Open Access Article
Open Access ArticlePreparation, characterization, and biological activity study of thymoquinone-cucurbit[7]uril inclusion complex
Lubna Alrawashdeh*a,
Khaleel I. Assaf *b,
Walhan Alshaer
*b,
Walhan Alshaer c,
Fadwa Odehd and
Suhair A. Bani-Attae
c,
Fadwa Odehd and
Suhair A. Bani-Attae
aDepartment of Chemistry, Faculty of Science, The Hashemite University, P.O. Box 330127, Zarqa, 13133, Jordan. E-mail: lubna.reyad@hu.edu.jo
bDepartment of Chemistry, Al-Balqa Applied University, Al-Salt, 19117, Jordan. E-mail: khaleel.assaf@bau.edu.jo
cCell Therapy Center, The University of Jordan, Amman, 11942, Jordan
dDepartment of Chemistry, School of Science, The University of Jordan, Amman, 11942, Jordan
eDepartment of Chemistry, Faculty of Science, University of Tabuk, 71491, Saudi Arabia
First published on 12th January 2022
Abstract
In this study, the formation of a host–guest inclusion complex between cucurbit[7]uril (CB[7]) and thymoquinone (TQ) was investigated in aqueous solution. The formation of a stable inclusion complex, CB[7]–TQ, was confirmed by using different techniques, such as 1H NMR and UV-visible spectroscopy. The aqueous solubility of TQ was clearly enhanced upon the addition of CB[7], which provided an initial indication for supramolecular complexation. The complexation stoichiometry and the binding constant of the inclusion complex were determined through a combination of two sets of titration methods, including UV-visible and fluorescence displacement titrations. Both methods suggested the formation of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry between CB[7] and TQ with moderate binding affinity of 3 × 103 M−1. Density functional theory (DFT) calculations were also performed to verify the structure of the resulted host–guest complex and to support the complexation stoichiometry. The theoretical calculations were in agreement with experimental results obtained by 1H NMR spectroscopy. Most importantly, the cytotoxic effect of the CB[7]–TQ complex was investigated against cancer and normal cell lines. The results showed that the anticancer activity of TQ against MDA-MB-231 cells was enhanced by the complexation with CB[7], while no significant effect was observed in MCF-7 cells. The results also confirmed the low toxicity of the CB[7] host molecule that supports the use of CB[7] as a drug carrier.
1 stoichiometry between CB[7] and TQ with moderate binding affinity of 3 × 103 M−1. Density functional theory (DFT) calculations were also performed to verify the structure of the resulted host–guest complex and to support the complexation stoichiometry. The theoretical calculations were in agreement with experimental results obtained by 1H NMR spectroscopy. Most importantly, the cytotoxic effect of the CB[7]–TQ complex was investigated against cancer and normal cell lines. The results showed that the anticancer activity of TQ against MDA-MB-231 cells was enhanced by the complexation with CB[7], while no significant effect was observed in MCF-7 cells. The results also confirmed the low toxicity of the CB[7] host molecule that supports the use of CB[7] as a drug carrier.
Introduction
Nigella sativa (also called black seed or black cumin) is a promising medicinal plant, especially in the Mediterranean region and Western Asian countries including India, Pakistan, and Afghanistan.1,2 It has been used to treat a range of health conditions for many years, such as lung diseases, arthritis, and hypercholesterolemia. The biological activity of nigella sativa is mainly related to one of its main active components, namely, thymoquinone (TQ, 2-isopropyl-5-methylbenzo-1,4-quinone, Fig. 1).3 Several studies have been established to investigate the therapeutic effect of TQ. These studies showed strong indications that TQ has a very interesting and significant therapeutic effect in cancer, inflammation, and as an antioxidant agent through different modes of action. For example, TQ is able to arrest tumor cells at different stages of their progression.2 Recently, TQ showed a potential inhibition activity against coronavirus infections.4,5Despite the promising therapeutic efficiency of TQ, the lack of bioavailability, pharmacokinetic parameters, and formulation problems deferred the usage of TQ in the clinical development.3 The bioavailability problem (for example, poor solubility in water) is mainly related to the fact that TQ is a hydrophobic molecule. In addition, it has been reported that TQ has stability problems in solution; in particular, TQ is very sensitive to heat and light.6 Many attempts have been done to overcome the bioavailability and stability difficulties. The most important attempt was established by encapsulating TQ within different nanomaterials (vehicles/carriers). For example, several drug delivery systems were used, such as liposomes,7 lipids,8 cyclodextrins (CDs),9,10 nanoparticles,11 etc. These studies showed that encapsulation of TQ within the new environments (carrier/vehicle) can improve its bioavailability, stability (thermal/light), and activity as an anti-cancer agent.
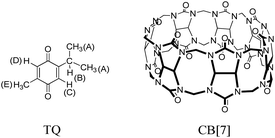
In this work, we are interested in introducing another and new host molecule to TQ, for the purpose of defining possible improvements in the performance of TQ pharmaceutical properties when associated with this host. Then we compare the obtained results with the previous studies that used different host molecules. Cucurbit[n]urils (CB[n], Fig. 1) are another attractive class of water soluble macrocyclic host molecules, which, like CDs and calixarenes, have a cavity that can accommodate different sizes of hydrophobic guest molecules.12 CBs are composed of a different number of glycoluril units (n = 5–10) joined by pairs of methylene bridges.13,14 The members of this family have a pumpkin shaped, highly symmetrical and rigid structure. They also possess a hydrophobic cavity and two identical partially-negatively charged carbonyl portals (hydrophilic).15 The binding of guest molecules to CB[n] can be driven by ion-dipole, and dipole–dipole interactions (suitable for cationic moiety parts), or through the hydrophobic effect (suitable to bind neutral and hydrophobic residues).16 Cucurbit[7]uril (CB[7], Fig. 1) is the most soluble member of the CB[n] family.14,17
Recent studies start to give notable interest to the CB[n] family in the biological and medicinal fields.18 Particular focus has been given to the CB[n] family in the drug delivery area, and many studies have been using CB[n]-type molecular hosts as a drug delivery vehicle for a large number of drugs.19 Based on the previous studies, it has been suggested that CB[n] showed significant advantages over the CD host molecule, which is the most common choice in the drug delivery area. For example, the binding constants (K) of CB[n]-guest complexes are higher than those of CDs in aqueous medium.20 Also, the encapsulation of drug molecules in CB[n] provides a method for slow drug release, facilitate drug targeting, improve chemical and thermal stability, reduction in toxicity, and enhance the aqueous solubility of poorly soluble drugs. Most importantly, CBs are a promising drug delivery system, being non-toxic and highly biocompatible.20,21
Herein, we report for the first time the formation of a stable inclusion complex between TQ and CB[7] in aqueous solution. The complexation is investigated by 1H NMR and UV-visible spectroscopy. The binding constant is determined by using UV-visible and fluorescence displacement titrations. In addition, a theoretical evaluation of the CB[7]–TQ inclusion complex stoichiometry and energy measurements are performed. Moreover, the antiproliferative effect of the CB[7], TQ, and CB[7]–TQ complex is investigated against two breast cancer cell lines, MDA-MB-231 and MCF-7, and compared to the effect against human dermal fibroblast cell line, HDF, to evaluate retaining toxicity of TQ after complexation with CB[7]. A previously reported βCD–TQ complex was used for comparison with CB[7]–TQ.
Experimental section
Chemicals and instruments
Thymoquinone, acridine orange base (AO), β-cyclodextrin (βCD) and D2O (99 atom % D) were purchased from Sigma-Aldrich (USA). Cucurbit[7]uril (CB[7]) was prepared according to literature.22 All reagents and chemicals were used without further treatment. 1H NMR spectra were performed on a Bruker AVANCE-III 400 MHz NanoBay FT-NMR spectrometer, in D2O and referenced in ppm with respect to a tetramethylsilane (TMS). The UV-visible absorption spectra were measured on Cary-100 Bio instrument. Fluorescence experiments were performed using Jasco spectrofluorometer (FP-6500).1H NMR titration of TQ and CB[7]
CB[7] stock solution (host) was prepared by dissolving 0.01 g of CB[7] in 0.7 mL D2O, heating and sonication were used to speed the dissolution; the CB[7] concentration was corrected to 80% content. The CB[7] solution was then filtrated using syringe filter (0.45 μm, nylon). Different aliquots of CB[7] stock solution were then added into a solution of TQ (9.7 mM) in D2O to obtain 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of CB[7]
1 ratio of CB[7]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TQ.
TQ.
UV-visible titration of TQ and CB[7]: complexation stoichiometry and binding study
Stock solution of CB[7] was prepared first (11 mM), different aliquots of CB[7] stock solution were added to a solution of TQ (1 mM). The UV-visible spectrum was recorded after each addition until getting a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of CB[7]
1 ratio of CB[7]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TQ. The complexation stoichiometry and the binding constant of CB[7]–TQ inclusion complex were determined using the Benesi–Hildebrand equation based on the data from UV-visible spectrophotometry. Eqn (1) and (2) were used for 1
TQ. The complexation stoichiometry and the binding constant of CB[7]–TQ inclusion complex were determined using the Benesi–Hildebrand equation based on the data from UV-visible spectrophotometry. Eqn (1) and (2) were used for 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 binding models.
2 binding models.
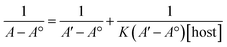 | (1) |
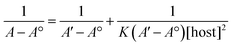 | (2) |
According to the design of Benesi–Hildebrand experiment, A° is absorbance of the free TQ, while A is absorbance at different CB[7] concentrations, A′ is absorbance at the maximum concentration of the host. The association constant (K) is calculated from the Benesi–Hildebrand equation, after plotting (1/A − A°) against (1/[host]) for 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (host
1 (host![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) guest stoichiometric ratio), or (1/[host]2) for 1
guest stoichiometric ratio), or (1/[host]2) for 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (host
2 (host![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) guest stoichiometric ratio). Binding constant is then calculated as:
guest stoichiometric ratio). Binding constant is then calculated as:
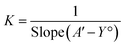 | (3) |
Fluorescence titration of CB[7] with AO dye and binding constant of CB[7]–AO inclusion complex
An aqueous solution contains CB[7] and AO (1 mM CB[7] and 1 μM AO dye) was prepared first. Different aliquots of this solution were added into a solution of AO (1 μM). The emission peak of AO at λem = 521 nm (λex = 470 nm) was followed to study the effect of adding CB[7] on the emission of AO. The additions were stopped after obtaining a plateau in the experimental fluorescence data. Binding constant of CB[7]–AO inclusion complex was obtained by fitting the titration data20,21 at λem = 510 nm by using Origin program.Displacement titration and binding constant of CB[7]–TQ inclusion complex
An aqueous solution contains 10 μM CB[7] and 1 μM AO dye, was prepared first. Then, a solution of 10 μM CB[7], 1 μM AO dye, and 1.6 mM TQ was prepared. For displacement titration, different aliquots of TQ solution were added into the first solution (CB[7]–AO). The emission peak of AO encapsulated species at λem = 510 nm (λex = 470 nm) was followed to study the effect of adding TQ on the emission. The additions were stopped after obtaining a plateau in the experimental fluorescence data. Binding constant of CB[7]–TQ inclusion complex was obtained by fitting the titration data22,23 at λem = 510 nm by using Origin program.Quantum chemical calculations
Density functional theory (DFT) calculations were performed using Gaussian 09.23 The dispersion-correct DFT method (wB97XD)24 was used with the 6-31G* basis set. All calculations were performed in the gas phase.Cells culture
Human breast adenocarcinoma (MDA-MB-231) and human dermal fibroblast (HDF) cell lines were cultured in Dulbecco's Modified Eagle Medium (DMEM) (Euroclone SpA, Italy), while Human breast cancer (MCF-7) cell line was cultured in RPMI-1640 medium (Euroclone SpA, Italy). Both DMEM and RPMI-1640 were supplemented with 10% (v/v) fetal bovine serum (FBS), 100 U mL−1 penicillin, 100 mg mL−1 streptomycin, and 2 mM L-glutamine, and maintained in an incubator (Memmert, Germany) at 37 °C under an atmosphere of 5% CO2 and 90% relative humidity. The cells were sub-cultivated approximately every 2 to 3 days using 0.05% (w/v) trypsin-EDTA.Cell viability assay
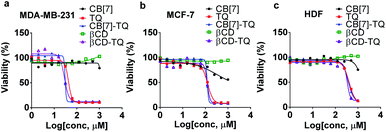
MDA-MB-231, MCF-7 and HDF (5 × 103 cells per well) were seeded in 96-well plates. After 24 h, the cells were treated with serial dilution of each of the CB[7], βCD, TQ, βCD–TQ complex, CB[7]–TQ complex in a concentration range of 0 to 1000 μM for 72 h (the host–guest solutions were prepared in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio). After 72 h of incubation at 37 °C, treatment was removed from the wells, followed by adding 15 μL of 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium Bromide (MTT) solution and 100 μL of the medium. After 3 h of incubation, the medium was removed, then 50 μL of dimethyl sulfoxide (DMSO) was added for dissolving the formazan. The absorbance was measured at a wavelength of 570 nm using Glomax microplate reader (Promega, USA).
1 molar ratio). After 72 h of incubation at 37 °C, treatment was removed from the wells, followed by adding 15 μL of 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium Bromide (MTT) solution and 100 μL of the medium. After 3 h of incubation, the medium was removed, then 50 μL of dimethyl sulfoxide (DMSO) was added for dissolving the formazan. The absorbance was measured at a wavelength of 570 nm using Glomax microplate reader (Promega, USA).
Results and discussion
CB[7]–TQ inclusion complex
The inclusion complex between TQ and CB[7] (structures shown in Fig. 1) was investigated using several spectroscopic methods. In particular, NMR is a very powerful technique for studying guest–host complexation since it is highly influenced by any changes in the microenvironment of the nucleus under study in addition to its chemical identity capabilities. Several NMR experiments are useful in gaining insights on type and location of interaction between molecules such as the chemical shift (Δδ), relaxation time (T1) and diffusion measurements (DOSY experiments). However, simple 1H NMR experiments to observe chemical shift changes upon complexation (or interaction) are very efficient. The 1H NMR spectrum of free TQ in D2O is shown in Fig. 2a, the protons' signals are broadened due to the low solubility of TQ in water, and the possible aggregation.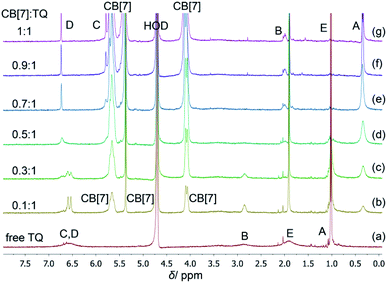 | ||
| Fig. 2 1H NMR titration of TQ and CB[7], (a) the spectrum of free TQ, (b–g) the spectra of CB[7]–TQ at different molar ratio, in D2O at ambient temperature. | ||
Upon the addition of different aliquots of CB[7] to TQ solution, the proton of the guest experienced complexation induced chemical shifts (Δδ), and the shape of the signals changed suggesting the formation of CB[7]–TQ inclusion complex. As shown in Table 1, a significant upfield shift was noticed in HA, HB and HC signals of TQ (+Δδ = 0.66, 0.89, 0.84 ppm, respectively), this indicated that these protons are accommodated deep inside the CB[7] cavity, and that the observed upfield shift is related to the shielding effect of the hydrophobic cavity of CB[7].16,25 On the other hand, protons HD and HE are positioned out of the cavity of CB[7], and close to the carbonyl rims, as indicated by the downfield shift (−Δδ).25 In addition, the shape of TQ protons in the 1H NMR spectra was largely affected by CB[7] additions. The signals (protons B, C, D and E) in D2O were broad (Fig. 2a), indicating their presence in an aggregated state and not as free hydrated TQ molecules. This behavior has been reported in literature regarding self-assembly of small organic molecules in aqueous solutions.26 Upon the addition of CB[7], an enhancement in solubility is achieved due to complexation and the aggregates start breaking giving sharp 1H NMR signals (Fig. 2b–g).27 No further shift in the TQ peaks was noticed after reaching 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mole ratio (host:guest), which indicated the formation of 1
1 mole ratio (host:guest), which indicated the formation of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stable inclusion complex of CB[7]–TQ. The gradual disappearance of some signals of free TQ and the appearance of CB[7]–TQ inclusion complex signals, suggested slow exchange on the NMR time scale.
1 stable inclusion complex of CB[7]–TQ. The gradual disappearance of some signals of free TQ and the appearance of CB[7]–TQ inclusion complex signals, suggested slow exchange on the NMR time scale.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CB[7]–TQ inclusion complex in D2O at ambient temperature
1 CB[7]–TQ inclusion complex in D2O at ambient temperature
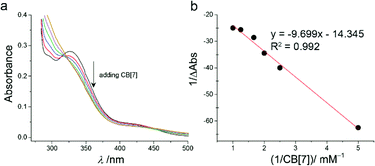
The formation of inclusion complex CB[7]–TQ was further investigated by using UV-visible spectroscopy. The free TQ showed two absorption peaks, at 334 nm and at 434 nm (as a shoulder) in aqueous medium. As shown in Fig. 3a, addition of CB[7] into the TQ solution (1 mM) resulted in gradually decrease of the absorption peak at ∼334 nm (λmax of the free TQ) until almost disappeared at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mole ratio of CB[7]
1 mole ratio of CB[7]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TQ. Also, a decrease in the absorbance of the shoulder peak of TQ (λ = 434 nm) with bathochromic shift (from 434 nm to 447 nm) was observed upon CB[7] addition. These changes confirmed the formation of host–guest inclusion complex (CB[7]–TQ complex).25
TQ. Also, a decrease in the absorbance of the shoulder peak of TQ (λ = 434 nm) with bathochromic shift (from 434 nm to 447 nm) was observed upon CB[7] addition. These changes confirmed the formation of host–guest inclusion complex (CB[7]–TQ complex).25
Complexation stoichiometry and binding constant determination
The UV-visible spectrophotometry was used to investigate the host–guest complex formation of CB[7]–TQ, the spectra obtained for successive additions of CB[7] to TQ in aqueous solution are shown in Fig. 3a. The complexation stoichiometry and the binding constant of CB[7]–TQ inclusion complex were determined using the Benesi–Hildebrand equation based on the titration data from UV-visible spectrophotometry.28 A linear dependence of the type 1/(A − A°) vs. 1/[CB7]n, with n = 1, indicated the presence of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry of the CB[7]
1 stoichiometry of the CB[7]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TQ complex. Fig. 3b represents the Benesi–Hildebrand plot for CB[7]–TQ inclusion complex using UV-visible spectrophotometric titration for 1
TQ complex. Fig. 3b represents the Benesi–Hildebrand plot for CB[7]–TQ inclusion complex using UV-visible spectrophotometric titration for 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry. A Good linear correlation was obtained for n = 1 (Fig. 3b), with a binding constant (K) of 2.5 × 103 M−1. Fitting the data according to nonlinear model also provided a K of 3 × 103 M−1.
1 stoichiometry. A Good linear correlation was obtained for n = 1 (Fig. 3b), with a binding constant (K) of 2.5 × 103 M−1. Fitting the data according to nonlinear model also provided a K of 3 × 103 M−1.
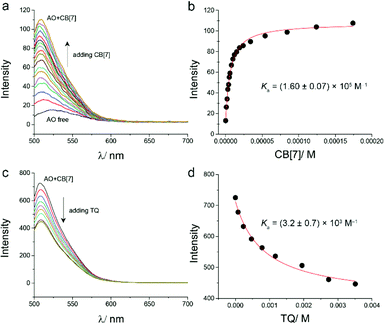
The binding of TQ and CB[7] was also investigated using optical dye displacement titration. This method relies on the competitive displacement of a fluorescent dye from a host molecule by the guest we intend to study.29,30 Different dyes were used in the indicator displacement titration with CB[7].31,32 In this study, acridine orange dye (AO) was used for this purpose. Upon the addition of CB[7] to aqueous solution of AO, a blue shift (from λem = 521 nm for free AO, to λem = 510 nm after adding CB[7]) and a large enhancement in the emission peak of AO were observed (Fig. 4a). The binding constant of CB[7]–AO inclusion was found to be 1.6 × 105 M−1 based on the titration experiment (Fig. 4b), which is in line with the reported value.15
For displacement titration, different aliquots of TQ solution were added into an aqueous solution of the encapsulated adduct CB[7]–AO, a continuous decrease in the emission spectrum of CB[7]–AO was observed (Fig. 4c). The addition of TQ was stopped after reaching a plateau in the experiment data, which confirmed reaching the equilibrium between the encapsulated and the free TQ. By fitting the data of fluorescence displacement titration experiment (adding TQ to CB[7]–AO solution), the value of the binding constant of CB[7] and TQ was obtained (K = 3.2 × 103 M−1) (Fig. 4d). This value was almost similar to the value obtained from UV-visible titration (2.5 × 103 M−1).
DFT calculations
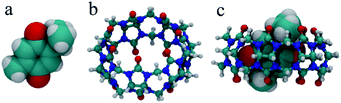
The optimized structures of the free TQ, CB[7], and the CB[7]–TQ complex are shown in Fig. 5. The cavity volume of CB[7] (242 Å3) can accommodate only one residue of TQ (size 161 Å3), resulting in a packing coefficient (PC% = guest size/cavity volume) of 67%, which verified the experimentally observed 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry. The calculations showed the formation of an inclusion complex was stabilized by −37 kcal mol−1 in the gas phase compared to the free components. The obtained structure of the CB[7]–TQ complex showed that the benzoquinone was fully encapsulated inside the hydrophobic cavity of CB[7], while the methyl substituent was positioned inside the cavity and close to the carbonyl rim. In contrast, the isopropyl substituent was partial excluded and more exposed to the surrounding. These structural results are in line with the 1H NMR data.
1 binding stoichiometry. The calculations showed the formation of an inclusion complex was stabilized by −37 kcal mol−1 in the gas phase compared to the free components. The obtained structure of the CB[7]–TQ complex showed that the benzoquinone was fully encapsulated inside the hydrophobic cavity of CB[7], while the methyl substituent was positioned inside the cavity and close to the carbonyl rim. In contrast, the isopropyl substituent was partial excluded and more exposed to the surrounding. These structural results are in line with the 1H NMR data.
Cytotoxicity
The cytotoxic effect of the CB[7]–TQ complex was investigated against breast cancer using MTT assay. Moreover, the toxicity of the CB[7], βCD, free TQ, and βCD–TQ complex were assessed and compared to CB[7]–TQ complex. To do so, triple-negative human breast cancer cell line (MDA-MB-231), estrogen receptor (ER)-positive human breast cancer cell line (MCF-7), and normal human dermal fibroblast cell line (HDF) were used. The three cell lines were treated with different concentrations of CB[7], βCD, free TQ, βCD–TQ complex, or CB[7]–TQ complex for 72 h. In general, the results revealed higher toxicity for free TQ, βCD–TQ complex, and CB[7]–TQ complex to both breast cancer cell lines (Fig. 6a, b) compared to normal fibroblast (Fig. 6c). Furthermore, based on IC50 values (Table 2), it was noticed that the toxicity of βCD–TQ and CB[7]–TQ complexes was higher than the free TQ in MDA-MB-231 cells compared to no significant differences in the cytotoxicity of TQ, βCD–TQ complex, and CB[7]–TQ complexes in MCF-7 cells (Table 2). Importantly, there was no significant toxicity against the three tested cell lines obtained from βCD and CB[7] treatment. All of these results confirmed that the anticancer activity of TQ was almost not affected by the kind of the host molecule that used to form the inclusion complexes (based on IC50 values, Table 2). An interesting finding in the current work was the similar antiproliferative effect of TQ when complexed with CB[7] or βCD counterparts. The similar effect is most likely due to the formation of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 inclusion complexes and the ability of both CB[7] and βCD to deliver and release of TQ into the cells in a similar manner. Moreover, an intensive and comparative study of uptake and release of TQ from CB[7] and βCD is of high interest for more understanding.
1 inclusion complexes and the ability of both CB[7] and βCD to deliver and release of TQ into the cells in a similar manner. Moreover, an intensive and comparative study of uptake and release of TQ from CB[7] and βCD is of high interest for more understanding.
| IC50 ± SD (μM, n = 3) | |||
|---|---|---|---|
| MDA-MB-231 | MCF-7 | HDF | |
| CB[7] | >1000 | >1000 | >1000 |
| βCD | >1000 | >1000 | >1000 |
| TQ | 42.5 ± 1.5 | 123.7 ± 11.3 | 407.9 ± 14.0 |
| CB[7]–TQ | 29.2 ± 1.7 | 118.3 ± 8.8 | 370.8 ± 33.3 |
| βCD–TQ | 28.6 ± 2.7 | 121.3 ± 6.2 | 332.3 ± 19.9 |
The antiproliferative activity of TQ loaded into different nanocarriers have been described by several studies and have been shown either higher or equal activity in comparison to free TQ.9,33–37 For instance, Bhattacharya et al. reported similar antiproliferative effect of TQ-loaded into polymeric nanoparticles against MCF7 when compared to free TQ.37 Moreover, Ganea et al. reveled twofold higher toxicity of TQ-loaded into PLGA nanoparticles in MDA-MB-231 cells compared to free TQ.35 In contrary, Odeh et al. showed that the encapsulation of TQ in βCD, enhanced its antiproliferative activity against MCF7.9 Encapsulation of TQ in liposomes also enhanced its solubility and hence its bioavailability against MCF7 and MDA-MB-231 cell lines.7
The TQ's mode of action on different cell lines is an important factor in deciding the suitable carrier aside from enhanced solubility and bioavailability. For example, Sunoqrot et al. reported the encapsulation of TQ in polymeric particles and tested their activity on various cell lines.38 Their results showed no enhancement of the antiproliferative activity of TQ-polymer compared to free TQ, but the selectivity index and in vivo bioavailability were more prominent. TQ, either free, encapsulated in a nanocarrier or in combination with other chemotherapeutics, is very promising against many cancers including cancer stem cells.39–41 Moreover, the results also confirmed the low toxicity of CB[7] that supports the use of CB[7] as a drug carrier. Previously described reports about toxicity and safety of CB[n] demonstrated the relative safety and acceptable toxicity in vitro and in vivo.42,43 For example, Oun et al. reported the lack of significant tissue specific toxicity of CB[6] and CB[7] using ex vivo rat model.42 Moreover, Hettiarachchi et al. revealed the very low toxicity of CB[5] and CB[7] and high cell tolerance at concentrations of up to 1 mM when tested against normal human cell lines originated from kidney, liver, and blood tissues.43
Conclusion
The inclusion complex CB[7]–TQ was successfully prepared in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mole ratio of CB[7]
1 mole ratio of CB[7]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TQ with moderate strength binding (K of 3 × 103 M−1) in aqueous solution. As a result of this inclusion process, the solubility of TQ was clearly enhanced (based on 1H NMR spectroscopy study), which is useful in increasing the bioavailability of TQ in aqueous solution. The experimental and computational theoretical results are consistent with inclusion of a single TQ molecule within CB[7] cavity. The cytotoxic effect of the CB[7]–TQ complex was investigated against cancer and normal cell lines, which revealed that the anticancer activity of TQ can be enhanced by forming a stable inclusion complex with CB[7]. Results showed also that the anticancer activity of TQ was not affected by the kind of the host molecule that used in the complexation process. Overall, the results highlight the potential implementation of supramolecular complexation in medicinal chemistry and drug delivery applications.
TQ with moderate strength binding (K of 3 × 103 M−1) in aqueous solution. As a result of this inclusion process, the solubility of TQ was clearly enhanced (based on 1H NMR spectroscopy study), which is useful in increasing the bioavailability of TQ in aqueous solution. The experimental and computational theoretical results are consistent with inclusion of a single TQ molecule within CB[7] cavity. The cytotoxic effect of the CB[7]–TQ complex was investigated against cancer and normal cell lines, which revealed that the anticancer activity of TQ can be enhanced by forming a stable inclusion complex with CB[7]. Results showed also that the anticancer activity of TQ was not affected by the kind of the host molecule that used in the complexation process. Overall, the results highlight the potential implementation of supramolecular complexation in medicinal chemistry and drug delivery applications.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
L.A. acknowledges the Deanship of Scientific Research at the Hashemite University for the financial support (Grant number: 58/2019).References
- C. C. Woo, A. P. Kumar, G. Sethi and K. H. B. Tan, Biochem. Pharmacol., 2012, 83, 443–451 CrossRef CAS PubMed.
- S. Banerjee, S. Padhye, A. Azmi, Z. Wang, P. A. Philip, O. Kucuk, F. H. Sarkar and R. M. Mohammad, Nutr. Cancer, 2010, 62, 938–946 CrossRef CAS PubMed.
- S. Darakhshan, A. Bidmeshki Pour, A. Hosseinzadeh Colagar and S. Sisakhtnezhad, Pharmacol. Res., 2015, 95–96, 138–158 CrossRef CAS.
- H. Xu, B. Liu, Z. Xiao, M. Zhou, L. Ge, F. Jia, Y. Liu, H. Jin, X. Zhu, J. Gao, J. Akhtar, B. Xiang, K. Tan and G. Wang, Infect. Dis. Ther., 2021, 10, 483–494 CrossRef PubMed.
- S. Elgohary, A. A. Elkhodiry, N. S. Amin, U. Stein and H. M. El Tayebi, Cells, 2021, 10, 302 CrossRef CAS PubMed.
- J. M. M. Salmani, S. Asghar, H. Lv and J. Zhou, Molecules, 2014, 19, 5925–5939 CrossRef PubMed.
- F. Odeh, S. I. Ismail, R. Abu-Dahab, I. S. Mahmoud and A. Al Bawab, Drug Delivery, 2012, 19, 371–377 CrossRef CAS PubMed.
- S. I. Abdelwahab, B. Y. Sheikh, M. M. E. Taha, C. W. How, R. Abdullah, U. Yagoub, R. El-Sunousi and E. E. M. Eid, Int. J. Nanomed., 2013, 8, 2163–2172 CrossRef PubMed.
- R. Abu-Dahab, F. Odeh, S. I. Ismail, H. Azzam and A. Al Bawab, Pharmazie, 2013, 68, 939–944 CAS.
- M. S. Al-Qubaisi, A. Rasedee, M. H. Flaifel, E. E. M. Eid, S. Hussein-Al-Ali, F. H. Alhassan, A. M. Salih, M. Z. Hussein, Z. Zainal, D. Sani, A. H. Aljumaily and M. I. Saeed, Eur. J. Pharm. Sci., 2019, 133, 167–182 CrossRef CAS PubMed.
- V. K. Kumar, R. R. Devi and E. Hemananthan, J. Dispersion Sci. Technol., 2020, 41, 243–256 CrossRef.
- H. Yin, D. Bardelang and R. Wang, Trends Chem., 2021, 3, 1–4 CrossRef CAS.
- A. L. Koner and W. M. Nau, Supramol. Chem., 2007, 19, 55–66 CrossRef CAS.
- K. I. Assaf and W. M. Nau, Chem. Soc. Rev., 2015, 44, 394–418 RSC.
- M. Shaikh, J. Mohanty, P. K. Singh, W. M. Nau and H. Pal, Photochem. Photobiol. Sci., 2008, 7, 408–414 CrossRef CAS PubMed.
- K. I. Assaf, M. Florea, J. Antony, N. M. Henriksen, J. Yin, A. Hansen, Z. W. Qu, R. Sure, D. Klapstein, M. K. Gilson, S. Grimme and W. M. Nau, J. Phys. Chem. B, 2017, 121, 11144–11162 CrossRef CAS PubMed.
- E. Masson, X. Ling, R. Joseph, L. Kyeremeh-Mensah and X. Lu, RSC Adv., 2012, 2, 1213–1247 RSC.
- H. S. El-Sheshtawy, S. Chatterjee, K. I. Assaf, M. N. Shinde, W. M. Nau and J. Mohanty, Sci. Rep., 2018, 8, 1–10 CrossRef CAS PubMed.
- K. I. Kuok, S. Li, I. W. Wyman and R. Wang, Ann. N. Y. Acad. Sci., 2017, 1398, 108–119 CrossRef CAS PubMed.
- D. Das, K. I. Assaf and W. M. Nau, Front. Chem., 2019, 7, 619 CrossRef CAS PubMed.
- A. I. Day and A. S. Atthar, in Cucurbiturils and Related Macrocycles, The Royal Society of Chemistry, 2020, pp. 238–282 Search PubMed.
- A. Day, A. P. Arnold, R. J. Blanch and B. Snushall, J. Org. Chem., 2001, 66, 8094–8100 CrossRef CAS PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci, G. A. Petersson, H. Nakatsuji, M. Caricato, X. Li, H. P. Hratchian, A. F. Izmaylov, J. Bloino, G. Zheng, J. L. Sonnenberg, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. Bearpark, J. J. Heyd, E. Brothers, K. N. Kudin, V. N. Staroverov, T. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, N. Rega, J. M. Millam, M. Klene, J. E. Knox, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, R. L. Martin, K. Morokuma, V. G. Zakrzewski, G. A. Voth, P. Salvador, J. J. Dannenberg, S. Dapprich, A. D. Daniels, O. Farkas, J. B. Foresman, and J. V. Ortiz, J. Cioslowski and D. J. Fox, Gaussian 09, Gaussian, Inc., Wallingford, CT, 2010 Search PubMed.
- T. C. Lee, E. Kalenius, A. I. Lazar, K. I. Assaf, N. Kuhnert, C. H. Grün, J. Jänis, O. A. Scherman and W. M. Nau, Nat. Chem., 2013, 5, 376–382 CrossRef CAS PubMed.
- G. H. Aryal, K. I. Assaf, K. W. Hunter, W. M. Nau and L. Huang, Chem. Commun., 2017, 53, 9242–9245 RSC.
- F. Odeh, A. Al-Bawab and Y. Li, J. Dispersion Sci. Technol., 2010, 31, 162–168 CrossRef CAS.
- A. Thangavel, I. A. Elder, C. Sotiriou-Leventis, R. Dawes and N. Leventis, J. Org. Chem., 2013, 78, 8297–8304 CrossRef CAS PubMed.
- J. S. Negi and S. Singh, Carbohydr. Polym., 2013, 92, 1835–1843 CrossRef CAS PubMed.
- K. I. Assaf, O. Suckova, N. Al Danaf, V. Von Glasenapp, D. Gabel and W. M. Nau, Org. Lett., 2016, 18, 932–935 CrossRef CAS PubMed.
- A. Praetorius, D. M. Bailey, T. Schwarzlose and W. M. Nau, Org. Lett., 2008, 10, 4089–4092 CrossRef CAS PubMed.
- K. I. Assaf and W. M. Nau, Supramol. Chem., 2014, 26, 657–669 CrossRef CAS.
- S. Gupta, Y. Zhao, R. Varadharajan and V. Ramamurthy, ACS Omega, 2018, 3, 5083–5091 CrossRef CAS.
- I. Fakhoury, W. Saad, K. Bouhadir, P. Nygren, R. Schneider-Stock and H. Gali-Muhtasib, J. Nanopart. Res., 2016, 18, 1–16 CrossRef CAS.
- H. Dehghani, M. Hashemi, M. Entezari and A. Mohsenifar, Iran. J. Pharm. Res., 2015, 14, 539–546 CAS.
- G. M. Ganea, S. O. Fakayode, J. N. Losso, C. F. Van Nostrum, C. M. Sabliov and I. M. Warner, Nanotechnology, 2010, 21, 285104 CrossRef PubMed.
- J. Ravindran, H. B. Nair, B. Sung, S. Prasad, R. R. Tekmal and B. B. Aggarwal, Biochem. Pharmacol., 2010, 79, 1640–1647 CrossRef CAS PubMed.
- S. Bhattacharya, M. Ahir, P. Patra, S. Mukherjee, S. Ghosh, M. Mazumdar, S. Chattopadhyay, T. Das, D. Chattopadhyay and A. Adhikary, Biomaterials, 2015, 51, 91–107 CrossRef CAS PubMed.
- S. Sunoqrot, M. Alfaraj, A. M. Hammad, V. Kasabri, D. Shalabi, A. A. Deeb, L. H. Ibrahim, K. Shnewer and I. Yousef, Pharmaceutics, 2020, 12, 1–16 CrossRef PubMed.
- F. R. Ballout and H. Gali-Muhtasib, Pharmacogn. Rev., 2020, 14, 155–159 CrossRef CAS.
- F. Odeh, R. Naffa, H. Azzam, I. S. Mahmoud, W. Alshaer, A. Al Bawab and S. Ismail, Heliyon, 2019, 5, e02919 CrossRef PubMed.
- M. M. Mehanna, R. Sarieddine, J. K. Alwattar, R. Chouaib and H. Gali-Muhtasib, Int. J. Nanomed., 2020, 15, 9557–9570 CrossRef CAS PubMed.
- R. Oun, R. S. Floriano, L. Isaacs, E. G. Rowan and N. J. Wheate, Toxicol. Res., 2014, 3, 447–455 CrossRef CAS PubMed.
- G. Hettiarachchi, D. Nguyen, J. Wu, D. Lucas, D. Ma, L. Isaacs and V. Briken, PLoS One, 2010, 5, 2–11 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2022 |