acute lymphoblastic leukemia cell line CCRF-CEM (IC50 = 0.49 μg mL−1). These values showed that 65 displayed strong cytotoxic activities, especially against human-derived carcinoma cells.109 Compounds 66–68 also showed cytotoxic activities against mouse leukemia cell line P388, with IC50 values of 15.4, 0.94, 12.1 μg mL−1, respectively.110 However, there were no significant cytotoxic effects for compounds 69–72. Interestingly, Matsuura et al. found that a toxic metabolite is responsible for rhabdomyolysis caused by R. subnigricans, and they revealed it to be an unstable compound cycloprop-2-ene carboxylic acid (see Section 2.8.).111
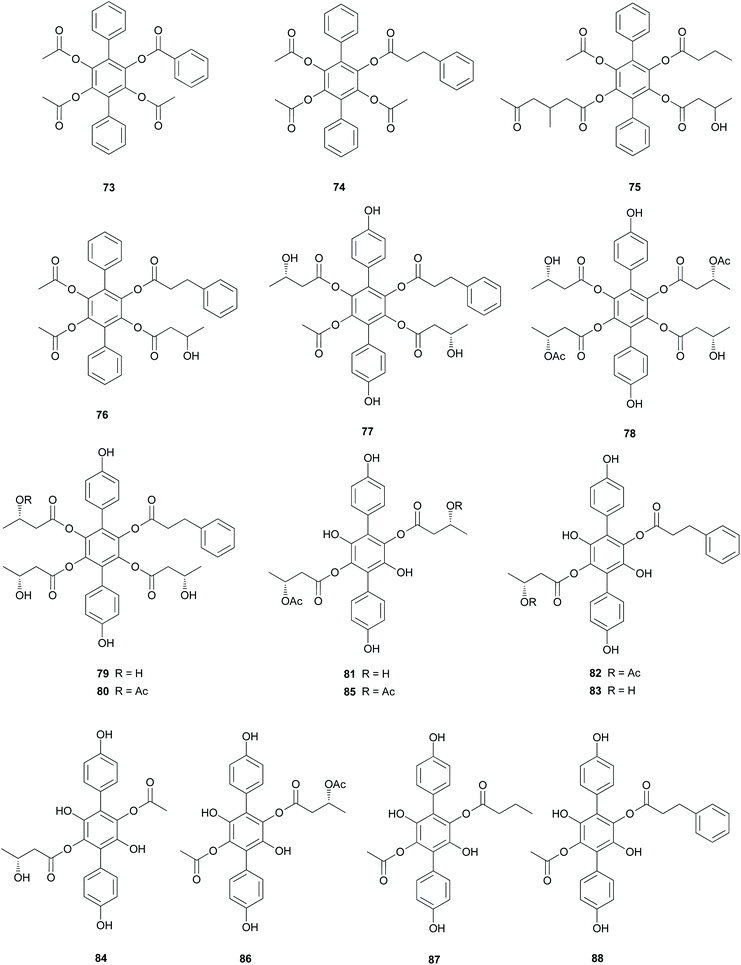
Curtisians D, E, and I (76, 77, and 81) were tested for iron-chelating ability using chrome azurol S assay, which is commonly used for the detection of siderophores from microorganisms.124 The tested compounds showed positive reaction results by demonstrating similar chelation ability compared to the controls, including a traditionally used iron chelator-deferrioxamine, an antioxidant-butylated hydroxyanisole (BHA), and Trolox.118 This result suggests that the catechol moiety of p-terphenyls can provide some significant iron chelation and that this can act as a key modulator in the biological degradation of wood for P. curtisii metabolism and growth.125
Compound 77 also functions as an antiplatelet agent for rat platelets collected from the abdominal aorta.126 It showed broad inhibitory effects on platelet aggregation induced by agonists, such as ADP, thrombin, and collagen (12.5–200.0 μM) via MAPK, PI-3k/Akt, and cAMP-mediated VASP phosphorylation.116 In addition, the antioxidant activity of 77 was evaluated using the DPPH, ABTS, and superoxide radical scavenging methods, but these approaches did not reveal any significant activity of this compound (IC50 >500 μM) compared with that of the control caffeic acid (IC50 = 21.11 ± 1.22, 5.37 ± 0.09, and 58.0 ± 4.3 μM, respectively).117
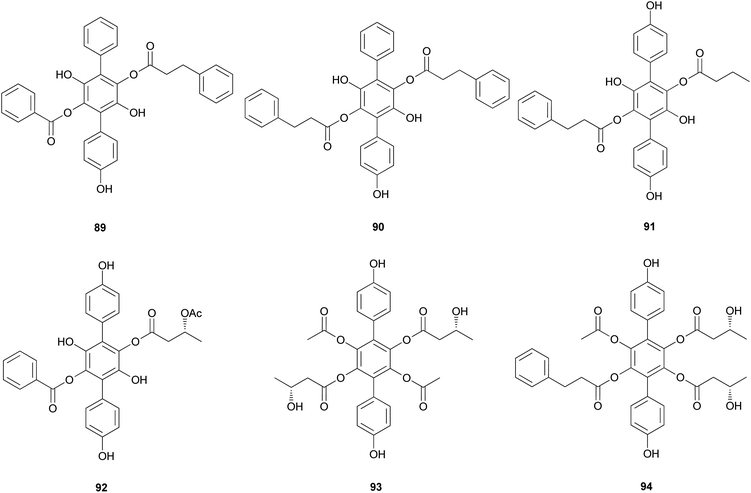
Curtisians H, J, P, and Q (80, 82, 88, and 89) were examined for their inhibition of LPS-induced NO production in the culture media of RAW 264.7 cells. The tested compounds showed NO inhibitory activities with IC50 values of 46.2, 51.1, 27.8, and 31.1 μM, respectively.127 Curtisians I–P (81–89) and R–V (90–94) were tested for antioxidant activity using the DPPH, ABTS, and superoxide radical scavenging methods. They showed antioxidant activity in the ABTS radical scavenging method with IC50 values of 60.41 ± 4.53, 13.45 ± 0.55, 11.30 ± 0.07, 18.64 ± 0.10, 8.15 ± 0.18, 5.50 ± 0.23, 7.11 ± 0.19, 6.44 ± 0.39, 8.02 ± 0.28, 8.16 ± 0.34, 8.43 ± 0.29, >500, and >500 μM, respectively, compared with the control caffeic acid (IC50 = 5.37 ± 0.09 μM). However, all tested compounds did not show significant activity in the DPPH and superoxide radical scavenging methods (IC50 >500 μM) compared with the control caffeic acid (IC50 = 21.11 ± 1.22 and 58.0 ± 4.3 μM, respectively).117 In contrast, in another study, compounds 81–84 showed antioxidant activity through free DPPH-radical scavenging activity, with IC50 values of 19.1, 24.0, 31.0, and 117.8 μM, respectively, compared with the control, ascorbic acid, which showed an IC50 value of 16.5 μM.120
Additionally, compounds 85–89 showed significant antioxidant activity through free DPPH-radical scavenging activity, with IC50 values of 45.9, 48.8, 58.7, 44.0, and 43.4 μM, respectively, compared with the control, ascorbic acid, which showed an IC50 value of 16.5 μM.119
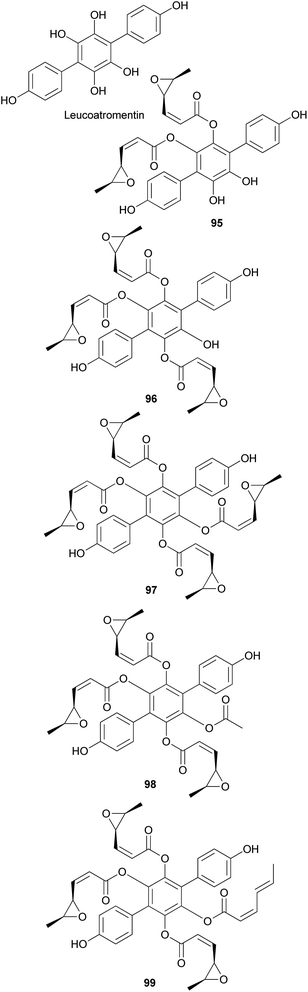
Other p-terphenyl compounds, such as leucomentin analogs, have also been isolated from toxic mushrooms. Similar to curtisians A–V (73–94), leucomentin analogs are derived by the substitution of esters of leucoatromentin with (2Z,4S,5S)-4,5-epoxy-2-hexenoic acid.112 Leucomentins-2, -3, -4, -5, and -6 (95–99) are novel compounds found in the toxic mushroom Paxillus panuoides.128–130 Adequate information is not available regarding the toxicity of P. panuoides; however, it is currently known as Tapinella panuoides. Compound 96 has also been isolated from P. atrotomentosus, and only the relative configurations of compounds 95–99 are confirmed.131 The neuroprotective effects of compounds 95 and 97–99 against glutamate, NMDA, and H2O2 have been tested in mouse cortical neurons.128 Against glutamate neurotoxicity, the tested metabolites 95 and 97–99 showed strong activities with IC50 values of 0.7, 0.9, 1.6, and 4.5 μM, respectively, compared with the control MK-801 (IC50 = 0.3 μM). Against NMDA neurotoxicity, they showed IC50 values of 6.6, 5.1, 5.5, and 5.6 μM, respectively, compared with the control MK-801 (IC50 = 0.4 μM). Finally, against H2O2 neurotoxicity, they showed IC50 values of 4.5, 3.3, 3.3, and 1.1 μM, respectively, compared with the control vitamin E (IC50 >50 μM). These metabolites were also tested for neuroprotective effects on a different cell line, N18-RE-105, which did not show any activity with all the compounds, with IC50 values of >100 μM compared with the control vitamin E (IC50 = 1.2 μM). In addition, free radical scavenging method was used to evaluate the inhibitory activity of compounds 95 and 97–99 against lipid peroxidation in rat liver microsomes. The tested metabolites, 95 and 97–99, showed strong lipid peroxidation inhibitory activities with IC50 values of 0.06, 0.11, 0.11, and 0.06 μg mL−1, respectively, compared with the control vitamin E (IC50 = 1.5 μg mL−1).130
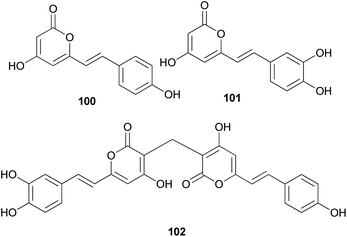
Both compounds 100 and 101 were able to scavenge DPPH, ABTS, and superoxide radicals in a concentration-dependent manner.132 In addition, 3,3-fused bis(styrylpyrone), squarrosidine (102) has been isolated as a novel compound from the toxic mushroom Pholiota squarrosa.133 A plausible biosynthetic pathway of 102 is proposed in Scheme 2. The formation of 102 can be rationalized by the nucleophilic vinylogous addition of 102 to the tautomeric form of a methylated bisnoryangonin derivative (or vice versa). The inhibitory effects of 102 on xanthine oxidase (XO) have been tested; XO catalyzes the crystallization of uric acid in the joints, causing gouty arthritis.134 Compound 102 showed strong inhibitory activity against XO with an IC50 value of 8.1 μM.
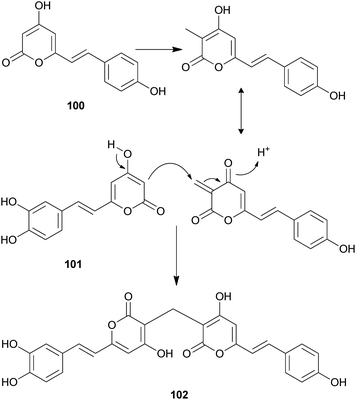 | ||
| Scheme 2 Plausible biosynthetic pathway of squarrosidine (102) proposed by Kemami Wangun et al.133 | ||
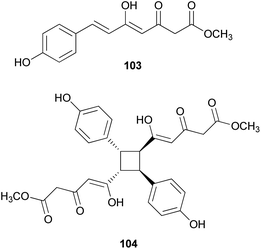
Other phenylpropanoid-derived metabolites found in toxic mushrooms include methyl 5-hydroxy-7-(p-hydroxyphenyl)-3-keto-4Z,6E-heptadienoate (103), and a novel phenyl heptadienoic acid dimer, gymnodimericin (104), isolated from the hallucinogenic mushroom G. spectabilis,135 whose scientific name was recently corrected as G. orientispectabilis.136 In particular, the structure of 104 has been identified as an achiral and a centrosymmetric meso compound via characteristic coupling constants and NOESY analysis. These metabolites, 103 and 104, were tested for their activities on lipid metabolism using 3T3-L1 adipocytes by determining the mRNA levels of the four related genes, namely Fabp4, Adipsin (biomarkers), ATGL (lipolytic gene), and SREBP1 (lipogenic gene). Compound 103 downregulated Adipsin and Fabp4 (both ∼0.6-fold), and compound 104 showed selective upregulation of Adipsin and Fabp4 (up to 2-fold). Metabolites 103 and 104 were also examined for their cytotoxic activities toward human cervical cancer (HeLa), human triple-negative breast cancer (MDA-MB-231), and human acute promyelocytic leukemia (HL-60) cells as well as their autophagic activities against the HeLa cells, which stably express a marker protein (mRFP-EGFP-LC3B) of autophagy; however, none of them were active. Plausible biosynthetic pathway interactions of 103 and 104 were proposed through the hydrolysis and methylation of triketide intermediate i. Light-driven [2 + 2] self-photocycloaddition in the olefinic group of 103 (gray-colored in Scheme 3) would lead to the formation of 104, a centrosymmetric photodimer.135,137
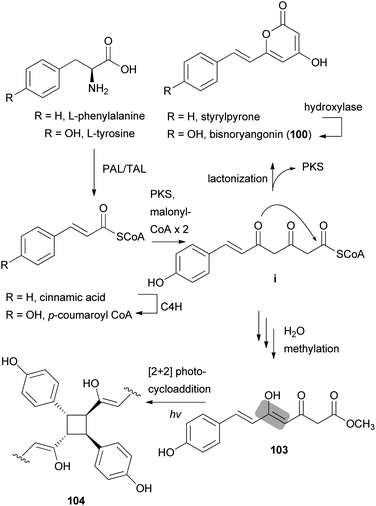 | ||
| Scheme 3 Plausible biosynthetic pathway of compounds 100, 103, and 104 proposed by Lee et al.135 (PAL, phenylalanine ammonia-lyase; TAL, tyrosine ammonia-lyase; PKS, polyketide synthase; and C4H, trans-cinnamate 4-hydroxylase). | ||
The proposed plausible biosynthetic pathway of 106 starts from orsellinic acid and undergoes methylation and oxidoreductive tailoring steps (Scheme 4).138 From the proposed pathway, benzofuran-derived compounds 107 and 108 were both isolated from P. squarrosa. Compound 106 was tested for its inhibitory activity against the serine protease, α-chymotrypsin, where it inhibited α-chymotrypsin with an IC50 value of 84 μM, thereby helping in digestion.140,141
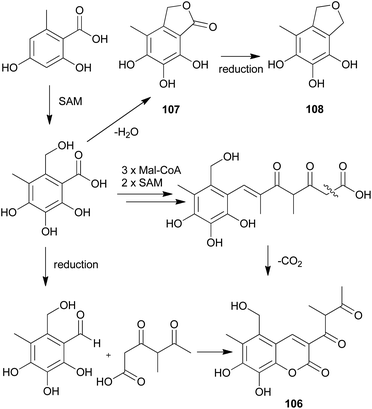 | ||
| Scheme 4 Plausible biosynthetic pathway proposed by Kemami Wangun et al.138 (SAM, S-adenosylmethionine; Mal-CoA, Malonyl-CoA). | ||
2.5 Terpenoids
Terpenoids are major secondary metabolites of fungi derived from the five-carbon unit isopentenyl diphosphate and its isomer dimethylallyl diphosphate that are sequentially coupled via prenyltransferase enzymes to produce long chains of prenyl diphosphates, which serve as precursors to terpene biosynthesis.146 Terpene synthases, which are also known as terpene cyclases, catalyze ionization of prenyl diphosphates to produce a carbocation, which is then quenched through intramolecular rearrangements and/or nucleophilic attack according to the active-site topology of the enzyme.147,148 The hydrocarbon terpene skeleton is produced and further derivatized by one or more enzymes, resulting in a broad variety of bioactive terpenoids.1462.5.1.1 Illudins. Illudins are predicted to be formed by a primary cyclization of FPP between C-1 and the distal C-10–C-11 double bond, forming humulene as the first cyclic precursor.156,157 Humulene is then protonated for further cyclization and rearrangement, producing the illudin skeleton.56 Illudins possess spirocyclopropane and α,β-unsaturated ketone, which constitute a bis-electrophile that is responsible for DNA damage.158
The identification of illudins has been reported in a member of the division Basidiomycetes, Lampteromyces japonicus, a bioluminescent toxic mushroom.159,160 This mushroom is known as one of the most notorious poisonous mushrooms in Japan, and it causes severe illnesses, including liver damage and hemorrhagic changes in the lungs, kidneys, and digestive organs.161 Illudin S (111), the first illudin identified from L. japonicus, is the major toxic substance present in this mushroom. The isolation of 111 has been reported by several groups,159,162–165 and its antibiotic and antitumor properties have also been studied.166–169 It strongly inhibits in vivo DNA synthesis from thymidine, presumably caused by DNA damage, as well as RNA synthesis.169 It produces a unique type of DNA lesion, that is largely ignored by global repair pathways.170 In one study, 111 was found to cross-link with DNA in the cell, but it did not react directly with the DNA, which is activated possibly by phosphorylation, converting 111 into a bifunctional alkylating agent at physiological pH.171 Compound 111 damage does not only affect the transcription complexes but also inhibits DNA replication, and topoisomerase inhibitors that are expected to comprise recombinational processing of stalled replication forks are known to sensitize cells to illudins.172,173 In relatively sensitive tumor cells, the uptake of 111 occurs through a saturable, energy-dependent mechanism, whereas in relatively resistant cells it occurs by passive diffusion.174
The mechanism underlying the toxicity and antitumor activity of illudins is explained in Scheme 5, where Michael-type addition to the α,β-unsaturated ketone produces a highly reactive cyclohexadiene intermediate, which undergoes cyclopropane ring-opening with an attack by nucleophiles.175
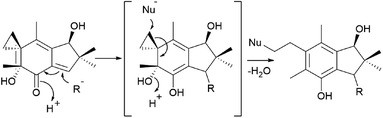 | ||
| Scheme 5 Mechanism underlying the toxicity and antitumor activity of illudins.175 | ||
Tanaka et al. found that cyclopropane ring cleavage occurs when 111 is orally administered to rats, ultimately excreting two metabolites, 111a and 111b (Scheme 6), which can be effectively used to identify poisoning caused by L. japonicus.161
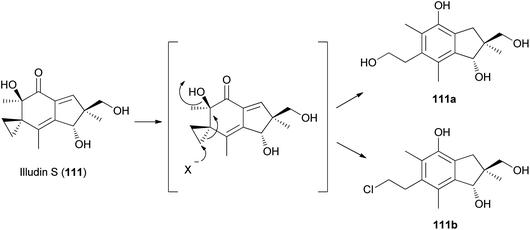 | ||
| Scheme 6 Proposed metabolism of illudin S by rat liver supernatant. X− is OH− or Cl−.161 | ||
Compound 111 is predicted to be biosynthetically formed from farnesyl pyrophosphate, forming a humulene-type sesquiterpene as an intermediate, as shown in Scheme 7.156,164 Further cyclization and rearrangement of the humulene intermediate results in the formation of the illudin skeleton. Subsequent dehydration and oxidative reactions produce 111.
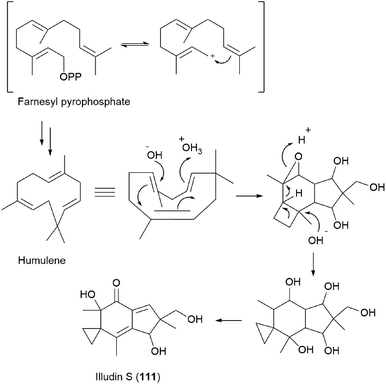 | ||
| Scheme 7 Proposed biosynthesis of illudin S (111).156,164 | ||
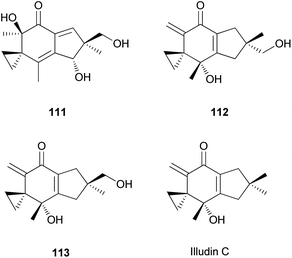
Illudins C2 (112) and C3 (113), which are new derivatives of illudin C, have been identified in the culture broth of Coprinus atramentarius, a poisonous mushroom known to produce coprine, a mycotoxin (see Section 2.7.1.2.).176 Both compounds have been evaluated for antimicrobial activity, and they exhibited minimum inhibitory concentrations of 12 and 25 μg mL−1 against Staphylococcus aureus 209 and S. aureus R209, respectively.176
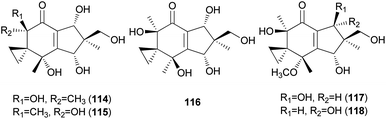
Neoilludins A (114) and B (115), which are structurally closely related to 111, have been isolated from L. japonicus, a bioluminescent toxic mushroom.177 Their antibacterial activity, along with that of 111, against Bacillus subtilis and S. aureus was tested, and all compounds exhibited antibacterial activity against these bacteria. Interestingly, compound 114 exhibited selective inhibitory activity against S. aureus at a concentration of 10 ppm compared with 111.177 Since neoilludins were less toxic than 111 against the murine leukemia cell line P388, they were considered as potential nontoxic antibacterial agents, although further study is necessary.177 Neoilludin C (116), a new epimer of 115, was recently identified from L. japonicus, along with new illudane sesquiterpenes 4-O-methylneoilludins A (117) and B (118).178 These compounds, along with 111, were evaluated for their growth-restoring activity against a mutant yeast strain, YNS17. It was found that compounds 111, 117 and 118 showed weak activity via Ca2+ signal transduction at 1.250, 2.500, and 0.625 μg per spot, respectively.178
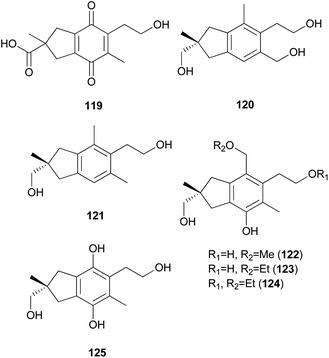
2.5.1.2 Illudalanes. Paraquinoic acid (119), a novel norilludalane sesquiterpene, was isolated from mycelial cultures of the Basidiomycetes Mycena pura, a poisonous mushroom.179 Its effects on the differentiation of HL-60 (human promyelocytic leukemia) and U-937 (human histiocytic lymphoma) cells were examined, and it was found that up to 30–40% of HL-60 cells were differentiated into granulocyte- or monocyte/macrophage-like cells at 380 μM and up to 10–15% of U-937 cells were differentiated at the same concentration.179
Russujaponols A–L are sesquiterpenes isolated from a toxic mushroom Russula japonica.180,181 Russujaponols E (120) and F (121) are novel illudalane sesquiterpenes from this source.180 Russujaponols I–L (122–125) are novel illudalane type sesquiterpenes isolated from the same source.181 Compounds 122–124 were evaluated for neurite outgrowth promoting activity, and they all induced neurite outgrowth of cultured rat cortical neurons at concentrations from 0.1–10 μM.181
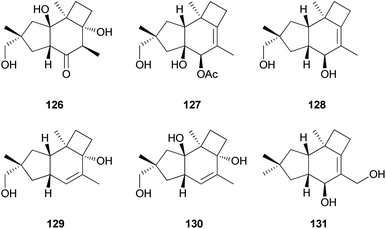
Russujaponols A–D (126–129) are novel protoilludalane sesquiterpenes identified from R. japonica.180 Compound 126 was tested for anti-invasive activity in human fibrosarcoma cell line (HT1080) as the invasion of tumor cells through basement membrane is one of the crucial steps for cancer metastasis. Compound 126 suppressed the invasion of HT1080 to reconstituted basement membrane (Matrigel) with 63% inhibition at 3.73 μM.180 Russujaponols G (130) and H (131) are also novel protoilludalane sesquiterpenes isolated from R. japonica.181
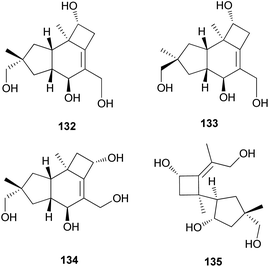
Tsukiyols A (132) and B (133) are new protoilludane sesquiterpenes, and 5-hydroxydichomitol (134) is the same type of compound isolated from L. japonicus, a bioluminescent poisonous mushroom.178 Tsukiyol C (135) is a new fomannosane sesquiterpene identified from L. japonicus. These compounds were evaluated for their growth-restoring activity against a mutant yeast strain, YNS17, and compound 135 showed activity via Ca2+ signal transduction at 1.25 μg per spot.178
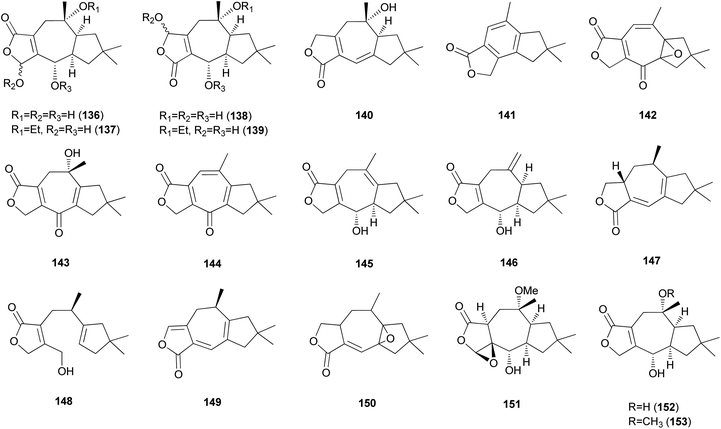
2.5.1.3 Lactaranes. Diverse types of lactarane sesquiterpenes have been identified from the Russulaceae family of fungi (genera Lactarius and Russula),182–189 particularly from the poisonous mushroom Lactarius scrobiculatus, which has an intensely pungent taste presumed to be responsible for the high toxicity of the mushroom. Among different types, sesquiterpene lactones account for the majority of lactarane-type lactones. Four new γ-hydroxybutenolide sesquiterpenes (136–139) were identified in L. scrobiculatus by de Bernardi et al.182 New sesquiterpene lactones, namely lactaroscrobiculide B (140),183 8-norlactaranelactone (141), 2,9-epoxylactarotropone (142), and 3,13-dihydroxy-8-oxo-2(9),6-lactaradien-5-oic acid γ-lactone (143), were isolated from the same source, along with known sesquiterpene lactones (144-146).184 The stereochemistry of 142 was not determined. Lactaroscrobiculide A (147),185 blennin C (148), a new triene-enolactone, chysorrhelactone (149),186 and a new sesquiterpene, epoxylactone (150),187 were also identified in L. scrobiculatus; however, the absolute configuration of 150 remains to be deduced. These lactarane sesquiterpene lactones have also been isolated from the toxic mushroom Russula emetica, from which a new sesquiterpene (151) with a β,γ-epoxy-γ-lactone moiety and a known sesquiterpene, lactarorufin A (152), were isolated.188 Compound 153 was isolated along with 152 from another Russula species, R. nigricans, which contains toxins that cause gastrointestinal upset.145,189
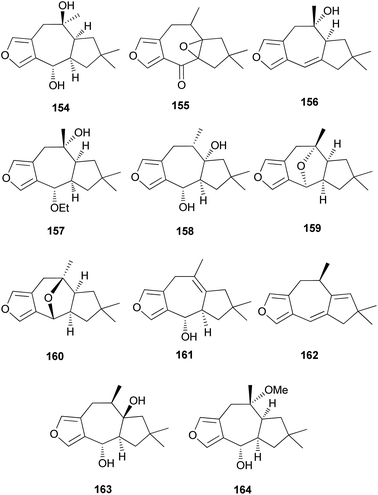
Furanosesquiterpenes are another major type of lactarane-type sesquiterpenes. 4-Epi-furandiol (154) was identified as a new compound from L. scrobiculatus.184 Furoscrobiculins A–D (155–158) and furanethers A and B (159 and 160) were also identified in L. scrobiculatus.183 The stereochemistry of 155 remains to be deduced, and 158 was identified in R. emetica.188 Additionally, other lactarane furans (161 and 162) were isolated,185,186 and the chemical structure of 158, which was reported in 1980,183 was corrected to 163 in 1994.190 Compound 163 was discovered in R. nigricans in 2017,189 and methoxyfuranalcohol (164) was identified from R. emetica.188
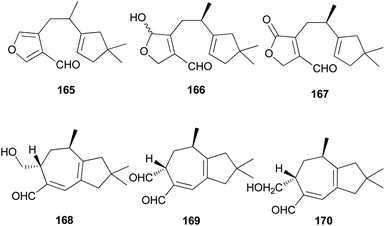
Several sesquiterpene aldehydes belonging to the lactarane-type sesquiterpenes have also been identified in L. scrobiculatus. Furthermore, lactaral (165) and lactardial (166), which are known sesquiterpene aldehydes, were isolated,183,185 and aldehydes, compound 167, and scrobicalol (168), which were not previously reported as natural products, were identified.185 A new dialdehyde, chrysorrhedial (169), and chrysorrheal (170) were also identified in L. scrobiculatus.186
2.5.1.4 Trichothecenes. Trichothecenes are produced by various fungal genera, including Fusarium, Trichothecium, Trichoderma, Myrothecium, and Stachybotrys.191 Their biosynthetic pathways have been extensively studied, which begin with trichodiene synthase catalyzing the conversion of FPP into sesquiterpene hydrocarbon (+)-trichodiene.192 This step includes the isomerization of FPP into tertiary allylic (E)-nerolidyl diphosphate and further cyclization to a bisabolyl cation through a 1Re attack of the vinylic double bond.193
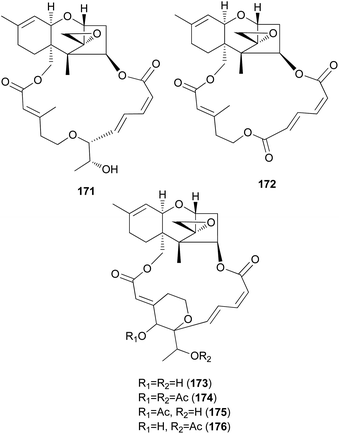
Podostroma cornu-damae, a rare deadly poisonous mushroom, is a great source for producing lethal macrocyclic trichothecene mycotoxins.194,195 Ingestion of only a small amount of this mushroom is known to cause fatal poisoning, and common symptoms include vomiting, dehydration, and diarrhea in the early stage, followed by changes in perception, hypotension, and disturbance of consciousness, ultimately leading to death with multiple organ failure.194,195 Saikawa et al. reported the isolation of macrocyclic trichothecenes, including roridin E (171), verrucarin J (172), and satratoxin H (173), from the culture broth of P. cornu-damae, and from the fruiting bodies of the same fungus, compound 173, satratoxin H 12′,13′-diacetate (174), satratoxin H 12′-acetate (175), and satratoxin H 13′-acetate (176) were isolated.196 All of the macrocyclic trichothecenes, except for 172, showed lethal effects on mice by at least 0.5 mg per capita.196
It has been reported that tri17 and tri18 participate in the biosynthetic pathways of macrocyclic trichothecenes, where tri17 is considered to act as a polyketide synthase gene and tri18 as an acyltransferase gene according to BLAST analyses and function prediction.197 The study reported the roles of tri17 and tri18 in the biosynthesis of macrocyclic trichothecenes in certain species as well as the phylogenetic trees for the two genes, providing information on gene homologs and the evolutionary relationship of the macrocyclic trichothecene-producing species; however, their genome information has not been reported in Podostroma species.197 Although genomic analysis regarding the biosynthesis of these trichothecenes still requires thorough investigation, possible biosynthetic pathways of macrocyclic trichothecenes have been predicted by a few studies, as shown in Scheme 8.197–199 The pathway begins with trichodermol reacting with polyketide chains through esterification. Further oxidation or substitution produces either verrol or trichoverrols, and the esterification of both compounds yields trichoverrins, which produce 171via dehydration, constructing a macrocycle. Compound 171 can then be modified to various other derivatives, including roridin F, roridin L2, verrucarin J, 12′-episatratoxin H, and satratoxin I.197
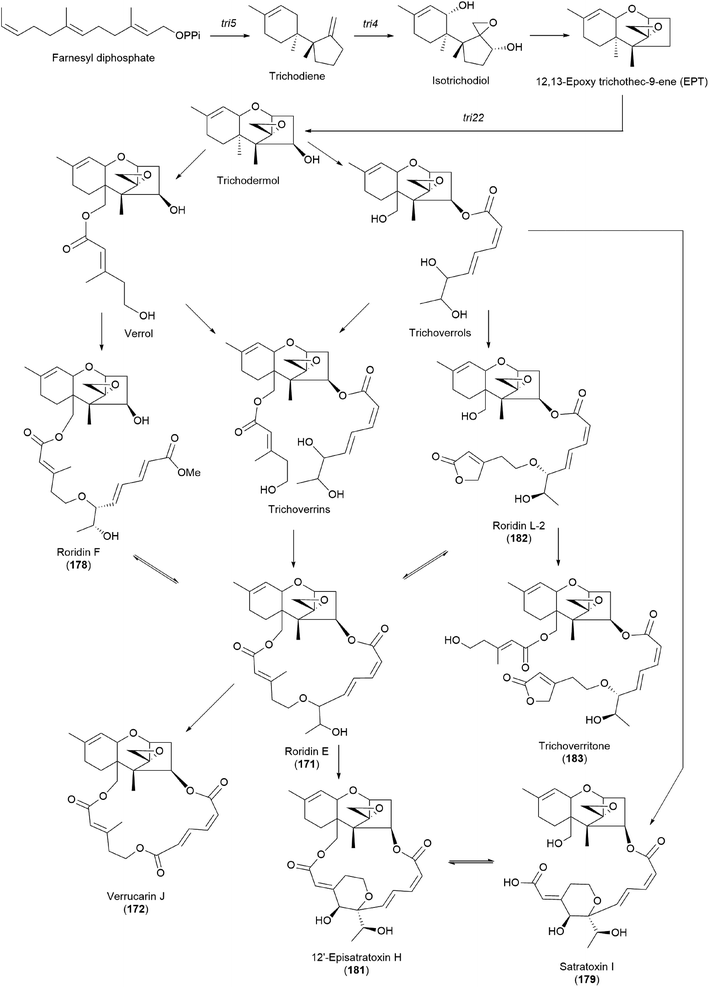 | ||
| Scheme 8 Plausible biosynthetic pathway of macrocyclic trichothecenes.197 | ||
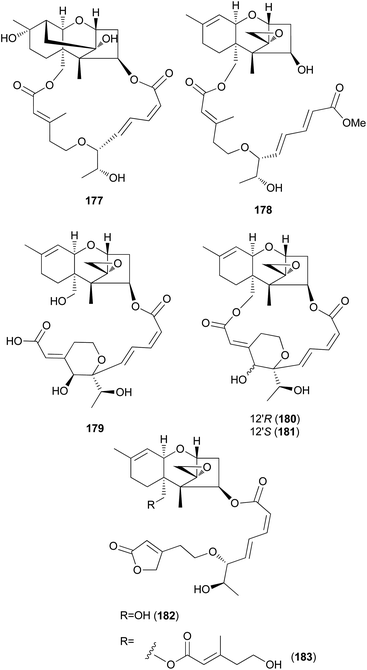
New trichothecenes, miophytocen D (177), roridin F (178), and satratoxin I (179), along with known compounds, satratoxin H (180), 12′-episatratoxin H (181), roridin L-2 (182), and trichoverritone (183), were isolated from P. cornu-damae.200 Compound 173 was previously reported without determination of its absolute configuration;196 however, Lee et al. reported the isolation of both 12′S (180) and 12′R (181) isomers.200 All the isolated compounds (177–183), including 171, were evaluated for cytotoxicity against four human breast cancer cell lines (Bt549, HCC70, MDA-MB-231, and MDA-MB-468), and 171, 181 and 183 exhibited strong cytotoxic effects against cancer cell lines, with IC50 values ranging from 0.02–80 nM, which indicated stronger effects than those of the positive control doxorubicin. Structure–activity relationship (SAR) analysis suggests that the 12,13-epoxy ring in trichothecenes plays a crucial role in toxicity, and in agreement with this, 177 was inactive against all tested cell lines (IC50 >10 μM).200
Furthermore, SAR analysis also proposed that the breakage between the macrocyclic part and the trichothecene unit can influence the potency by decreasing the toxicity, and in the case of compound 183, despite the cleavage, the presence of an additional substituent might have contributed to the cytotoxicity. Furthermore, the difference in the cytotoxicities of compounds 180 and 181 suggests the possible role of 12′-OH in potency.200
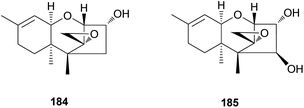
Trichothecene sesquiterpenes have also been reported from Gymnopilus junonius, formerly known as G. spectabilis, a hallucinogenic toxic mushroom.201 Isotrichodermol (184), a well-known trichothecene, was isolated along with a new trichothecene derivative, isotrichojunol (185). These compounds (184 and 185) were evaluated for their cytotoxic effects on human lung and prostate cancer cell lines A549 and PC3, respectively; both showed strong cytotoxicity, similar to that of the control doxorubicin, with IC50 values of 0.424 and 1.572 μM, respectively, against A549 cell lines, and 0.630 μM and 1.711 μM, respectively, against PC3 cell lines.201
The biosynthetic pathway of 184 has been determined in Fusarium sporotrichioides, a fungal plant pathogen responsible for damaging crops, and in several studies, the core trichothecene ring was discovered to be formed from the cyclization of farnesyl pyrophosphate by trichodiene synthase encoded by the gene Tri5 and subsequent multiple oxygenations by the enzyme TRI4.202–204
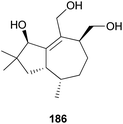
2.5.1.5 Tremulane. Tremulanes are rare sesquiterpenes, and only one type of tremulane has been reported from G. junonius, a hallucinogenic toxic mushroom.201 Tremujunol (186) was isolated as a new compound along with the trichothecenes 184 and 185 and evaluated for its cytotoxic effects against human lung and prostate cancer cell lines. The results showed that unlike trichothecenes, compound 186 did not have any significant cytotoxicity against the tested cancer cell lines at 10 μM.201
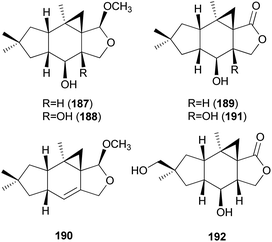
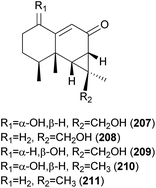
2.5.1.6 Marasmanes. Lactapiperanol E (187) was isolated as a new marasmane-type sesquiterpene from the Basidiomycete R. foetens, a poisonous mushroom possessing gastrointestinal irritants, along with a known compound lactapiperanol A (188).205 In the following study, Wang et al. described the isolation of two new marasmane-type sesquiterpenes, 8α,13-dihydroxy-marasm-5-oic acid γ-lactone (189) and 13-hydroxymarasm-7(8)-en-5-methoxy γ-acetal (190), together with a known sesquiterpene, 7α,8α,13-trihydroxy-marasm-5-oic acid γ-lactone (191).206 Another new marasmane-type sesquiterpene, russulfoen (192), along with 189 and 191, was reported from the same source by Kim et al.207 The cytotoxic activities of 189, 191, and 192 were evaluated against A549, SK-OV-3, SK-MEL-2, and HCT-15 human tumor cell lines in vitro. All three sesquiterpenes showed moderate cytotoxicity against all four cell lines tested; compound 191 exhibited cytotoxicity, with IC50 values ranging from 15.4–28.2 μM, against A549, SK-OV-3, and SK-MEL-2 cell lines (IC50 >30 μM against HCT-15); 189 exhibited cytotoxicity, with IC50 values ranging from 20.9–25.7 μM, against A549 and HCT-15 cell lines (IC50 >30 μM against SK-OV-3 and SK-MEL-2); and 192 showed cytotoxicity, with IC50 values ranging from 22.5–29.4 μM, against A549, SK-OV-3, and SK-MEL-2 cell lines (IC50 >30 μM against HCT-15).207
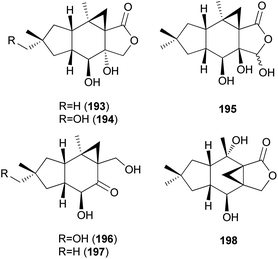
Three new marasmane sesquiterpenes, russulanigrins A–C (193–195), and a new normarasmane, russulanigrin D (196), were identified in R. nigricans, a toxic mushroom, along with known sesquiterpenes, 191, 197, and isolactarorufin (198).189 Compounds 196–198 were evaluated for antibacterial (Bacillus cereus and Enterococcus faecium), antimycobacterial (Mycobacterium tuberculosis H37Ra), antiplasmodial (Plasmodium falciparum K1), and antifungal (Candida albicans) activities as well as cytotoxicity against KB, MCF-7, and NCI-H187 cancer cell lines. However, all were found to be inactive in all assays.189
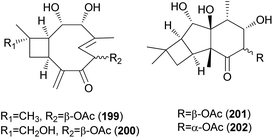
2.5.1.7 Caryophyllanes. Naematolin (199) and naematolin B (200) were reported as two new caryophyllane-type sesquiterpenes from culture filtrates of Naematoloma fasciculare (Fr.) Karst, a bitter poisonous mushroom.208 In a subsequent study, Doi et al. reported the isolation of two new additional metabolites, naematolins C (201) and G (202), which are the first examples of tetramethyltricyclo[5.4.0.02,3]undecane skeleton in naturally occurring sesquiterpenoids.209
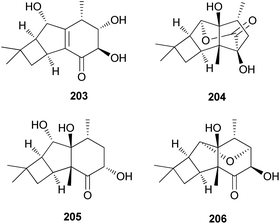
2.5.1.8 Tricyclo[5.4.0.02,5]undecanes. Fascicularones A (203) and B (204), novel tricyclo[5.4.0.02,5]undecane-type sesquiterpenoids, were isolated from the mycelial culture of N. fasciculare.210 In a subsequent study, Shiono et al. reported the identification of fascicularones C (205) and D (206).211 The structures of fascicularones resemble those of 201 and 202, all derived from N. fasciculare. The biological activity of 203–206 was examined in lettuce seedlings, wherein they accelerated radicle growth to 148%, 184%, 170%, and 188% of the control, promoting radicle elongation of lettuce seedlings.211
2.5.1.9 Aristolanes. The crude extract of Ramaria formosa, a poisonous coral fungus that causes acute gastrointestinal symptoms of nausea, vomiting, and diarrhea, has been previously reported to have antioxidant, antitumor, and cellulose activities.212–214 Kim et al. discovered that the MeOH extract of this fungus exhibits a considerable human neutrophil elastase (HNE)-inhibitory activity (IC50 = 40.6 μg mL−1), and carried out the first study on the secondary metabolites of this mushroom.215 As a result, two new aristolane-type sesquiterpenes, ramarins A (207) and B (208), were isolated, together with three known compounds, nambinone A (209), axinysone A (210), and ent-aristolone (211). Compounds 207–211 were evaluated for their inhibitory activity against HNE in vitro, and all were found to inhibit HNE by 30-35% at a concentration of 100 μM, while epigallocatechin gallate, the positive control, exhibited 60% inhibition at the same concentration.215
2.5.2.1 Labdanes. Labdane-related diterpenoids are characterized by the use of class II diterpene cyclases in their biosynthesis.217 These class II diterpene cyclases catalyze protonation-initiated bicyclization of GGDP, forming the eponymous labdadienyl diphosphate intermediate. Subsequent deprotonation of this intermediate produces copalyl diphosphate (CPP) via CPP synthases, which are the backbone of labdane diterpenes.218
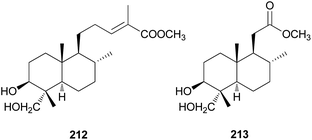
Diterpenes are uncommon in toxic mushrooms. Two new labdane-type diterpenes (212 and 213) were identified from the fruiting bodies of R. formosa, a poisonous coral fungus.219 Compounds 212 and 213 were evaluated for their inhibitory activity against human neutrophil elastase (HNE) in vitro, where both showed moderate activity, with IC50 values of 36.4 ± 1.2 μM and 40.8 ± 1.5 μM, respectively. The HNE inhibitory behavior was further characterized through a kinetic study involving different concentrations of 213 and substrate, and it was found that 213 inhibited HNE through a mixed-type noncompetitive inhibition mechanism, with a Ki of 41.5 ± 1.8 μM.219
2.5.3.1 Lanostanes. The abovementioned biosynthetic pathway simply includes the formation of lanostanoids.222–224 After OS adopts a pre-organized chair–boat–chair conformation, protonation provokes a cascade of ring-forming reactions to the protosterol cation. Subsequent rearrangement of this intermediate via a series of 1,2-hydride and 1,2-methyl groups shifts along with deprotonation, eventually producing lanosterol, which is the backbone of lanostane-type triterpenes.227
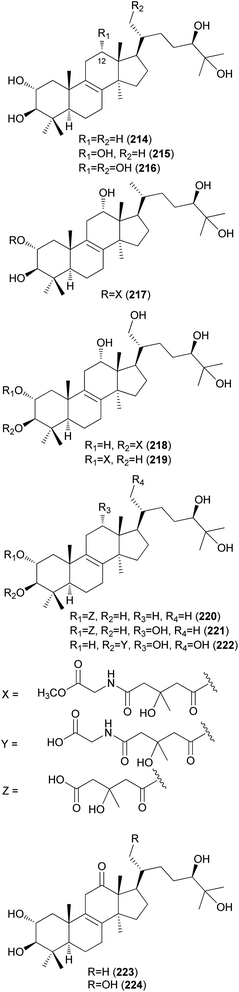
Fasciculols and fasciculic acids are lanostane-type triterpenes identified from N. fasciculare, a bitter poisonous mushroom. Fasciculols A–D (214–217) were isolated from the fruiting bodies of N. fasciculare;228,229 fasciculols B and C (215 and 216) exhibited inhibitory activity against Chinese cabbage seedlings, while fasciculol A (214) only exhibited one-fourth the potency of the inhibitory activity of the other two fasciculols.228 It was assumed that the presence of the C12-hydroxyl group would play an important role in the biological activity. Suzuki et al. identified fasciculols E and F (218 and 219) as mycotoxins of N. fasciculare, which caused paralysis and death in mice with LD50 values of 50 mg kg−1 and 168 mg kg−1, respectively.230 Compounds 215, 216, and 219 were isolated through biological activity-guided separation, and all were found to possess calmodulin inhibitory activity, with IC50 values of 21, 400, and 70 μg mL−1, respectively.231 Compound 216 also exhibited antitumor activity against MCF-7 cell line proliferation in vitro with an IC50 value of 1.54 μg mL−1, and against tumor growth in H22 implanted mice in vivo with an inhibition rate of 39.13% at a dose of 100 mg per kg per day.232 Fasciculic acids A–C (220–222) as new lanostane-type triterpenes, were isolated along with 216–218 from the same source by Takahashi et al.233 All of the isolated compounds were tested for their calmodulin antagonistic activity, using calmodulin-sensitive and calmodulin-insensitive phosphodiesterases (PDE). Fasciculic acid B (221) exhibited the strongest inhibitory activity on both PDEs with an IC50 value of 6 μM, and fasciculic acid A (220) specifically inhibited the activity of calmodulin-sensitive PDE with an IC50 value of 10 μM. The inhibitory activity of 221 was approximately 10 times stronger than that of a well-known calmodulin antagonist.233 Compounds 216, 217, and 222 also showed inhibition but with low selectivities.233 Until 1993, the chirality of the 3-hydroxy-3-methylglutaric acid (HMGA) moiety of 220 was not determined. By comparing the 1H NMR spectral analysis of L-alanylamides prepared from the condensation of 220 and synthetic HMGA, the absolute configuration at C-3′ of the HMGA moiety was established as S, whereas that of synthetic HMGA was found to be R.234 Based on its biosynthesis as well as previous reports, naturally occurring HMGA has the S configuration.235 Fasciculols H and I (223 and 224) were identified from N. fasciculare in 2011, along with 222,236 where compounds 222 and 223 weakly inhibited the growth of human glioma cell line U87 with inhibition rates of 24.5% and 13.3%, respectively.236
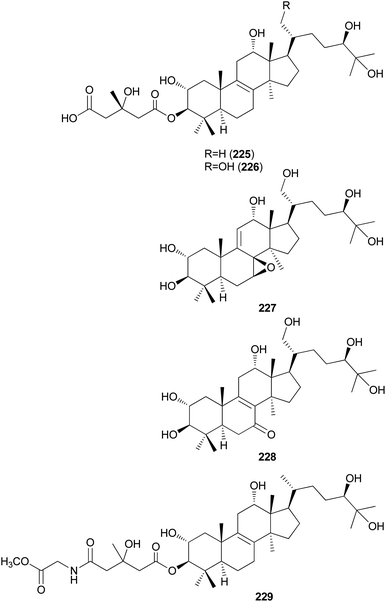
Kim et al. isolated fasciculols J–M (225–228) from N. fasciculare in 2013 as new lanostane-type triterpenes, and tested their antiproliferative activities against four human cancer cell lines, A549, SK-OV-3, SK-MEL-2, and HCT-15.237 Fasciculol G (229) was also isolated and tested for antiproliferative activities, and its identification was first reported by Ikeda et al.,238 but without full NMR data assignment. Compounds 227, 228, and 229 showed cytotoxic activity against all four human cancer cell lines, and 227 and 228 exhibited significant cytotoxic activity against A549, SK-OV-3, SK-MEL-2, and HCT-15 cell lines (IC50 = 6.59, 7.08, 8.26, and 8.53 μM, and IC50 = 3.99, 7.36, 4.77, and 8.50 μM, respectively). Compound 229 exhibited selective cytotoxicity against the SK-MEL-2 cell line, with an IC50 value of 8.60 μM.237
2.5.3.2 Cucurbitanes. The biosynthesis of cucurbitane-type triterpenoids also begins with OS (2,3-oxidosqualene) as in lanostane; however, through the catalysis of the enzyme cucurbitadienol synthase, OS forms the cucurbitadienol triterpenoid skeleton, where the major difference with lanostanes includes the shift of a methyl group from 10-position to 9-position. Various modification steps result in the production of a wide variety of cucurbitane derivatives.239
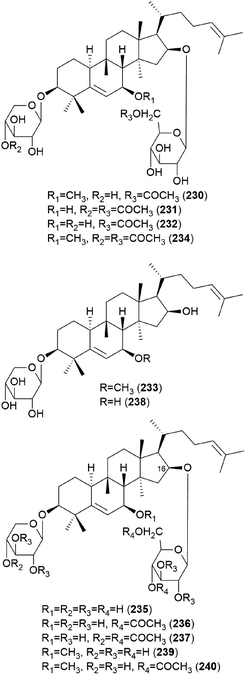
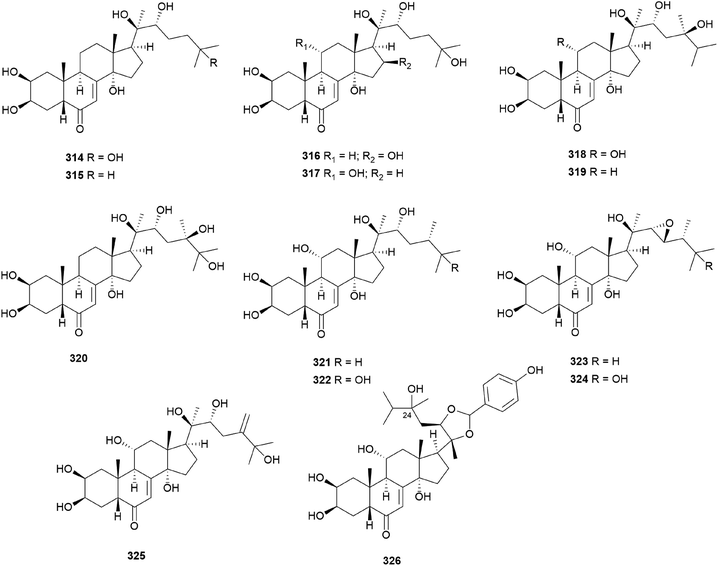
In 1986, hebevinosides I–V (230–234) were isolated as new toxic cucurbitane-type triterpenes from Hebeloma vinosophyllum, a poisonous mushroom that is neurotropically and lethally toxic to mice.240 Intraperitoneal administration of 50 mg kg−1 of 230 and 234, or 100 mg kg−1 of 231 and 232, to mice led to death after paralysis within 72 h. However, compound 233 did not cause paralytic death in mice at a dose of 200 mg kg−1.240 In the following study in 1987, Fujimoto et al. reported the identification of hebevinosides VI–XI (235–240) in H. vinosophyllum.241 The administration of 100 mg kg−1 of hebevinoside VI (235) to mice resulted in death after paralysis; however, administration of 50 mg kg−1 of 235 led to survival of the mice. The LD50 value of 235 after intraperitoneal administration was 66 mg kg−1. The presence of a glucosyl moiety at C-16 was revealed to be indispensable for the toxicity of the compound, and compounds with methoxyhebevinogenin aglycone were proven to be artifacts produced from the use of methanol for extraction.241
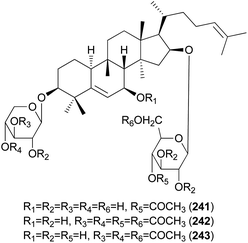
Hebevinosides XII–XIV (241–243) were isolated from H. vinosophyllum in 1991, along with 231, 232, and 235–237.242 These compounds were studied for their productivity at three growth stages: mycelium (stage I), mycelium with primordia (stage II), and mycelium with fruit-bodies (stage III). Compounds with acetyl groups at O-4 in glucosyl and/or O-3 in the xylosyl moieties (236, 237, and 241–243) decreased and those with acetyl groups at O-6 in the glucosyl and/or O-4 in the xylosyl moieties (231 and 232) increased overtime throughout stages I-III. In contrast, compounds with no acetyl group (233) were independent of the growth stages.242
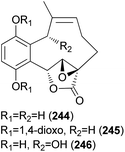
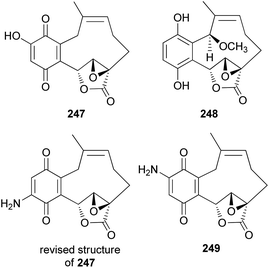
2.5.4.1 Hydroquinone and benzoquinone meroterpenoids. Clavilactones A–C (244–246) have been isolated as new meroterpenoids having a 10-membered carbocycle connected to a hydroquinone and an α,β-epoxy/unsaturated lactone obtained from cultures of the toxic mushroom C. clavipes.248 A possible biosynthetic origin of 244 was predicted to start with geranylhydroquinone, which requires several oxidation steps (Scheme 9), including the oxidation of benzylic C-6 and 8-Me, oxidation of the C-7–C-8 double bond, and activation of the terminal methyl group followed by lactonization and ring closure. Compound 245 exhibited antibacterial activity against Bacillus subtilis, B. cereus, Sarcinea lutea, and Saccharomyces cerevisiae (50 μg per disc), while 244 was only active against B. subtilis at 100 μg mL−1. Compound 245 also inhibited the growth of Lepidium sativum. Compounds 244–246 were found to have antifungal activity against Cladosporium cladosporioides and C. cucumerinum, with concentrations as low as 50 μg per plate.248
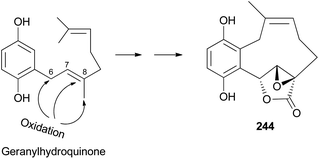 | ||
| Scheme 9 Plausible biosynthetic pathway of clavilactone A (244).248 | ||
New meroterpenoid analogs, clavilactones D and E (247 and 248), were also isolated from the poisonous mushroom C. clavipes, and 247 was found to be a strong specific inhibitor of tyrosine kinases, such as epidermal growth factor receptor (EGFR), with an IC50 value of 5.5 μM in the kinase assay.249 In addition, it exhibited selective antiproliferative activity against the A431 cell line in another study.250
Cassinelli et al. performed kinase assays using compounds 244, 245, and 247.251 All three compounds were also tested for antiproliferative effects in human cell lines A431, IGROV-1, and SKOV-3, and 247 was found to be the strongest inhibitor, with IC50 values of 0.1 μM, 0.8 μM, and 1.0 μM, respectively, against each cell line. Compounds 244, 245, and 247 were further evaluated for their ability to inhibit the activity of protein tyrosine kinases against Ret/ptc1 and EGFR tyrosine kinases, and 247 was again found to be the strongest inhibitor, with IC50 values of 2.8 μM and 5.5 μM, respectively.251 Additionally, 245 was revealed to be a noncompetitive inhibitor of EGFR, which does not compete with ATP or polyGAT substrate (IC50 = 27 μM). Cassinelli et al. ultimately reported the discovery of clavilactones A, B, and D, representing prototypes of a new structural class of tyrosine kinase inhibitors.251 The total synthesis process of the EGFR tyrosine kinase inhibitors, 244, 245, and 247, as well as the revision of the structure of 247, was reported by the Takao group.252 Another new meroterpenoid analog, clavilactone F (249), was isolated from C. clavipes in 2019 by Sun et al., along with 244 and 247.253 They were evaluated for antiproliferative activities against human cancer cells HepG2 and A549, as well as against normal cell line GES-1; however, they did not show any significant activity (249: IC50 = 18.22–33.27 μM, 244: IC50 = 32.14 μM against HepG2, IC50 >50 μM and >100 μM against A549 and GES-1, respectively, and 247: IC50 = 9.32 μM–38.13 μM).253
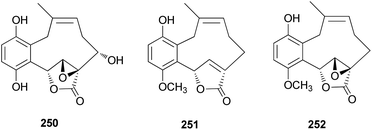
From the fruiting bodies of the Basidiomycete C. clavipes, new meroterpenoids with the same skeleton, clavilactones G–I (250–252), were identified in 2019 by Sun et al. All the meroterpenoids were evaluated for their cytotoxic activity against three human tumor cell lines, HeLa, SGC-7901, and SHG-44.254 Clavilactone H (251) showed moderate cytotoxicity against HeLa and SGC-7901 cell lines, with IC50 values of 23.5 and 14.5 μM, respectively.254
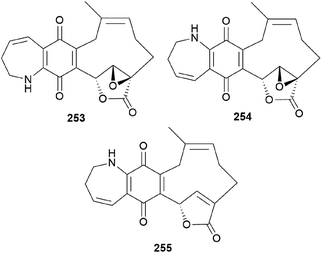
Clavipines A–C (253–255) composed of a benzoquinone fused to an azepine ring and a ten-membered carbocycle with α,β-epoxy/unsaturated-γ-lactone were isolated from the poisonous mushroom C. clavipes along with 244, 247, and 249 by the Sun group.253 Clavipine A (253) showed significant cytotoxic activity against A549 and HepG2 cells with IC50 values of 7.49 ± 0.41 and 4.28 ± 0.26 μM, respectively.253
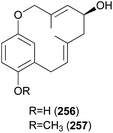
Clavipols A and B (256 and 257) were identified as new meroterpenoids with a 12-membered ether ring from the same mushroom, along with 250–252, and were also tested against HeLa, SGC-7901, and SHG-44 tumor cell lines. Compounds 256 and 257 exhibited weak cytotoxicity against HeLa (IC50 = 63.2 ± 0.43 μM and 38.6 ± 1.3 μM, respectively) and SGC-7901 cell lines (IC50 = 44.1 ± 0.5 μM and 51.2 ± 0.7 μM, respectively).254
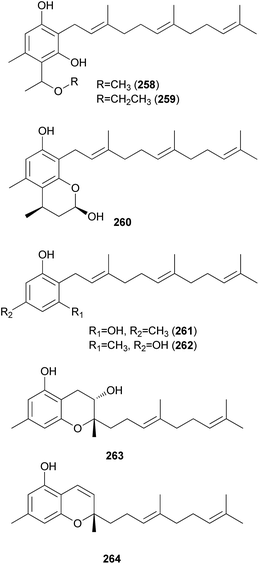
2.5.4.2 Phenolic and chromene meroterpenoids. Grifolin derivatives (258–260), another type of meroterpenoid, were isolated as new compounds from the poisonous mushroom B. pseudocalopus, along with known analogs (261–264), including grifolin (261) and neogrifolin (262).143 All of the derivatives except 263 exhibited anticancer activities against human lung carcinoma A549 and mouse melanoma B16F1 cell lines, with IC50 values in the range of 3.5–11.0 μg mL−1. Additionally, all compounds (258–264) showed moderate radical-scavenging activities, with IC50 values in the range of 11.1–75.9 μg mL−1.143
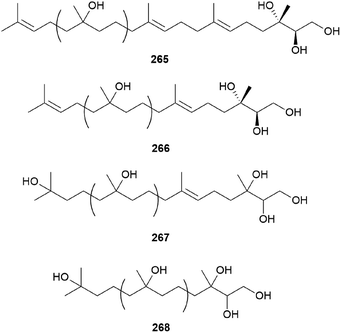
2.5.5 Miscellaneous terpenes. Gymnoprenols A and B (265 and 266) were identified from the hallucinogenic mushroom G. spectabilis, as a novel type of polyisoprenepolyols with 45–60 carbon atoms.255 Gymnoprenols D and E (267 and 268) were also isolated from the same mushroom in the following year by the same group.256 The structures of these polyisoprenepolyols were found to occur as a mixture of isoprene homologs, and the stereochemistry of the gymnoprenols were not determined at the time of isolation. However, the correct structure of 265 and the absolute configurations of 265 and 266 were determined in a later study.257,258
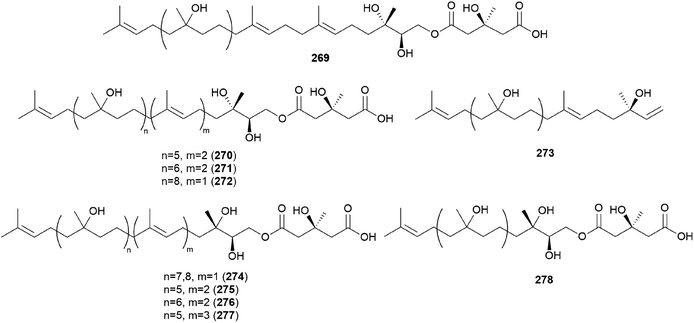
Gymnopilin (269), a polyisoprenoid with a 3-hydroxy-3-methylglutaric acid unit, was identified as a bitter principle from the hallucinogenic mushroom G. spectabilis by the Nozoe group.259 In the same year, gymnopilins A9, A10, and B11 (270–272) were also identified as the main bitter principles of G. spectabilis by the Aoyagi group.260 Gymnopilene (273), a novel polyisoprenol, was isolated in 1991,67 and a new series of gymnopilins 274–277 were identified in 1993 by Tanaka et al.261 Gymnopilin K (278), a new cytotoxic gymnopilin, was isolated from G. spectabilis in 2012 by the Kim group together with 265, 270, 271, and 273.257 The isolated compounds (265, 270, 271, 273, and 278) were investigated for their cytotoxic activities against four human cancer cell lines, including SK-OV-3, SK-MEL-2, A549, and HCT-15, and all the compounds exhibited activity against proliferation of the tested cell lines with IC50 values ranging from 3.26–26.30 μM.257 Kim et al. also reported the structure–activity relationship (SAR), assuring the contribution of the methyl (S)-3-hydroxy-3-methylglutarate moiety to the cytotoxicity of a molecule, based on compounds with this moiety (270, 271, and 278) having more significant cytotoxicity compared to those without the moiety (265 and 273).257
2.6 Steroids
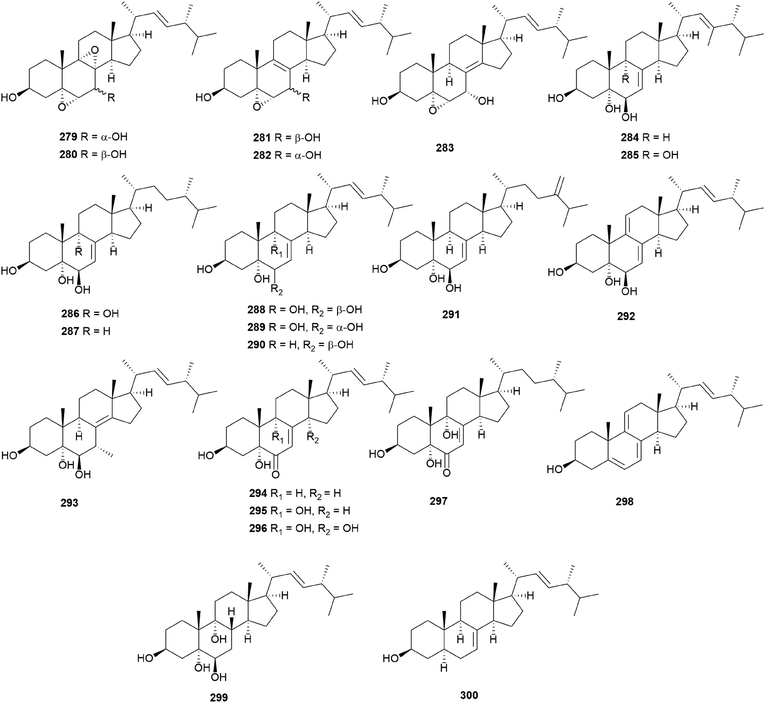
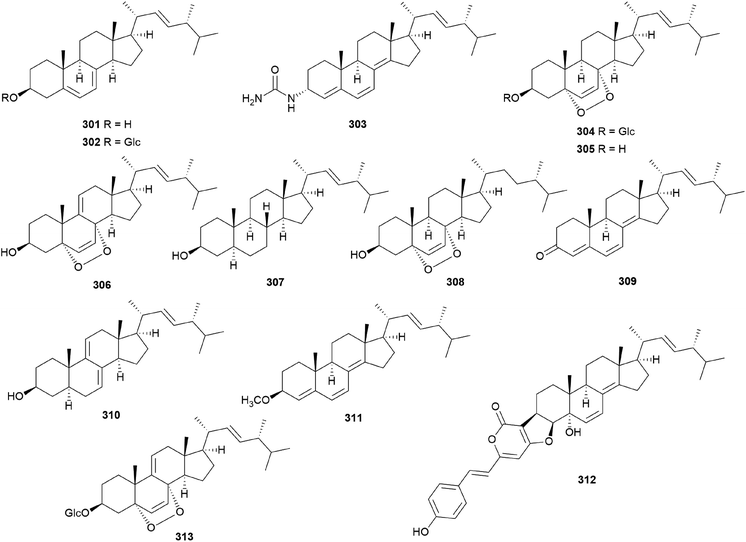
In eukaryotes, steroids are formed through the mevalonate pathway, where acetyl-CoA is used as a building block. Biosynthetically, they are derived from triterpenes, and their characteristic is the lack of methyl groups at 28- and 29-positions in the lanostane and cycloartane backbones.262 In the first step of the pathway, acetyl-CoA undergoes Claisen-type condensation mediated by acetoacetyl-CoA thiolase to form acetoacetyl-coenzyme A. This reaction is followed by the action of (S)-hydroxy-3-methylglutaryl-coenzyme A synthase (HMGS), yielding HMG-CoA, which is converted into mevalonic acid via a reduction reaction mediated by (S)-hydroxy-3-methylglutaryl-CoA reductase (HMGR). The enzymes mevalonate kinase, mevalonate-5-phosphate kinase, and mevalonate-5-phosphate decarboxylase convert mevalonic acid to isopentenyl pyrophosphate (IPP), which is then converted into the allylic isomer dimethylallyl pyrophosphate (DMAPP). The continuous ‘head-to-tail’ 1′4-condensation of IPP and DMAPP by farnesyl pyrophosphate synthase yields farnesyl pyrophosphate (FPP), which acts as a precursor for steroidogenesis.263,264Ergosterol (301) as well as cerevisterol (290) were isolated from the poisonous mushrooms Russula foetens and Paxillus involutus.95,207 Additionally, 290, 301, ergosterol 3-O-β-D-glucopyranoside (302), (22E,24R)-3α-ureido-ergosta-4,6,8(14),22-tetraene (303), 5α,8α-epidioxy-(22E,24R)-ergosta-6,22-diene-3β-ol 3-O-β-D-glucopyranoside (304), and ergosterol peroxide (305) were isolated from Chlorophyllum molybdites,271,272 which was previously known as Lepiota morgana or the “green parasol” mushroom273 and is known to cause poisoning with symptoms such as nausea, vomiting, diarrhea, and abdominal pain showing in a couple of hours.274,275 Interestingly, compound 303 consists of an unusual urea junction at C-3, and both 303 and 304 were new compounds isolated from the toxic mushroom C. molybdites. Compound 305, one of the most abundant ergostane-type steroids in mushrooms, has been isolated from several toxic mushrooms, including Russula subnigricans,268Russula foetens,95,207 and Gymnophilus spectabilis.49,132,135,276 It was also reported to be isolated from the toxic mushroom Amanita subjunquillea, together with 9,11-dehydroergosterol peroxide (306),269 and the ergostane-type steroids 305 and 306 were isolated from Macrolepiota neomastoidea, along with compound 283 and brassicastanol (307).277 Extensive data regarding the toxicity of M. neomastoidea have not been reported, but there have been cases where ingesting the mushroom caused nausea, vomiting, diarrhea, and abdominal pain.278,279 The ergostane-type steroids 305 and 306, as well as their related analog 308, were also identified in Amanita pantherina.265
Other ergostane-type steroids 309-312 were isolated from the hallucinogenic mushroom Gymnophilus spectabilis, recently revised to be G. orientispectabilis, along with 290, 301, and 308.49,132,135,276 Interestingly, the styrylpyrone-fused ergosterol derivative ergopyrone (312) was isolated and identified as a novel compound, which is composed of a unique hexacyclic 6/5/6/6/6/5 skeleton that is formed from ergosterol and styrylpyrone precursors via [3 + 2] cycloaddition. The theoretical biosynthesis of 312 was proposed to involve the unique conjunction of bisnoryangonin (100) and ergosterol (301) (Scheme 10). Compound 301 accepts a molecular oxygen at the C-5 position to produce hydroperoxide intermediate i, which is further oxidized to synthesize ergosterol peroxide derivatives. Peroxide degradation of i produces C-5 hydroxylated intermediate ii, and subsequent dehydration at C-3 and C-4 yields C-3/C-4 unsaturated intermediate iv. Another pathway leading from 301 to ivvia simple oxidation and isomerization is also possible. The enol moiety in 100 and the olefinic functionality at C-3 and C-4 in iv yield carbon–carbon/carbon–oxygen-coupled product 312via the [3 + 2] cycloaddition reaction. The sterol 310 is produced by simple isomerization step(s) from 301, whereas sterols 309 and 311 are generated by oxidation and methylation, respectively, from iii, the oxidation product of 301. Another ergostane-type steroid, (22E,24R)-5α,8α-epidioxyergosta-6,9,22-triene-3β-ol 3-O-β-D-glucopyranoside (313), along with 288, 290, 301, 304, and 305, were isolated from the toxic mushroom Naematoloma fasciculare.232,280–282
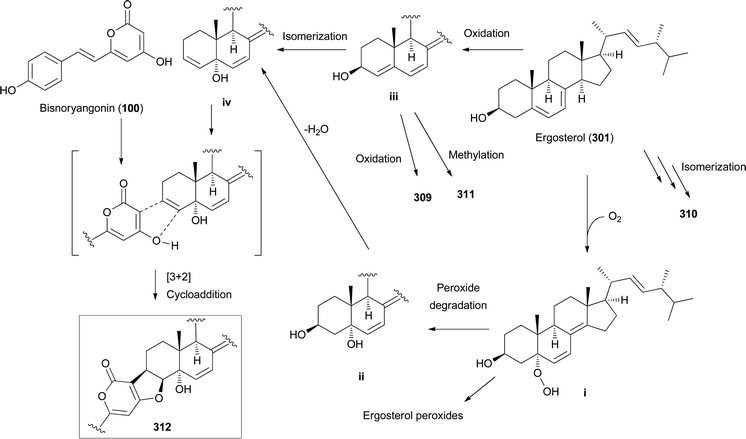 | ||
| Scheme 10 Plausible biosynthetic pathway of ergopyrone (312) and other sterols 309–311, as proposed by Lee et al.135 | ||
Regarding the bioactivities of the reported ergostane-type steroids from toxic mushrooms, the metabolites 283 and 305–308 were tested for their cytotoxicity against cancer cell lines A549, SK-OV-3, SK-MEL-2, and HCT-15.277 Compound 283 exerted cytotoxic effects on A549, SK-OV-3, SK-MEL-2, and HCT-15, with IC50 values of >30.0, 16.5, >30.0, and >30.0 μM, respectively. Compounds 305–308 showed cytotoxicity against A549, SK-OV-3, SK-MEL-2, and HCT-15 (305: IC50 = 14.0, 17.9, 12.7, and 10.0 μM, respectively; 306: IC50 = 25.6, 17.5, 11.8, and 17.1 μM, respectively; 307: IC50 >30.0, >30.0, >30.0, and 28.3 μM, respectively; 308: IC50 >30.0, 27.20, 23.49, and >30.0 μM, respectively), compared to the control doxorubicin (IC50 = 0.16, 0.38, 0.04, and 0.82 μM, respectively for A549, SK-OV-3, SK-MEL-2, and HCT-15).277 In addition, 301 and 305 displayed a strong anticomplementary activity in the classical pathway, with IC50 values of 0.1 and 5.0 μM, respectively.280 The ergostane-type steroids 288, 304, and 313 were also tested for their cytotoxicity toward four cancer cell lines, namely SK-OV-3, SK-MEL-2, A549, and HCT-15. Compound 288 showed cytotoxicity against SK-OV-3, SK-MEL-2, A549, and HCT-15 with IC50 values of 12.16, 10.39, 10.83, and 13.20 μM, respectively. Compounds 304 and 313 showed moderate cytotoxicity against A549, SK-OV-3, SK-MEL-2, and HCT-15 (304: IC50 = 14.21, 15.11, 9.01, and 17.49 μM, respectively; 313: IC50 = 15.42, 18.26, 12.96, and 19.32 μM, respectively), compared to the control doxorubicin (IC50 = 0.02, 0.01, 0.01, and 0.13 μM, respectively).281 The ergostane-type steroids 290, 301, 302, 304, and 305 were tested for their cytotoxicity against two human cancer cell lines, A549 and Panc-28; however, all the tested sterols did not show any activities (GI50 >50 μM).271 The ergostane-type steroids 309–312 were investigated for their effect on lipid metabolism using 3T3-L1 adipocytes by the measurement of mRNA levels of the four related genes, namely Fabp4, Adipsin (biomarkers), ATGL (lipolytic gene), and SREBP1 (lipogenic gene). Compounds 309, 310, and 312 showed selective upregulation among the tested genes (up to 2-fold), while 311 showed upregulated expression of all the tested genes at concentrations of 2.5 μM and 10 μM, dose-dependently (∼5.5-fold for Fabp4 and Adipsin, ∼3.2-fold for ATGL, and ∼1.5-fold for SREBP1); this suggested that the metabolite 311 can promote the adipogenesis of 3T3-L1 preadipocytes.135 The ergostane-type steroids 303–305 were examined for their cytotoxic activities against gastrocarcinoma-causing Kato III cells, where all showed significant cytotoxicity against gastrocarcinoma with IC50 values of 7.60, 14.27, and 2.69 μg mL−1, respectively (positive control, hinokitiol: IC50 = 0.6 μg mL−1).272 In addition, 305 and 308 were tested for their inhibitory effects on NO production in LPS-activated BV-2 cells, with IC50 values of 2.73 and 23.95 μM, respectively, compared to the control NMMA (IC50 = 18.29 μM).49
2.7 Alkaloids
Alkaloids were first defined as alkaline compounds from plants exhibiting various biological activities.289 This definition has been amended several times to include secondary metabolites from other natural sources (outside the plant kingdom), which share similar biosynthetic genes with alkaloids. Considering their broad definition, alkaloids are nitrogen-containing metabolites synthesized as a result of secondary or specialized metabolism. Alkaloids, usually referred to as structurally complex alkaloids, are compounds with nitrogen atoms originating from amino acids and sometimes have heterocyclic moieties. The biosynthetic precursors of alkaloids are mainly derived from amino acids, including lysine, ornithine, tyrosine, phenylalanine, tryptophan, aspartic acid, histidine, and anthranilic acid. In addition, some building blocks derived from terpenes or the acetate pathway are frequently incorporated into the structures of alkaloids. A variety of alkaloid biosynthesis pathways have been identified so far, but they remain complex and unclear.Some non-protein amino acids often provide toxic substances to mushrooms; the toxins are then used to protect the mushrooms against predation. The amino acid biosynthetic pathway in fungi is known to be identical or similar to that in bacteria and plants.
2.7.1.1 Isoxazole amino acids. Amanita muscaria is a hallucinogenic and poisonous mushroom that causes symptoms including nausea, drowsiness, auditory and visual distortions, mood changes, euphoria, relaxation, and ataxia depending on the amount ingested per body weight.290 Ibotenic acid (327) and muscimol (328) from A. muscaria were identified as the main components, structurally containing the isoxazole moieties.290 The synthesis of 328, a major active hallucinogenic compound, from 327, is easily triggered by low pH, high temperature, exposure to light, and digestion and during the purification of 327.33 Although the structures of compounds 327 and 328 were elucidated by the analysis of their physical and spectroscopic data in 1964, their biosynthetic pathway and origin were uncovered. Based on the co-isolation of compounds, the Eugster group assumed that 3-hydroxyglutamate is a biosynthetic precursor for the formation of 327.291 However, 3-hydroxyglutamate has not been isolated and identified from A. muscaria. Thus, the biosynthetic pathway and origin of 327 and 328 remain ambiguous.
Recently, Muller et al. investigated the biosynthesis of 327 and 328 from at least three Amanita species and found the ibo genes to be responsible for the production of two toxic metabolites (Scheme 11).292 A glutamate hydroxylase gene is surrounded by six biosynthetic genes; the genes are linked to each other and are involved in the production of 327 and 328, as demonstrated by recent genomic and transcriptomic data (Scheme 11).292
 | ||
| Scheme 11 Proposed biosynthetic pathway of ibotenic acid (327) and muscimol (328). IboH: hydroxylase; IboA: adenylating; IboG: PLP; IboC: cytochrome P450; IboF: FMO; IboD: decarboxylase.292 | ||
A. muscaria has been reported to exert mycoatropinic effects; symptoms of poisoning include dizziness, confusion, tiredness, and auditory esthesia. These symptoms are attributable to 327 and 328 because their chemical structures are highly similar to those of glutamic acid and γ-aminobutyric acid (GABA), respectively.293,294 Compound 327 mainly acts as an agonist of N-methyl-D-aspartate (NMDA) and 1-aminocyclopentane-trans-1,3-dicarboxylic acid receptors in the central nervous system; thus, the symptoms related to ibotenic acid poisoning are associated with control and perception.293,294
The binding of compound 327 leads to the accumulation of Ca2+ in neuronal cells, which results in cell death. Additionally, increased concentrations of Ca2+ stimulate the phosphorylation of multiple kinases in neuronal cells. The activated kinases start to produce reactive oxygen species, which profoundly impair surrounding tissues.293,294 Compound 328 is a strong agonist of the receptor for the principal inhibitory neurotransmitter of the brain, GABAA.295 The receptors are widely dispersed in multiple brain regions, such as the cerebellum, cerebral cortex, and hippocampus. Administration of 328 alters various neuronal responses, leading to depressant effects, sedative effects, euphoria, and delirium.295
Muscazone (329) is another related metabolite isolated from European specimens of A. muscaria.296 It is synthesized via the breakdown of 327 by UV radiation. Compound 329 exhibits relatively minor physiological activity, compared with 327 and 328.297
2.7.1.2 Cyclopropyl amino acids. Coprine (330) was identified as a mycotoxin, which harbors the first discovered cyclopropanone group in natural products, from Coprinopsis atramentaria in 1975.46,47Coprinopsis atramentaria, commonly known as inky cap, is an edible mushroom that can be poisonous when combined with alcohol. Since the poisoning of C. atramentaria is similar to that of disulfiram, it was anticipated for a long time that disulfiram was the main active compound in C. atramentaria.
The mechanism of action of toxicity was investigated in 1979.298 If consumed with alcohol, 330 hampers ethanol metabolism in liver and induces the elevation of acetaldehyde in the blood, resulting in symptoms of coprine intoxication, including facial reddening/flushing, nausea, vomiting, agitation, headache, and salivation.299 Compound 330 is not a direct inhibitor of acetaldehyde dehydrogenase, considering its in vitro experimental results. When administered, 330 undergoes hydrolysis to produce glutamic acid and 1-aminocyclopropanol, which is quickly converted into cyclopropanone hydrate (Scheme 12).298,300 The cyclopropanone hydrate easily forms a covalent bond with the thiol group of acetaldehyde dehydrogenase, which causes the deactivation of dehydrogenase activity. The concentration of acetaldehyde in the body increases if ethanol is consumed. However, the symptoms of coprine poisoning can be alleviated if no more alcohol is ingested because the covalent bond between cyclopropanone hydrate and acetaldehyde dehydrogenase is reversible.298,300
 | ||
| Scheme 12 Mechanism of action for coprine (330) intoxification.298,300 | ||
2-Amino-3-cyclopropyl-butanoic acid (331) was identified from the toxic mushroom Amanita castanopsidis Hongo,301 widely distributed in China, Japan, and Korea in deciduous forests. It displayed substantial inhibitory effects on root elongation of lettuce (Lactuca sativa) at 10−4 M. The inhibition of root elongation by the treatment of 331 was mainly due to the reduction in the mechanical extensibility of cell walls.302 In 2002, Shibata et al. determined the absolute configuration of 331 to be (2S,3S) via racemic and enantioselective synthesis coupled with chelate-enolate Claisen rearrangement.303
2.7.1.3 Alkyne-containing amino acids. Since 1978, more than 400 healthy people in Yunnan Province, located in the south of China, died suddenly without manifest causes. Approximately 90% of the sudden unexplained death (SUD) situations in Yunnan Province occurred from June to September.304 In 2005, epidemiologic research suggested that mushroom picking and eating were the main risk factors; investigation from 2006–2009 indicated consumption of an unidentified mushroom, which was determined to be Trogia venenata, as the cause of the SUD incidents.304 This is similar to the symptoms of rhabdomyolysis.304 Liu et al. isolated and structurally elucidated 2R-amino-4S-hydroxy-5-hexynoic acid (332) from the fruits of T. venenata and proposed that metabolite 332 could be a reason for the SUD incidents.305 They also synthesized metabolite 332 for toxicological studies. Mice administered with the compound showed an increase of 1.1–1.6-fold in serum creatinine kinase level, and analysis of a victim of the Yunnan SUD incident confirmed the existence of 332 in the blood.305 Additionally, the crude extract of T. venenata exhibited severe hypoglycemia (median 0.66 mmol L−1 glucose) in the blood of mice after injection via oral exposure within 2 h. Hypoglycemia triggers the depletion of ATP and neuronal cell death, which may lead to death in humans and experimental animals.306,307
Another fatal poisoning incident of Amanita abrupta was found in Japanese forests in autumn, although its symptoms and mechanism of action have not been documented. Amanita abrupta grows on the ground, typically solitary, in mixed conifer and deciduous forests, usually during autumn. In 1986, Hashimoto et al. described some toxic effects of an aqueous extract derived from the fruiting bodies of A. abrupta in mice,308 where the extract was intraperitoneally injected into male mice; after 6 h, it was found that concentrations of serum glucose and liver glycogen decreased to 60% and 10%, respectively, compared with those of the control levels. In addition, enzymatic activities of serum glutamic oxaloacetic transaminase and glutamic pyruvic transaminase were elevated 3-and 8 folds, respectively. Hashimoto et al. identified 2-amino-4-pentynoic acid (333) from A. abrupta and reported that metabolite 333 showed toxic effects similar to those associated with extracts of A. abrupta. They hypothesized that 333 may affect the inhibitory effect of gluconeogenesis and/or enhancement of glycolysis because of the inhibition of β-oxidation of fatty acids.
2.7.1.4. Chloride-containing amino acids. An interesting chloride-containing amino acid, 2-amino-5-chloro-4-pentenoic acid (334), was isolated from the toxic mushroom A. castanopsidis along with 331.301 Compound 334 was considered to exist as a racemic mixture based on its zero optical rotation value; thus, its absolute configuration has not been identified.301 In the same way as compound 331, metabolite 334 at 10−4 M substantially inhibited root elongation, while it had no effect on hypocotyl elongation in the growth experiments of L. sativa.301 In 1987, Nozoe et al. discovered physiologically active compounds from A. abrupta, which led to the isolation of another chlorohydrin amino acid, (2S,4Z)-2-amino-5-chloro-hydroxy-4-hexenoic acid (335). The bioactivity of metabolite 335 has not yet been investigated.309
2.7.1.5 Heterocyclohexanone-containing amino acids. The structurally rare amino acids with 6-oxo-pyran-2-carboxylic acid, L-stizolobic acid (336), and L-stizolobinic acid (337), were isolated from the fruits of the poisonous mushroom, Clitocybe acromelalga, which belongs to the basidiomycete fungus distributed in Japan and found to be a toxic mushroom in 1918.310 The mushroom was known to cause severe allodynia and burning pain that is noticeable by the presence of reddish edema at the terminal parts of hands and feet although these are not fatal on the human body. The structurally related metabolites, acromelic acids A (338), B (339), and C (340) were also isolated from C. acromelalga, and the metabolites 338–340 were recognized as the main toxic metabolites because of their strong neuroexcitatory and neurotoxic properties.311,312
The co-isolation of the metabolites 336–340 from C. acromelalga strongly indicated that the pyridone group of compounds 338–340 was biosynthetically synthesized from L-DOPA.310Scheme 13 shows the proposed biosynthetic pathway for the formation of 338. Compound 337 is converted into a pyridine amino acid intermediate and then condensed with glutamic acid, followed by cyclization and decarboxylation to produce 338.310
 | ||
| Scheme 13 Biosynthetic pathway of acromelic acid A (338).310 | ||
Compounds 338–340 containing the pyrrolidine dicarboxylic acid moiety found in kainic acid displayed strong depolarizing and neurotoxic activities by affecting the kainite receptors.313 Rats treated with a single dose administration of the metabolites 338 exhibited various abnormal behaviors, including a tonic extension of the hind limbs, severe tonic/clonic seizures, temporary flaccid paraplegia, and strong spastic paraplegia for a long time.314 These behavioral symptoms of 338 in rats may clarify the allodynia in humans by the consumption of C. acromelalga.
2.7.1.6 Cyano-containing amino acids. Two rare α-amino acids with a cyano group, β-cyano-L-alanine (341) and N-(γ-L-glutamyl)-β-cyano-L-alanine (342), were identified from the toxic mushroom C. acromelalga.315 Compound 341 has been isolated in Vicia species, and C. acromelalga is the first example of the isolation of 341 from a mushroom.315 The metabolite 341 has been recognized as a neurolathyrogen, exhibiting irreversible tremors, hyperactivity, and rigidity.315 Considering these bioactivities, the poisonous symptoms caused by the ingestion of C. acromelalga might be attributable to 338–340 as well as cyano-containing amino acid (341). In 2000, Hiroshi et al. reported the identification of 341 from Vibrio sp.316 They found that 341 inhibited the growth of some cyanobacteria at a concentration of 0.4–25 μg mL−1.316
2.7.1.7 Thiol-containing amino acids. In 1992, Shigeo et al. isolated a diastereomeric mixture of 2-amino-3-(1,2-dicarboxyethylthio)propanoic acid (343) containing a thiol group from the toxic mushroom Amanita pantherine, and metabolite 343 showed an antagonistic effect on NMDA-sensitive glutamate receptors in rat brain and spinal motoneurons.317 Shigeo et al. isolated four diastereomeric mixtures (L-A, L-B, L-C, and L-D) of the metabolite 343 using a Dowex 50 W column equilibrated with pH 2.7, ammonia formate buffer, and high-performance liquid column chromatography.317
The mushroom Cortinarius orellanus first came to public attention in 1957 when there were 19 deaths, resulting from a large poisoning of 135 people in Poland.319,320 Many severe and fatal poisoning incidents in humans have been constantly reported since people often confuse the toxic mushroom C. orellanus with edible and nontoxic mushroom, Cantharellus tubaeformis and Cantharellus cibarius in North America and Europe.321 In 1962, Stanisław Grzymala successfully isolated orellanine (344) from the methanolic extract of the fruits of C. orellanus and confirmed that crude extract of the mushroom exhibited strong nephrotoxicity.322 After this successful purification of 344, its chemical structure was elucidated using X-ray crystallography of the orellanine-trifluoroacetic acid complex in 1979.320 Structurally, 344 has bipyridines with positively charged nitrogen atoms, which is already known as the toxic functional group. Compound 344 is predicted to be biosynthesized from anthranilic acid as a precursor. Oxidative ring opening and a new ring closure result in the production of 3,4-dihydroxy-pyridine-2-carboxylic acid, and the formation of 4-O-glucoside takes place enabling the regioselective coupling to orelline-4,4′-di-β-D-glucopyranoside. After N-oxidation, compound 344 is generated by hydrolysis of the diglucoside.17
When humans consume 344 or C. orellanus, the distinctive nephrotoxicity of 344 has a long latency. After a few days or even longer, 344 triggers acute kidney failure, often extending to severe kidney injury, and kidney transplantation is the only option for the treatment of the symptoms. In mice, cats, and pigs, toxin 344 causes histological changes in the liver and spleen, and the physiological symptoms are identical to those caused by the consumption of C. orellanus and its methanolic extract.322,323 The biochemical mechanism of nephrotoxicity of 344 has been investigated using electron spin resonance research, which suggested that 344 was oxidized to form the radical via a tyrosinase/O2 system and visible light. During this reaction, many superoxide radicals were formed, which induced severe oxidative damage to DNA, RNA, and proteins.324–326 This mechanism of oxidative stress by 344 may be relevant to the depletion of glutathione and ascorbate, which are important defence mechanisms against oxidative stress in humans.324–326 In 2007, it was found that 344 targets both primary and metastatic renal cell carcinoma (RCC) in vitro and in vivo.327 In nude rats carrying human RCC xenografts, treatment with 344 eliminated more than 90% of viable tumor mass compared to control rats.
The phosphorylated indole derivative, psilocybin (345), is widely known as a naturally occurring psychedelic compound mainly derived from the Psilocybe genus. After consuming 345, the fungal metabolite is quickly dephosphorylated to form psilocin (346), which displays mind-altering effects similar to those of psychedelic hallucinogens, lysergic acid diethylamide, mescaline, and N,N-dimethyltryptamine. The effects include misperception, euphoria, visual and mental hallucinations, nausea, and panic. Compounds 345 and 346 were first identified in Psilocybe mexicana, known as a psychedelic mushroom by the natives of North and Central America over 2000 years ago. Their analogs, baeocystin (347) and norbaeocystin (348) were isolated from Psilocybe baeocystis, which is known as a psilocybin mushroom of the family Hymenogastraceae.331–334
Recently, Hoffmeister et al. demonstrated the enzymatic foundation for the biosynthesis of 345 by identifying four psilocybin biosynthetic enzymes. In the biosynthetic route of 345, L-tryptophan is converted into tryptamine via decarboxylation and consecutive oxidation, phosphorylation, and methylation of tryptamine leads to the formation of 345 (Scheme 14).335
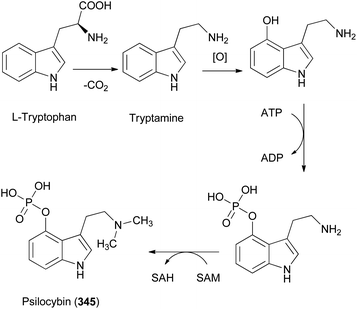 | ||
| Scheme 14 Biosynthetic pathway for the production of psilocybin (345).335 | ||
Although compound 345 has no activity in vivo, compound 346, an active form converted from 345via dephosphorylation, exhibited psychoactive effects by changing neurotransmission through serotonin receptors, especially binding with a strong affinity to 5-HT2A receptors and increasing the level of dopamine in the basal ganglia.336 The above evidence suggests that serotonin and indirect dopaminergic contribute to the psychoactive effects of 346 in humans. Interestingly, 345 may be utilized in clinical environments for the treatment of compulsive behavior, headache, and severe depression, although pharmacokinetic and pharmacological studies in humans are required for the application of 345 as a drug.336 In a short-duration Phase II trial, psilocybin treatment was at least as effective as escitalopram, a commonly prescribed antidepressant as a selective serotonin reuptake inhibitor treatment.337
The toxic mushroom Leucocoprinus birnbaumii is known as flowerpot parasol and plantpot dapperling, and has attracted much public attention due to its intense yellow color and special shape. Until recently, no secondary metabolites have been reported in this mushroom. In 2005, Steglich et al. first reported the isolation and structural identification of the yellowish compounds, birnbaumins A and B (349 and 350) from the fruiting bodies of L. birnbaumii.338 These compounds have several distinctive structural features including N-hydroxyoxamidines and 7-hydroxyindoles. Unusual structural moieties are rarely found in natural products. Although the chemical structures of 349 and 350 have been determined, their biosynthetic pathway and biological activities have not been investigated to date. It was speculated that the starting products for the biosynthesis of 349 and 350 might be L-tryptophan, citrulline, glycine, and nitrite.338
Macrolepiota neomastoidea is a poisonous mushroom widely distributed in Europe and East Asia. This mushroom is known to cause severe gastrointestinal symptoms including nausea, vomiting, diarrhea, and abdominal pain.339,340 In 2009, a new indole alkaloid, macrolepiotin (351), was isolated and structurally identified from fruiting bodies of M. neomastoidea.341 The structure of 351 was elucidated using NMR and MS data analyses, but its absolute configuration has not been assigned yet. The cytotoxic activity of 351 was evaluated against four human tumor cell lines (A549, SK-OV-3, SK-MEL-2, and HCT15), which did not exhibit significant activity in any of the cell lines (IC50 >30.0).341
Gyromitra esculenta, distributed across Europe and North America, has been widely consumed as food. However, G. esculenta is suspected to be toxic to the human body because of poisoning incidents related to the consumption of raw mushrooms.28,345 Even if it is often preboiled before preparation, scientific evidence indicates that this practice is not useful for safe consumption. The main symptoms of poisoning are gastrointestinal and neurological. Initially, nausea, vomiting, and diarrhea occur, followed by ataxia, vertigo, tremor, lethargy, fever, and headaches develop soon.28,345 As the main mycotoxin, acetaldehyde N-methyl-N-formylhydrazone, namely gyromitrin (352), was isolated from G. esculenta and its derivatives, N-methyl-N-formyl hydrazones of pentanal (353), 3-methylbutanal (354), and hexanal (355) were also identified from the mushroom.346 Compound 352 is a volatile water-soluble and heat-durable hydrazine compound, but it is hydrolyzed in the human body into monomethylhydrazine and N-methyl-N-formlhydrazin (MFH).347,348 Monomethylhydrazine reacts with pyridoxal-5-phosphate to form a hydrazone. This affects the reduction of the neurotransmitter GABA by decreasing the activity of glutamic acid decarboxylase, which leads to the occurrence of neurological symptoms.347,348 In addition, MFH is oxidized by cytochrome P450 into reactive nitrosamide intermediates, which leads to the production of various radicals that affect liver necrosis.347,348
Epicoccum is a genus widely distributed as cosmopolitan saprophytic fungi and is pathogenic to various plants, insects, and human skin. In 2003, an unusual tetramic acid derivative, epicoccamide, was first isolated by König et al. from the fungus Epicoccum purpurascens (also known as Epicoccum nigrum) derived from the inner tissue of the jellyfish Aurelia aurita.354 Additionally, Hertweck et al. reported the purification and identification of epicoccamide A (356), originally termed epicoccamide, and epicoccamides B–D (357–359) from an Epicoccum sp. growing within the fruiting body of the tree mushroom Pholiota squarrosa,355 which causes poisoning symptoms, such as vomiting and diarrhea approximately 10 h later when consumed with alcohol.139 Compounds 356–359 have biosynthetically distinctive features of 3-acyltetramic acids bearing β-mannosylated fatty acid chains. Unfortunately, the absolute configurations of 356–359 have not been elucidated to date owing to the difficulties of diastereomeric purification of enantiomeric mixtures. Among the isolated epicoccamides, compound 359 exhibited cytotoxicity against HeLa cell lines (CC50 = 17.0 μM) and antiproliferative activities toward mouse fibroblast (L-929) and human leukemia cell lines (K-562) with GI50 values of 50.5 and 33.3 μM, respectively.355
2.8 Miscellaneous secondary metabolites
Russula subnigricans has been recognized as a toxic mushroom distributed in China, Japan, and Taiwan because its poisoning incidents showed vomiting and diarrhea initially, followed by severe indications in late steps, such as chronic convulsions, loss of consciousness, backaches, acute renal failure, and respiratory failure.356 Although many poisoning incidents from R. subnigricans have been reported, there have been no investigations of the responsible toxins until 2009.111 Nakata et al. reported the isolation and structural elucidation of the small, highly strained cycloprop-2-ene carboxylic acid (360) from the fruiting bodies of R. subnigricans.111 Since unstable, volatile, and easy polymerization of 360, it was difficult to isolate and perform toxicity assessments. To verify the structure and toxicity, 360 was also synthesized by Nakata and Fox et al.111,357 In mice, the oral LD50 of 360 was 2.5 mg kg−1 and the serum creatine phosphokinase activity increased by 360.111In 2016, Hashimoto et al. identified unique naturally occurring compounds, including cyclopropylacetyl-(R)-carnitine (361) and 3-hydroxybaikiain (362) from R. subnigricans, along with 360, where it was proposed that 361 is specific to R. subnigricans since the upfielded proton signals corresponding to the cyclopropane ring are easily detectable in the 1H NMR spectrum of its crude extract.356 In fact, 361 is very stable under normal isolation procedures and it can be useful as a chemical marker for the identification of genuine R. subnigricans.
A quaternary ammonium compound, muscarine (363), was identified in Amanita muscaria for the first time and was also found in some mushrooms, particularly in Inocybe, Clitocybe, Entoloma, and Mycena species. After identifying the structure of 363, Eugster et al. investigated the biosynthesis of 363 in the mycelial culture of Clitocybe rivulosa utilizing the incorporation of several 14C-labeled simple compounds into the culture system. It was found that 363 was formed from the combination of pyruvate and glutamate.358 The colorless and odorless 363 stimulates the parasympathetic nervous system of animals. Because the structure of compound 363 is almost similar to that of the natural neurotransmitter acetylcholine in the muscarinic part of the cholinergic nervous system, 363 binds to acetylcholine receptors and causes poisonous symptoms, including central nervous system disorder, hallucination-induced excitation, sweating, tearing, and bradycardia.359–361 The lethal dose of 363 in humans is projected to be 180–300 mg, therefore, consumption of a mushroom containing 0.33% of 363 can be lethal.362 In contrast, by utilizing the mode of action of 363, muscarinic agonists are used as drugs in clinics for the treatment of glaucoma, postoperative ileus, congenital megacolon, urinary retention, and xerostomia.
3 Conclusions and future perspectives
Mushroom-forming fungi belong to the phyla Ascomycota or Basidiomycota, and they are proficient producers of structurally diverse and unique secondary metabolites that mediate a wide range of bioactivities and potential applications in drug discovery and development by providing core template structures and pharmacophores. However, the secondary metabolites sometimes simultaneously make mushrooms poisonous, turning them into life-threatening toadstools. These secondary metabolites thus contribute to the utility of mushrooms as prolific producers of lead compounds for drug discovery and development, as well as mushroom poisoning, resulting in serious illness and even death. However, the secondary metabolites of poisonous mushrooms have rarely been summarized and discussed systematically for their chemical information, including biosynthesis. This review summarizes the current information on 363 secondary metabolites from poisonous mushrooms from the chemistry, bioactivity, and biosynthesis-based points of view. Since the investigation of toxic substances from poisonous mushrooms began in the early 19th century, more than 300 secondary metabolites, including toxic compounds, have been characterized from poisonous mushrooms, and some specific mushroom toxins, such as cyclopeptides, mainly phallotoxins, amatoxins, and virotoxins have been extensively studied for structural determination synthesis, toxicology, diagnosis and detection from blood or urine, emergency medical treatment, and pharmacological activities. A variety of reviews on toxic mushrooms have been published, but they have focused on toxic mushrooms themselves or their toxicity and summarized the chemistry, biosynthesis, and toxicity of the above-mentioned cyclopeptides. Thus, in the present review article, we cover only non-peptide secondary metabolites from poisonous mushrooms and summarize their chemistry by structural classification, bioactivity, and biosynthesis.In summary, poisonous mushrooms produce a large number of structurally diverse secondary metabolites that are unique to natural products found in other natural resources. Phenolic compounds with diverse carbon skeletons, ergostane-type steroids, and terpenoids, including meroterpenoids, are the major bioactive metabolites. Poisonous mushrooms are particularly prolific in creating sesquiterpenoid diversity; diverse sesquiterpenoid skeletons have been reported from the poisonous mushrooms including illudin-type, illudalane-type, lactarane-type, trichothecene-type, tremulane-type, marasmane-type, caryophyllene-type, tricyclo[5.4.0.02,5]undecane-type, and aristolane-type sesquiterpenoids. The mycotoxins of poisonous mushrooms are mainly composed of alkaloids described in this review, and the mycotoxins also include toxic cyclopeptides, although they are not covered. Table 1 summarizes the key mycotoxins of poisonous mushrooms from the total metabolites described in this review. Based on the reported bioactivities of the secondary metabolites from poisonous mushrooms, the most promising approach for the medicinal development of secondary metabolites includes antitumor and anticancer agents. For example, Illudin S (111), the first illudin-type sesquiterpenoid identified from Lampteromyces japonicus, a bioluminescent toxic mushroom as a major toxic substance, is currently used in clinical programs for the development of anticancer drugs through structural modifications to reduce its toxicity and improve its low therapeutic index. In addition, muscarine (363), a quaternary ammonium compound identified from a poisonous mushroom Amanita muscaria is currently used as a drug for muscarinic agonists in the clinic for the treatment of some diseases such as glaucoma, postoperative ileus, congenital megacolon, urinary retention, and xerostomia. The potential of psilocybin (345) identified from the psychedelic mushroom Psilocybe mexicana, for the treatment of compulsive behavior, headache, and severe depression has been noticed. Moreover, the biosynthetic pathways of many secondary metabolites, but not all, are presented in this review. Many aspects of the biosynthetic pathway of secondary metabolites from poisonous mushrooms still need to be investigated.
| Mushroom | Mushroom poisoning | Metabolites | Types of metabolites | Toxicity of metabolite | References |
|---|---|---|---|---|---|
| Tricholoma ustale | Gastrointestinal poisoning | Ustalic acid (33) | Phenolic compound | Inhibition of Na+/K+-ATPase activity | 83 |
| Lampteromyces japonicus | Cytotoxic poisoning | Illudin S (111) | Sesquiterpene | Inhibition of DNA and RNA synthesis | 169 and 170 |
| Podostroma cornu-damae | Gastrointestinal and cytotoxic poisoning | Roridin E (171), verrucarin J (172), and satratoxin H (173) | Sesquiterpene | Lethal effects on mice | 196 |
| Naematoloma fasciculare | Gastrointestinal poisoning | Fasciculols E and F (218 and 219) | Triterpene | Paralytic death in mice | 230 |
| Hebeloma vinosophyllum | Neurotoxic poisoning | Hebevinosides I–XI (230–240) | Triterpene | Paralytic death in mice | 240 and 241 |
| Amanita muscaria | Neurotoxic poisoning | Ibotenic acid (327) | Alkaloid (amino acid) | Agonist of N-methyl-D-aspartate (NMDA) and 1-aminocyclopentane-trans-1,3-dicarboxylic acid receptors | 290, 293 and 294 |
| Amanita muscaria | Neurotoxic poisoning | Muscimol (328) | Alkaloid (amino acid) | Agonist of the receptor for GABAA | 290 and 295 |
| Coprinopsis atramentaria | Gastrointestinal poisoning | Coprine (330) | Alkaloid (amino acid) | Inhibition of acetaldehyde dehydrogenase | 45, 298 and 300 |
| Trogia venenata | Cytotoxic poisoning | 2R-Amino-4S-hydroxy-5-hexynoic acid (332) | Alkaloid (amino acid) | Increasing serum creatinine kinase in mice | 305 |
| Clitocybe acromelalga | Neurotoxic poisoning | Acromelic acids A–C (338–340) | Alkaloid (amino acid) | Neurotoxic activity by affecting the kainite receptors | 312–314 |
| Cortinarius orellanus | Cytotoxic poisoning | Orellanine (344) | Alkaloid (pyridine alkaloid) | Oxidative damage to DNA, RNA, and proteins | 322–324 |
| Psilocybe mexicana | Neurotoxic poisoning | Psilocybin (345) and psilocin (346) | Alkaloid (indole alkaloid) | Increasing the level of dopamine in the basal ganglia by binding to 5-HT2A receptors | 331 and 336 |
| Gyromitra esculenta | Gastrointestinal and neurotoxic poisoning | Gyromitrin (352) | Alkaloid (hydrazine alkaloid) | Inhibition of glutamic acid decarboxylase | 346–348 |
| Russula subnigricans | Gastrointestinal and cytotoxic poisoning | Cycloprop-2-ene carboxylic acid (360) | Alkaloid | Induction of serum creatine phosphokinase | 111 and 357 |
| Amanita muscaria | Neurotoxic poisoning | Muscarine (363) | Alkaloid | Muscarinic agonist binding to acetylcholine receptors | 359–361 |
The goal of this review is to provide chemical information (chemistry and biosynthesis) and bioactivity of the reported secondary metabolites from poisonous mushrooms, and ultimately to switch the perspective that poisonous mushrooms are always harmful into a new aspect that the secondary metabolites from poisonous mushrooms can be useful for advances in chemistry, biochemistry, and pharmacology, which can lead to potential lead structures for drug discovery and development. Compared to the extensive and comprehensive studies of Amanita toxic cyclopeptides in a variety of interdisciplinary research, other toxins or secondary metabolites from poisonous mushrooms still require further research for their poisoning mechanism, protein targets, and potential medical applications beyond their chemistry. Furthermore, the secondary metabolites of most poisonous mushrooms remain uninvestigated. This review may inspire natural product chemists or related researchers to investigate the secondary metabolites of toxic mushrooms, which are still unexplored. The chemical information will be utilized by clinicians or relevant staff to diagnose and treat mushroom poisoning, train the public on how to prevent it, as well as provide potential lead structures for drug discovery. In addition, many aspects of poisonous mushrooms that evolve the unique metabolic pathway are still obscure, and thus a better understanding of the biosynthetic pathway of secondary metabolites will also be helpful for the discovery of new bioactive secondary metabolites. Consequently, close collaborative mushroom natural product research on chemistry/mycology/molecular biology/toxicology and mutual assistance is necessary not only to deepen our knowledge of poisonous mushrooms but also to provide new nature-inspired approaches for drug development.
4 Author contributions
Conceptualization, K. H. K.; formal analysis, S. L., J. S. Y., and S. R. L.; investigation, S. L., J. S. Y., and S. R. L.; writing—original draft preparation, S. L., J. S. Y., S. R. L., and K. H. K.; writing—review and editing, S. L. and K. H. K.; visualization, S. L., J. S. Y., S. R. L., and K. H. K.; supervision, K. H. K.; project administration, K. H. K.; funding acquisition, K. H. K. All authors have read and agreed to the published version of the manuscript.5 Conflicts of interest
There are no conflicts to declare.6 Acknowledgements
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (2019R1A5A2027340 and 2021R1A2C2007937). This study was also supported by a biological toxicity research grant (KNA 1-3-2, 19-5) of the National Institute of Forest Science affiliated with the Korea Forest Service.7 References
- M. Blackwell, Am. J. Bot., 2011, 98, 426–438 CrossRef
.
- P. Spiteller, Chem.–Eur. J., 2008, 14, 9100–9110 CrossRef CAS
.
- P. Spiteller, Nat. Prod. Rep., 2015, 32, 971–993 RSC
.
- A. Fleming, Br. J. Exp. Pathol., 1929, 10, 226 CAS
.
- D. A. Dias, S. Urban and U. Roessner, Metabolites, 2012, 2, 303–336 CrossRef CAS
.
- A. Schueffler and T. Anke, Nat. Prod. Rep., 2014, 31, 1425–1448 RSC
.
- M. S. Butler, J. Nat. Prod., 2004, 67, 2141–2153 CrossRef CAS
.
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2012, 75, 311–335 CrossRef CAS
.
- P. L. Schiff Jr, Am. J. Pharm. Educ., 2006, 70(5), 98 CrossRef PubMed
.
-
S.-T. Chang, Mushrooms as functional foods, 2008, vol. 260 Search PubMed
.
- H.-C. Lin, R. T. Hewage, Y.-C. Lu and Y.-H. Chooi, Org. Biomol. Chem., 2019, 17, 1027–1036 RSC
.
- Z.-Y. Zhou and J.-K. Liu, Nat. Prod. Rep., 2010, 27, 1531–1570 RSC
.
- S. Rittenhouse, S. Biswas, J. Broskey, L. McCloskey, T. Moore, S. Vasey, J. West, M. Zalacain, R. Zonis and D. Payne, Antimicrob. Agents Chemother., 2006, 50, 3882–3885 CrossRef CAS
.
- H. Sauter, W. Steglich and T. Anke, Angew. Chem., Int. Ed., 1999, 38, 1328–1349 CrossRef
.
- H. E. Hallen, H. Luo, J. S. Scott-Craig and J. D. Walton, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 19097–19101 CrossRef CAS PubMed
.
- H. Luo, H. E. Hallen-Adams, J. S. Scott-Craig and J. D. Walton, Fungal Genet. Biol., 2012, 49, 123–129 CrossRef
.
- P. Spiteller, M. Spiteller and W. Steglich, Angew. Chem., Int. Ed., 2003, 42, 2864–2867 CrossRef CAS
.
-
P. G. Miles and S.-T. Chang, Mushrooms: cultivation, nutritional value, medicinal effect, and environmental impact, CRC press, 2004 Search PubMed
.
- M. P. Barbato, Med. J. Aust., 1993, 158, 842–848 CrossRef CAS
.
- J. White, S. A. Weinstein, L. De Haro, R. Bédry, A. Schaper, B. H. Rumack and T. Zilker, Toxicon, 2019, 157, 53–65 CrossRef CAS PubMed
.
- A. D. Lima, R. C. Fortes, M. G. Novaes and S. Percário, Nutr. Hosp., 2012, 27, 402–408 CAS
.
- W.-S. Jo, M. A. Hossain and S.-C. Park, Mycobiology, 2014, 42, 215–220 CrossRef
.
- J. H. Diaz, Wilderness Environ. Med., 2018, 29, 111–118 CrossRef PubMed
.
- T. Wieland, Int. J. Pept. Protein Res., 1983, 22, 257–276 CrossRef CAS
.
- E. Munekata, H. Faulstich and T. Wieland, Justus Liebigs Ann. Chem., 1978, 9, 1758–1765 Search PubMed
.
- H. Faulstich, A. Buku, H. Bodenmueller and T. Wieland, Biochemistry, 1980, 19, 3334–3343 CrossRef CAS
.
- J. Vetter, Toxicon, 1998, 36, 13–24 CrossRef CAS PubMed
.
- C. Karlson-Stiber and H. Persson, Toxicon, 2003, 42, 339–349 CrossRef CAS
.
- I. Riede, Am. J. Biomed. Res., 2013, 1, 93–107 CrossRef CAS
.
- J. Garcia, V. M. Costa, A. Carvalho, P. Baptista, P. G. de Pinho, M. de Lourdes Bastos and F. Carvalho, Food Chem. Toxicol., 2015, 86, 41–55 CrossRef CAS PubMed
.
-
J. H. WONG and T. Ng, in Handbook of Biologically Active Peptides, Elsevier, 2006, pp. 131–135 Search PubMed
.
- D. Michelot and L. M. Melendez-Howell, Mycol. Res., 2003, 107, 131–146 CrossRef CAS
.
- X. Yin, A.-A. Yang and J.-M. Gao, J. Agric. Food Chem., 2019, 67, 5053–5071 CrossRef CAS PubMed
.
- H. S. Bukhari, R. P. Jakob and T. Maier, Structure, 2014, 22, 1775–1785 CrossRef CAS
.
- M. Reich, C. Göbel, A. Kohler, M. Buée, F. Martin, I. Feussner and A. Polle, New Phytol., 2009, 950–964 CrossRef CAS
.
-
A. D. McNaught and A. Wilkinson, Compendium of chemical terminology, Blackwell Science Oxford, 1997 Search PubMed
.
- S. Jenni, M. Leibundgut, D. Boehringer, C. Frick, B. Mikolásek and N. Ban, Science, 2007, 316, 254–261 CrossRef CAS
.
- F. Wieland, E. A. Siess, L. Renner, C. Verfürth and F. Lynen, Proc. Natl. Acad. Sci. U. S. A., 1978, 75, 5792–5796 CrossRef CAS PubMed
.
- J. K. Stoops, S. J. Kolodziej, J. P. Schroeter, J.-P. Bretaudiere and S. J. Wakil, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 6585–6589 CrossRef CAS
.
- S. J. Kolodziej, P. A. Penczek and J. K. Stoops, J. Struct. Biol., 1997, 120, 158–167 CrossRef CAS PubMed
.
- S. Jenni, M. Leibundgut, T. Maier and N. Ban, Science, 2006, 311, 1263–1267 CrossRef CAS
.
- F. Lynen, H. Engeser, E. C. Foerster, J. L. Fox, S. Hess, G. B. Kresze, T. Schmitt, T. Schreckenbach, E. Siess and F. Wieland, Eur. J. Biochem., 1980, 112, 431–442 CrossRef CAS
.
- M. Leibundgut, T. Maier, S. Jenni and N. Ban, Curr. Opin. Struct. Biol., 2008, 18, 714–725 CrossRef CAS
.
- H. Kawagishi, T. Miyazawa, H. Kume, Y. Arimoto and T. Inakuma, J. Nat. Prod., 2002, 65, 1712–1714 CrossRef CAS PubMed
.
- P. Lindberg, R. Bergman and B. Wickberg, J. Chem. Soc., Perkin Trans. 1, 1977, 1, 684–691 RSC
.
- P. Lindberg, R. Bergman and B. Wickberg, J. Chem. Soc., Chem. Commun., 1975, 946–947 RSC
.
- G. Hatfield and J. Schaumberg, Lloydia, 1975, 38, 489–496 CAS
.
- H. M. So, S. Lee, K.-H. Baek, H.-S. Roh, S. Kim, M. S. Jo, S. C. Baek, S. Seok, R. Ryoo and K. H. Kim, Nat. Prod. Res., 2021, 35, 649–654 CrossRef CAS
.
- W. S. Suh, S. R. Lee, C. S. Kim, E. Moon, S. Y. Kim, S. U. Choi, K. S. Kang, K. R. Lee and K. H. Kim, J. Chem. Res., 2016, 40, 156–159 CrossRef CAS
.
-
E. Wong, Cells: molecules and mechanisms, Axolotl Academic Publishing, 2009 Search PubMed
.
- G. Carta, E. Murru, S. Lisai, A. Sirigu, A. Piras, M. Collu, B. Batetta, L. Gambelli and S. Banni, PLoS One, 2015, 10, e0120424 CrossRef PubMed
.
- G. Carta, E. Murru, S. Banni and C. Manca, Front. Physiol., 2017, 8, 902 CrossRef PubMed
.
- C. S. Kim, E. Moon, S. U. Choi, S. Y. Kim, K. R. Lee and K. H. Kim, J. Antibiot., 2015, 68, 414–416 CrossRef CAS
.
- K. H. Kim, S. U. Choi and K. R. Lee, Lipids, 2012, 47, 593–599 CrossRef CAS
.
- S. R. Lee, S. A. Yi, K. H. Nam, R. Ryoo, J. Lee and K. H. Kim, J. Nat. Prod., 2019, 82, 3489–3493 CrossRef CAS
.
- T. Kokta, A. Strat, M. Papasani, J. Szasz, M. Dodson and R. Hill, Animal, 2008, 2, 92–99 CrossRef CAS
.
- Y. Guo, T. C. Walther, M. Rao, N. Stuurman, G. Goshima, K. Terayama, J. S. Wong, R. D. Vale, P. Walter and R. V. Farese, Nature, 2008, 453, 657–661 CrossRef CAS
.
-
D. Kahveci, N. Zhong and X. Xu, in Ionic Liquids in Lipid Processing and Analysis, Elsevier, 2016, pp. 251–278 Search PubMed
.
- R. E. Minto and B. J. Blacklock, Prog. Lipid Res., 2008, 47, 233–306 CrossRef CAS
.
- R. Negri, Fitoterapia, 2015, 106, 92–109 CrossRef CAS
.
- J. D. Bu'Lock and G. N. Smith, J. Chem. Soc. C, 1967, 332–336 RSC
.
- I. Fleming and J. Harley-Mason, J. Chem. Soc. C, 1963, 4771–4778 RSC
.
- M. T. Hearn, E. R. Jones, M. G. Pellatt, V. Thaller and J. L. Turner, J. Chem. Soc., Perkin Trans. 1, 1973, 1, 2785–2788 RSC
.
- G. Kusano, Y. Koike, H. Inoue and S. Nozoe, Chem. Pharm. Bull., 1986, 34, 3465–3470 CrossRef CAS PubMed
.
- M. Hashimoto, M. Yanagiya, T. Okuno and H. Shirahama, Bull. Chem. Soc. Jpn., 1989, 62, 2751–2752 CrossRef CAS
.
- I.-K. Lee, S.-M. Cho, S.-J. Seok and B.-S. Yun, Mycobiology, 2008, 36, 55–59 CrossRef CAS PubMed
.
- J. A. Findlay and Z.-q. He, J. Nat. Prod., 1991, 54, 184–189 CrossRef CAS
.
- R. M. Bell, Y. A. Hannun and A. Merrill Jr, Adv. Lipid Res., 1993, 26, 384 Search PubMed
.
- J. Patton, B. Srinivasan, R. Dickson and R. Lester, J. Bacteriol., 1992, 174, 7180–7184 CrossRef CAS PubMed
.
- R. C. Dickson, Annu. Rev. Biochem., 1998, 67, 27 CrossRef CAS PubMed
.
- B. Zanolari, S. Friant, K. Funato, C. Sütterlin, B. J. Stevenson and H. Riezman, EMBO J., 2000, 19, 2824–2833 CrossRef CAS PubMed
.
- R. C. Dickson and R. L. Lester, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2002, 1583, 13–25 CrossRef CAS
.
- M. Fornarotto, L. Xiao, Y. Hou, K. A. Koch, E. Chang, R. M. O'Malley, T. A. Black, M. B. Cable and S. S. Walker, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2006, 1761, 52–63 CrossRef CAS
.
- R. Mashima, T. Okuyama and M. Ohira, Future Sci. OA, 2019, 6, FSO434 CrossRef PubMed
.
- Y. Yaoita, R. Kohata, R. Kakuda, K. Machida and M. Kikuchi, Chem. Pharm. Bull., 2002, 50, 681–684 CrossRef CAS PubMed
.
- J. M. Gao, A. L. Zhang, C. L. Zhang and J. k. Liu, Helv. Chim. Acta, 2004, 87, 1483–1487 CrossRef CAS
.
- K. H. Kim, S. U. Choi, K. M. Park, S. J. Seok and K. R. Lee, Arch. Pharmacal Res., 2008, 31, 579–586 CrossRef CAS PubMed
.
- P. G. Anantharaju, P. C. Gowda, M. G. Vimalambike and S. V. Madhunapantula, Nutr. J., 2016, 15, 99 CrossRef PubMed
.
- P. M. Dewick, Nat. Prod. Rep., 1998, 15, 17–58 RSC
.
- A. R. Knaggs, Nat. Prod. Rep., 2003, 20, 119–136 RSC
.
- M. Gill, Nat. Prod. Rep., 2003, 20, 615–639 RSC
.
- M. Gill and W. Steglich, Fortschr. Chem. Org. Naturst., 1987, 51, 1–317 CrossRef CAS
.
- Y. Sano, K. Sayama, Y. Arimoto, T. Inakuma, K. Kobayashi, H. Koshino and H. Kawagishi, Chem. Commun., 2002, 13, 1384–1385 RSC
.
- K. H. Kim, H. J. Noh, S. U. Choi, K. M. Park, S. J. Seok and K. R. Lee, J. Antibiot., 2010, 63, 575–577 CrossRef CAS PubMed
.
- F. Rineau, D. Roth, F. Shah, M. Smits, T. Johansson, B. Canbäck, P. B. Olsen, P. Persson, M. N. Grell and E. Lindquist, Environ. Microbiol., 2012, 14, 1477–1487 CrossRef CAS PubMed
.
- F. Bschor and H. Mallach, Arch. Toxikol., 1963, 20, 82–95 CrossRef CAS
.
- H. Mlodecki, Acta Pol. Pharm., 1967, 24, 71–76 Search PubMed
.
- S. Musselius, A. Ryk, A. Lebedev, G. Pakhomova, P. Golikov, B. Davydov, L. Donova, L. Zimina, G. Platonova and I. Selina, Anesteziologiia i reanimatologiia, 2002, 30–35 CAS
.
- J. Schmidt, W. Hartmann, A. Würstlin and H. Deicher, Dtsch. Med. Wochenschr., 1971, 96, 1188–1191 CrossRef CAS
.
- M. Winkelmann, F. Borchard, W. Stangel and B. Grabensee, Dtsch. Med. Wochenschr., 1982, 107, 1190–1194 CrossRef CAS
.
- M. Winkelmann, W. Stangel, I. Schedel and B. Grabensee, Klin. Wochenschr., 1986, 64, 935–938 CrossRef CAS
.
- R. Antkowiak, W. Z. Antkowiak, I. Banczyk and L. Mikolajczyk, Can. J. Chem., 2003, 81, 118–124 CrossRef CAS
.
- R. Edwards, G. Elsworthy and N. Kale, J. Chem. Soc. C, 1967, 405–409 RSC
.
- L. Mikolajczyk and W. Z. Antkowiak, Heterocycles, 2009, 79, 423–426 CrossRef CAS
.
- J.-X. Zhang, J.-H. Lv, L.-Q. Zhao, X.-X. Shui, J. Zhang and L.-A. Wang, Nat. Prod. Res., 2020, 34, 1246–1249 CrossRef CAS
.
- S. Kim, H. M. So, H.-S. Roh, J. Kim, J. S. Yu, S. Lee, S. Seok, C. Pang, K.-H. Baek and K. H. Kim, RSC Adv., 2017, 7, 35297–35304 RSC
.
- S. A. Yi, K. H. Nam, S. Kim, H. M. So, R. Ryoo, J. W. Han, K. H. Kim and J. Lee, Genes, 2019, 11, 18 CrossRef
.
- C. J. Duncan, M. Cuendet, F. R. Fronczek, J. M. Pezzuto, R. G. Mehta, M. T. Hamann and S. A. Ross, J. Nat. Prod., 2003, 66, 103–107 CrossRef CAS PubMed
.
- D. N. Quang, T. Hashimoto, M. Nukada, I. Yamamoto, M. Tanaka, S. Takaoka and Y. Asakawa, Chem. Pharm. Bull., 2003, 51, 330–332 CrossRef CAS
.
- L. Zhang, F. Wang and Z.-J. Dong, Heterocycles, 2006, 68, 1455–1458 CrossRef CAS
.
- H. Besl, A. Bresinsky, G. Geigenmüller, R. Herrmann, C. Kilpert and W. Steglich, Liebigs Ann. Chem., 1989, 1989, 803–810 CrossRef
.
- M. S. Buchanan, T. Hashimoto, S. Takaoka and Y. Asakawa, Phytochemistry, 1995, 40, 1251–1257 CrossRef CAS
.
- Z. Béni, M. Dékány, B. Kovács, B. Csupor-Löffler, Z. P. Zomborszki, E. Kerekes, A. Szekeres, E. Urbán, J. Hohmann and A. Ványolós, Molecules, 2018, 23, 1082 CrossRef PubMed
.
- A. Bresinsky, M. Jarosch, M. Fischer, I. Schönberger and B. Wittmann-Bresinsky, Plant Biol., 1999, 1, 327–333 CrossRef CAS
.
- M. Matsuura, S. Kato, Y. Saikawa, M. Nakata and K. Hashimoto, Chem. Pharm. Bull., 2016, 64, 602–608 CrossRef CAS
.
- J. T. Cho and J. H. Han, J. Korean Med. Sci., 2016, 31, 1164–1167 CrossRef CAS PubMed
.
- S. Lin, M. Mu, F. Yang and C. Yang, Wilderness & Environmental Medicine, 2015, 26, 380–383 Search PubMed
.
- T. Ohta, A. Takahashi, M. Matsuda, S.-y. Kamo, T. Agatsuma, T. Endo and S. Nozoe, Tetrahedron Lett., 1995, 36, 5223–5226 CrossRef CAS
.
- A. Takahashi, T. Agatsuma, M. Matsuda, T. Ohta, T. Nunozawa, T. Endo and S. Nozoe, Chem. Pharm. Bull., 1992, 40, 3185–3188 CrossRef CAS PubMed
.
- A. Takahashi, T. Agatsuma, T. Ohta, T. Nunozawa, T. Endo and S. Nozoe, Chem. Pharm. Bull., 1993, 41, 1726–1729 CrossRef CAS
.
- M. Matsuura, Y. Saikawa, K. Inui, K. Nakae, M. Igarashi, K. Hashimoto and M. Nakata, Nat. Chem. Biol., 2009, 5, 465–467 CrossRef CAS
.
- J. K. Liu, Chem. Rev., 2006, 106, 2209–2223 CrossRef CAS
.
- W. Li, X.-B. Li and H.-X. Lou, J. Asian Nat. Prod. Res., 2018, 20, 1–13 CrossRef
.
- V. Jirawongse, E. Ramstad and J. Wolinsky, J. Pharm. Sci., 1962, 51, 1108–1109 CrossRef CAS PubMed
.
- J. Burton and B. Cain, Nature, 1959, 184, 1326–1327 CrossRef CAS
.
- S. M. Kamruzzaman, T. Yayeh, H. D. Ji, J. Y. Park, Y. S. Kwon, I. K. Lee, S. Kim, S. H. Oh, S. D. Kim, S. S. Roh, B. S. Yun and M. H. Rhee, Vasc. Pharmacol., 2013, 59, 83–89 CrossRef CAS
.
- I. K. Lee, J. Y. Jung, Y. S. Kim, M. H. Rhee and B. S. Yun, Bioorg. Med. Chem., 2009, 17, 4674–4680 CrossRef CAS PubMed
.
- I. K. Lee, D. W. Ki, S. E. Kim, M. S. Lee, J. G. Song and B. S. Yun, J. Microbiol. Biotechnol., 2013, 23, 652–655 CrossRef CAS
.
- D. N. Quang, T. Hashimoto, M. Nukada, I. Yamamoto, M. Tanaka and Y. Asakawa, Chem. Pharm. Bull., 2003, 51, 1064–1067 CrossRef CAS
.
- D. N. Quang, T. Hashimoto, M. Nukada, I. Yamamoto, M. Tanaka and Y. Asakawa, Planta Med., 2003, 69, 1063–1066 CrossRef CAS
.
- D. N. Quang, T. Hashimoto, M. Nukada, I. Yamamoto, M. Tanaka and Y. Asakawa, Phytochemistry, 2003, 64, 649–654 CrossRef CAS
.
- B.-S. Yun, I.-K. Lee, J.-P. Kim and I.-D. Yoo, J. Antibiot., 2000, 53, 114–122 CrossRef CAS
.
- I.-K. Lee, B.-S. Yun, J.-P. Kim, W.-G. Kim, I.-J. Ryoo, S. Oh, Y.-H. Kim and I.-D. Yoo, Planta Med., 2003, 69, 513–517 CrossRef CAS
.
- B. Schwyn and J. Neilands, Anal. Biochem., 1987, 160, 47–56 CrossRef CAS
.
- B. Goodell, J. Jellison, J. Liu, G. Daniel, A. Paszczynski, F. Fekete, S. Krishnamurthy, L. Jun and G. Xu, J. Biotechnol., 1997, 53, 133–162 CrossRef CAS
.
- D.-H. Lee, H.-J. Cho, H.-Y. Kang, M. H. Rhee and H.-J. Park, J. Ginseng Res., 2012, 36, 40 CrossRef CAS
.
- D. N. Quang, L. Harinantenaina, T. Nishizawa, T. Hashimoto, C. Kohchi, G.-I. Soma and Y. Asakawa, J. Nat. Med., 2006, 60, 303–307 CrossRef CAS
.
- I. K. Lee, B. S. Yun, J. P. Kim, I. J. Ryoo, Y. H. Kim and I. D. Yoo, Biosci., Biotechnol., Biochem., 2003, 67, 1813–1816 CrossRef CAS PubMed
.
- B.-S. Yun, I.-K. Lee, J.-P. Kim and I.-D. Yoo, J. Microbiol. Biotechnol., 2000, 10, 233–237 CAS
.
- B. S. Yun, I. K. Lee, J. P. Kim and I. D. Yoo, J. Antibiot., 2000, 53, 711–713 CrossRef CAS
.
- T. Hashimoto, T. Arakawa and M. Tanaka, Heterocycles, 2002, 56, 581–588 CrossRef CAS
.
- I. K. Lee, S. M. Cho, S. J. Seok and B. S. Yun, Mycobiology, 2008, 36, 55–59 CrossRef CAS PubMed
.
- H. V. Kemami Wangun and C. Hertweck, Eur. J. Org. Chem., 2007, 3292–3295 CrossRef
.
- G. Rastelli, L. Costantino and A. Albasini, J. Am. Chem. Soc., 1997, 119, 3007–3016 CrossRef CAS
.
- S. Lee, C. S. Kim, J. S. Yu, H. Kang, M. J. Yoo, U. J. Youn, R. Ryoo, H. Y. Bae and K. H. Kim, Org. Lett., 2021, 23, 3315–3319 CrossRef CAS PubMed
.
- R. G. Thorn, D. W. Malloch, I. Saar, Y. Lamoureux, E. Nagasawa, S. A. Redhead, S. Margaritescu and J.-M. Moncalvo, Botany, 2020, 98, 293–315 CrossRef
.
- S. Poplata, A. Tröster, Y.-Q. Zou and T. Bach, Chem. Rev., 2016, 116, 9748–9815 CrossRef CAS
.
- H. V. Kemami Wangun, K. Ishida and C. Hertweck, Eur. J. Org. Chem., 2008, 3781–3784 CrossRef CAS
.
- R. L. Shaffer, Mycologia, 1965, 57, 318–319 CrossRef CAS PubMed
.
- R. J. Cannell, S. J. Kellam, A. M. Owsianka and J. M. Walker, Planta Med., 1988, 54, 10–14 CrossRef CAS PubMed
.
- D. Leung, G. Abbenante and D. P. Fairlie, J. Med. Chem., 2000, 43, 305–341 CrossRef CAS PubMed
.
- S. Kawamura, T. Horii, T. Nakabayashi, Y. Abe and M. Chubachi, Annu. Rep. Radiat. Cent. Osaka Prefect., 1979, 20, 109–114 CAS
.
- J. Song, M. M. Manir and S. S. Moon, Chem. Biodiversity, 2009, 6, 1435–1442 CrossRef CAS PubMed
.
- J. W. Tan, J. B. Xu, Z. J. Dong, D. Q. Luo and J. K. Liu, Helv. Chim. Acta, 2004, 87, 1025–1028 CrossRef CAS
.
-
O. K. Miller and H. H. Miller, North American Mushrooms: A Field Guide to Edible and Inedible Fun, Falcon Guide, Globe Pequot Press, 2006 Search PubMed
.
- G. T. Wawrzyn, S. E. Bloch and C. Schmidt-Dannert, Methods Enzymol., 2012, 515, 83–105 CAS
.
- D. W. Christianson, Curr. Opin. Chem. Biol., 2008, 12, 141–150 CrossRef CAS PubMed
.
- E. M. Davis and R. Croteau, Biosynthesis, 2000, 53–95 CAS
.
- M. B. Quin, C. M. Flynn, G. T. Wawrzyn, S. Choudhary and C. Schmidt-Dannert, ChemBioChem, 2013, 14, 2480–2491 CrossRef CAS
.
- W.-R. Abraham, Curr. Med. Chem., 2001, 8, 583–606 CrossRef CAS
.
- B. Fraga, Nat. Prod. Rep., 2011, 28, 1580–1610 RSC
.
- D. W. Christianson, Chem. Rev., 2006, 106, 3412–3442 CrossRef CAS PubMed
.
- D. J. Miller and R. K. Allemann, Nat. Prod. Rep., 2012, 29, 60–71 RSC
.
- C. A. Lesburg, J. M. Caruthers, C. M. Paschall and D. W. Christianson, Curr. Opin. Struct. Biol., 1998, 8, 695–703 CrossRef CAS
.
- Y. Yamada, D. E. Cane and H. Ikeda, Methods Enzymol., 2012, 515, 123–162 CAS
.
- A. P. W. Bradshaw, J. R. Hanson and M. Siverns, J. Chem. Soc., Chem. Commun., 1978, 303–304 RSC
.
- J. R. Hanson, T. Marten and R. Nyfeler, J. Chem. Soc., Perkin Trans. 1, 1976, 1, 876–880 RSC
.
- A. Padwa, E. A. Curtis and V. P. Sandanayaka, J. Org. Chem., 1997, 62, 1317–1325 CrossRef CAS
.
- K. Nakanishi, M. Ohashi, N. Suzuki, M. Tada, Y. Yamada and S. Inagaki, Yakugaku Zasshi, 1963, 83, 377–380 CrossRef CAS
.
- K. Tanaka, T. Inoue, S. Kadota and T. Kikuchi, Xenobiotica, 1990, 20, 671–681 CrossRef CAS PubMed
.
- K. Tanaka, T. Inoue, M. Kanai and T. Kikuchi, Xenobiotica, 1994, 24, 1237–1243 CrossRef CAS PubMed
.
- M. Tada, Y. Yamada, N. S. Bhacca, K. Nakanishi and M. Ohashi, Chem. Pharm. Bull., 1964, 12, 853–855 CrossRef CAS
.
- K. Nakanishi, M. Tada and Y. Yamada, Chem. Pharm. Bull., 1964, 12, 856–857 CrossRef CAS PubMed
.
- K. Nakanishi, M. Ohashi, M. Tada and Y. Yamada, Tetrahedron, 1965, 21, 1231–1246 CrossRef CAS
.
- T. Matsumoto, H. Shirahama, A. Ichihara, Y. Fukuoka, Y. Takahashi, Y. Mori and M. Watanabe, Tetrahedron, 1965, 21, 2671–2676 CrossRef CAS PubMed
.
- T. ITANO, Okayama Igakkai Zasshi, 1979, 91, 453–460 CrossRef CAS
.
- S. Shinozawa, K. Tsutsui and T. Oda, Experientia, 1979, 35, 1102–1103 CrossRef CAS
.
- M. J. Kelner, T. C. McMorris, W. T. Beck, J. M. Zamora and R. Taetle, Cancer Res., 1987, 47, 3186–3189 CAS
.
- J. Walser and P. Heinstein, Antimicrob. Agents Chemother., 1973, 3, 357–363 CrossRef CAS PubMed
.
- N. G. Jaspers, A. Raams, M. J. Kelner, J. M. Ng, Y. M. Yamashita, S. Takeda, T. C. McMorris and J. H. Hoeijmakers, DNA Repair, 2002, 1, 1027–1038 CrossRef CAS
.
- T. C. McMorris, M. J. Kelner, W. Wang, S. Moon and R. Taetle, Chem. Res. Toxicol., 1990, 3, 574–579 Search PubMed
.
- M. Kelner, T. McMorris, L. Estes, K. Samson, N. Trani and J. MacDonald, Leukemia, 2000, 14, 136–141 CrossRef CAS PubMed
.
- L. Hammond, S. Hilsenbeck, S. Eckhardt, J. Marty, G. Mangold, J. MacDonald, E. Rowinsky, D. Von Hoff and S. Weitman, Eur. J. Cancer, 2000, 36, 2430–2436 CrossRef CAS
.
- M. J. Kelner, T. C. McMorris and R. Taetle, J. Natl. Cancer Inst., 1990, 82, 1562–1565 CrossRef CAS
.
- T. C. McMorris, Q. Cong and M. J. Kelner, J. Org. Chem., 2003, 68, 9648–9653 CrossRef CAS PubMed
.
- I.-K. Lee, C.-Y. Jeong, S.-M. Cho, B.-S. Yun, Y.-S. Kim, S.-H. Yu, H. Koshino and I.-D. Yoo, J. Antibiot., 1996, 49, 821–822 CrossRef CAS
.
- M. Kuramoto, T. Tsukihara and N. Ono, Chem. Lett., 1999, 28, 1113–1114 CrossRef
.
- S. Aoki, T. Aboshi, Y. Shiono, K.-i. Kimura, T. Murata, D. Arai, Y. Iizuka and T. Murayama, Chem. Pharm. Bull., 2020, 68, 436–442 CrossRef CAS PubMed
.
- U. Becker, G. Erkel, T. Anke and O. Sterner, Nat. Prod. Lett., 1997, 9, 229–236 CrossRef CAS
.
- K. Yoshikawa, A. Kaneko, Y. Matsumoto, H. Hama and S. Arihara, J. Nat. Prod., 2006, 69, 1267–1270 CrossRef CAS
.
- K. Yoshikawa, Y. Matsumoto, H. Hama, M. Tanaka, H. Zhai, Y. Fukuyama, S. Arihara and T. Hashimoto, Chem. Pharm. Bull., 2009, 57, 311–314 CrossRef CAS PubMed
.
- M. de Bernardi, G. Fronza, G. Mellerio, G. Vidari and P. Vita-Finzi, Phytochemistry, 1979, 18, 293–298 CrossRef CAS
.
- R. Battaglia, M. De Bernardi, G. F. G. Mellerio, G. Vidari and P. Vita-Finsi, J. Nat. Prod., 1980, 43, 319–328 CrossRef CAS
.
- A. Bosetti, G. Fronza, G. Vidari and P. Vita-Finzi, Phytochemistry, 1989, 28, 1427–1431 CrossRef CAS
.
- Z. Pang, F. Bocchio and O. Sterner, Tetrahedron Lett., 1992, 33, 6863–6866 CrossRef CAS
.
- M. De Bernardi, L. Garlaschelli, L. Toma, G. Vidari and P. Vita-Finzi, Tetrahedron, 1993, 49, 1489–1504 CrossRef CAS
.
- G. Vidari and D. Bernardi, Tetrahedron Lett., 1975, 22, 1773–1774 CrossRef
.
- K. Kobata, S. Kano and H. Shibata, Biosci., Biotechnol., Biochem., 1995, 59, 316–318 CrossRef CAS
.
- M. Isaka, A. Yangchum, S. Wongkanoun and S. Kongthong, Phytochem. Lett., 2017, 21, 174–178 CrossRef CAS
.
- L. Garlaschelli, L. Toma, G. Vidari and D. Colombo, Tetrahedron, 1994, 50, 1211–1216 CrossRef CAS
.
- J. S. Dickschat, N. L. Brock, C. A. Citron and B. Tudzynski, ChemBioChem, 2011, 12, 2088–2095 CrossRef CAS PubMed
.
- S. Nozoe and Y. Machida, Tetrahedron Lett., 1970, 11, 2671–2674 CrossRef
.
- D. E. Cane and H. J. Ha, J. Am. Chem. Soc., 1986, 108, 3097–3099 CrossRef CAS
.
- J. Y. Ahn, S. J. Seok, J. E. Song, J. H. Choi, S. H. Han, J. Y. Choi, C. O. Kim, Y. G. Song and J. M. Kim, Yonsei Med. J., 2013, 54, 265 CrossRef PubMed
.
- H. N. Kim, H. H. Do, J. S. Seo and H. Y. Kim, Clin. Exp. Emerg. Med., 2016, 3, 186 CrossRef PubMed
.
- Y. Saikawa, H. Okamoto, T. Inui, M. Makabe, T. Okuno, T. Suda, K. Hashimoto and M. Nakata, Tetrahedron, 2001, 57, 8277–8281 CrossRef CAS
.
- M. Zhu, Y. Cen, W. Ye, S. Li and W. Zhang, Toxins, 2020, 12, 417 CrossRef CAS PubMed
.
- S. Trapp, T. Hohn, S. McCormick and B. Jarvis, Mol. Gen. Genet., 1998, 257, 421–432 CrossRef CAS PubMed
.
- T. Degenkolb, R. Dieckmann, K. F. Nielsen, T. Gräfenhan, C. Theis, D. Zafari, P. Chaverri, A. Ismaiel, H. Brückner and H. Von Döhren, Mycol. Prog., 2008, 7, 177–219 CrossRef
.
- S. R. Lee, S. Seok, R. Ryoo, S. U. Choi and K. H. Kim, J. Nat. Prod., 2018, 82, 122–128 CrossRef
.
- S. Lee, R. Ryoo, J. H. Choi, J.-H. Kim, S.-H. Kim and K. H. Kim, Arch. Pharmacal Res., 2020, 43, 214–223 CrossRef CAS
.
- G. S. Garvey, S. P. McCormick, N. J. Alexander and I. Rayment, Protein Sci., 2009, 18, 747–761 CAS
.
- T. M. Hohn and P. D. Beremand, Gene, 1989, 79, 131–138 CrossRef CAS
.
- S. P. McCormick, N. J. Alexander and R. H. Proctor, Can. J. Microbiol., 2006, 52, 636–642 CrossRef CAS
.
- X.-N. Wang, F. Wang, J.-C. Du, H.-M. Ge, R.-X. Tan and J.-K. Liu, Zeitschrift für Naturforschung B, 2005, 60, 1065–1067 CrossRef CAS
.
- X.-N. Wang, J.-H. Shen, J.-C. Du and J.-K. Liu, J. Antibiot., 2006, 59, 669–672 CrossRef CAS PubMed
.
- K. H. Kim, H. J. Noh, S. U. Choi, K. M. Park, S.-J. Seok and K. R. Lee, J. Antibiot., 2010, 63, 575–577 CrossRef CAS
.
- S. Tsuboyama, T. Sakurai, K. Tsuboyama and K. Doi, Bull. Chem. Soc. Jpn., 1986, 59, 1921–1924 CrossRef CAS
.
- K. Doi, T. Shibata, N. Yokoyama, H. Terasawa, O. Matsuda and S. Kashino, J. Chem. Soc., Chem. Commun., 1990, 725–726 RSC
.
- Y. Shiono, R. Matsuzaka, H. Wakamatsu, K. Muneta, T. Murayama and M. Ikeda, Phytochemistry, 2004, 65, 491–496 CrossRef CAS
.
- Y. Shiono, H. Wakamatsu, T. Murayama and M. Ikeda, Zeitschrift für Naturforschung B, 2004, 59, 119–123 CrossRef CAS
.
- C. Ramesh and M. G. Pattar, Pharmacogn. Res., 2010, 2, 107 CrossRef CAS
.
- I.-S. Yoo, M.-S. Woo, E.-C. Choi and B.-K. Kim, Korean J. Mycol., 1982, 10, 165–171 CAS
.
- A. Ginterová, O. Janotková and E. Findová, Folia Microbiol., 1981, 26, 133–136 CrossRef
.
- K.-C. Kim, I.-S. Lee, I.-D. Yoo and B.-J. Ha, Chem. Pharm. Bull., 2015, 63, 554–557 CrossRef
.
- T. Toyomasu, Biosci., Biotechnol., Biochem., 2008, 72, 1168–1175 CrossRef CAS PubMed
.
- R. J. Peters, Nat. Prod. Rep., 2010, 27, 1521–1530 RSC
.
- M. Xu, M. L. Hillwig, M. S. Tiernan and R. J. Peters, J. Nat. Prod., 2017, 80, 328–333 CrossRef CAS
.
- I.-S. Lee, K.-C. Kim, I.-D. Yoo and B.-J. Ha, Biosci., Biotechnol., Biochem., 2015, 79, 1921–1925 CrossRef CAS
.
- J. K. Volkman, Org. Geochem., 2005, 36, 139–159 CrossRef CAS
.
- Y.-L. Lin, Y.-R. Lee, N.-W. Tsao, S.-Y. Wang, J.-F. Shaw and F.-H. Chu, J. Nat. Prod., 2015, 78, 1556–1562 CrossRef CAS PubMed
.
- I. Abe, M. Rohmer and G. D. Prestwich, Chem. Rev., 1993, 93, 2189–2206 CrossRef CAS
.
- I. Abe, Nat. Prod. Rep., 2007, 24, 1311–1331 RSC
.
- R. Xu, G. C. Fazio and S. P. Matsuda, Phytochemistry, 2004, 65, 261–291 CrossRef CAS PubMed
.
- K.-H. Hsu, Y.-R. Lee, Y.-L. Lin and F.-H. Chu, Int. J. Med. Mushrooms, 2011, 13, 513–523 CrossRef CAS PubMed
.
- C.-H. Lee, K.-H. Hsu, S.-Y. Wang, T.-T. Chang, F.-H. Chu and J.-F. Shaw, J. Agric. Food Chem., 2010, 58, 4800–4807 CrossRef CAS
.
- R. Thoma, T. Schulz-Gasch, B. D'Arcy, J. Benz, J. Aebi, H. Dehmlow, M. Hennig, M. Stihle and A. Ruf, Nature, 2004, 432, 118–122 CrossRef CAS PubMed
.
- M. Ikeda, G.-i. Niwa, K. Tohyama, T. Sassa and Y. Miura, Agric. Biol. Chem., 1977, 41, 1803–1805 CAS
.
- M. Ikeda, H. Watanabe, A. Hayakawa, K. Sato, T. Sassa and Y. Miura, Agric. Biol. Chem., 1977, 41, 1543–1545 CAS
.
- K. Suzuki, H. Fujimoto and M. Yamazaki, Chem. Pharm. Bull., 1983, 31, 2176–2178 CrossRef CAS PubMed
.
- I. Kubo, A. Matsumoto, M. Kozuka and W. F. Wood, Chem. Pharm. Bull., 1985, 33, 3821–3825 CrossRef CAS PubMed
.
- Y. Ding, H. Y. Bao, T. Bau, Y. Li and Y.-H. Kim, J. Microbiol. Biotechnol., 2009, 19, 1135–1138 CAS
.
- A. Takahashi, G. Kusano, T. Ohta, Y. Ohizumi and S. Nozoe, Chem. Pharm. Bull., 1989, 37, 3247–3250 CrossRef CAS PubMed
.
- S. Nozoe, A. Takahashi and T. Ohta, Chem. Pharm. Bull., 1993, 41, 1738–1742 CrossRef CAS
.
- C. Seeka, P. Sutthivaiyakit, J. Youkwan, N. Hertkorn, M. Harir, P. Schmitt-Kopplin and S. Sutthivaiyakit, Phytochemistry, 2016, 127, 38–49 CrossRef CAS PubMed
.
- X. W. Shi, X. J. Li, J. M. Gao and X. C. Zhang, Chem. Biodiversity, 2011, 8, 1864–1870 CrossRef CAS
.
- K. H. Kim, E. Moon, S. U. Choi, S. Y. Kim and K. R. Lee, J. Nat. Prod., 2013, 76, 845–851 CrossRef CAS PubMed
.
- M. Ikeda, Y. Sato, T. Sassa and Y. Miura, Tennen Yuki Kagobutsu Toronkai Koen Yoshishiu, 1978, 21, 584–591 Search PubMed
.
- J. Zhang, L. Dai, J. Yang, C. Liu, Y. Men, Y. Zeng, Y. Cai, Y. Zhu and Y. Sun, Plant Cell Physiol., 2016, 57, 1000–1007 CrossRef CAS PubMed
.
- H. Fujimoto, K. Suzuki, H. Hagiwara and M. Yamazaki, Chem. Pharm. Bull., 1986, 34, 88–99 CrossRef CAS PubMed
.
- H. Fujimoto, H. Hagiwara, K. Suzuki and M. Yamazaki, Chem. Pharm. Bull., 1987, 35, 2254–2260 CrossRef CAS PubMed
.
- H. Fujimoto, K. Maeda and M. Yamazaki, Chem. Pharm. Bull., 1991, 39, 1958–1961 CrossRef CAS
.
- Y. Matsuda and I. Abe, Nat. Prod. Rep., 2016, 33, 26–53 RSC
.
- J.-M. Molina, M. Tourneur, C. Sarfati, S. Chevret, A. de Gouvello, J.-G. Gobert, S. Balkan and F. Derouin, N. Engl. J. Med., 2002, 346, 1963–1969 CrossRef CAS
.
- S. Liu, J. Widom, C. W. Kemp, C. M. Crews and J. Clardy, Science, 1998, 282, 1324–1327 CrossRef CAS PubMed
.
- R. Geris and T. J. Simpson, Nat. Prod. Rep., 2009, 26, 1063–1094 RSC
.
- Y. Matsuda, T. Awakawa, T. Mori and I. Abe, Curr. Opin. Chem. Biol., 2016, 31, 1–7 CrossRef CAS
.
- S. ValdoáMeille, J. Chem. Soc., Perkin Trans. 1, 1994, 2165–2168 Search PubMed
.
- L. Merlini, G. Nasini, L. Scaglioni, G. Cassinelli and C. Lanzi, Phytochemistry, 2000, 53, 1039–1041 CrossRef CAS
.
- G. Cassinelli, C. Lanzi, D. Polizzi, R. Gambetta, T. Pensa, L. Merlini, G. Nasini and F. Zunino, Ann. Oncol., 1998, 9, 109 Search PubMed
.
- G. Cassinelli, C. Lanzi, T. Pensa, R. A. Gambetta, G. Nasini, G. Cuccuru, M. Cassinis, G. Pratesi, D. Polizzi and M. Tortoreto, Biochem. Pharmacol., 2000, 59, 1539–1547 CrossRef CAS PubMed
.
- K.-i. Takao, K. Mori, K. Kasuga, R. Nanamiya, A. Namba, Y. Fukushima, R. Nemoto, T. Mogi, H. Yasui and A. Ogura, J. Org. Chem., 2018, 83, 7060–7075 CrossRef CAS
.
- Z. Sun, N. Zhu, M. Zhou, X. Huo, H. Wu, Y. Tian, J. Yang, G. Ma, Y.-L. Yang and X. Xu, Org. Chem. Front., 2019, 6, 3759–3765 RSC
.
- Z. Sun, X. Xu, H. Liang, X. Xia, G. Ma and L. Shi, Molecules, 2019, 24, 4015 CrossRef CAS PubMed
.
- S. Nozoe, Y. Koike, E. Tsuji, G. Kusano and H. Seto, Tetrahedron Lett., 1983, 24, 1731–1734 CrossRef CAS
.
- S. Nozoe, Y. Koike, N. Ito and G. Kusano, Chem. Lett., 1984, 13, 1001–1002 CrossRef
.
- K. H. Kim, S. U. Choi and K. R. Lee, J. Antibiot., 2012, 65, 135–137 CrossRef CAS
.
- S. Nozoe, T. Ohta, Y. Koike and G. Kusano, Tetrahedron Lett., 1984, 25, 4023–4024 CrossRef CAS
.
- S. Nozoe, Y. Koike, G. Kusano and H. Seto, Tetrahedron Lett., 1983, 24, 1735–1736 CrossRef CAS
.
- F. Aoyagi, S. Maeno, T. Okuno, H. Matsumoto, M. Ikura, K. Hikichi and T. Matsumoto, Tetrahedron Lett., 1983, 24, 1991–1994 CrossRef CAS
.
- M. Tanaka, K. Hashimoto, T. Okuno and H. Shirahama, Phytochemistry, 1993, 34, 661–664 CrossRef CAS
.
-
L. P. Sandjo and V. Kuete, in Medicinal plant research in Africa, Elsevier, 2013, pp. 135–202 Search PubMed
.
- G. D. Brown, Nat. Prod. Rep., 1998, 15, 653–696 RSC
.
- D. M. Harrison, Nat. Prod. Rep., 1990, 7, 459–484 RSC
.
- Y. Yaoita, M. Endo, Y. Tani, K. Machida, K. Amemiya, K. Furumura and M. Kikuchi, Chem. Pharm. Bull., 1999, 47, 847–851 CrossRef CAS
.
- L. Satora, D. Pach, K. Ciszowski and L. Winnik, Toxicon, 2006, 47, 605–607 CrossRef CAS PubMed
.
- A. Vendramin and M. Brvar, Toxicon, 2014, 90, 269–272 CrossRef CAS
.
- Q.-F. Gong, Y.-M. Zhang, N.-H. Tan and Z.-H. Chen, Nat. Prod. Res. Dev., 2007, 19, 436–438 CAS
.
- K. H. Kim, S. U. Choi, K. M. Park, S. J. Seok and K. R. Lee, Arch. Pharmacal Res., 2008, 31, 579–586 CrossRef CAS
.
- H. J. Rho, J. H. Kim, H. R. Kang, M. K. Lee, S. H. Hyun, Y. M. Kang, J. M. Lee and N. S. Kim, Korean J. Med., 2000, 58, 453–461 Search PubMed
.
- Z. Su, P. Wang, W. Yuan and S. Li, Nat. Prod. Commun., 2013, 8, 1227–1228 CAS
.
- K. Yoshikawa, M. Ikuta, S. Arihara, E. Matsumura and S. Katayama, Chem. Pharm. Bull., 2001, 49, 1030–1032 CrossRef CAS PubMed
.
- D. Blayney, E. Rosenkranz and A. Zettner, West. J. Med., 1980, 132, 74 CAS
.
- P. F. Lehmann and U. Khazan, Mycopathologia, 1992, 118, 3–13 CrossRef CAS PubMed
.
- P. H. Stenklyft and W. Lynn Augenstein, J. Toxicol., Clin. Toxicol., 1990, 28, 159–168 CrossRef CAS PubMed
.
- G. Kusano, Y. Koike, H. Inoue and S. Nozoe, Chem. Pharm. Bull., 1986, 34, 3465–3470 CrossRef CAS PubMed
.
- K. H. Kim, K. M. Park, S. U. Choi and K. R. Lee, J. Antibiot., 2009, 62, 335–338 CrossRef CAS PubMed
.
- S. Y. Kim, Y.-H. Baek, S. Y. Han, S. W. Lee, Y. H. Roh, K. W. Kim, S. H. Kang and J. S. Jeong, Korean J. Gastroenterol., 2018, 71, 94–97 CrossRef PubMed
.
- C. Chan, H. Lam, S. Chiu, M. Tse and F. Lau, Hong Kong Med. J., 2016, 22, 124–130 CAS
.
- D. S. Kim, N.-I. Baek, S. R. Oh, K. Y. Jung, J. H. Kim and H.-K. Lee, Arch. Pharmacal Res., 1997, 20, 201–205 CrossRef CAS
.
- K. H. Kim, S. U. Choi, H. J. Noh, O. Zee and K. R. Lee, Nat. Prod. Sci., 2014, 20, 76–79 CAS
.
- J. Herbich, K. Lohwag and R. Rotter, Arch. Toxicol., 1966, 21, 310–320 CAS
.
-
R. Lafont, Archives of Insect Biochemistry and Physiology, Published in Collaboration with the Entomological Society of America, 1997, vol. 35, pp. 3–20 Search PubMed
.
-
R. Lafont and D. Horn, Ecdysone from chemistry to mode of action, ed. J. Koolman, Thieme Medical Publishers, New York, 1989, pp. 39–63 Search PubMed
.
- Y. Miyata, T. Diyabalanage, C. D. Amsler, J. B. McClintock, F. A. Valeriote and B. J. Baker, J. Nat. Prod., 2007, 70, 1859–1864 CrossRef CAS
.
- D. J. Faulkner, Nat. Prod. Rep., 2001, 18, 1R–49R RSC
.
- K. Vokáč, M. Buděšnský, J. Harmatha and J. Kohoutová, Phytochemistry, 1998, 49, 2109–2114 CrossRef
.
- K. Vokáč, M. Buděšínský, J. Harmatha and J. Píš, Tetrahedron, 1998, 54, 1657–1666 CrossRef
.
- L. Zhang, O. E. Fasoyin, I. Molnár and Y. Xu, Nat. Prod. Rep., 2020, 37, 1181–1206 RSC
.
- M. J. Moss and R. G. Hendrickson, Clin. Toxicol., 2019, 57, 99–103 CrossRef CAS
.
- T. Matsumoto, W. Trueb, R. Gwinner and C. Eugster, Helv. Chim. Acta, 1969, 52, 716–720 CrossRef CAS
.
- S. Obermaier and M. Müller, Angew. Chem., Int. Ed., 2020, 59, 12432–12435 CrossRef CAS
.
- W. C. Zinkand, W. C. Moore, C. Thompson, A. I. Salama and J. Patel, Mol. Chem. Neuropathol., 1992, 16, 1–10 CrossRef CAS PubMed
.
- M. B. Hermit, J. R. Greenwood, B. Nielsen, L. Bunch, C. G. Jørgensen, H. T. Vestergaard, T. B. Stensbøl, C. Sanchez, P. Krogsgaard-Larsen and U. Madsen, Eur. J. Pharmacol., 2004, 486, 241–250 CrossRef CAS
.
- D. Chandra, F. Jia, J. Liang, Z. Peng, A. Suryanarayanan, D. Werner, I. Spigelman, C. Houser, R. Olsen and N. Harrison, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 15230–15235 CrossRef CAS
.
- H. Fritz, A. Gagneux, R. Zbinden, J. Geigy, S. Basle and C. Eugster, Tetrahedron Lett., 1965, 6, 2075–2076 CrossRef
.
- P. Catalfomo and C. H. Eugster, Bull. Narc., 1970, 22, 33 CAS
.
- J. S. Wiseman and R. H. Abeles, Biochemistry, 1979, 18, 427–435 CrossRef CAS
.
- B. Haberl, R. Pfab, S. Berndt, C. Greifenhagen and T. Zilker, Clin. Toxicol., 2011, 49, 113–114 CrossRef PubMed
.
- O. Tottmar and P. Lindberg, Acta Pharmacol. Toxicol., 1977, 40, 476–481 CrossRef CAS
.
- H. Yoshimura, K. Takegami, M. Doe, T. Yamashita, K. Shibata, K. Wakabayashi, K. Soga and S. Kamisaka, Phytochemistry, 1999, 52, 25–27 CrossRef CAS
.
- K. Wakabayashi, K. Soga, T. Hoson, S. Kamisaka, H. Yoshimura and K. Shibata, Plant Growth Regul., 2001, 33, 169–173 CrossRef CAS
.
- Y. Morimoto, M. Takaishi, T. Kinoshita, K. Sakaguchi and K. Shibata, Chem. Commun., 2002, 42–43 RSC
.
- R. Stone, Science, 2012, 335, 1293 CrossRef CAS PubMed
.
- Z. Y. Zhou, G. Q. Shi, R. Fontaine, K. Wei, T. Feng, F. Wang, G. Q. Wang, Y. Qu, Z. H. Li and Z. J. Dong, Angew. Chem., 2012, 124, 2418–2420 CrossRef
.
- P. E. Cryer, J. Clin. Invest., 2007, 117, 868–870 CrossRef CAS PubMed
.
- G.-Q. Shi, W.-L. Huang, J. Zhang, H. Zhao, T. Shen, R. E. Fontaine, L. Yang, S. Zhao, B.-L. Lu and Y.-B. Wang, PLoS One, 2012, 7, e35894 CrossRef CAS PubMed
.
- Y. Yamaura, M. Fukuhara, E. Takabatake, N. Ito and T. Hashimoto, Toxicology, 1986, 38, 161–173 CrossRef CAS PubMed
.
- T. Ohta, S. Nakajima, S.-I. Hatanaka, M. Yamamoto, Y. Shimmen, S. Nishimura, Z. Yamaizumi and S. Nozoe, Phytochemistry, 1987, 26, 565–566 CrossRef CAS
.
- S. Fushiya, S. Sato and S. Nozoe, Phytochemistry, 1992, 31, 2337–2339 CrossRef CAS
.
- K. Konno, K. Hashimoto, Y. Ohfune, H. Shirahama and T. Matsumoto, J. Am. Chem. Soc., 1988, 110, 4807–4815 CrossRef CAS
.
- S. Fushiya, S. Sato, T. Kanazawa, G. Kusano and S. Nozoe, Tetrahedron Lett., 1990, 31, 3901–3904 CrossRef CAS
.
- B. E. Cantrell, D. M. Zimmerman, J. A. Monn, R. K. Kamboj, K. H. Hoo, J. P. Tizzano, I. A. Pullar, L. N. Farrell and D. Bleakman, J. Med. Chem., 1996, 39, 3617–3624 CrossRef CAS PubMed
.
- K. Shin, A. Hitoshi and I. Michiko, Exp. Neurol., 1992, 116, 145–155 CrossRef CAS
.
- S. Fushiya, S. Sato, G. Kusano and S. Nozoe, Phytochemistry, 1993, 33, 53–55 CrossRef CAS
.
- K. Yoshikawa, K. Adachi, M. Nishijima, T. Takadera, S. Tamaki, K.-i. Harada, K. Mochida and H. Sano, Appl. Environ. Microbiol., 2000, 66, 718 CrossRef CAS PubMed
.
- S. Fushiya, Q. Q. Gu, K. Ishikawa, S. Funayama and S. Nozoe, Chem. Pharm. Bull., 1993, 41, 484–486 CrossRef CAS PubMed
.
- Y. Aizenberg-Gershtein, I. Izhaki, R. Santhanam, P. Kumar, I. T. Baldwin and M. Halpern, Sci. Rep., 2015, 5, 1–11 Search PubMed
.
- E. V. Dehmlow and H.-J. Schulz, Tetrahedron Lett., 1985, 26, 4903–4906 CrossRef CAS
.
- M. Tiecco, M. Tingoli, L. Testaferri, D. Chianelli and E. Wenkert, Tetrahedron, 1986, 42, 1475–1485 CrossRef CAS
.
- L. C. Wyatt, A. Moshnikova, T. Crawford, D. M. Engelman, O. A. Andreev and Y. K. Reshetnyak, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, E2811–E2818 CrossRef CAS PubMed
.
- R. J. Dinis-Oliveira, M. Soares, C. Rocha-Pereira and F. Carvalho, Hum. Exp. Toxicol., 2016, 35, 1016–1029 CrossRef CAS PubMed
.
- C. J. Deutsch and D. Swallow, Bmj, 2012, 345 CrossRef PubMed
.
- J.-M. Richard, D. Cantin-Esnault and A. Jeunet, Free Radicals Biol. Med., 1995, 19, 417–429 CrossRef CAS PubMed
.
- J.-M. Richard, E. E. Creppy, J.-L. Benoit-Guyod and G. Dirheimer, Toxicology, 1991, 67, 53–62 CrossRef CAS PubMed
.
- D. Cantin-Esnault, H. Oubrahim and J.-M. Richard, Free Radical Res., 2000, 33, 129–137 CrossRef CAS PubMed
.
- L. Buvall, H. Hedman, A. Khramova, D. Najar, L. Bergwall, K. Ebefors, C. Sihlbom, S. Lundstam, A. Herrmann and H. Wallentin, Oncotarget, 2017, 8, 91085 CrossRef PubMed
.
- N. K. Kaushik, N. Kaushik, P. Attri, N. Kumar, C. H. Kim, A. K. Verma and E. H. Choi, Molecules, 2013, 18, 6620–6662 CrossRef CAS PubMed
.
- R. G. Mehta, J. Liu, A. Constantinou, C. F. Thomas, M. Hawthorne, M. You, C. Gerhäuser, J. M. Pezzuto, R. C. Moon and R. M. Moriarty, Carcinogenesis, 1995, 16, 399–404 CrossRef CAS PubMed
.
- A. Duflos, A. Kruczynski and J.-M. Barret, Curr. Med. Chem.: Anti-Cancer Agents, 2002, 2, 55–70 CrossRef CAS
.
- A. Hofmann, R. Heim, A. Brack and H. Kobel, Experientia, 1958, 14, 107–109 CrossRef CAS PubMed
.
- P. ZILKA, Experientia, 1958, 14, 397–399 CrossRef PubMed
.
- A. Hofmann, R. Heim, A. Brack, H. Kobel, A. Frey, H. Ott, T. Petrzilka and F. Troxler, Helv. Chim. Acta, 1959, 42, 1557–1572 CrossRef CAS
.
- A. Leung and A. Paul, J. Pharm. Sci., 1968, 57, 1667–1671 CrossRef CAS PubMed
.
- J. Fricke, F. Blei and D. Hoffmeister, Angew. Chem., Int. Ed., 2017, 56, 12352–12355 CrossRef CAS PubMed
.
- T. Passie, J. Seifert, U. Schneider and H. M. Emrich, Addict. Biol., 2002, 7, 357–364 CrossRef CAS PubMed
.
- R. Carhart-Harris, B. Giribaldi, R. Watts, M. Baker-Jones, A. Murphy-Beiner, R. Murphy, J. Martell, A. Blemings, D. Erritzoe and D. J. Nutt, N. Engl. J. Med., 2021, 384, 1402–1411 CrossRef CAS PubMed
.
- A. Bartsch, M. Bross, P. Spiteller, M. Spiteller and W. Steglich, Angew. Chem., Int. Ed., 2005, 44, 2957–2959 CrossRef CAS PubMed
.
- S. Y. Kim, Y. H. Baek, S. Y. Han, S. W. Lee, Y. H. Roh, K. W. Kim, S. H. Kang and J. S. Jeong, Korean J. Gastroenterol., 2018, 71, 94–97 CrossRef PubMed
.
- T. Ohta, H. Inoue, G. Kusano and Y. Oshima, Heterocycles, 1998, 47, 883–891 CrossRef CAS
.
- K. H. Kim, K. M. Park, S. U. Choi and K. R. Lee, J. Antibiot., 2009, 62, 335–338 CrossRef CAS PubMed
.
- M. Strous and M. S. Jetten, Annu. Rev. Microbiol., 2004, 58, 99–117 CrossRef CAS PubMed
.
- E. G. Johnson, J. P. Sparks, B. Dzikovski, B. R. Crane, D. M. Gibson and R. Loria, Chem. Biol., 2008, 15, 43–50 CrossRef CAS PubMed
.
- T. Sawa, T. Akaike and H. Maeda, J. Biol. Chem., 2000, 275, 32467–32474 CrossRef CAS
.
- K. F. Lampe, Annu. Rev. Pharmacol. Toxicol., 1979, 19, 85–104 CrossRef CAS PubMed
.
- H. Pyysalo, 1975.
- R. Braun, U. Greeff and K. Netter, Xenobiotica, 1980, 10, 557–564 CrossRef CAS PubMed
.
- R. Braun, U. Greeff and K. J. Netter, Toxicology, 1979, 12, 155–163 CrossRef CAS PubMed
.
- J. W. Sims, J. P. Fillmore, D. D. Warner and E. W. Schmidt, Chem. Commun., 2005, 186–188 RSC
.
- T. B. Kakule, D. Sardar, Z. Lin and E. W. Schmidt, ACS Chem. Biol., 2013, 8, 1549–1557 CrossRef CAS
.
- P.-K. Chang, B. W. Horn and J. W. Dorner, Fungal Genet. Biol., 2009, 46, 176–182 CrossRef CAS PubMed
.
- M. Tokuoka, Y. Seshime, I. Fujii, K. Kitamoto, T. Takahashi and Y. Koyama, Fungal Genet. Biol., 2008, 45, 1608–1615 CrossRef CAS PubMed
.
- Q. Yue, L. Chen, Y. Li, G. F. Bills, X. Zhang, M. Xiang, S. Li, Y. Che, C. Wang, X. Niu, Z. An and X. Liu, mBio, 2015, 6, e00703–e00715 CrossRef CAS PubMed
.
- A. D. Wright, C. Osterhage and G. M. König, Org. Biomol. Chem., 2003, 1, 507–510 RSC
.
- H. V. K. Wangun, H.-M. Dahse and C. Hertweck, J. Nat. Prod., 2007, 70, 1800–1803 CrossRef CAS PubMed
.
- M. Matsuura, S. Kato, Y. Saikawa, M. Nakata and K. Hashimoto, Chem. Pharm. Bull., 2016, 64, 602–608 CrossRef CAS PubMed
.
- N. Yan, X. Liu, M. K. Pallerla and J. M. Fox, J. Org. Chem., 2008, 73, 4283–4286 CrossRef CAS PubMed
.
- K. Nitta, R. Stadelmann and C. Eugster, Helv. Chim. Acta, 1977, 60, 1747–1753 CrossRef CAS
.
- M. Malone, R. Robichaud, V. Tyler and L. Brady, Lloydia, 1962, 25, 231–237 CAS
.
- P. Catalfomo and C. H. Eugster, Helv. Chim. Acta, 1970, 53, 848–851 CrossRef CAS
.
- L. Gurevich and E. Nezdoiminogo, Mikol. Fitopatol., 1992, 26, 88–97 CAS
.
- J. Patocka, R. Wu, E. Nepovimova, M. Valis, W. Wu and K. Kuca, Int. J. Mol. Sci., 2021, 22, 2218 CrossRef CAS
.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |