Integration of a capacitor to a 3-D DNA walker and a biofuel cell-based self-powered system for ultrasensitive bioassays of microRNAs†
Fu-Ting
Wang
b,
Ke-Jing
Huang
 *ab,
Yang-Yang
Hou
b,
Xuecai
Tan
*ab,
Yang-Yang
Hou
b,
Xuecai
Tan
 *a,
Xu
Wu
*b,
Xin-Meng
Yu
b and
Xin
Zhou
b
*a,
Xu
Wu
*b,
Xin-Meng
Yu
b and
Xin
Zhou
b
aKey Laboratory of Chemistry and Engineering of Forest Products, Guangxi Key Laboratory of Chemistry and Engineering of Forest Products, Key Laboratory of Guangxi Colleges and Universities for Food Safety and Pharmaceutical Analytical Chemistry, School of Chemistry and Chemical and Engineering, Guangxi University for Nationalities, Nanning 530008, China. E-mail: kejinghuang@163.com; gxuntan@126.com
bCollege of Chemistry and Chemical Engineering, Xinyang Normal University, Xinyang 464000, China. E-mail: xwu@xynu.edu.cn
First published on 6th December 2021
Abstract
A self-powered microRNA biosensor with triple signal amplification systems was assembled through the integration of three-dimensional DNA walkers, enzymatic biofuel cells and a capacitor. The DNA walker is designed from an enzyme-free target triggered catalytic hairpin assembly of modified gold nanoparticles. When triggered by the target microRNA, the DNA walker will move along the catalytic hairpin track, resulting in a payload release of glucose oxidase. The enzymatic biofuel cell contains the glucose oxidase bioanode and a bilirubin oxidase biocathode that bring a dramatic open circuit voltage to realize the self-powered bioassays of microRNA. A capacitor is further coupled with the enzymatic biofuel cell to further amplify the electrochemical signal, and the sensitivity increases 28.82 times through optimizing the matching capacitor. Based on this design, the present biosensor shows high performance, especially for detection limit and sensitivity. Furthermore, the present biosensor was successfully applied for serum samples, directly demonstrating its good application in clinical biomedicine and disease diagnosis.
Introduction
MicroRNA (miRNA) is an endogenous, small non-coding regulatory molecule, and it plays an important role in the expression of genes.1,2 MiRNA-21 plays an essential role in a plethora of biological functions and diseases including development, cancer, cardiovascular diseases and inflammation.3–6 So developing effective detection strategies for miRNA-21 is crucial for many clinical diagnoses in biological processes and early disease screening. Recently, various DNA nanomachines, such as robots, DNA gears and DNA walkers have been developed for use in biosensor platforms.7–12 In particular, DNA walkers which can precisely control the specified tracks at the nanoscale have shown great potential for the sensitive detection of miRNAs.13–15 For instance, Jung et al.16 constructed a sensing platform using a catalytic hairpin assembly (CHA) to drive walking on the surface of the DNA-coated microparticles and realizing the sensitive detection of miRNA. Peng et al.17 designed a DNA zyme nanomachine, activated by a specific intracellular target and initiated autonomous walking on AuNPs for miRNA imaging in living cells. However, the common method of energy input for DNA walkers is to cleave the DNA substrates along the tracks via ribozymes18 or protein enzymes.19 The addition of extra reactants will simultaneously release and increase the detection complexity. Thus, efficient target-triggered, enzyme-free and self-powered strategies are still required for DNA walkers to detect miRNAs.Realizing the rapid payload release during the DNA walker movement is another advantage of DNA nanomachines.20–26 The electrical output of enzymatic biofuel cells (EBFCs) can provide a convenient way for realizing self-powered sensing, which is crucial for portable and on-site bioassays.27–29 However, the low current/power of EBFCs limits the sensitivity of the self-powered biosensor. Coupling EBFCs with capacitors can be an effective route, since the capacitor can accumulate the charges and deliver high power pulse outputs.30 Recently, Huang et al. have introduced capacitors into EBFCs, and the sensitivity increased by 18.4 times.31 Another work from Slaughter also demonstrated the successful utilization of a capacitor as a transducing element to convert the biochemical energy of glucose into electrical power.32 Au nanoparticles (AuNPs) have been mostly recommended because they can markedly facilitate electron transport due to excellent conductive ability and immobilization of more biomolecules on the electrode by the Au–S bond, and have been broadly applied to develop electrochemical biosensors.1–6
Based on the above discussion, herein, we propose an ultrasensitive and easy-to-use bioassay of miRNAs based on the combination of 3-D DNA walkers, EBFCs and a capacitor. As shown in Scheme 1, the EBFCs were constructed from a bilirubin oxidase (BOD)/AuNPs/carbon paper (CP) biocathode and a glucose oxidase (GOD)/3D DNA walker/CP bioanode. The local CHA reaction based 3-D DNA walker was constructed by assembling biotin-labeled and polyA-mediated hairpin DNA strands (H1, H2) on the surface of gold nanoparticles (AuNPs). Once the target miRNA was encountered, the recognition strand DNA H1 will hybridize with miRNA and open the hairpin structure. Then, the exposed part of H1 can spontaneously hybridize with the H2 and release miRNA. During this process, each step released two biotins, and the GOD will subsequently combine with the biotin by interacting with biotin. The miRNA will act as a catalyst to walk within one AuNP or even between particles to fix more GOD. After the GOD appeared at the surface of the bioanode, glucose oxidation will take place and the lost electrons will transfer to the cathode to drive the digital multimeter (DMM), realizing the self-powered biosensing of miRNA. In addition, when capacitor is added in the system, the electrons from the EBFCs can be first stored in the capacitor, and then the capacitor give an amplified transient current. Thus, through the integration of 3-D DNA walkers, EBFCs and a capacitor, a target-triggered, enzyme-free and self-powered biosensing strategy with triple signal amplification systems for miRNA detection was developed.
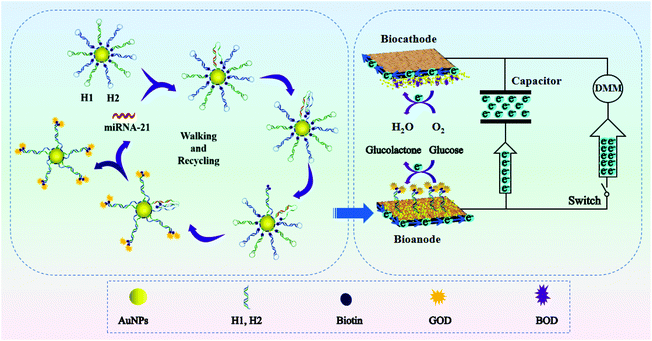 | ||
| Scheme 1 Schematic illustration of enzyme-free 3-D DNA walking nanomachine based self-powered biosensing with triple signal amplification systems. | ||
Results and discussion
Fabrication of self-powered miRNA biosensor
The self-powered miRNA biosensor was assembled on the breadboard, consisting of EBFCs, a capacitor and a DMM-joined electronic circuit (Fig. 1). The EBFC was operated at room temperature. To test the sensing performance, 30 μL of different concentrations of miRNA-21 was incubated with 30 μL of H1, H2-AuNPs at 4 °C for 150 min, and added to 5 mL of a supporting electrolyte to test the EOCV. The input voltage from the EBFC is converted to stepped-up power and charged to the capacitor. Thereafter, the capacitor is automatically shorted under the control of a switch and provides instantaneous current, which can be conveniently determined using DMM.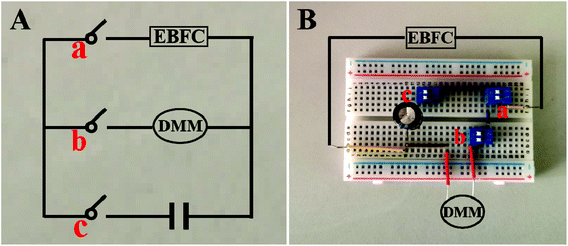 | ||
| Fig. 1 Photos of circuit diagram (A) and breadboards used for measuring the instantaneous current of the device (B). | ||
Characterization of AuNP substrate electrode
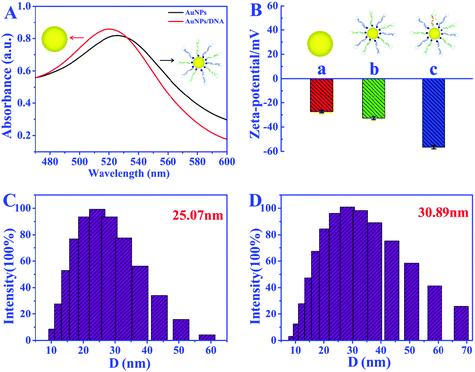
AuNPs were used as carriers for poly A mediated hairpin DNA strands. The SEM images in Fig. S1A and S1B† and the TEM image in Fig. S1C† show the even distribution of AuNPs with average diameter of 15 nm. As shown in the HRTEM image in Fig. S1D,† the stripe spacing of 0.238 nm is in accordance with the (111) crystal planes of Au, further demonstrating the successful synthesis of AuNPs.The preparation of the 3-D DNA walker was verified through UV–vis absorption spectroscopy, zeta potential analysis, and dynamic light scattering (DLS) (Fig. 2C and D). From the results of the UV–vis absorption spectroscopy shown in Fig. 2A, the red-shift (from 520 to 528 nm) of the absorption peak of AuNPs confirms the successful loading of H1 and H2 DNA strands. The increased zeta potentials of H1, H2-AuNPs and 3D DNA walker in Fig. 2B demonstrate the remarkable alterations of the surface charges during the assembly process. In Fig. 2C and D, the DLS analysis results show that the average hydrodynamic diameter was increased from 25.07 nm (AuNPs) to 30.89 nm (3D DNA walker nanodevice). The above results suggest the successful formation of the 3-D DNA walker.
Assembly and characterization of bioanode and biocathode
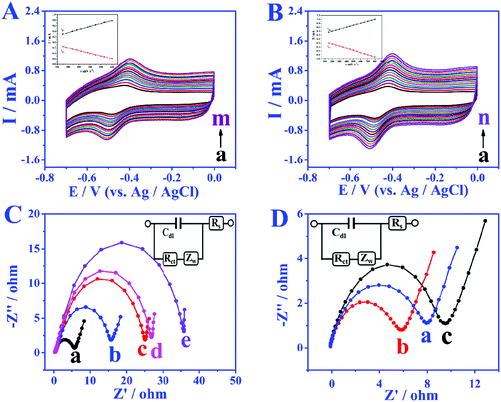
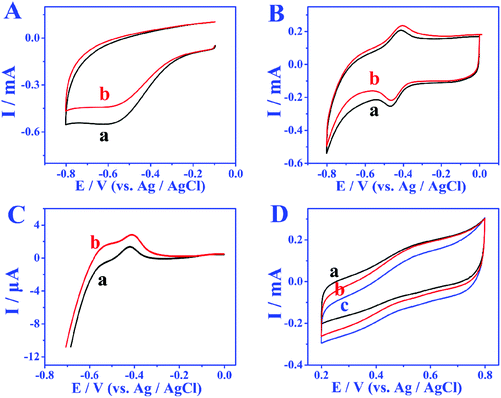
In order to study the electron transfer mode and electron transfer rate between the immobilized enzyme and electrode the GOD modified electrode was taken as an example. The cyclic voltammetry (CV) curves of different modified electrodes at different sweep rates were recorded and the enzyme loadings were calculated through the laviron equation.33 From the CV curves in Fig. 3A and B, the redox peak spacings are all lower than 20 mV with the increase of scan rates, indicating the high reversibility of the redox process of GOD active sites. The apparent surface coverage (Γ × cat) of GOD on the miRNA/3D DNA walker/CP electrode (Fig. 3B) was estimated to be 1.0638 × 10−9 mol cm−2, which was 2.3 times the value of the GOD/3D DNA walker/CP electrode (Fig. 3A). The higher value was mainly because the miRNA can open the hairpin structure, and the exposed biotin part of H1 and H2 can spontaneously interact with GOD to enhance the electron transfer. The assembly processes of the bioanodes and biocathodes were demonstrated through electrochemical impedance spectroscopy (EIS). The EIS results of different bioanodes are shown in Fig. 3C. The CP electrode (curve a) has minimal charge-transfer resistance (Ret). The Ret significantly increased after the modification of H1, H2-AuNPs (curve b), an inert blocking layer being formed by the biomolecules that block electron transfer. Because of the steric-hindrance effects, the Ret values are stepwise increased after the loading of MCH (curve c) and the assembling of miRNA (curve d) and GOD (curve e). As shown in Fig. 3D, different biocathodes also exhibit different EIS results. The EIS result of bare CP is displayed as curve a, and the semicircle decreases after the assembling of AuNPs (curve b), thus the enhancement of matrix conductivity by AuNPs was proved. The Ret significantly increased after the loading of BOD (curve c). These results clearly illustrate the assembly and electrochemical kinetics of the bioelectrodes.The bioelectrocatalytic behavior and target miRNA capture of bioanodes were revealed by CV test (Fig. 4A). When no miRNA was present, there were almost no peak currents in phosphate buffer solution without glucose (curve a) and phosphate buffer solution containing 5 mmol L−1 glucose (curve b), which is attributed to biotinylated-H1, and H2 not being able to fix more GOD, leading to the blocking of the redox probes. When miRNA is present, compared with the CV result of GOD/MCH/H1, H2-AuNPs/CP electrode (Fig. 4A), the GOD/miRNA/MCH/H1, the H2-AuNPs/CP (Fig. 4B) bioanode shows obvious redox peaks, thus demonstrating the successful modification of GOD. After the employment of miRNA and 5 mM glucose (curve b in Fig. 4B), as GOD is fixed, the peak intensity will become greater. The signal enhancement of glucose was also demonstrated by the linear sweep voltammetry (LSV) test. In Fig. 4C, when curve b (with 5 mmol L−1 glucose) is compared with curve a (without glucose), higher oxidation peaks were observed. The BOD/AuNPs/CP biocathode (Fig. 4D) in N2 (curve a) saturated solution, air (curve b) saturated solution, and O2 (curve c) saturated solution were also investigated by CV test. From the comparison, a pair of redox peaks at about 520 mV caused by the redox process of BOD can still be found with the absence of O2, confirming that BOD is the first electron acceptor. These results successfully demonstrated the effective assembly of bioanodes and biocathodes.
Optimization
To achieve the high performance self-powered biosensing system, a series of experimental conditions were optimized. The number of poly A was first optimized. As shown in Fig. S2A,† poly A with a number of 10 is used, since electrochemical response signal is the strongest under this value. Then the molar ratio ranges of AuNPs, H1 and H2 were optimized to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to approach the peak intensity saturation (Fig. S2B†). On the other hand, the hybridization reaction between AuNPs and H1, H2 was simultaneously implemented, so the effect of hybridization reaction time on the electrochemical signal was studied. As shown in Fig. S2C,† when the incubation time increased to 16 h, the peak reaches a plateau, thus the optimized time is chosen as 16 h. In addition, a hybridization reaction time of 150 min between the target miRNA and H1 is applied in the following experiments to achieve the maximum peak current (Fig. S2D†).
1 to approach the peak intensity saturation (Fig. S2B†). On the other hand, the hybridization reaction between AuNPs and H1, H2 was simultaneously implemented, so the effect of hybridization reaction time on the electrochemical signal was studied. As shown in Fig. S2C,† when the incubation time increased to 16 h, the peak reaches a plateau, thus the optimized time is chosen as 16 h. In addition, a hybridization reaction time of 150 min between the target miRNA and H1 is applied in the following experiments to achieve the maximum peak current (Fig. S2D†).
Analytical performance of the MiRNA biosensor
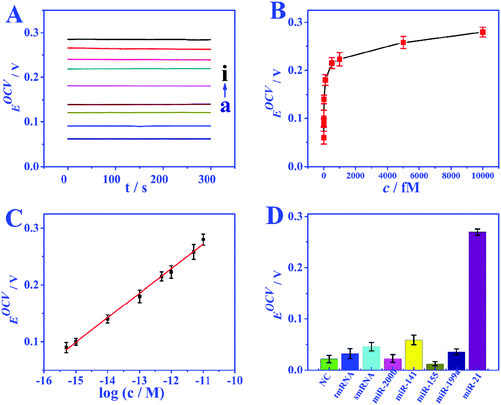
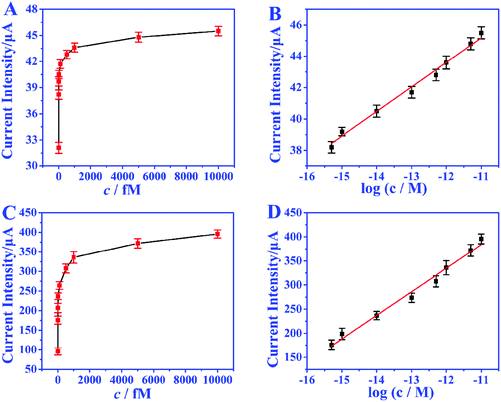
Under the optimized conditions, the analytical performance of the biosensor was tested. The feasibility of the biosensor is investigated through the response of EOCV to different miRNA concentrations. In the absence of miRNA, nearly no GOD molecules can bind to biotin to provide an electron donor, and the EOCV is only 0.06 V. Nevertheless, when miRNAs are present, the recognition strand H1 will hybridize with miRNAs, and this hybridization process will walk within one particle or even between particles to continuously immobilize GOD. Fig. 5A and B show the EOCV of different miRNA-21 concentrations. There is a linear relationship between the EOCV and the logarithm of miRNA-21 concentration in the range of 0.5–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fmol L−1 (Fig. 5C). The linear equation is EOCV = 0.75 + 0.04
000 fmol L−1 (Fig. 5C). The linear equation is EOCV = 0.75 + 0.04![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) c (correlation coefficient R2 = 0.996), and the detection limit of 0.17 fmol L−1 (S/N = 3). These analytical performances are superior to miRNA biosensors based on other detection techniques, especially the detection limit (Table S3†).
c (correlation coefficient R2 = 0.996), and the detection limit of 0.17 fmol L−1 (S/N = 3). These analytical performances are superior to miRNA biosensors based on other detection techniques, especially the detection limit (Table S3†).
Selectivity, reproducibility and stability
The specificity of the proposed EBFC-based biosensor was studied by possible interfering substances (miRNA-141, miRNA-200b, miRNA-199a, and miRNA-155) and base mismatched sequences (NC, smRNA and tmRNA). The analytical performances on the above target molecules are compared in Fig. 5D, and the result clearly indicates that the developed biosensor has high selectivity. The reproducibility was studied by repeatedly measuring six freshly fabricated electrodes under the same experimental conditions, and a low relative standard deviation (RSD) of less than 5.8% indicating the high reproducibility. In addition, the stability was further evaluated. The performance of the bioanode and biocathode stored under 4 °C for 20 days was compared with the freshly prepared ones. A high signal intensity retention of over 95.4% demonstrates excellent stability.Real sample analysis
The feasibility of self-powered miRNA biosensor was verified by human serum sample (from the Xinyang Central Hospital), and an additional standard recovery experiment was conducted. Samples 1 and 2 were obtained from the serum of healthy persons, and samples 3 and 4 were got from the serum of patients. The test results are provided in Table S4,† recoveries of 98.33–103.34% and RSDs of 2.56–5.88% were obtained after the addition of miRNA. These testing data show the high promise of this self-powered biosensor for miRNA determination in human serum samples.Optimization of capacitor matching mechanism
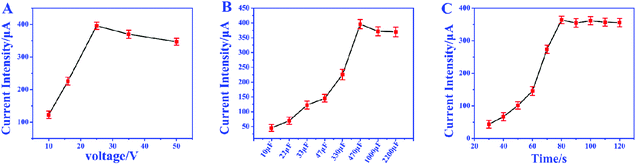
In order to achieve the highest amplification, the matching capacitor was carefully selected. The detecting currents upon commercial capacitors with different nominal voltages were tested. As shown in Fig. 6A, the highest instantaneous current is achieved with the 25 V capacitor. Therefore, 25 V was chosen as the nominal voltage of the matching capacitor. Then 25 V capacitors with different nominal capacitances were further evaluated in the biosensor. From Fig. 6B, we can find that the instantaneous currents increase with higher capacitance and reach the top at 470 μF. So, the suitable nominal parameters of the matching capacitor should be 25 V and 470 μF. The instantaneous current delivered under different charging times was also studied to ensure the complete electron storage. In Fig. 6C, the current density reaches the highest value after charging the capacitor for 80 s, which means the electrons transferred from the biosensor were completely stored in the capacitor. Thus, the instantaneous current obtained after charging the capacitor (25 V, 470 μF) for 80 s was recorded as the current response for each investigation.The amplification enhancement of the capacitor was then demonstrated. Fig. 7A and B show the linear relationship between the corresponding EOCV and the logarithm of miRNA-21 concentrations. After the capacitor was introduced, the current signals were significantly increased (Fig. 7C) and a good linear relationship was obtained (Fig. 7D). The sensitivity of the biosensor can reach 8.07 μA nM−1, which is 28.82 times higher than with no capacitor being added (0.28 μA nM−1). Then a digital multimeter (DMM) was further introduced to display the current response. Fig. 8A is the instantaneous current value of the uncharged capacitor, Fig. 8B is the instantaneous current value when charging, and Fig. 8C is the instantaneous current value after charging. The highly increased current directly reveals the amplification effect of the capacitor. The above analysis demonstrates that the capacitor can significantly enhance the detection signals, and high-performance biosensors can be developed by combining the capacitor, biological fuel cells and biological amplification technologies.
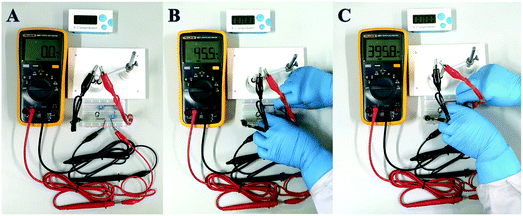 | ||
| Fig. 8 The optical photographs of uncharged capacitor (A), after incubation with 1 nM miRNA (B), and after charging (C). | ||
Conclusion
In summary, a triple signal amplification self-powered ultrasensitive electrochemical biosensing device has been developed for the selective detection of miRNAs by combining a 3D DNA nanomachine, EBFCs and a capacitor. The CHA reaction-based enzyme-free 3D DNA walker can effectively avoid enzyme distraction and significantly enhance the reaction efficiency to output high detection signals. The EBFC biosensor constructed from a BOD/AuNPs/CP biocathode and GOD/3D DNA walker/CP bioanode shows a wide linear range and low detection limit, with high selectivity, reproducibility and stability. In addition, the sensitivity of the biosensor can be dramatically increased to 28.82 times through coupling suitable capacitors, and the detecting signals can be directly read out by a portable digital multimeter (DMM). This work provides an ultrasensitive and easy-to-use detection strategy for miRNAs and shows great potential for accurate early cancer diagnosis.Data availability
Additional data are available in the ESI.†Author contributions
F. T. Wang and Y. Y. Hou: most of the experiments and writing of the manuscript. X. M. Yu and X. Zhou: prepared materials. X. C. Tan: supervision of experiments, discussion, and writing of the manuscript. K. J. Huang and X. Wu: conceptualization, funding acquisition, and supervision; writing, review, and editing of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the financial support from the Natural Science Foundation of China (No. 22074130, 51902280, 21365004), Zhongyuan Thousand Talents Plan – Science and Technology Innovation Leading Talents Project (No. 204200510030), Natural Science Foundation of Henan (No. 202300410329), the Key Research and Development Project of Guangxi (AB18126048), Guangxi Innovation-Driven Development Special Fund Project (AA18118013-10), and Innovation Project of Guangxi Graduate Education (JGY2019062). The authors also acknowledge the great support from the Analysis Testing Center of Xinyang Normal University for materials characterization.References
- Y. X. Chen, K. J. Huang and K. X. Niu, Biosens. Bioelectron., 2018, 99, 612–624 CrossRef CAS.
- H. L. Shuai, K. J. Huang, Y. X. Chen, L. X. Fang and M. P. Jia, Biosens. Bioelectron., 2017, 89, 989–997 CrossRef CAS PubMed.
- F. T. Wang, Y. H. Wang, J. Xu, K. J. Huang, Z. H. Liu, Y. F. Lu, S. Y. Wang and Z. W. Han, Nano Energy, 2020, 68, 104310 CrossRef CAS.
- C. C. Gu, P. P. Gai, X. K. Kong, T. Hou and F. Li, Anal. Chem., 2020, 92, 5426–5430 CrossRef CAS.
- J. F. Chang, W. X. Lv, J. H. Wu, H. Y. Li and F. Li, Chin. Chem. Lett., 2021, 32, 775–778 CrossRef CAS.
- H. L. Shuai, K. J. Huang, L. L. Xing and Y. X. Chen, Biosens. Bioelectron., 2016, 86, 337–345 CrossRef CAS PubMed.
- P. Zhang, Z. Lin and Y. Zhuo, Anal. Chem., 2017, 89, 1338–1345 CrossRef CAS PubMed.
- Y. Chang, Z. Wu and Q. Sun, Anal. Chem., 2019, 91, 8123–8128 CrossRef CAS PubMed.
- F. Luo, F. Chen and Y. Xiong, Anal. Chem., 2021, 93, 4506–4512 CrossRef CAS PubMed.
- Y. Qi, Y. Zhai and W. Fan, Anal. Chem., 2020, 93, 1620–1626 CrossRef PubMed.
- N. Wu, K. Wang and Y. T. Wang, Anal. Chem., 2020, 92, 11111–11118 CrossRef CAS PubMed.
- T. Kang, J. Zhu and X. Luo, Anal. Chem., 2021, 93, 2519–2526 CrossRef CAS PubMed.
- M. M. Lv, J. W. Liu, R. Q. Yu and J. H. Jiang, Chem. Sci., 2020, 11, 10361–10366 RSC.
- L. Wang, P. Liu and Z. Liu, ACS Sens., 2020, 5, 3584–3590 CrossRef CAS PubMed.
- H. Y. Lv, A. Y. Chen, W. Q. Cheng, L. S. Kong, M. Zhao, S. J. Ding, H. X. Ju and W. Cheng, Anal. Chem., 2020, 92, 15624–15631 CrossRef CAS PubMed.
- C. Jung, P. Allen and A. Ellington, Nat. Nanotechnol., 2016, 11, 157–163 CrossRef CAS PubMed.
- H. Peng, X. F. Li and H. Zhang, Nat. Commun., 2017, 8, 14378 CrossRef PubMed.
- B. Y. Huo, Y. L. Hu, Z. X. Gao and G. K. Li, Talanta, 2021, 222, 121565 CrossRef CAS PubMed.
- J. Chen, Z. W. Luo, C. J. Sun, Z. J. Huang, C. Zhou, S. Yin, Y. X. Duan and Y. X. Li, TrAC, Trends Anal. Chem., 2019, 120, 115626 CrossRef CAS.
- T. Yan, L. Zhu and H. Ju, Anal. Chem., 2018, 90(24), 14493–14499 CrossRef CAS PubMed.
- Y. Yin, G. Chen and L. Gong, Anal. Chem., 2020, 92, 9247–9256 CrossRef CAS PubMed.
- H. Yang, M. Xiao and W. Lai, Anal. Chem., 2020, 92, 4990–4995 CrossRef CAS PubMed.
- X. Peng, Z. B. Wen and P. Yang, Anal. Chem., 2019, 91, 14920–14926 CrossRef CAS PubMed.
- N. Liao, M. C. Pan and L. Wang, Anal. Chem., 2021, 93, 4051–4058 CrossRef CAS PubMed.
- P. Miao and Y. G. Tang, Anal. Chem., 2019, 91, 15187–15192 CrossRef CAS PubMed.
- E. Xiong, D. Zhen and L. Jiang, Anal. Chem., 2019, 91, 15317–15324 CrossRef CAS PubMed.
- J. Xu, Y. H. Wang and Z. Wei, Sens. Actuators, B, 2021, 327, 128933 CrossRef CAS.
- F. T. Wang, Y. H. Wang and J. Xu, J. Mater. Chem. B, 2020, 8, 1389–1395 RSC.
- C. C. Gu, L. P. Bai, L. Pu, P. P. Gai and F. Li, Biosens. Bioelectron., 2021, 176, 112907 CrossRef CAS PubMed.
- H. Liu, Y. Xiang, Y. Lu and R. M. Crooks, Angew. Chem., Int. Ed., 2012, 51, 6925–6928 CrossRef CAS PubMed.
- Y. H. Wang, F. T. Wang, Z. W. Han, K. J. Huang, X. M. Wang, Z. H. Liu, S. Y. Wang and Y. F. Lu, Sens. Actuators, B, 2020, 304, 127418 CrossRef CAS.
- A. Baingane, S. Narayanan and G. Slaughter, Sens. Bio-Sens. Res., 2018, 20, 41–46 CrossRef.
- L. L. Wang, H. H. Shao, W. J. Wang, J. R. Zhang and J. J. Zhu, Nano Energy, 2018, 44, 95–102 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1nr06271a |
| This journal is © The Royal Society of Chemistry 2022 |