Open Access Article
Open Access ArticleCold atmospheric pressure plasma for attenuation of SARS-CoV-2 spike protein binding to ACE2 protein and the RNA deactivation
Rakesh Ruchel Khanikar†
a,
Monalisa Kalita†b,
Parismita Kalitaa,
Bhaswati Kashyapb,
Santanu Dasb,
Mojibur R. Khanb,
Heremba Bailung*a and
Kamatchi Sankaranarayanan *a
*a
aPhysical Sciences Division, Institute of Advanced Study in Science and Technology, (An Autonomous Institute Under DST, Govt. of India), Vigyan Path, Paschim Boragaon, Garchuk, Guwahati, Assam 781035, India. E-mail: hbailung@yahoo.com; kamatchi.sankaran@gmail.com
bLife Sciences Division, Institute of Advanced Study in Science and Technology, (An Autonomous Institute Under DST, Govt. of India), Vigyan Path, Paschim Boragaon, Garchuk, Guwahati, Assam 781035, India
First published on 25th March 2022
Abstract
Cold atmospheric pressure (CAP) plasma has a profound effect on protein–protein interactions. In this work, we have highlighted the deactivation of the Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) spike protein by CAP plasma treatment. Complete deactivation of spike protein binding to the human ACE2 protein was observed within an exposure time of 5 minutes which is correlated to the higher concentration of hydrogen peroxide formation due to the interaction with the reactive oxygen species present in the plasma. On the other hand, we have established that CAP plasma is also capable of degrading RNA of SARS-CoV-2 virus which is also linked to hydrogen peroxide concentration. The reactive oxygen species is produced in the plasma by using noble gases such as helium, in the absence of any other chemicals. Therefore, it is a green process with no chemical waste generated and highly advantageous from the environmental safety prospects. Results of this work could be useful in designing plasma-based disinfection systems over those based on environmentally hazardous chemical-based disinfection and biomedical applications.
1. Introduction
The novel coronavirus disease (Covid-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) which is a positive sense single stranded RNA virus.1 The genome of the virus is packed in an envelope comprising three proteins namely, membrane, spike, and envelope proteins. It is reported that the sustained interactions in SARS-CoV-2 and the major infection spread is caused by the binding of spike protein with the human Angiotensin Converting Enzyme 2 (hACE2) protein for the virus entry through the receptor binding domain (RBD) region.2,3 The major route of the spread is air-borne or surfaces wherein the virus floats or sits as respiratory droplets of infected patients. On entry into the nasal cavity, the spike protein of the virus binds to the ACE2 protein in the cell wall to make way for the virus entry.4 The second wave has hit hard in most of the developing countries especially India (especially due to the delta variant of the SARS-CoV-2). With the rapid increase in the Covid-19 infections worldwide (∼280 million) and deaths reaching ∼5.4 million as of December 2021 and it is need of the hour to look for additional approaches to minimize the risk of SARS-CoV-2 transmission.The interaction of cold atmospheric pressure (CAP) plasma with biological materials like living tissues, cells, bacteria, viruses is an emerging interdisciplinary field of research.5–7 The CAP plasma contains various active species such as electrons, ions, reactive oxygen and nitrogen species (RONS) and UV-photons making it useful in biomedical field specially for deactivation of bacteria, fungi, spores and viruses.8–11 CAP plasma has also been found effective for inactivation or modification of several proteins such as lysozyme,12 myoglobin and haemoglobin,13 NADPH oxidase (NOX) enzymes14 and RNAse.15 Recently, few reports have shown that CAP plasma is effective in inactivation of SARS-CoV-2 virus.16–19 Efforts have also been made to utilize plasma activated water to study the spike protein deactivation.17
SARS-CoV-2 virus remains stable as aerosols for longer periods of time and, to stop their spread, it is crucial to treat not only surfaces but also the air inside hospital/laboratory as well as living/office rooms. Traditional chemical disinfectants used in fogging have serious limitations due to its toxicity for human as well as material body. UV based disinfection can effectively deactivate the genetic material of the virus, however, has several associated issues with respect to affecting the human skin.20 In view of environmental protection, novel environmentally friendly decontamination methods are a need of the hour. In this work, we explored the process of deactivation of the spike protein of the Covid-19 virus using cold atmospheric pressure plasma and its effect on the RNA of the virus.
2. Materials and methods
SARS-CoV-2 (2019-nCoV) spike (S1 + S2 ECD-His Recombinant) protein, Anti-His tag HRP Antibody, human ACE2 protein (mFc Tag), TMB highly Sensitive HRP Substrate were purchased from Krishgen Biosystems, India and used without further purification. BSA, materials for buffer preparation were purchased from Sigma Aldrich, India. The reagents and buffers were prepared in Millipore water. RNA samples of SARS-CoV-2 virus extracted from samples of Covid-19 positive infected patients for Covid-19 testing (RT-PCR) were used. RNA extraction was done following standard protocols. Abundance of the viral RNA before and after CAP treatment was is evaluated in RT-PCR. A sample volume of 100 μl is taken in the wells of a 24 well plate for plasma treatment for different exposure times. Each experiment has been performed in triplicate.2.1 Experimental setup
A cold atmospheric pressure plasma jet is used for the experiment. It is a modified form of the plasma jet described elsewhere with a modified electrode arrangement as shown in Fig. 1(a).21 It consists of a glass tube with an inner diameter of 3 mm and an outer diameter of 7 mm. The live electrode is made of stainless steel rod (1 mm in diameter) and placed inside the glass tube. The grounded electrode is copper strip wrapped around the outer surface of the glass tube. The glass tube and the live electrode assembly is fitted into a Teflon housing with a gas inlet. Plasma is generated in between the electrodes applying sinusoidal voltages (2.5 kV and 5.0 kV) at frequency (30 kHz). Helium and a mixture of helium with compressed air have been used as the plasma forming gases. The gas flow has been measured using a variable area flowmeter (Cole Parmer). For He plasma, a helium gas flow of 1000–3000 sccm (standard cubic cm per minute) has been maintained to obtain a stable plasma. For He–air plasma, air flow of 50–200 sccm is mixed with helium in order to enhance the RONS level in the plasma jet. The plasma produced between the two electrodes comes out into the open air with the gas flow. The sample for plasma treatment is placed underneath (as shown in Fig. 1) for different exposure times.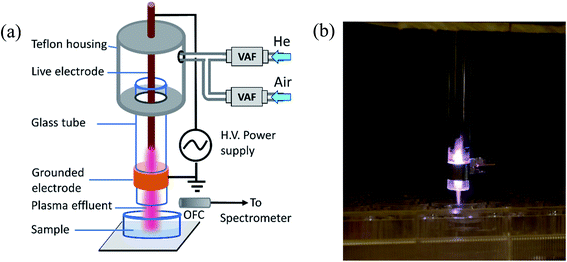 | ||
| Fig. 1 (a) Schematic diagram of the experimental set up. VAF: variable area flowmeter, OFC: optical fiber cable. (b) Photograph of the CAP plasma plume coming out of the glass tube. | ||
Further, the Teflon housing can be extended to cover the whole electrode arrangement up to the end through which the plasma comes out into open air. It will provide insulation from high voltage and then it will be possible to use the device as a handheld portable one that can be operated by a user without having a strong scientific background by just switching on the power supply and the gas flow. The device presented here is a prototype one for scientific study. For large area applications, it will be possible to upscale the treatment capacity by designing an array of multi-jet structure.
To investigate the various reactive species, present in the plasma, an emission spectrum of the plasma jet was obtained by using an optical emission spectrometer (Andor-SR-303i-A). The light emitted from the plasma was collected by an optical fiber fitted to the spectrometer equipped with three gratings consisting of 300, 1200 and 2400 grooves per mm and the slit width can be varied from 10–2500 μm. In this experiment, a slit width of 50 μm was used to achieve a fair resolution along with sufficient intensity. The survey spectrum was recorded using 1200 groves per mm grating in the wavelength range 250–900 nm, whereas the OH emission is recorded using 2400 groves per mm grating in the wavelength range 306–310 nm to achieve high resolution. The spectrometer's wavelength calibration is performed using a standard NIST traceable calibration lamp (StellarNet Inc).
2.2 Enzyme-linked immunosorbent assay (ELISA)
The hACE2 protein was diluted with a coating buffer PBS (136.9 mM NaCl, 10.1 mM Na2HPO4, 2.7 mM KCl, 1.8 mM KH2PO4, 0.2 μm filtered) adjusted to pH 7.4 to 2 μg ml−1 and added to the wells of 96-well plates (100 μl per well). The plates were incubated at 4 °C overnight and washed with water. Then, the wells were blocked with blocking buffer PBS-T (0.05% Tween20 in PBS) containing 2% BSA for 60 min and washed. Next, a series of diluted SARS-CoV-2 spike protein from 20 μg ml−1 was prepared in the diluting buffer (0.1% BSA in PBS-T) and incubated for 60 min in the well plates. To study the binding of spike proteins with hACE2, various concentrations of the spike protein viz., 2.5, 5, 10, 12.5, 15, 17.5, 20 μg ml−1 were prepared and subjected to plasma treatment with two different feed gases He and He–air mixture. The wells were washed and incubated with 100 μl of HRP detection antibody for 60 min. After washing, TMB substrate (Thermo Fisher) and 2 M H2SO4 were sequentially added to the wells. Then, the optical density was measured using a microplate reader (Thermo Scientific Varioskan Flash) at 450 nm. The optical density was normalized to 100% for the binding of ACE2 with the untreated SARS-CoV 2 spike protein and the further compared with the binding of ACE2 to the CAP treated spike proteins.2.3 Protein carbonyl content estimation
The carbonyl content of the native and plasma treated spike protein was determined by 2,4-dinitrophenylhydrazine (DNPH) assay.22 Briefly, 200 μl protein samples were properly mixed with 800 μl of freshly prepared 0.1% 2,4-dinitrophenylhydrazine in 2.5 M HCL and incubated for 1.5 h at room temperature. Protein was precipitated by addition of 1000 μl of 20% chilled trichloroacetic acid (TCA) followed by centrifugation at 4 °C. The pellet was then re-suspended and washed with chilled 10% TCA followed by washing twice with ice-cooled 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 (v/v) acetate and ethanol. Finally, the pellet was re-suspended in 500 μl of 6 M guanidium chloride and absorption spectra were recorded at 370 nm with a molar absorptivity of 22
50 (v/v) acetate and ethanol. Finally, the pellet was re-suspended in 500 μl of 6 M guanidium chloride and absorption spectra were recorded at 370 nm with a molar absorptivity of 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1.
000 M−1 cm−1.
3. Results and discussion
3.1 Optical diagnostics and gas temperature of the CAP plasma
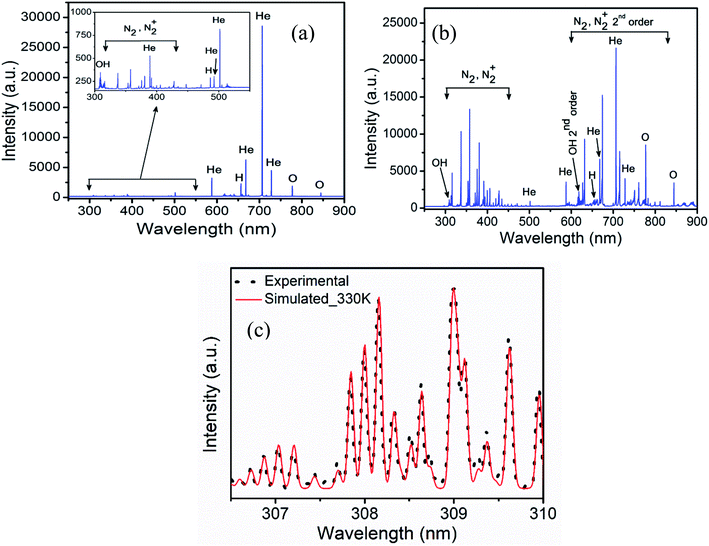
Fig. 2 shows emission spectra obtained from the He and He–air plasma jet. The spectrum covers wavelength from 250 nm to 900 nm. The spectral lines are identified using NIST atomic spectra database28 and ref. 29. The presence of various reactive species in the plasma jet can be seen from the spectrum, which includes molecular bands such as OH (A–X), N2 (C–B), N2+ (B–X) and atomic lines of helium and singlet oxygen etc. These reactive species diffuse into the liquid media thereby forming reactive oxygen and nitrogen species (RONS) in the sample solution. The presence of the molecular bands in the UV region of the emission spectrum indicates the energetic UV photon emission from the plasma jet. In Fig. 2(a), the OES spectrum corresponding to the He plasma jet is shown where very weak emission lines from reactive oxygen and nitrogen species are seen (enlarged in the inset). In He–air plasma, the concentrations of reactive oxygen and nitrogen species are significantly increased. As a result, we observed a manifold increase in the RONS concentrations in the He–air plasma jet spectrum as shown in Fig. 2(b). The mixing flow rate of air with helium has been optimized to achieve such optimized concentration of RONS in the plasma jet. Slight enhancement in UV radiation has also been observed for the He–air plasma. The reactive species in the plasma are known to be generated by electron impact phenomena, which cause excitation, ionization and dissociation of the molecules. The plasma generated excited/ionized species and metastables strike the water molecules on the liquid media surface leading to formation of different ROSs such as hydroxyl radical, hydrogen peroxide etc30.It is essential to know the gas temperature in a cold atmospheric pressure plasma jet accurately specially for biological applications. One of the most popular gas temperature determination methods in low temperature humid atmospheric plasmas is from the measurement of rotational temperature from OH (A–X) emission band around 306–310 nm. The rotational temperature is determined from the comparison of the experimental spectrum with a simulated theoretical spectrum obtained from spectral simulation software like LIFBASE31 and SPECAIR.32 The models assume Boltzmann distribution of rotational levels. The rotational temperature obtained from them is a good measure of gas temperature since the translational and rotational degrees of freedom remain in equilibrium in atmospheric pressure plasmas due to large number of collisions.33,34 Fig. 2(c) shows the best fit the OH molecular band between the recorded and the simulated spectrum from the software LIFBASE. The temperature of both He plasma jet and He–air plasma jet is found to be similar and estimated to be 330 ± 20 K in the active region of plasma between the electrodes. While at the point of application, where the plasma plume comes out into open air, the gas temperature remains at room temperature. This is also verified by the fact that the temperature of the sample remains unchanged after plasma treatment.
3.2 ELISA characterization of the spike protein binding to the ACE2 proteins
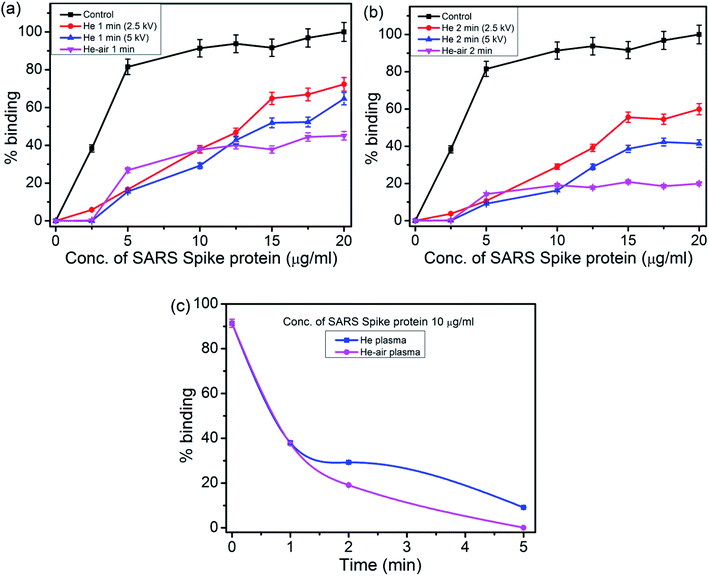
We performed the Enzyme-linked Immunosorbent Assay to characterize the binding of the ACE2 proteins to the spike proteins before and after subjecting to the plasma exposure. The results are presented in Fig. 3. As mentioned earlier, CAP plasma was produced with two different feed gases, He and He-air mixed gas with different discharge voltages (2.5 kV and 5 kV, 30 kHz) and very short interaction time ∼2 minutes (1 minute in step), which will be relevant for practical applications in large scale.The binding of ACE2 protein to the spike protein is the most important factor behind the Covid-19 infection. To model such binding, in our study, we had taken various concentration of the spike protein from 2.5 μg ml−1 to 20 μg ml−1 (Fig. 3(a)). The binding efficiency of the various concentration of the spike proteins with ACE2 was evaluated after treating the spike protein with the CAP plasma jet. The native spike protein binds to ACE2 with ∼100% efficiency (without plasma treatment). He plasma reduces the binding efficiency to about 70% and 60% relative to the control, for the voltages of 2.5 kV and 5 kV within 1 min for a concentration of 20 μg ml−1 (Fig. 3(a)). The binding further reduces to 40% with He–air plasma for same treatment time 1 min and for the same concentration 20 μg ml−1. Further increase in treatment time to 2 min reduces the binding efficiency more effectively and maximum reduction to ∼18% has been estimated for He–air plasma treatment (Fig. 3(b)) for the spike protein concentration 20 μg ml−1. In both the cases the He–air plasma was operated at 5 kV of discharge voltage.
A similar treatment of the spike protein with the CAP plasma using both Helium and He–air mixed feed gas, for a particular concentration 10 μg ml−1 is presented in Fig. 3(c). Complete deactivation of the spike protein with the longer exposure time of 5 minutes is clearly evident for He–air plasma. The superior effect of the He–air mixed feed gas plasma can be attributed to the increase in the RONS production as observed from the optical emission spectra.
Further, the role of the reactive oxygen species (ROS) in the deactivation of the spike protein was determined using the DNPH assay. ROS can have major interactions with some amino acid residues such as Pro, Arg, Lys, and Thr leading to an oxidative cleavage of the protein backbone.35,36 Protein-bound carbonyls are the most often used as markers to estimate protein oxidation. Earlier reports have shown that plasma treatment can modify or oxidize a variety of proteins, including horse radish peroxidase37 and such interactions can be tuned by adjusting the RONS.38 In this experiment with DNPH assay our estimation indicate protein-bound carbonyls for the plasma treated spike proteins enhancement from 2.1% for the control to 17.2% and 36.1% for the He and He–air mixed plasma respectively for the 2 min plasma exposure time. The various modifications of the protein backbone due to the increased carbonyl content in the protein could be primarily responsible for the deactivation of the spike protein.
Additionally, we performed RT-PCR analysis of the extracted RNA after plasma treatment. The relative abundance of the nucleocapsid protein encoding N-gene and the open reading frame (ORF1ab) gene of the plasma treated samples were compared with the control and presented in Fig. 4. The N-gene encodes the structural proteins in SARS-CoV-2, whereas the ORF gene encodes the nonstructural proteins.39 It is reported that by detecting the sole N-gene, a positive rate similar to that of detecting both N-gene and ORF1ab gene can be obtained.40 From Fig. 4, it is evident that the influence of the He and He–air plasma are dissimilar on the abundance of the N-gene and ORF-gene. Treatment with He plasma caused increase in the relative abundance of both the genes with respect to the untreated controls (239% for ORF1ab gene and 442% for N-gene; p < 0.05). On the other hand, treatment with He–air plasma caused a significant decrease in the abundance of the both the genes (23% for ORF1ab gene and 35% for N-gene; p < 0.05). This could be due to the non-interacting neutrals present in the plasma. The contrasting effects of the He and He–air plasma could be due to their differential effects on protein and RNA with relative abundance of RONS in the plasma jets. This is possible if the plasma treatment causes degradation/deformation of structural proteins of the virus particles releasing RNA. While He–air plasma treatment as explained earlier in Fig. 2, induces more OH radicals which can interfere with the RNA structure causing the reduction in abundance of the gene. However, to have a clear understanding we have to quantify the role of different active radicals produced in the He and He–air plasma.
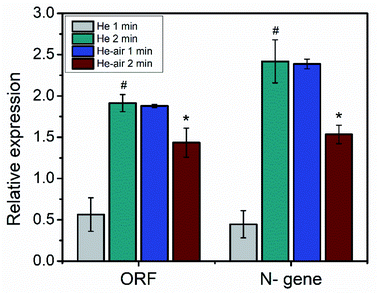 | ||
| Fig. 4 Relative abundance of ORF-gene and N-gene upon plasma treatment (*is significant compared to #). | ||
To better understand the role of the different radicals in the deactivation of the spike protein as well as the gene abundance, we quantified the different active radicals OH, H2O2 and nitrates in the liquid media using different assays (Table 1). The striking feature in this work is that with He plasma we had a high concentration of H2O2 for a treatment time of 1 min, which decreases nearly 5 times when treated for 2 min. This change is reflected in the increase of OH radicals (8 μM to 100 μM in 2 min) which could be due to the dissociation of the H2O2 to OH radicals upon interacting with higher energy electrons obtained from the He plasma.41 In the case of He–air plasma, the concentration of H2O2 increases from 28.6 μM to 37.2 μM in 2 min. The concentration of nitrate remains almost the same for He–air plasma whereas the nitrite concentration increases for both He and He–air plasma with the treatment time. The presence of nitrite and its increasing concentration with treatment time may be responsible for the interaction of these nitrites with the spike protein for further reactions. Such conversion of nitrites to nitrogen is reported for several amino acids and proteins interactions.42
| Samples/time | Hydroxyl radicals (μM) | H2O2 (μM) | Nitrate (μM) | Nitrite (μM) |
|---|---|---|---|---|
| He plasma 1 min | 7.29 ± 0.23 | 29.4 ± 0.45 | 21 ± 0.53 | 54.2 ± 0.9 |
| He plasma 2 min | 100 ± 1.2 | 6.1 ± 0.06 | 61.5 ± 1.2 | 97.4 ± 1.56 |
| He–air plasma 1 min | 1.56 ± 0.05 | 28.6 ± 0.57 | 10.1 ± 0.18 | 22.1 ± 0.29 |
| He–air plasma 2 min | 3.11 ± 0.08 | 37.2 ± 0.53 | 10.6 ± 0.24 | 39.6 ± 0.65 |
Our results suggest that the spike protein deactivation is possibly due to the amount of hydrogen peroxide produced in the sample. Such reports on deactivation of proteins in the presence of H2O2 have been earlier reported.43,44 We also see differential peroxide production in different plasma such as He and He–air plasma which could lead to the protein deactivation (Fig. 3) as well as the change in the gene abundance (Fig. 4). With respect to RNA, earlier reports suggest that the oxidative stress induced by the hydrogen peroxide can lead to the oxidative modification of RNA further affecting the structure of the RNA, which results in the lower gene abundance.45–47
4. Conclusions
In this study we have conducted a systematic evaluation of CAP plasma-based decontamination of SARS-Cov-2 virus. We observed complete inactivation of the spike protein in the presence of the He–air mixed feed gas plasma compared to the He-plasma. DNPH assay suggests the carbonyl modification of the spike protein due to the reactive oxygen species (ROS) induced in the plasma which inhibits the binding with hACE2 protein. On the other hand, direct exposure of the extracted RNA to the He and He–air plasma has shown evidence on modification on the gene abundance in the RT-PCR experiment. We have shown the hydrogen peroxide from the CAP plasma mediates the deactivation of the spike protein and influences the gene abundance. The reactive oxygen and nitrogen species are produced in the plasma by using noble gases such as helium as plasma forming gas. The transfer of chemical reactivity to the sample takes place from the gas phase of the plasma interacting with normal air, in the absence of any other chemicals. Hence, it is a green process and no chemical waste is generated in this method with remarkable advantage from the environmental safety prospects. Therefore, CAP plasma disinfection is definitely a better alternative compared to the current chemical-based decontamination and can be utilized for medical devices, surfaces and in air purification and optimal tuning of the reactive species of CAP could be an energy efficient and dry method for SARS-CoV-2 virus inactivation.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
Authors would like to thank National Health Mission, Govt. of Assam, Guwahati Medical College and Hospital, and Covid Testing and Research Laboratory, IASST, Guwahati for providing infrastructure facility and technical support. Authors KS and HB thank the IASST in-house grant. Author RRK thanks DST, Govt. of India for the support under DST-INSPIRE Fellowship Scheme. Author MK, PK thanks IASST for the research fellowship. The authors thank SAIC, IASST for the analytical facilities.References
- C. Y. Chen, C. ke Chang, Y. W. Chang, S. C. Sue, H. I. Bai, L. Riang, C. D. Hsiao and T. huang Huang, J. Mol. Biol., 2007, 368, 1075–1086 CrossRef CAS PubMed.
- J. Yang, S. J. L. Petitjean, M. Koehler, Q. Zhang, A. C. Dumitru, W. Chen, S. Derclaye, S. P. Vincent, P. Soumillion and D. Alsteens, Nat. Commun., 2020, 11, 4541 CrossRef CAS PubMed.
- S. Sakkiah, W. Guo, B. Pan, Z. Ji, G. Yavas, M. Azevedo, J. Hawes, T. A. Patterson and H. Hong, Front. Chem., 2021, 8, 622632 CrossRef PubMed.
- L. Morawska, J. W. Tang, W. Bahnfleth, P. M. Bluyssen, A. Boerstra, G. Buonanno, J. Cao, S. Dancer, A. Floto, F. Franchimon, C. Haworth, J. Hogeling, C. Isaxon, J. L. Jimenez, J. Kurnitski, Y. Li, M. Loomans, G. Marks, L. C. Marr, L. Mazzarella, A. K. Melikov, S. Miller, D. K. Milton, W. Nazaroff, P. V. Nielsen, C. Noakes, J. Peccia, X. Querol, C. Sekhar, O. Seppänen, S. ichi Tanabe, R. Tellier, K. W. Tham, P. Wargocki, A. Wierzbicka and M. Yao, Environ. Int., 2020, 142, 105832 CrossRef CAS PubMed.
- A. Lin, Y. Gorbanev, J. De Backer, J. Van Loenhout, W. Van Boxem, F. Lemière, P. Cos, S. Dewilde, E. Smits and A. Bogaerts, Adv. Sci., 2019, 6, 1802062 CrossRef PubMed.
- M. Moreau, N. Orange and M. G. J. Feuilloley, Biotechnol. Adv., 2008, 26, 610–617 CrossRef CAS PubMed.
- A. Filipić, I. Gutierrez-Aguirre, G. Primc, M. Mozetič and D. Dobnik, Trends Biotechnol., 2020, 38, 1278–1291 CrossRef PubMed.
- T. Bernhardt, M. L. Semmler, M. Schäfer, S. Bekeschus, S. Emmert and L. Boeckmann, Oxid. Med. Cell. Longevity, 2019, 2019, 3873928 Search PubMed.
- M. H. Lee, B. J. Park, S. C. Jin, D. Kim, L. Han, J. Kim, S. O. Hyun, K. H. Chung and J. C. Park, New J. Phys., 2009, 11, 115022 CrossRef.
- M. Zashev, D. Donchev, I. Ivanov and R. Gornev, J. Theor. Appl. Phys., 2020, 14, 41–49 CrossRef.
- N. Hojnik, M. Modic, Y. Ni, G. Filipič, U. Cvelbar and J. L. Walsh, Environ. Sci. Technol., 2019, 53, 1893–1904 CrossRef CAS PubMed.
- S. Choi, P. Attri, I. Lee, J. Oh, J. H. Yun, J. H. Park, E. H. Choi and W. Lee, Sci. Rep., 2017, 7, 1027 CrossRef PubMed.
- P. Attri, N. Kumar, J. H. Park, D. K. Yadav, S. Choi, H. S. Uhm, I. T. Kim, E. H. Choi and W. Lee, Sci. Rep., 2015, 5, 8221 CrossRef CAS PubMed.
- P. Attri, J. H. Park, J. De Backer, M. Kim, J. H. Yun, Y. Heo, S. Dewilde, M. Shiratani, E. H. Choi, W. Lee and A. Bogaerts, Int. J. Biol. Macromol., 2020, 163, 2405–2414 CrossRef CAS.
- J. W. Lackmann, S. Baldus, E. Steinborn, E. Edengeiser, F. Kogelheide, S. Langklotz, S. Schneider, L. I. O. Leichert, J. Benedikt, P. Awakowicz and J. E. Bandow, J. Phys. D: Appl. Phys., 2015, 48, 494003 CrossRef.
- L. Guo, Z. Yao, L. Yang, H. Zhang, Y. Qi, L. Gou, W. Xi and D. Liu, Chem. Eng. J., 2021, 421, 127742 CrossRef CAS PubMed.
- Z. Chen, G. Garcia, V. Arumugaswami and R. E. Wirz, Phys. Fluids, 2020, 32, 111702 CrossRef CAS PubMed.
- A. Bisag, P. Isabelli, R. Laurita, C. Bucci, F. Capelli, G. Dirani, M. Gherardi, G. Laghi, A. Paglianti, V. Sambri and V. Colombo, Plasma Processes Polym., 2020, 17, e2000154 CrossRef.
- T. Jin, Y. Xu, C. Dai, X. Zhou, Q. Xu and Z. Wu, AIP Adv., 2021, 11, 085019 CrossRef CAS PubMed.
- V. K. Patra, S. N. Byrne and P. Wolf, Front. Microbiol., 2016, 7, 1235 Search PubMed.
- R. R. Khanikar, P. J. Boruah and H. Bailung, Plasma Res. Express, 2020, 2, 045002 CrossRef CAS.
- R. L. Levine, J. A. Williams, E. R. Stadtman and E. Shacter, Methods Enzymol., 1994, 233, 346–357 CAS.
- S. Shen, T. H. P. Tan and Y.-J. Tan, in Methods in Molecular Biology, ed. R. J. Sugrue, Humana Press, 2007, vol. 379, pp. 127–135 Search PubMed.
- L. Du, Y. He, Y. Zhou, S. Liu, B. J. Zheng and S. Jiang, Nat. Rev. Microbiol., 2009, 7, 226–236 CrossRef CAS PubMed.
- X. Xiao and D. S. Dimitrov, Cell. Mol. Life Sci., 2004, 61, 2428–2430 CrossRef CAS PubMed.
- A. K. Nalla, A. M. Casto, A. M. Casto, M. L. W. Huang, G. A. Perchetti, R. Sampoleo, L. Shrestha, Y. Wei, H. Zhu, K. R. Jerome, K. R. Jerome and A. L. Greninger, J. Clin. Microbiol., 2020, 58, e00557–20 CrossRef.
- T. D. Schmittgen and K. J. Livak, Nat. Protoc., 2008, 3, 1101–1108 CrossRef CAS PubMed.
- A. Kramida, Y. Ralchenko, J. Reader and NIST ASD Team, NIST Atomic Spectra Database (version 5.8), [Online], https://physics.nist.gov/PhysRefData/ASD/lines_form.html, accessed 6 June 2021 Search PubMed.
- R. W. B. Pearse and A. G. Gaydon, The Identification of Molecular Spectra, Chapman & Hall Ltd, London, 2nd edn, 1950 Search PubMed.
- M. Magureanu, D. Piroi, F. Gherendi, N. B. Mandache and V. Parvulescu, Plasma Chem. Plasma Process., 2008, 28, 677–688 CrossRef CAS.
- J. M. Luque and D. R. Crosley, SRI Int. Rep. MP Search PubMed.
- C. O. Laux, Radiation and Nonequilibrium Collisional-Radiative Models, von Karman Institute Lecture Series 2002–07, Physico-Chemical Modeling of High Enthalpy and Plasma Flows, ed. D. Fletcher, J. M. Charbonnier, G. S. R. Sarma and T. Magin, Rhode-Saint-Genese, Belgium, 2002 Search PubMed.
- C. O. Laux, T. G. Spence, C. H. Kruger and R. N. Zare, Plasma Sources Sci. Technol., 2003, 12, 125–138 CrossRef CAS.
- S. Hofmann, A. F. H. Van Gessel, T. Verreycken and P. Bruggeman, Plasma Sources Sci. Technol., 2011, 20, 065010 CrossRef.
- P. A. Grimsrud, H. Xie, T. J. Griffin and D. A. Bernlohr, J. Biol. Chem., 2008, 283, 21837–21841 CrossRef CAS PubMed.
- E. R. Stadtman, Curr. Med. Chem., 2012, 11, 1105–1112 CrossRef PubMed.
- Z. Ke and Q. Huang, Plasma Processes Polym., 2013, 10, 731–739 CrossRef CAS.
- J. H. Park, M. Kim, M. Shiratani, A. E. Cho, E. H. Choi and P. Attri, Sci. Rep., 2016, 6, 35883 CrossRef CAS PubMed.
- Y. Huang, C. Yang, X. feng Xu, W. Xu and S. wen Liu, Acta Pharmacol. Sin., 2020, 41, 1141–1149 CrossRef PubMed.
- X. Zhang, M. Li, B. Zhang, T. Chen, D. Lv, P. Xia, Z. Sun, X. Shentu, H. Chen, L. Li and W. Qian, Clin. Chim. Acta, 2020, 511, 291–297 CrossRef CAS PubMed.
- L. Lyu, L. Zhang, G. He, H. He and C. Hu, J. Mater. Chem. A, 2017, 5, 7153–7164 RSC.
- T. Kato and K. Kikugawa, Food Chem. Toxicol., 1992, 30, 617–626 CrossRef CAS PubMed.
- J. M. Denu and K. G. Tanner, Biochemistry, 1998, 37, 5633–5642 CrossRef CAS PubMed.
- R. Drodz and J. W. Naskalski, Folia Histochem. Cytobiol., 1993, 31, 71–75 Search PubMed.
- Q. Ding, E. Dimayuga and J. N. Keller, Free Radical Res., 2007, 41, 903–910 CrossRef CAS.
- Q. Kong and C. L. Lin, Cell. Mol. Life Sci., 2010, 67, 1817–1829 CrossRef CAS PubMed.
- R. T. Sapio, C. J. Burns and D. G. Pestov, Front. Mol. Biosci., 2021, 8, 678488 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |